Abstract
Background
We have previously identified associations between major histocompatibility complex (MHC) class I and resistance towards bacterial and viral pathogens in Atlantic salmon. To evaluate if only MHC or also closely linked genes contributed to the observed resistance we ventured into sequencing of the duplicated MHC class I regions of Atlantic salmon.
Results
Nine BACs covering more than 500 kb of the two duplicated MHC class I regions of Atlantic salmon were sequenced and the gene organizations characterized. Both regions contained the proteasome components PSMB8, PSMB9, PSMB9-like and PSMB10 in addition to the transporter for antigen processing TAP2, as well as genes for KIFC1, ZBTB22, DAXX, TAPBP, BRD2, COL11A2, RXRB and SLC39A7. The IA region contained the recently reported MHC class I Sasa-ULA locus residing approximately 50 kb upstream of the major Sasa-UBA locus. The duplicated class IB region contained an MHC class I locus resembling the rainbow trout UCA locus, but although transcribed it was a pseudogene. No other MHC class I-like genes were detected in the two duplicated regions. Two allelic BACs spanning the UBA locus had 99.2% identity over 125 kb, while the IA region showed 82.5% identity over 136 kb to the IB region. The Atlantic salmon IB region had an insert of 220 kb in comparison to the IA region containing three chitin synthase genes.
Conclusion
We have characterized the gene organization of more than 500 kb of the two duplicated MHC class I regions in Atlantic salmon. Although Atlantic salmon and rainbow trout are closely related, the gene organization of their IB region has undergone extensive gene rearrangements. The Atlantic salmon has only one class I UCA pseudogene in the IB region while trout contains the four MHC UCA, UDA, UEA and UFA class I loci. The large differences in gene content and most likely function of the salmon and trout class IB region clearly argues that sequencing of salmon will not necessarily provide information relevant for trout and vice versa.
Background
Major histocompatibility complex (MHC) class I and class II molecules are vital parts of the cellular immune system presenting self and/or foreign peptides to CD8 positive and CD4 positive T cells. Both classes of genes reside in a 4 Mb gene dense region on human chromosome 6 shared with many other immune genes [1].
Atlantic salmon and rainbow trout genomes encode one major MHC class I locus designated UBA in addition to the major MHC class II alpha and beta genes designated DAA and DAB respectively [2-4]. For UBA, the main polymorphism resides in the alpha 1 and alpha 2 domains with up to 60% sequence divergence between these antigen binding domains. Added variability for UBA is produced by shuffling of exon 2 onto different exon 3 and downstream regions through recombination occurring in intron 2 [4]. Additional class I loci and lineages have been described in both Atlantic salmon as well as in rainbow trout. The majority of reported salmonid MHC class I molecules are classified into a U-lineage consisting of both UBA as well as non-classical MHC molecules [5,6]. Two other described MHC class I-like lineages are ZE described by Miller et al. [5] and L described by Dijkstra et al. [7].
In all teleosts studied so far including salmonids the MHC class I and class II regions are unlinked [3,8]. Sequence data on the MHC class I region is available from zebrafish [9], fugu [10], medaka [11,12] and rainbow trout [6]. A general feature of these four MHC class I regions is a core region containing genes for the proteasome components (PSMBs) and the transporter for antigen processing (TAP2) being flanked by various numbers of MHC class I loci in addition to many other genes also residing in the human MHC region located on chromosome 6. Data from medaka and zebrafish indicate that other fish orthologs of the mammalian MHC-encoded genes are dispersed on several different chromosomes [13-16], similar to the paralogue MHC regions described on human chromosomes 1, 9 and 19 [17]. Salmonids are seen as partially tetraploid with a unique whole genome duplication occurring between 25 and 125 million years ago (mya) with remnants of tetraploidy visible also today [18-20]. Shiina et al. [6] sequenced two duplicated core MHC regions of rainbow trout. Based on sequence divergence they estimated the duplication event to have taken place approx. 60 mya, in agreement with the salmonid whole genome duplication theory. The classical or IA region contained the major expressed classical MHC class I UBA locus while the duplicated region denoted IB contained the four Onmy-UCA, -UDA, -UEA and -UFA class I loci. Based on expression and polymorphism data, Onmy-UCA, -UDA and -UEA were defined as non-classical loci and -UFA as a pseudogene due to an incapacitating mutation in exon 3 [6].
Data is rapidly emerging on associations between MHC and resistance to salmonid pathogens. In Atlantic salmon, UBA genotypes have been found to provide resistance towards Aeromonas salmonicida and Infectious Salmon Anaemia Virus [21,22]. Class IB, but not class IA was found associated with susceptibility towards infectious hematopoietic necrosis virus (IHNV) in Atlantic salmon and towards infectious pancreatic necrosis virus (IPNV) in rainbow trout [23,24].
Both trout and salmon are main aquaculture species and understanding their immune systems will improve our understanding of how these regions influence disease resistance and thus improve our breeding schemes for the trait. Atlantic salmon and rainbow trout are estimated to have split approx. 20 mya [25]. As Atlantic salmon is a major aquaculture species and displays some differences in response to pathogens when compared to rainbow trout [26], we ventured into sequencing of the two duplicated MHC class I regions of Atlantic salmon. Here we describe the gene organization of these two MHC class I regions comprising approx. 500 kb each and compare our results to data from other teleosts.
Results and discussion
The aim of this study was to characterize the gene organization and identify new genes potentially contributing to disease resistance in the two MHC class I regions of Atlantic salmon.
Characterization and sequencing of BAC clones
Sasa-UBA and TAP2 probes hybridized to 74 BAC clones, where 18 clones were positive for both probes. The 74 BAC clones were ordered into three contigs using restriction fragment analysis together with GRASP HindIII fingerprint information [27].
The two contigs that were positive for UBA, TAP2, PSMB9 and PSMB8 by southern hybridization, were tested for presence of a polymorphic dinucleotide repeat located in the 3'UTR of the UBA locus [3]. Only BAC clones from one of the two contigs gave PCR-products, thus this contig was defined as the IA region, and the other contig remained a candidate for the duplicated IB region. The BAC clones in the third contig hybridized to the UBA probe as well as a mixed UBA exon 2 probe. These clones also tested positive for a U-lineage ULA locus that has previously been found closely linked to UBA [5].
Three BACs were sequenced from the IA region. The BAC clones 92I04 and 714P22 indicated allelic variants based on variation in the UBA 3'UTR marker (data not shown) with 523M19 as a continuation of 714P22. From the duplicated IB region we chose 8I14, 424M17, 15L20 and 189M18 for sequencing. 30C23 was chosen as a candidate from the third contig and was extended 5 kb with the sequence of 868O01. The selected BAC clones were subcloned, sequenced, and assembled into continuous sequences. The Atlantic salmon IA region consisted of the BAC clones 30C23, 868O01, 92I04, 714P22 and 523M19 covering 502869 bp, while the IB region consisted of 8I14, 424M17, 15L20 and 189M18 totaling 522617 bp.
Gene organization of the Atlantic salmon MHC class I regions
We have adopted the nomenclature described by Shiina et al. [6] with IA covering the UBA locus region and IB for the duplicated region. Thus the genes identified in the regions will be named accordingly; the IA proteasome subunits are given an extension of a (PSMB9a) and the IB genes have an extension of b (PSMB9b). The previous symbol ABCB3 has been withdrawn for the transporter for antigen processing 2, so we have used the current symbol TAP2 [28].
The gene organization of the IA and IB MHC regions is shown in Fig. 1. A core region was identified in both regions which included MHC class I genes, together with the proteasome subunits genes PSMB8 (LMP7), PSMB10 (MECL-1), PSMB9-like (LMP2-δ), PSMB9 (LMP2) and TAP2. The gene order and orientation of the Atlantic salmon PSMBs and TAP2 was very similar to that found in rainbow trout and other teleost (Fig. 2). For the IA region, the main difference between Atlantic salmon and rainbow trout is that the rainbow trout PSMB8a gene is a pseudogene.
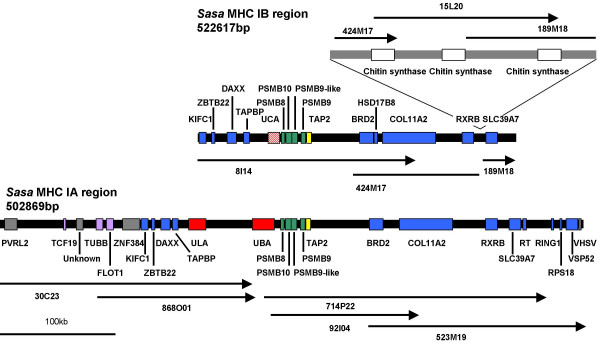
Figure 1.
