Abstract
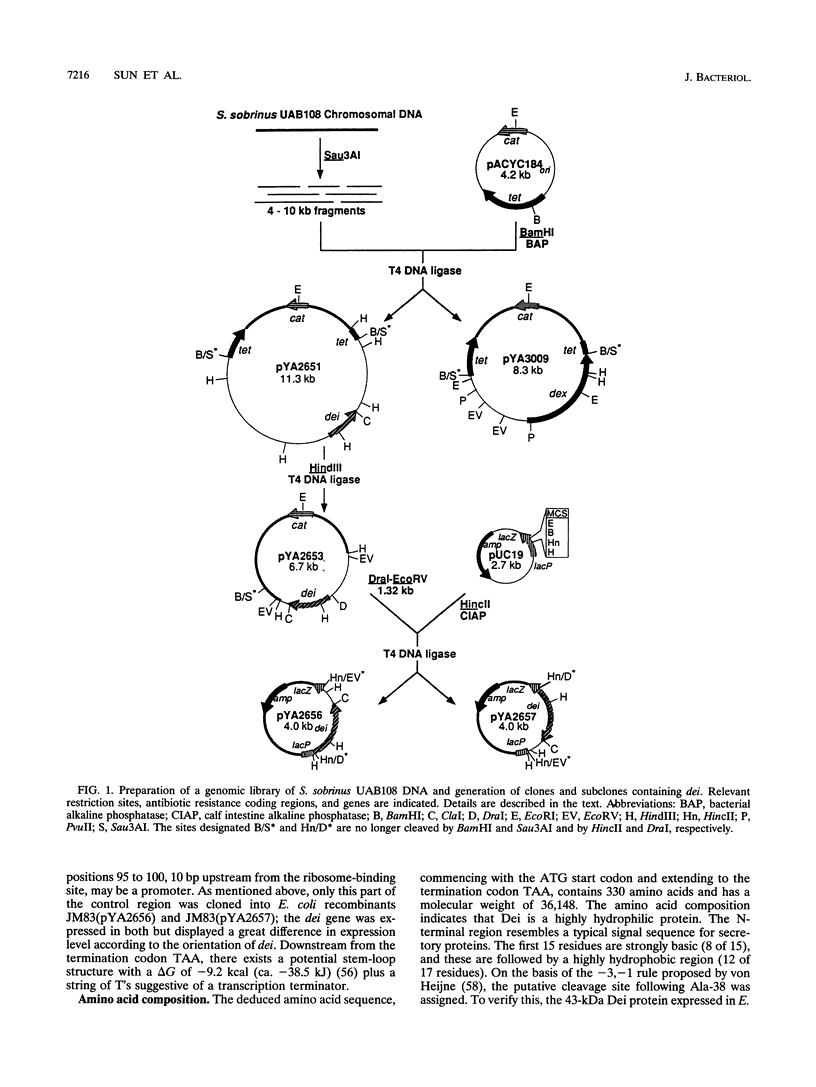
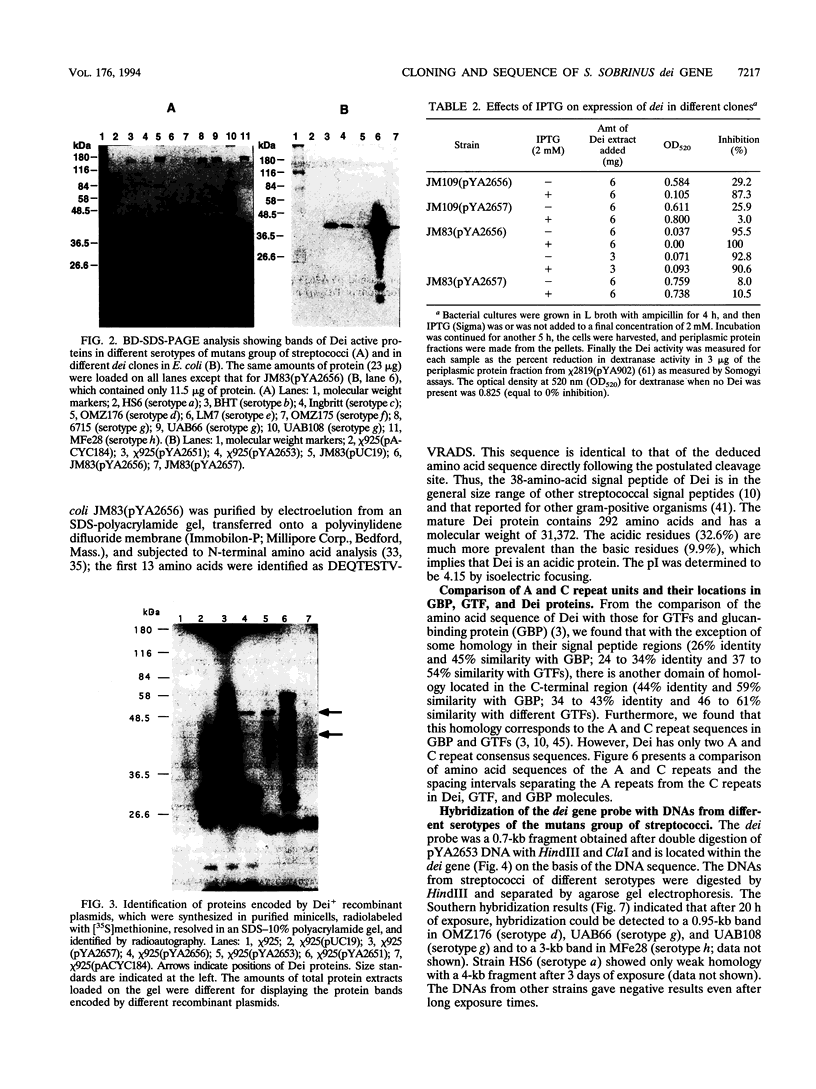
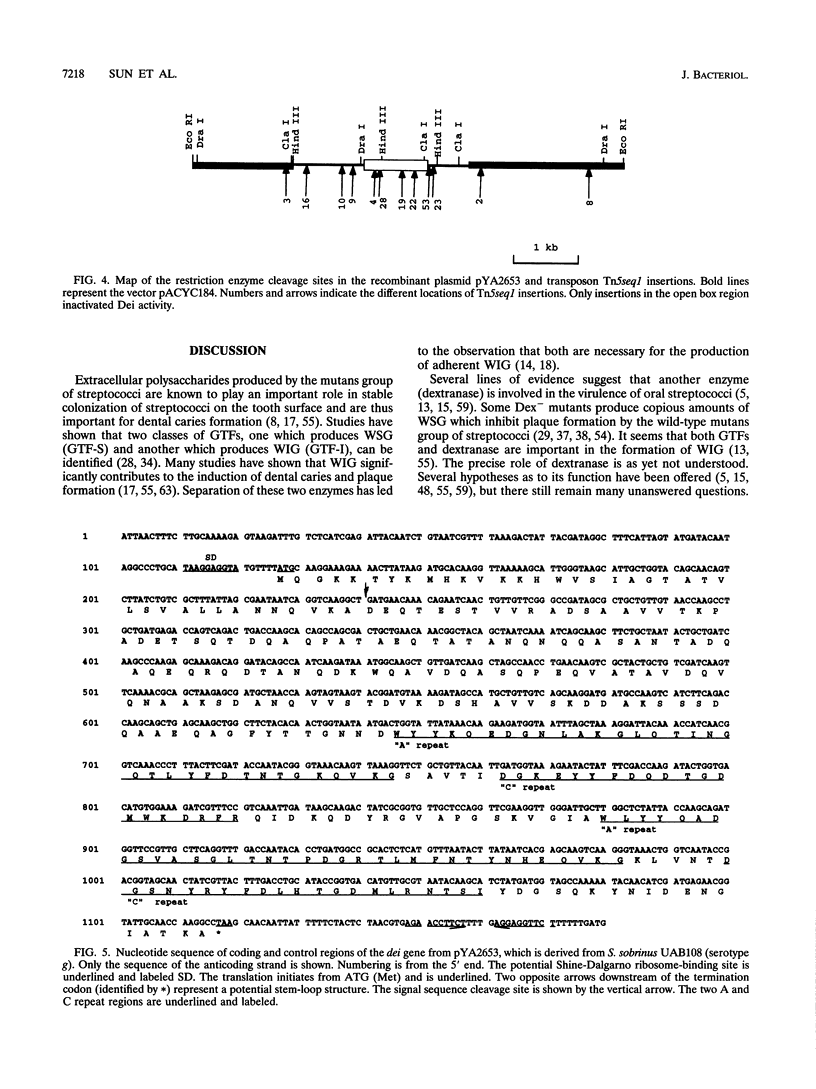
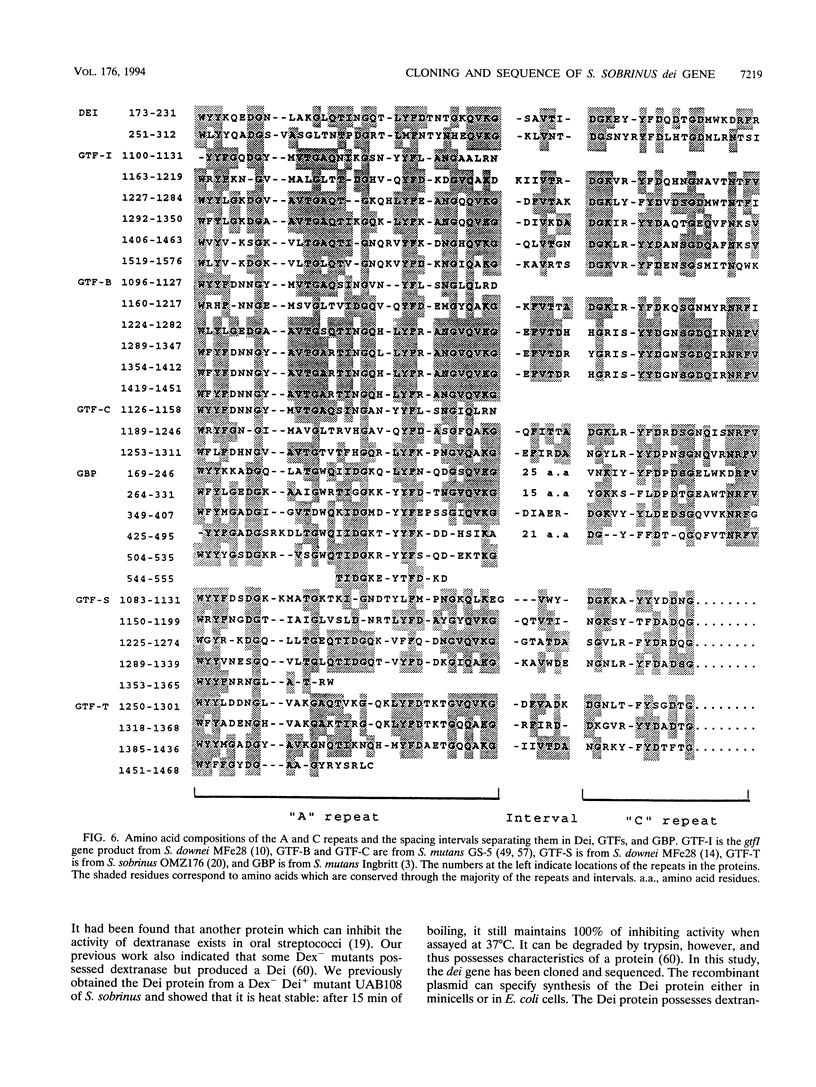
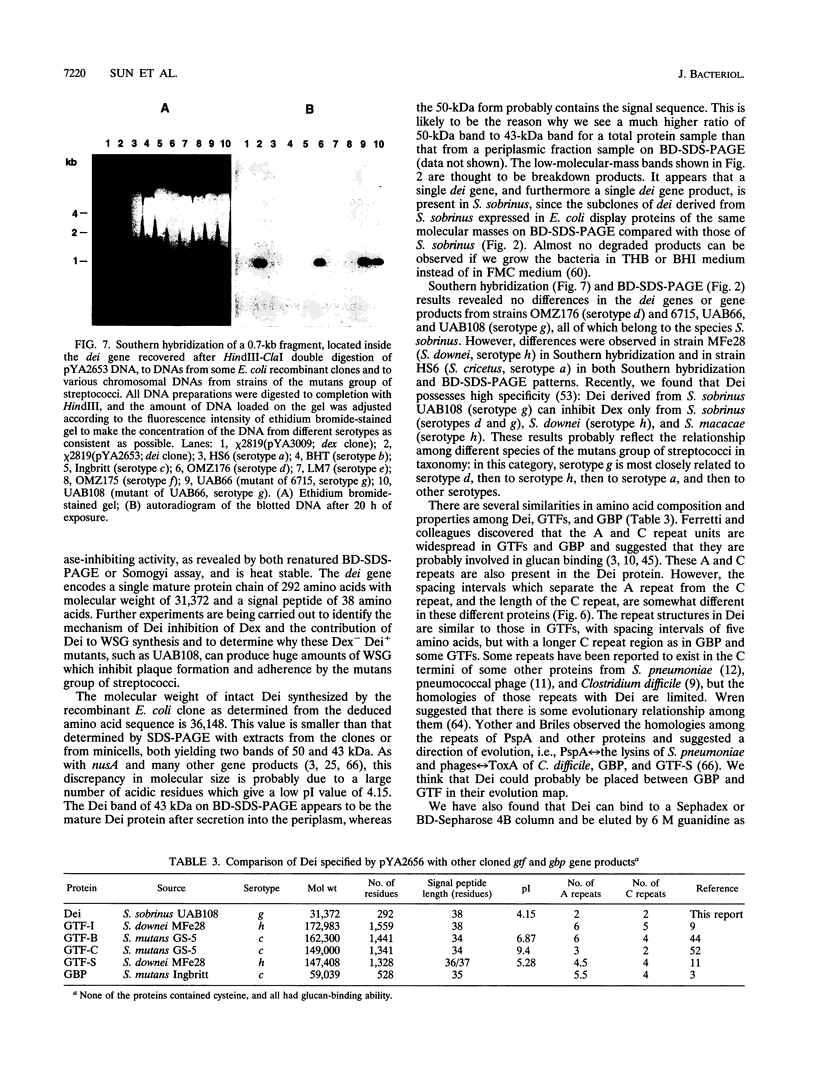
Some dextranase-deficient (Dex-) mutants of Streptococcus sobrinus UAB66 (serotype g) synthesize a substance which inhibits dextranase activity (S.-Y. Wanda, A. Camilli, H. M. Murchison, and R. Curtiss III, J. Bacteriol. 176:7206-7212, 1994). This substance produced by the Dex- mutant UAB108 was designated dextranase inhibitor (Dei) and identified as a protein. The Dei gene (dei) from UAB108 has been cloned into pACYC184 to yield pYA2651, which was then used to generate several subclones (pYA2653 to pYA2657). The DNA sequence of dei was determined by using Tn5seq1 transposon mutagenesis of pYA2653. The open reading frame of dei is 990 bp long. It encodes a signal peptide of 38 amino acids and a mature Dei protein of 292 amino acids with a molecular weight of 31,372. The deduced amino acid sequence of Dei shows various degrees of similarity with glucosyltransferases and glucan-binding protein and contains A and C repeating units probably involved in glucan binding. Southern hybridization results showed that the dei probe from UAB108 hybridized to the same-size fragment in S. sobrinus (serotype d and g) DNA, to a different-size fragment in S. downei (serotype h) and S. cricetus (serotype a), and not at all to DNAs from other mutans group of streptococci.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abo H., Matsumura T., Kodama T., Ohta H., Fukui K., Kato K., Kagawa H. Peptide sequences for sucrose splitting and glucan binding within Streptococcus sobrinus glucosyltransferase (water-insoluble glucan synthetase). J Bacteriol. 1991 Feb;173(3):989–996. doi: 10.1128/jb.173.3.989-996.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki H., Shiroza T., Hayakawa M., Sato S., Kuramitsu H. K. Cloning of a Streptococcus mutans glucosyltransferase gene coding for insoluble glucan synthesis. Infect Immun. 1986 Sep;53(3):587–594. doi: 10.1128/iai.53.3.587-594.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banas J. A., Russell R. R., Ferretti J. J. Sequence analysis of the gene for the glucan-binding protein of Streptococcus mutans Ingbritt. Infect Immun. 1990 Mar;58(3):667–673. doi: 10.1128/iai.58.3.667-673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J. F., Barrett T. A., Curtiss R., 3rd Purification and partial characterization of the multicomponent dextranase complex of Streptococcus sobrinus and cloning of the dextranase gene. Infect Immun. 1987 Mar;55(3):792–802. doi: 10.1128/iai.55.3.792-802.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J. F., Curtiss R., 3rd Renaturation of dextranase activity from culture supernatant fluids of Streptococcus sobrinus after sodium dodecylsulfate polyacrylamide gel electrophoresis. Anal Biochem. 1986 Nov 1;158(2):365–370. doi: 10.1016/0003-2697(86)90562-2. [DOI] [PubMed] [Google Scholar]