Gene organization of Atlantic salmon MHC IA and IB regions. Upper panel is the IB region represented by the BACs 8I14, 424M17, 15L20 and 189M18. Lower panel is the IA region represented by the BACs 30C23, 868O01, 92I04, 714P22 and 523M19. Locus designation is based on sequence identity to matching ESTs and human nomenclature is used. The regions are drawn to scale.
Figure 2.
Comparison of the human, Atlantic salmon, rainbow trout, medaka, zebrafish and fugu MHC class I. Color code: red is MHC class I genes, orange is MHC class II genes, yellow is TAP genes, green is proteasome genes, blue is human extended MHC class II region genes, purple is human class I region genes, grey is non-human class I region genes, black is genes unique to the medaka HN1 strain [12]. Pseudogenes are striped. Human class III region genes are not shown. References are: zebrafish [9,16], fugu [10], medaka [11,12], rainbow trout [6] and human [1].
The IA region contained the major MHC class I Sasa-UBA locus and the recently reported Sasa-ULA locus residing approximately 50 kb upstream. The duplicated class IB region contained an MHC class I locus resembling the rainbow trout UCA locus, but although transcribed it was a pseudogene. No other MHC class I-like genes were detected in the two duplicated regions.
Outside the core region we found 12 Atlantic salmon orthologs of genes residing in the extended human MHC class II region. Alternative nomenclature for these genes is described in Table 1. The following genes were found in both the IA and IB regions; KIFC1, ZBTB22, DAXX, TAPBP, BRD2, COL11A2, RXRB and SLC39A7. Three orthologs found in the IA region only were RING1, RPS18 and VPS52. For other teleosts, the gene organization of the extended MHC class I region is partly known for zebrafish [9,16], fugu [10,15] and medaka [11,12]. The TAPBP, DAXX, ZBTB22 and KIFC1 genes are conserved in the same order in both fish and human (Fig. 2). As described in medaka we also found a gene for ZNF384 in the IA region, which is located on Chromosome 12 in human.
Table 1.
EST match to genes in the Atlantic salmon MHC IA and IB regions
| GENE | Abbreviation | Alias | EST/cDNA match IA | EST/cDNA match IB |
| Poliovirus receptor like2 | PVRL2 | CA342790 | n.i. | |
| Transcription factor 19 | TCF19 | SC1 | ? | n.i. |
| UNKNOWN | ? | DW569240 | n.i. | |
| Tubulin | TUBB | OK/SW-cl.56 | DW589685 | n.i. |
| Flotillin | FLOT1 | DY703577 | n.i. | |
| Zinc finger protein 384 | ZNF384 | CIZ | CB506768 | n.i. |
| Kinesin family member C1 | KIFC1 | KNSL2/HSET | DW541824 | BX313539 |
| Zinc finger and BTB domain | ZBTB22 | ZNF297/BING1 | BX911712 | BX911712 |
| Death-associated protein 6 | DAXX | DY736372 | GRASP cluster 76574 | |
| Tapasin | TAPBP | DY735080 | DW580568 | |
| MHC class I | ULA | DY699730 | n.i. | |
| MHC class I | UCA | n.i. | DW563256 | |
| MHC class I | UBA | UBA*0201/*0601 | n.i. | |
| Proteasome subunit, beta type, 8 | PSMB8 | LMP7 | DY733578 | AF184938 |
| Proteasome subunit, beta type, 10 | PSMB10 | MECL1 | DY740375 | DY734168 |
| Proteasome subunit, beta type, 9-like | PSMB9-like | LMP2-delta | DW574810 | CN442539 |
| Proteasome subunit, beta type, 9 | PSMB9 | LMP2 | AF184935 | AF184934 |
| Transporter 2, ATP-binding cassette, sub-family B | TAP2 | ABCB3 | DW540744 | Z83328 |
| Bromodomain containing 2 | BRD2 | RING3 | CA349460 | CA349460 |
| Hydroxysteroid (17-beta) dehydrogenase 8 | HSD17B8 | KE6, FABGL | n.i. | CK880913 |
| Collagen, type XI, alpha 2 | COL11A2 | ? | ? | |
| Retinoid × receptor, beta | RXRB | CK879311 | CK357003 | |
| Chitin synthase | n.i. | DW550858 | ||
| Solute carrier family 39 (zinc transporter), member 7 | SLC39A7 | KE4, FABGL | DW552689 | DW563433 |
| Reverse transcriptase | RT | DW561676 | n.i. | |
| Ring finger protein 1 | RING1 | RNF1 | DY732022 | n.i. |
| Ribosomal protein S18 | RPS18 | KE3 | DY729361 | n.i. |
| Vacuolar protein sorting 52 | VSP52 | SAC2 | DY740244 | n.i. |
| VHSV induced gene | VHSV | DW547400 | n.i. |
Mostly one match is listed per gene. Current abbreviation vs. older nomenclature is listed for each gene. All EST/cDNA sequences are from Genbank (? denotes unidentified EST, n.i. denotes not identified).
HSD17B8, which resides in between SLC39A7 and RING1 in the extended human class II region, was found in the IB region only and showed more than 81% identity towards counterparts in tilapia [Genbank:AAV74184], zebrafish [Genbank:CAK04961] and medaka [Genbank:BAB83840]. HSD17B8 has thus been deleted from the Atlantic salmon IA region as it is also present in other fish MHC class I regions (Fig. 2).
Three orthologs of genes located in the human class I region were identified in the IA region; TCF19, TUBB and FLOT1. Atlantic salmon tubulin is highly conserved and showed more than 94% identity towards mammalian counterparts. Another highly conserved gene is RPS18, which showed 98% identity towards mammalian sequences.
A gene that was predicted by DIGIT in the IA region had one EST match [Genbank:DW569240], but no homology to annotated proteins and is thus denoted unknown in Fig. 1. However, some sequence identity was found towards a protein in zebrafish located on chromosome 19 [Genbank:XP_001344849] as well as to a tetraodon nigroviridis protein [Genbank:CAF97811], which could indicate a molecule unique to teleosts.
In addition to the genes described above we identified genes for PVRL2, RT, VSHV-induced gene and a novel gene similar to a non-vertebrate chitin synthase protein that are not MHC linked in humans. The human PVRL2 is located on chromosome 19 (19q13.2-q13.4). A homologue of this gene is also found on zebrafish chromosome 19 [Genbank:XP_689425]. A 220 kb insertion was found in the IB region in between the RXRB and SLC39A7 genes containing three copies of a chitin synthase gene approx. 45 kb apart (Figs. 1 and 3). Chitin synthase is involved in the synthesis of chitin, which is a main structural component of the fungal cell wall. A similar protein has also been identified in zebrafish [Genbank:CAK04859]. No chitin synthase genes were present in the IA region nor are chitin synthase genes found in any other teleost MHC regions suggestive of a single insertion of this gene in the IB region with two subsequent duplications (Fig. 2).
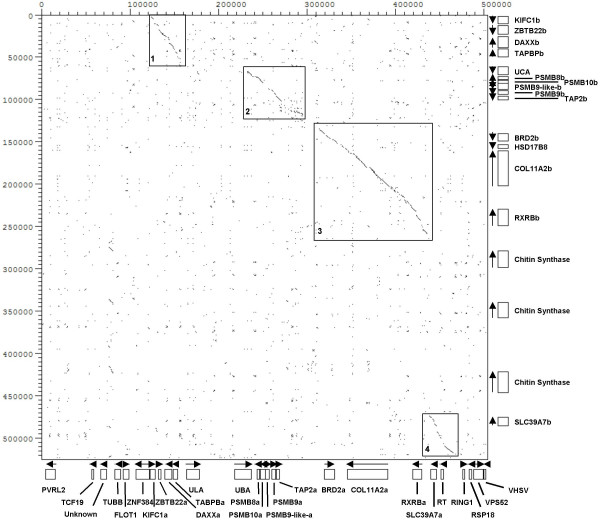
Figure 3.
Dot-plot analysis of the Atlantic salmon MHC IA and IB regions. The IA region is a collection of the BAC clones 30C23, 868O01, 714P22 and 523M19. The IB region is a summary of the BAC sequences 8I14, 424M17, 15L20 and 189M18. Position of genes is shown on the right (IB) and below (IA), and direction of transcription is marked with arrows. Genes found in both regions have an a or b extension.