- Clark-Curtiss J. E., Curtiss R., 3rd Analysis of recombinant DNA using Escherichia coli minicells. Methods Enzymol. 1983;101:347–362. doi: 10.1016/0076-6879(83)01026-5. [DOI] [PubMed] [Google Scholar]
- Curtiss R., 3rd Genetic analysis of Streptococcus mutans virulence. Curr Top Microbiol Immunol. 1985;118:253–277. doi: 10.1007/978-3-642-70586-1_14. [DOI] [PubMed] [Google Scholar]
- Dove C. H., Wang S. Z., Price S. B., Phelps C. J., Lyerly D. M., Wilkins T. D., Johnson J. L. Molecular characterization of the Clostridium difficile toxin A gene. Infect Immun. 1990 Feb;58(2):480–488. doi: 10.1128/iai.58.2.480-488.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti J. J., Gilpin M. L., Russell R. R. Nucleotide sequence of a glucosyltransferase gene from Streptococcus sobrinus MFe28. J Bacteriol. 1987 Sep;169(9):4271–4278. doi: 10.1128/jb.169.9.4271-4278.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García E., García J. L., García P., Arrarás A., Sánchez-Puelles J. M., López R. Molecular evolution of lytic enzymes of Streptococcus pneumoniae and its bacteriophages. Proc Natl Acad Sci U S A. 1988 Feb;85(3):914–918. doi: 10.1073/pnas.85.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García P., García J. L., García E., López R. Nucleotide sequence and expression of the pneumococcal autolysin gene from its own promoter in Escherichia coli. Gene. 1986;43(3):265–272. doi: 10.1016/0378-1119(86)90215-5. [DOI] [PubMed] [Google Scholar]