Most genes in both regions are supported by matching cDNAs apart from TCF19 and COL11A2 where no match has been found so far (Table 1). Other open reading frames were also identified, but were associated with transposon related repetitive elements.
Comparison of the IA and IB regions
The two allelic BACs 92I04 and 714P22 had an overall sequences identity of 99.2% over 124574 bp, with similar exon intron organization for all genes. The major differences between the two allelic regions resided in the UBA α2 and α3 exons and in differences in number of repeats (data not shown). Dotplot analysis of 714P22 or 92I04 against themselves showed no extended regions of local similarity, with the exception of the TAP2 region which showed similarity due to a duplicated TAP2 exon 11 (data not shown).
A dot plot analysis of more than 500 kb of the IA and IB regions showed four regions with high sequence similarity consisting of subregion one containing genes for KIFC1 to TAPBP, subregion two ranging from PSMB8 to TAP2, subregion three covering BRD2 to RXRB and subregion four containing SLC39A7 (Fig. 3). The conserved regions in IA and IB have 82.5% identity over 136104 bp. In total, repeats constituted approximately 24% of the sequence in both regions, and 17% of the repeats were fish-specific DNA elements.
MHC Class I genes
Sasa-UBA
The promoter, leader and α1 exons of Sasa-UBA were identified in 30C23/868O01, while the remaining exons of Sasa-UBA were found in 92I04 and 714P22. The leader and α1 exons found in 30C23/868O01 were identical to the PCR amplified UBA*0201 allele [Genbank:AF504023] as well as to the leader and α1 exons of another salmon full-length cDNA [Genbank:DY698957]. Together with the α2 and α3 exons of 92I04 they collectively provide a bona fida UBA*0201 allele. The UBA α2 exon and downstream sequences of the two allelic BACs 92I04 and 714P22 have complete sequence identity to the Sasa-UBA*0201/*0301 and Sasa-UBA*0601 alleles respectively. UBA*0201 and UBA*0301 are prime examples of the recombination shown to occur within intron 2 of salmonid UBA alleles [4] showing complete sequence identity in the α2 and downstream regions, but highly divergent α1 exons. The predicted amino acid sequences of UBA, ULA and the two open reading frames of UCAψ were aligned for comparison of the MHC class I genes encoded in the two regions (Fig. 4).
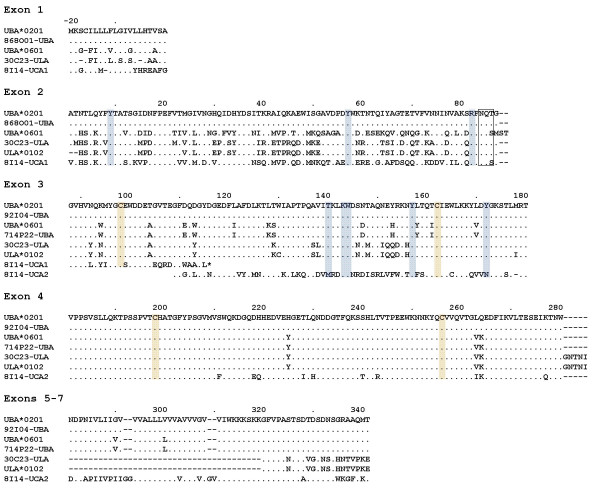
Figure 4.
Alignment of Atlantic salmon UBA, ULA and UCA alleles. Comparison of Atlantic salmon UBA, ULA and the two reading frames of UCA sequences found in the BACs. Also shown is UBA*0201 [Genbank:AAN75117] with exon 1 sequence from [Genbank:DY698957], UBA*0601 [Genbank:AAN75107] with exon 1 sequence from [Genbank: DW579225] and ULA*0102 [Genbank:AAZ73115]. Residues critical for binding of peptide anchors are in blue, and disulfide-bridge cysteins are in orange while the glycosylation site is boxed. Dots indicate identity while dashes indicate missing residues.
Analysis of the promoter sequence of UBA*0201 in 30C23/868O01 showed high similarity to a rainbow trout UBA*1501 promoter [29], both containing similar regulatory elements typical for MHC class I promoters such as an interferon stimulated response element (ISRE), W/S-box and enhancer B (enhB) (Fig. 5). The UBA*0201 promoter contains a potential site α element according to the core sequence (TGACGC) [30] while a sequence more resembling an X2-box has been found in rainbow trout (TGAGGCA). Both the site α and the homologous X2-box found in mammalian MHC class I and MHC class II promoters respectively, are involved in regulation of transcription and bind ATF/CREB family transcription factors [31]. A potential TATA-box was also identified in the promoter sequence of UBA*0201 [32]. A salmon UBA*0301 promoter published by Jorgensen et al. [33] had lower sequence identity to the UBA*0201 promoter, but both promoters are supported by complete identity to 5'UTR cDNA sequences of bona fida UBA alleles suggesting that Atlantic salmon UBA alleles have different promoters. The functional consequences of these differences are being investigated.
Figure 5.
Comparison of UBA promoter elements. Comparison of the SasaUBA*0201 promoter region with other reported UBA promoters in Atlantic salmon and rainbow trout; SasaUBA*0301 [Genbank:DQ243891], OnmyUBA*0501 [Genbank:AB162342] and OnmyUBA*1501 [Genbank:AF441856]. Promoter elements are boxed [31,32] and the genomic sequences with complete identity to 5'UTRs of UBA cDNA alleles are indicated in italics. Dots indicate identity while dashes indicate gaps.
The 30C23/868O01 and 92I04 BACs jointly have an intron sequence of 7 kb while in rainbow trout, the intron between the UBA α1 and α2 exons is 18 kb [6] suggesting we lack approximately 11 kb to have a continuous genomic sequence of the entire UBA region. PCR and cloning of the gap was performed multiple times, but despite successful PCR amplification no fragments covering the gap have been cloned suggestive of an unclonable region. The amplified products support an intron sequence of approx. 18 kb. Unfortunately no mRNA or cDNA is available from the BAC library fish, preventing verification of expressed UBA alleles in this animal. To verify the linkage between 92I04 and 30C23 fluorescent in situ hybridization was undertaken and showed that both BACs hybridized to the same region of one of the smallest chromosomes, potentially chromosome 27 (Fig. 6). The close linkage described by Miller et al. [5] between ULA and UBA also supported 30C23/868O01 being an extension of the IA region.
Figure 6.
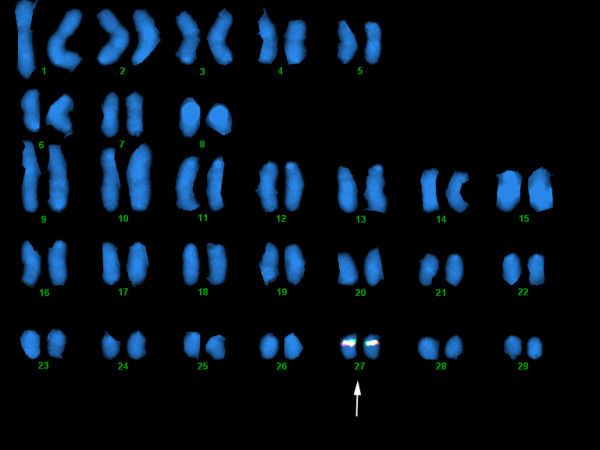
FISH mapping of BAC clones. Both 92I04 (in red) and 30C23 (in green) hybridized to one of the smallest chromosome, potentially chromosome 27 defining the Atlantic salmon IA region.
Sasa-ULA
The ULA locus residing approximately 50 kb upstream of the UBA locus matched a partial ULA*0102 sequence [Genbank:DQ091800] described by Miller et al. [5]. Another EST in the cGRASP database [34,35] provided us with a full-length match [Genbank:DY699730]. The exon encoding the transmembrane domain is missing, suggestive of a secreted MHC class I molecule (Fig. 4). Similar secretory class I molecules are also found for human class I molecules and the potential role of secretory HLA-G is currently being deciphered and holds promise for an interesting function. The 30C23 ULA gene has an α1 exon with highest sequence identity to UBA*0301 while α2 and downstream exons have highest identity to UBA*0801. No ESTs for ULA have been identified in rainbow trout, and a negative PCR-based survey for this gene in rainbow trout by Miller et al. [5] suggest this gene may be unique to Atlantic salmon.
Sasa-UCAψ
Only one MHC class I locus was identified in the four BACs representing the IB region. This locus found in 8I14 showed highest sequence identity to the Onmy UCA*0301 allele and was thus denoted Sasa-UCA. Multiple salmon ESTs with a polymorphic pattern resembling that of Onmy-UCA sequences were found in databases. However, both the 8I14 UCA ORF sequence as well as matching ESTs (Table 1), contained an internal stop codon in exon 3 making Sasa-UCA an expressed pseudogene. The exon intron organization of the UBA, ULA and UCAψ loci are quite similar apart from the enlarged first intron in ULA, the even larger second intron of UBA and the missing transmembrane exon of ULA (Fig. 7).
Figure 7.
Exon intron organization of the Atlantic salmon UBA, UCA ψ and ULA loci. The exons are boxed with the sizes in bp above sequence and intron sizes below.
Antigen presenting genes
Previously reported cDNAs for TAP2, which were assumed to reside in the IA and IB region and denoted TAP2B [Genbank:Z83329] and TAP2A [Genbank:Z83328] [36] respectively, indeed did match the TAP2 in the IA and IB BAC sequences. To avoid nomenclature confusion we hereby rename our IA UBA-linked TAP2 locus to TAP2a and the IB UCAψ linked TAP2 locus to TAP2b. We adapted a similar nomenclature for the reported rainbow trout TAP2 sequences, and suggest that TAP2 sequences without locus identification should be denoted TAP2 only (Fig. 8). Atlantic salmon IA and IB TAP2 sequences have more than 90% aa sequence identity and a similar identity to the rainbow trout IA and IB TAP2 sequences described by Shiina et al. [6].
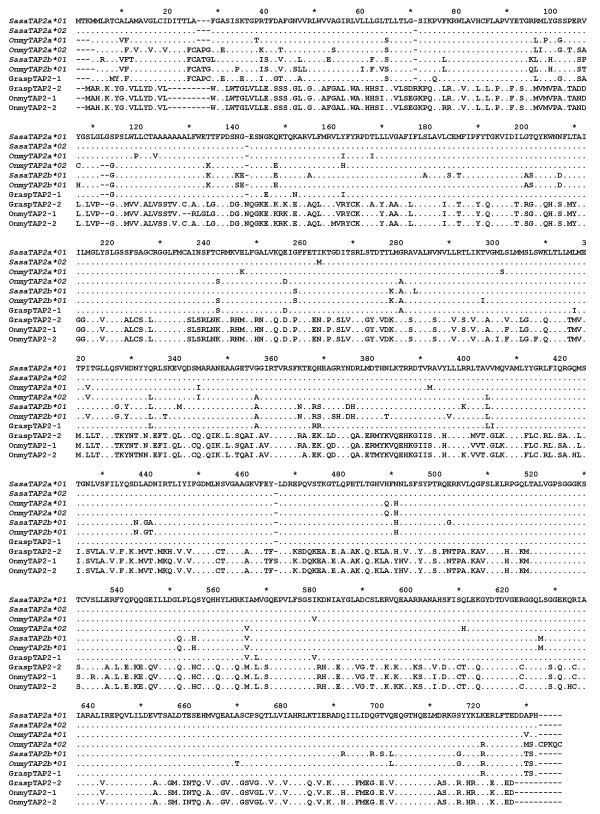
Figure 8.
TAP2 sequence alignment. A comparison of the amino acid sequences for TAP2s in Atlantic salmon and rainbow trout. SasaTAP2a*01 and SasaTAP2a*02 are from the allelic BACs 714P22 and 92I04 respectively while SasaTAP2b*01 is from the IB region BAC 8I14. GraspTAP2-1 represents an assembly of GRASP EST-clones [Genbank:DW551454, Genbank:DW563627, Genbank:DW563628, Genbank:DW580394 and Genbank:DY730930]), as does GraspTAP2-2 [Genbank:DW544931, Genbank:DW577600, Genbank:DW577601, Genbank:DW580644]. The TAP2 sequences from rainbow trout are OnmyTAP2a*01 [Genbank:AAD53035], OnmyTAP2-1 [Genbank:AAB62237], OnmyTAP2-2 [Genbank:AAD53034], OnmyTAP2a*02 [Genbank:BAD89546] and OnmyTAP2b*01 [Genbank:BAD89558].
Other TAP2 ESTs were also found in databases, which were difficult to define as TAP2a or TAP2b variants such as the GraspTAP2-1 in Fig. 8. Attempts to decipher locus origin including rainbow trout information shows that the TAP2a and TAP2b sequences described by Shiina et al. [6] resembles the TAP2b sequence identified in Atlantic salmon containing for instance the characteristic FCA motif at position 25 and the two aa deletion at position 110 (Fig. 8). A rainbow trout TAP2a (previously denoted TAP2B) [Genbank:AAD53035] sequence described by Hansen et al. [37], shown by in situ hybridization to reside in the IA region [8], resembles the Atlantic salmon IA TAP2a sequence and does not contain these motifs mentioned above. Thus, rainbow trout has a polymorphic TAP2a locus and the confusing sequence identities between the two TAP2 loci may suggest that these genes are exposed to recombination or gene conversion mechanisms. Locus designation of either salmon or trout TAP2 sequences therefore can not be performed on sequence alone, but must be verified by linkage mapping. Other more divergent Atlantic salmon TAP2 ESTs [Genbank:DW580644 and Genbank:DW577601] have approx. 50% sequence identity to all above described IA and IB TAP2 sequences (GraspTAP2-2 in Fig. 8), but has 94% sequence identity to a rainbow trout TAP2 variant described by Hansen et al. [37] (previously denoted TAP2A) [Genbank:AF115537]. If these sequences represent an additional TAP2 locus, i.e. a TAP2c locus, or are allelic variants of the TAP2a/b loci is currently unknown. Ancient lineages of divergent MHC class I, TAP1, TAP2 and LMP7 haplotypes have been described in Xenopus where the sequence identity between allelic TAP2s was less than 76% [38]. Similar ancient lineages of UBA and TAP2a may also exist in salmonids, where we were unfortunate enough to sequence allelic variants belonging to similar lineages.
All PSMBs genes in the salmon IA and IB regions were fully intact, as opposed to the IA region in rainbow trout where PSMB8 was a pseudogene, lacking the first three exons. The core region PSMB8, PSMB9, PSMB9-like, PSMB10 and TAP2 loci were found to be organized in a similar fashion in both regions and also displayed a high amount of sequence identity (Table 2). Some nucleotide differences were found between the allelic BACs 92I04 and 714P22 genes that resulted in amino acid changes; PSMB10a*01 and PSMB10a*02 had 2 aa differences located in the propeptide at position 12 (T vs. S) and 17 (E vs. K). The proteasome subunits therefore seem to be non-restrictive in providing peptides for UBA molecules as opposed to Xenopus where PSMB8 (LMP7) segregates in lineages with MHC class I, TAP1 and TAP2 [39].
Table 2.
Sequence comparison between antigen presenting genes in the MHC IA and IB regions in Atlantic salmon and rainbow trout
| GENE | % nt Sasa IA/IB | % aa Sasa IA/IB | % nt IA Sasa/Onmy | % aa IA Sasa/Onmy | % nt IB Sasa/Onmy | % aa IB Sasa/Onmy |
| PSMB8 | 95 | 95 | - | 98 | 96 | 98 |
| PSMB10 | 94 | 92 | 94 | 92 | 97 | 98 |
| PSMB9-like | 96 | 99 | 97 | 97 | 98 | 99 |
| PSMB9 | 97 | 100 | 96 | 98 | 98 | 99 |
| TAP2 | 92 | 90 | 94 | 93 | 96 | 95 |
The table shows comparison of % nucleotide (nt) and % amino acid (aa) identity between the genes found respectively in the Atlantic salmon IA vs IB region and in the IA and IB region of Atlantic salmon versus rainbow trout [6].
Tapasin (TAPBP) is a key member of MHC class I antigen-loading complexes, linking the class I molecule to the TAP. A full-length cDNA [Genbank:DQ451008] recently described by Jorgensen et al. [40] matched the TAPBPa locus in IA and another EST matched the TAPBPb locus in IB. As opposed to Atlantic salmon, the rainbow trout TAPBP in the IB region was described as a pseudogene both by Shiina et al. [6] as well as by Landis et al. [41] due to a deletion of the last 3 or 4 exons respectively. Landis et al. did however find transcripts of the first 4 exons. Different rainbow trout strains were used in the two studies, potentially accounting for the observed differences in deleted exons.