- Germaine G. R., Harlander S. K., Leung W. L., Schachtele C. F. Streptococcus mutans dextransucrase: functioning of primer dextran and endogenous dextranase in water-soluble and water-insoluble glucan synthesis. Infect Immun. 1977 May;16(2):637–648. doi: 10.1128/iai.16.2.637-648.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore K. S., Russell R. R., Ferretti J. J. Analysis of the Streptococcus downei gtfS gene, which specifies a glucosyltransferase that synthesizes soluble glucans. Infect Immun. 1990 Aug;58(8):2452–2458. doi: 10.1128/iai.58.8.2452-2458.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guggenheim B., Burckhardt J. J. Isolation and properties of a dextranase from streptococcus mutans OMZ 176. Helv Odontol Acta. 1974 Oct;18(2):101–113. [PubMed] [Google Scholar]
- Gulig P. A., Curtiss R., 3rd Cloning and transposon insertion mutagenesis of virulence genes of the 100-kilobase plasmid of Salmonella typhimurium. Infect Immun. 1988 Dec;56(12):3262–3271. doi: 10.1128/iai.56.12.3262-3271.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S., Koga T., Ooshima T. Virulence factors of Streptococcus mutans and dental caries prevention. J Dent Res. 1984 Mar;63(3):407–411. doi: 10.1177/00220345840630031001. [DOI] [PubMed] [Google Scholar]