The core IA region in Atlantic salmon, ranging from the UBA α2 exon and downstream including TAP2, shows 87.6% sequence identity over 20289 bp to the same region in rainbow trout. A comparison of the salmon and rainbow trout IB region sequences from PSMB8 to TAP2 show 91.4 % identity over 20331 bp. This would be in accordance with the general perception that UBA lineages are ancient while the polymorphism of the duplicated IB region has evolved after the duplication event.
Salmonid MHC evolution and function
In the Atlantic salmon IB region we found only one MHC class I pseudo locus denoted UCAΨ, which is still being transcribed and shows a polymorphic pattern similar to that of rainbow trout UCA and UDA [42]. The rainbow trout IB region contained four MHC class I loci denoted UCA, UDA, UEA and UFAΨ [6]. As suggested by Shiina et al. [6] there has been a primordial salmonid MHC region containing three MHC class I loci (UCA-, UEA – and UBA-like) where UEA and UBA have been deleted from the Atlantic salmon IB region and UCA and UEA have been deleted from the Atlantic salmon IA region. The trout IB UDA locus is a duplication of UCA that occured in trout only. Once the extended trout IA region is sequenced we will see if the UBA to ULA duplication occurred in both species and if the UCA and UEA homologues have been retained in this region of trout.
The salmonid whole-genome duplication was estimated to have occurred between 25 and 125 mya [18] while the study of Shiina et al. [6] estimate the duplication to have occurred 60 mya based on sequence identity of the MHC class I regions. Evolving from a tetraploid to a diploid state includes not only accumulation of mutations, but also random rearrangements and recombinations as exemplified by the multiple deletions that have occurred in the Atlantic salmon IA and IB regions. With a sequence identity between the Atlantic salmon IA and IB regions of approximately 82 percent, recombination may even be occurring between the two duplicates today. Salmonids are also known for using recombination within the second intron of the UBA locus to generate "new" alleles using exons already tested for functionality [2-4]. As recombination was not observed in 800 siblings [43] the recombination frequency is probably low. One way of reducing the risk of recombination between duplicates may be insertions such as the 220 kb insertion with three copies of chitin synthase genes in the IB region.
Another example of differences between Atlantic salmon and rainbow trout is the chromosomal location of the IA region. In both species, the IB region is located on chromosome 14 [6,8] (data not shown for salmon), while the IA region is located on chromosome 18 in rainbow trout and on one of the smaller chromosomes, potentially chromosome 27, in Atlantic salmon (Fig. 6) [8,44]. In Atlantic salmon, the IA and IB regions map to linkage groups 15 and 3 respectively [45], while in rainbow trout they map to linkage groups 16 and 3 [20] supporting the differences. Atlantic salmon and rainbow trout have diploid chromosome numbers ranging from 58 to 64 [46,47]. Most likely, different centric fusions have occurred in the diploidization processes of Atlantic salmon and rainbow trout leading to IA residing on one arm of a metacentric chromosome in rainbow trout while on an acrocentric chromosome in Atlantic salmon.
Why the Atlantic salmon IB region has undergone more deletions than trout is unknown, but it has functional consequences. The IB region has been identified as a major QTL for resistance towards IHNV in Atlantic salmon and IPNV in rainbow trout where the polymorphic UCA, UDA or UEA loci were suggested as prime candidates for the observed effects [23,24]. As our study indicates that the Atlantic salmon IB region only contains a UCA pseudolocus, there must either be other genes flanking our BACs which contribute to resistance or there could be haplotype variation in number of class I loci between Norwegian and Canadian Atlantic salmon.
The IA region was not found associated with resistance towards IHNV in Atlantic salmon nor IPNV in rainbow trout. Atlantic salmon UBA genotypes have however been shown to provide resistance towards the viral pathogen causing Infectious Salmon Anaemia (ISA) [21,22]. An ongoing study will identify the role of Atlantic salmon IA and IB in providing resistance towards IPNV, enabling us to decipher between differences in pathogens versus genetic organization. Apart from the potential TAP2a and UBA lineages, limited polymorphism in PSMBs and other linked loci suggest that the observed linkage between Sasa-UBA and disease resistance in Norwegian Atlantic salmon [21,22] is caused by Sasa-UBA alleles or genotypes and not closely linked genes. However, the PSMBs and TAP2 molecules residing in the IB region might still influence the overall peptide repertoire available for presentation by UBA alleles. Due to the pseudo status of Sasa-UCA, the PSMBs and TAP2B in the IB region will most likely devolve over time.
Conclusion
We have characterized the gene organization of more than 500 kb of the two duplicated MHC regions in Atlantic salmon. Although Atlantic salmon and rainbow trout are closely related, the gene organization of their IB region has undergone extensive gene rearrangements. The Atlantic salmon had only one identified MHC class I UCA pseudo gene in the IB region while this region in trout contained the four MHC class I loci UCA, UDA, UEA and UFAψ . The Atlantic salmon IB region also contained a 220 kb insertion as compared to the IA region potentially limiting recombination between the two regions. The large difference in gene content and most likely function of salmon and trout class IB regions clearly argues that sequencing of salmon will not necessarily provide information relevant for trout and vice versa.
Methods
Screening of the BAC library
An Atlantic salmon BAC library (CHORI214) was obtained from CHORI [48]. The library consisted of approximately 300.000 recombinant clones with an average insert size of 190 kb, representing 18-fold genome coverage [49]. All filters in the library were hybridized with two probes containing conserved exons of the Sasa-UBA and Sasa-TAP2 loci or using over go's as described by Han et al. [50]. Both probes were PCR amplified (primers listed in Table 3) from cDNA clones and gel purified, and then radioactively labeled with the Rediprime Random Labelling Kit (Amersham), including spermine precipitation of labeled DNA. Filter hybridizations were performed as described by CHORI. Probed BAC library filters were visualized using a Typhoon Phospho Image Scanner (Amersham).
Table 3.
Primers used for probes, southern hybridization and PCR
| Sequence (5'-3') | Position | Comments | |
| UBA 540F | |||
| UBA TmR | TCTTCTTCCAAATGACGACCCC | Exon 5 | cDNA amplification Sasa-UBA |
| TAP2.550F | GCGGGACACCGTCAGGGCAGT | Exon 5 | cDNA amplification Sasa-TAP2a/b |
| TAP2.850R | CGGCCCCACCAGAGCAGTCAG | Exon 8 | cDNA amplification Sasa-TAP2a/b |
| PSMB8.50F | GACTTCGTGGGCAGATTCTT | Exon 2 | cDNA amplification Sasa-PSMB8 |
| PSMB8.350R | TCAGCAGCACTACCAGACAT | Exon 3 | cDNA amplification Sasa-PSMB8 |
| PSMB9b4.590F | GAGGACTGCCAACAGTTTGTT | Exon 4 | cDNA clone (2B4) amplification Sasa-PSMB9 |
| PSMB9b4.876R | ATATCATGCTGGCACAATGTT | 3' UTR | cDNA clone (2B4) amplification Sasa-PSMB9 |
| UBAlead1.F | CTGGGAATAGGCCTTCTACAT | Exon 1 | Sasa-UBA*0201,*0801,*0901 specific |
| UBAlead2.F | AGCCCTACATTCTTCATCTGC | Exon 1 | Sasa-UBA*0301,*1001 specific |
| UBA02.285R | GTTTGATTGAAGCGGGATTTC | Exon 2 | Sasa-UBA*0201 specific |
| UBA03.281R | TCTGATTGAAGCGCTGCTTGG | Exon 2 | Sasa-UBA*0301 specific |
| UBA08.280R | CTGATTGAAGCGCTGCTTGAC | Exon 2 | Sasa-UBA*0801 specfic |
| UBA01.286R | GTTTGATTGAAACGTGTCTG | Exon 2 | Sasa-UBA*0901 specific |
| UBA10.272R | TTGAAAGATTGTGAGGTGCT | Exon 2 | Sasa-UBA*1001 specific |
| ssal.phc-004.039-2 | CTCTCCAGTGACCTGCCACGCTACAGGTTTCTATCCCAGTGGAGTCATGGTGTTCTGGCAGAAAGATGGA | MHC Non-classical | |
| ssal.rgb-532.282-r | GCCAATGTGCTCATGGCTATAGTCATCATTGTGTCTGTAGTCTTGATACTCACTGTCCTATTCAAGTATT | MHC Non-classical | |
| ssal.rgb-505.101 | GCTACCATCGGCTCAAAGAGGAACTCTTTGAGGACTGATTAGGAATCACACAGCTGTCAGAGAGAGAGAC | TAP2 | |
| ssal.rgb-516.329 | TCAACCTGTACCACATGCAGGAGGACGGCTGGATAAAGGTGTGTAAGGAGGACGTTTCAGAGCTGATC | PSMB8 | |
| ssal.rgb-523.298 | GTTGATGGACAGGAAGGGGAGCTACTACAAACTAAGAGAGAGACTGTTCACGGAAGACGACACGTCACAT | TAP2 | |
| ssal.rgb-550.202 | TTCATTGGCTATGAGTCGAGATGGTTCCAGCGGAGGCGTGGCCTACCTTGTCACTATTGATGAAAAGGGT | PSMB9 | |
| 868O01_F1 | GGCTTGCGGAGTAGAACACTTGAAAAAGAA | GAP-primers | |
| 868O01_F2 | GCAACCAATACACTGCAGTATTTCTACACGGCCACTTCTGGTATAGATAAC | GAP-primers | |
| 868O01_F3 | TCAATCAAACTGGAGGTGAGTAGAGACAGA | GAP-primers | |
| 714P22_R1 | CAGCGGAAACGCAACAAACACAAGAATAACTTACTAACAAATTAGAATCA | GAP-primers | |
| 714P22_R2 | AGCATACAACCATGCCGACAACCATC | GAP-primers | |
| 92I04_R1 | AACATTTCAAGGTGGTGAAACTATTCACAT | GAP-primers | |
| 92I04_R2 | AATTAGTTGTTGTTCACTATGTTAATTAGT | GAP-primers |
Characterization of BACs
MHC class I positive BAC clones were ordered into contigs using restriction fragment analysis together with GRASP HindIII fingerprint information [27]. Southern blot analysis of NotI (NEB) and NruI (NEB) digested BAC DNA was performed to characterize the clones. The digested DNA was electrophoresed for 16 h and then transferred to Hybond membranes (Amersham). The MHC class I and TAP2 probes described earlier together with probes for PSMB8 and PSMB9 (unpublished data), were used for hybridization to the southern blots. A mixed probe containing 5 UBA leader to alpha1 exons amplified from the alleles UBA*0201, *0301, *0801, *0901 and *1001 cDNAs (primers listed in Table 3) was also used. Hybridization with end-labeled Sp6 and T7 oligos were used to orient end-fragments of BAC inserts.