- Hamada S., Slade H. D. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol Rev. 1980 Jun;44(2):331–384. doi: 10.1128/mr.44.2.331-384.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelik R. M., McCabe M. M. An endodextranase inhibitor from batch cultures of streptococcus mutans. Biochem Biophys Res Commun. 1982 Jun 15;106(3):875–880. doi: 10.1016/0006-291x(82)91792-2. [DOI] [PubMed] [Google Scholar]
- Hanada N., Isobe Y., Aizawa Y., Katayama T., Sato S., Inoue M. Nucleotide sequence analysis of the gtfT gene from Streptococcus sobrinus OMZ176. Infect Immun. 1993 May;61(5):2096–2103. doi: 10.1128/iai.61.5.2096-2103.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J. B., Abiko Y., Curtiss R., 3rd Characterization of the Streptococcus mutans plasmid pva318 cloned into Escherichia coli. Infect Immun. 1981 Mar;31(3):1034–1043. doi: 10.1128/iai.31.3.1034-1043.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer G. L., Harayama S. Mutants in transmission of chemotactic signals from two independent receptors of E. coli. Cell. 1979 Mar;16(3):617–625. doi: 10.1016/0092-8674(79)90035-7. [DOI] [PubMed] [Google Scholar]
- Holt R. G., Abiko Y., Saito S., Smorawinska M., Hansen J. B., Curtiss R., 3rd Streptococcus mutans genes that code for extracellular proteins in Escherichia coli K-12. Infect Immun. 1982 Oct;38(1):147–156. doi: 10.1128/iai.38.1.147-156.