Blots were prehybridized at 65°C for 30 minutes in hybridization buffer (5× SSC, 5× Denhardt's solution and 1% SDS) with. This was followed by replacement with fresh, preheated (65°C) hybridization buffer and the addition of the radio labeled probes. Hybridization was allowed to proceed overnight. Following hybridization, the membranes were washed three times with 20 ml of 2× SSC, 0.1% SDS at 65°C for 30 min. Prehybridization, hybridization and wash conditions were the same for all probes. To further characterize the BACs we used primers spanning a polymorphic (CA)n repeat located in the 3'UTR of the UBA locus [3] both on individual BAC DNA as well as on genomic DNA from the animal the library was made from. PCR on genomic DNA from the BAC library animal was performed with GAP-primers (Table 3) with Herculase Enhanced polymerase (Stratagene) according to protocol. Amplified products were ligated into the three different vectors using TOPO-TA Cloning Kit with pCR2.1-TOPO (Invitrogen), TOPO-XL PCR Cloning Kit with pCR-XL-TOPO (Invitrogen) and CloneSmart LCKan Blunt Cloning Kit with pSMART LCKan (Lucigen Corporation) and subsequently transformed into XL-10 Gold cells (Stratagene).
Sequencing strategy
The selected BACs were subjected to a shotgun sequencing approach. Briefly, BAC DNA was purified by Nucleobond BAC Maxi Kit (BD Biosciences ClonTech). Isolated BAC DNA was nebulized (Invitrogen) (20PSI/15s) and fragments in size range 2–4 kb were purified from agarose gel and blunt-ended with Mung Bean Nuclease, T4 DNA polymerase and Klenow (NEB). Fragments were ligated into a pUC19 vector (Fermentas) cut with SmaI, and transformed into XL10-Gold (Stratagene). The sequencing templates were prepared by standard alkaline lysis, and sequencing reactions were run on an ABI3100 or ABI3700 DNA sequencer (Applied Biosystems). Bases were called using Phred [51,52]. High quality sequencing reads were assembled using Phrap, and viewed and edited using Consed [53]. Autofinish [54] was used for closing gaps by designing gap-closing primers with subsequent direct sequencing on BAC DNA or PCR amplification and PCR product sequencing. The BAC sequences were submitted to Genbank and given the following accession numbers: 8I14 (188042 bp, [Genbank:EF427379]), 15L20 (145959 bp [Genbank:EF427378]), 30C23 (218410 bp, [Genbank:EF427381]), 92I04 (128344 bp, [Genbank:EF427384]), 189M18 (170847 bp, [Genbank:EF427377]), 424M17 (163489 bp, [Genbank:EF427382]), 523M19 (188299 bp [Genbank:EF427383]), 714P22 (244579bp, [Genbank:EF210363]), and 868O01 (140046 bp, [Genbank:EF441211]).
Bioinformatics
DIGIT [55] and GENSCAN [56] were used to predict novel genes and to identify open reading frames. Dotter [57] was used to compare the BAC sequence to itself as well as to other BACs and to identify duplicated regions. Vista was used for sequence comparisons [58]. Blast searches identified possible functions of predicted genes [59]. Sim4 [60] and Spidey [61] were used to adjust exon and intron boundaries aligning EST/cDNA sequences to the BAC sequences. Repeatmasker [62] were used to identify repeats. Multiple sequence alignments of the assumed or verified expressed exons were done using ClustalX [63] followed by manual inspection.
In situ hybridization and karyotyping
Blood was cultured from the Norwegian strain of Atlantic salmon using standard methods [64]. DNA was isolated from three BAC clones (8I14, 30C23 and 92I04) from the CHORI library using the Qiagen Midi-Preparation kit. These clones were labeled with either Spectrum Orange (Vysis, Inc.) using a nick translation kit (Vysis, Inc.) or digoxigenin according to manufacturers instructions. Human placental DNA (0.2 μg) and Cot-1 DNA (1 μg, prepared from Atlantic salmon) were added to the probe mixture for blocking. Hybridizations were carried out at 37°C overnight and post-hybridization washes were as recommended by the manufacturer (Vysis, Inc.) with minor modifications [65]. Secondary antibodies to Spectrum Orange (Molecular Probes) were used to amplify the signal in some cases. Slides were counter-stained with 4'6'-diamidino-2-phenylindole (DAPI) at a concentration of 125 ng DAPI in 1 ml antifade solution. Images were captured with a Sensys camera and analyzed with Cytovision Genus (Applied Imaging, Inc.) software.
Authors' contributions
MFL: Performed sequencing, sequence data analysis, annotations and drafted the manuscript.
HH: Performed library screening, sequencing and BAC restriction mapping.
UG contributed to project design, library screening, sequencing and revision of manuscript.
HGB, MBS, GAC and LR: Performed library screening and sequencing.
RBP: Labelled BAC clones and performed fluorescence in situ hybridization on salmon chromosomes.
KMM: Identified 30C23 containing ULA, performed some sequence annotation
WSD contributed to the project planning and directions.
BFK contributed to the planning, design, direction and analysis.
All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
The present study was supported by The National Programme for Research in Functional Genomics in Norway (FUGE), The Research Council of Norway, and by NSERC, Genome Canada, Genome BC, and the Province of British Columbia.
Contributor Information
Morten F Lukacs, Email: morten.lukacs@veths.no.
Håvard Harstad, Email: havard.harstad@veths.no.
Unni Grimholt, Email: unni.grimholt@veths.no.
Marianne Beetz-Sargent, Email: marianbs@uvic.ca.
Glenn A Cooper, Email: gac@uvic.ca.
Linda Reid, Email: linda.l.reid@gmail.com.
Hege G Bakke, Email: hege.bakke@veths.no.
Ruth B Phillips, Email: phllipsr@vancouver.wsu.edu.
Kristina M Miller, Email: millerk@pac.dfo-mpo.gc.ca.
William S Davidson, Email: wdavidso@sfu.ca.
Ben F Koop, Email: bkoop@uvic.ca.
References
- Shiina T, Inoko H, Kulski JK. An update of the HLA genomic region, locus information and disease associations: 2004. Tissue Antigens. 2004;64:631–649. doi: 10.1111/j.1399-0039.2004.00327.x. [DOI] [PubMed] [Google Scholar]
- Aoyagi K, Dijkstra JM, Xia C, Denda I, Ototake M, Hashimoto K, Nakanishi T. Classical MHC class I genes composed of highly divergent sequence lineages share a single locus in rainbow trout (Oncorhynchus mykiss) J Immunol. 2002;168:260–273. doi: 10.4049/jimmunol.168.1.260. [DOI] [PubMed] [Google Scholar]
- Grimholt U, Drablos F, Jorgensen SM, Hoyheim B, Stet RJM. The Major Histocompatibility Class I locus in Atlantic salmon (Salmo salar L.): Polymorphism, linkage analysis and protein modelling. Immunogenetics. 2002;54:570–581. doi: 10.1007/s00251-002-0499-8. [DOI] [PubMed] [Google Scholar]
- Shum BP, Guethlein L, Flodin LR, Adkison MA, Hedrick RP, Nehring RB, Stet RJ, Secombes C, Parham P. Modes of salmonid MHC class I and II evolution differ from the primate paradigm. J Immunol. 2001;166:3297–3308. doi: 10.4049/jimmunol.166.5.3297. [DOI] [PubMed] [Google Scholar]
- Miller KM, Li S, Ming TJ, Kaukinen KH, Schulze AD. The salmonid MHC class I: more ancient loci uncovered. Immunogenetics. 2006;58:571–589. doi: 10.1007/s00251-006-0125-2. [DOI] [PubMed] [Google Scholar]
- Shiina T, Dijkstra JM, Shimizu S, Watanabe A, Yanagiya K, Kiryu I, Fujiwara A, Nishida-Umehara C, Kaba Y, Hirono I, Yoshiura Y, Aoki T, Inoko H, Kulski JK, Ototake M. Interchromosomal duplication of major histocompatibility complex class I regions in rainbow trout (Oncorhynchus mykiss), a species with a presumably recent tetraploid ancestry. Immunogenetics. 2005;56:878–893. doi: 10.1007/s00251-004-0755-1. [DOI] [PubMed] [Google Scholar]
- Dijkstra JM, Katagiri T, Hosomichi K, Yanagiya K, Inoko H, Ototake M, Aoki T, Hashimoto K, Shiina T. A third broad lineage of major histocompatibility complex (MHC) class I in teleost fish; MHC class II linkage and processed genes. Immunogenetics. 2007;59:305–321. doi: 10.1007/s00251-007-0198-6. [DOI] [PubMed] [Google Scholar]
- Phillips RB, Zimmerman A, Noakes MA, Palti Y, Morasch MR, Eiben L, Ristow SS, Thorgaard GH, Hansen JD. Physical and genetic mapping of the rainbow trout major histocompatibility regions: evidence for duplication of the class I region. Immunogenetics. 2003;55:561–569. doi: 10.1007/s00251-003-0615-4. [DOI] [PubMed] [Google Scholar]
- Michalova V, Murray BW, Sultmann H, Klein J. A contig map of the Mhc class I genomic region in the zebrafish reveals ancient synteny. J Immunol. 2000;164:5296–5305. doi: 10.4049/jimmunol.164.10.5296. [DOI] [PubMed] [Google Scholar]
- Clark MS, Shaw L, Kelly A, Snell P, Elgar G. Characterization of the MHC class I region of the Japanese pufferfish (Fugu rubripes). Immunogenetics. 2001;52:174–185. doi: 10.1007/s002510000285. [DOI] [PubMed] [Google Scholar]
- Matsuo M, Asakawa S, Shimizu N, Kimura H, Nonaka M. Nucleotide sequence of the MHC class I genomic region of a teleost, the medaka (Oryzias latipes). Immunogenetics. 2002;53:930–940. doi: 10.1007/s00251-001-0427-3. [DOI] [PubMed] [Google Scholar]
- Tsukamoto K, Hayashi S, Matsuo M, Nonaka M, Kondo M, Shima MI, Asakawa S, Shimizu N, Nonaka M. Unprecedented intraspecific diversity of the MHC class I region of a teleost medaka, Oryzias latipes. Immunogenetics. 2005;57:420–431. doi: 10.1007/s00251-005-0009-x. [DOI] [PubMed] [Google Scholar]
- Bingulac-Popovic J, Figueroa F, Sato A, Talbot WS, Johnson SL, Gates M, Postlethwait JH, Klein J. Mapping of mhc class I and class II regions to different linkage groups in the zebrafish, Danio rerio. Immunogenetics. 1997;46:129–134. doi: 10.1007/s002510050251. [DOI] [PubMed] [Google Scholar]
- Naruse K, Fukamachi S, Mitani H, Kondo M, Matsuoka T, Kondo S, Hanamura N, Morita Y, Hasegawa K, Nishigaki R, Shimada A, Wada H, Kusakabe T, Suzuki N, Kinoshita M, Kanamori A, Terado T, Kimura H, Nonaka M, Shima A. A detailed linkage map of medaka, Oryzias latipes: comparative genomics and genome evolution. Genetics. 2000;154:1773–1784. doi: 10.1093/genetics/154.4.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook JG, Russel R, Umrania Y, Edwards YJK, Campbell RD, Elgar G, Clark MS. Fugu orthologues of human major histocompatibility complex genes: a genome survey. Immunogenetics. 2002;54:367–380. doi: 10.1007/s00251-002-0478-0. [DOI] [PubMed] [Google Scholar]
- Sambrook JG, Figueroa F, Beck S. A genome-wide survey of Major Histocompatibility Complex (MHC) genes and their paralogues in zebrafish. BMC Genomics. 2005;6:152–162. doi: 10.1186/1471-2164-6-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M. Genome dynamics of the major histocompatibility complex: insights from genome paralogy. Immunogenetics. 1999;50:134–145. doi: 10.1007/s002510050589. [DOI] [PubMed] [Google Scholar]
- Allendorf FW, Thorgaard GH. Tetraploidy and the evolution of salmonid fishes. In: Turner BJ, editor. Evolutionary Genetics of Fishes. New York, Plenum; 1984. pp. 1–53. [Google Scholar]
- Arratia G. Basal teleosts and teleostean phylogeny. Palaeo Ichthyologica. 1997;7:5–168. [Google Scholar]
- Phillips RB, Nichols KM, Dekoning JJ, Morasch MR, Keatley KA, Rexroad C, III, Gahr SA, Danzmann RG, Drew RE, Thorgaard GH. Assignment of rainbow trout linkage groups to specific chromosomes. Genetics. 2006;174:1661–1670. doi: 10.1534/genetics.105.055269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjoglum S, Larsen S, Bakke HG, Grimholt U. How specific MHC class I and class II combinations affect disease resistance against infectious salmon anaemia in Atlantic salmon (Salmo salar) Fish Shellfish Immunol. 2006;21:431–441. doi: 10.1016/j.fsi.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Grimholt U, Larsen S, Nordmo R, Midtlyng P, Kjoeglum S, Storset A, Saebo S, Stet RJ. MHC polymorphism and disease resistance in Atlantic salmon (Salmo salar); facing pathogens with single expressed major histocompatibility class I and class II loci. Immunogenetics. 2003;55:210–219. doi: 10.1007/s00251-003-0567-8. [DOI] [PubMed] [Google Scholar]
- Miller KM, Winton JR, Schulze AD, Purcell MK, Ming TJ. Major histocompatibility complex loci are associated with susceptibility of Atlantic salmon to infectious hematopoietic necrosis virus. Environ Biol Fishes. 2004;69:307–316. doi: 10.1023/B:EBFI.0000022874.48341.0f. [DOI] [Google Scholar]
- Ozaki A, Sakamoto T, Khoo S, Nakamura K, Coimbra MR, Akutsu T, Okamoto N. Quantitative trait loci (QTLs) associated with resistance/susceptibility to infectious pancreatic necrosis virus (IPNV) in rainbow trout (Oncorhynchus mykiss) Mol Genet Genomics. 2001;265:23–31. doi: 10.1007/s004380000392. [DOI] [PubMed] [Google Scholar]
- McKay SJ, Devlin RH, Smith MJ. Phylogeny of Pacific salmon and trout based on growth hormone type-2 and mitocondrial NADH dehydrogenase subunit 3 DNA sequences. Can J Fish Aquatic Sci. 1996;53:1165–1168. doi: 10.1139/cjfas-53-5-1165. [DOI] [Google Scholar]
- Kibenge FS, Kibenge MJ, Groman D, McGeachy S. In vivo correlates of infectious salmon anemia virus pathogenesis in fish. J Gen Virol. 2006;87:2645–2652. doi: 10.1099/vir.0.81719-0. [DOI] [PubMed] [Google Scholar]
- Ng SH, Artieri CG, Bosdet IE, Chiu R, Danzmann RG, Davidson WS, Ferguson MM, Fjell CD, Hoyheim B, Jones SJ, de Jong PJ, Koop BF, Krzywinski MI, Lubieniecki K, Marra MA, Mitchell LA, Mathewson C, Osoegawa K, Parisotto SE, Phillips RB, Rise ML, von Schalburg KR, Schein JE, Shin H, Siddiqui A, Thorsen J, Wye N, Yang G, Zhu B. A physical map of the genome of Atlantic salmon, Salmo salar. Genomics. 2005;86:396–404. doi: 10.1016/j.ygeno.2005.06.001. [DOI] [PubMed] [Google Scholar]
- HUGO Gene Nomenclature Committee. Internet. 2007. http://www.genenames.org
- Dijkstra JM, Yoshiura Y, Kiryu I, Aoyagi K, Kollner B, Fischer U, Nakanishi T, Ototake M. The promoter of the classical MHC class I locus in rainbow trout (Oncorhynchus mykiss) Fish & Shellfish Immunology. 2003;14:177–185. doi: 10.1006/fsim.2002.0431. [DOI] [PubMed] [Google Scholar]
- Gobin SJ, Peijnenburg A, Keijsers V, van den Elsen PJ. Site alpha is crucial for two routes of IFN gamma-induced MHC class I transactivation: the ISRE-mediated route and a novel pathway involving CIITA. Immunity. 1997;6:601–611. doi: 10.1016/S1074-7613(00)80348-9. [DOI] [PubMed] [Google Scholar]
- van den Elsen PJ, Peijnenburg A, Van Eggermond MC, Gobin SJ. Shared regulatory elements in the promoters of MHC class I and class II genes. Immunol Today. 1998;19:308–312. doi: 10.1016/S0167-5699(98)01287-0. [DOI] [PubMed] [Google Scholar]
- Patikoglou GA, Kim JL, Sun L, Yang SH, Kodadek T, Burley SK. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 1999;13:3217–3230. doi: 10.1101/gad.13.24.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen SM, Lyng-Syvertsen B, Lukacs M, Grimholt U, Gjoen T. Expression of MHC class I pathway genes in response to infectious salmon anaemia virus in Atlantic salmon (Salmo salar L.) cells. Fish Shellfish Immunol. 2006;21:548–560. doi: 10.1016/j.fsi.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Consortium for Genomics Research on All Salmon. Internet. 2007. http://web.uvic.ca/cbr/grasp/
- Rise ML, von Schalburg KR, Brown GD, Mawer MA, Devlin RH, Kuipers N, Busby M, Beetz-Sargent M, Alberto R, Gibbs AR, Hunt P, Shukin R, Zeznik JA, Nelson C, Jones SR, Smailus DE, Jones SJ, Schein JE, Marra MA, Butterfield YS, Stott JM, Ng SH, Davidson WS, Koop BF. Development and application of a salmonid EST database and cDNA microarray: data mining and interspecific hybridization characteristics. Genome Res. 2004;14:478–490. doi: 10.1101/gr.1687304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimholt U. Transport-associated proteins in Atlantic salmon (Salmo salar) Immunogenetics. 1997;46:213–221. doi: 10.1007/s002510050264. [DOI] [PubMed] [Google Scholar]
- Hansen JD, Strassburger P, Thorgaard GH, Young WP, Du PL. Expression, linkage, and polymorphism of MHC-related genes in rainbow trout, Oncorhynchus mykiss. J Immunol. 1999;163:774–786. [PubMed] [Google Scholar]
- Ohta Y, Powis SJ, Lohr RL, Nonaka M, Pasquier LD, Flajnik MF. Two highly divergent ancient allelic lineages of the transporter associated with antigen processing (TAP) gene in Xenopus: further evidence for co-evolution among MHC class I region genes. Eur J Immunol. 2003;33:3017–3027. doi: 10.1002/eji.200324207. [DOI] [PubMed] [Google Scholar]
- Ohta Y, Haliniewski DE, Hansen J, Flajnik MF. Isolation of transporter associated with antigen processing genes, TAP1 and TAP2, from the horned shark Heterodontus francisci. Immunogenetics. 1999;49:981–986. doi: 10.1007/s002510050582. [DOI] [PubMed] [Google Scholar]
- Jorgensen SM, Grimholt U, Gjoen T. Cloning and expression analysis of an Atlantic salmon (Salmo salar L.) tapasin gene. Dev Comp Immunol. 2007;31:708–719. doi: 10.1016/j.dci.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Landis ED, Palti Y, Dekoning J, Drew R, Phillips RB, Hansen JD. Identification and regulatory analysis of rainbow trout tapasin and tapasin-related genes. Immunogenetics. 2006;58:56–69. doi: 10.1007/s00251-005-0070-5. [DOI] [PubMed] [Google Scholar]
- Dijkstra JM, Kiryu I, Yoshiura Y, Kumanovics A, Kohara M, Hayashi N, Ototake M. Polymorphism of two very similar MHC class Ib loci in rainbow trout (Oncorhynchus mykiss) Immunogenetics. 2006;58:152–167. doi: 10.1007/s00251-006-0086-5. [DOI] [PubMed] [Google Scholar]
- Kiryu I, Dijkstra JM, Sarder RI, Fujiwara A, Yoshiura Y, Ototake M. New MHC class Ia domain lineages in rainbow trout (Oncorhynchus mykiss) which are shared with other fish species. Fish & Shellfish Immunology. 2005:243–254. doi: 10.1016/j.fsi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Fujiwara A, Kiryu I, Dijkstra JM, Yoshiura Y, Nishida-Umehara C, Ototake M. Chromosome mapping of MHC class I in rainbow trout (Oncorhynchus mykiss) Fish & Shellfish Immunology. 2003;14:171–175. doi: 10.1006/fsim.2002.0426. [DOI] [PubMed] [Google Scholar]
- Salmon Genome Project. Internet. 2007. http://www.salmongenome.no/cgi-bin/sgp.cgi
- Phillips R, Rab P. Chromosome evolution in the Salmonidae (Pisces): an update. Biol Rev Camb Philos Soc. 2001;76:1–25. doi: 10.1017/S1464793100005613. [DOI] [PubMed] [Google Scholar]
- Hartley SE. The chromosomes of salmonid fishes. Biol Rev Camb Philos Soc. 1987;62:197–214. [Google Scholar]
- Children's Hospital Oakland Research Institute (CHORI) Internet. 2007. http://bacpac.chori.org/
- Thorsen J, Zhu B, Frengen E, Osoegawa K, de Jong PJ, Koop BF, Davidson WS, Hoyheim B. A highly redundant BAC library of Atlantic salmon (Salmo salar): an important tool for salmon projects. BMC Genomics. 2005;6:50–60. doi: 10.1186/1471-2164-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han CS, Sutherland RD, Jewett PB, Campbell ML, Meincke LJ, Tesmer JG, Mundt MO, Fawcett JJ, Kim UJ, Deaven LL, Doggett NA. Construction of a BAC contig map of chromosome 16q by two-dimensional overgo hybridization. Genome Res. 2000;10:714–721. doi: 10.1101/gr.10.5.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Gordon D, Desmarais C, Green P. Automated finishing with autofinish. Genome Res. 2001;11:614–625. doi: 10.1101/gr.171401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digit Web Server. Internet. 2007. http://digit.gsc.riken.go.jp/
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Durbin R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene. 1995;167:GC1–10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelan SJ, Church DM, Ostell JM. Spidey: a tool for mRNA-to-genomic alignments. Genome Res. 2001;11:1952–1957. doi: 10.1101/gr.195301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repeatmasker. Internet. 2007. http://www.repeatmasker.org/
- Thomson JD, Higgins DG, Gibson TJ. ClustalW: improving the sensitivity of progressive multiple sequence alignments through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed KM, Phillips RB. Molecular cytogenetic analysis of the double-CMA3 chromosome of lake trout, Salvelinus namaycush. Cytogenet Cell Genet. 1995;70:104–107. doi: 10.1159/000134002. [DOI] [PubMed] [Google Scholar]
- Phillips RB, Reed KM. Localization of repetitive DNAs to zebrafish (Danio rerio) chromosomes by fluorescence in situ hybridization (FISH) Chromosome Res. 2000;8:27–35. doi: 10.1023/A:1009271017998. [DOI] [PubMed] [Google Scholar]