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M., Koga T., Sato S., Hamada S. Synthesis of adherent insoluble glucan by the concerted action of the two glucosyltransferase components of Streptococcus mutans. FEBS Lett. 1982 Jun 21;143(1):101–104. doi: 10.1016/0014-5793(82)80282-2. [DOI] [PubMed] [Google Scholar]
- Ishii S., Ihara M., Maekawa T., Nakamura Y., Uchida H., Imamoto F. The nucleotide sequence of the cloned nusA gene and its flanking region of Escherichia coli. Nucleic Acids Res. 1984 Apr 11;12(7):3333–3342. doi: 10.1093/nar/12.7.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs W. R., Barrett J. F., Clark-Curtiss J. E., Curtiss R., 3rd In vivo repackaging of recombinant cosmid molecules for analyses of Salmonella typhimurium, Streptococcus mutans, and mycobacterial genomic libraries. Infect Immun. 1986 Apr;52(1):101–109. doi: 10.1128/iai.52.1.101-109.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramitsu H. K. Characterization of extracellular glucosyltransferase activity of Steptococcus mutans. Infect Immun. 1975 Oct;12(4):738–749. doi: 10.1128/iai.12.4.738-749.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramitsu H. K., Wondrack L. Insoluble glucan synthesis by Streptococcus mutans serotype c strains. Infect Immun. 1983 Nov;42(2):763–770. doi: 10.1128/iai.42.2.763-770.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Larrimore S., Murchison H., Shiota T., Michalek S. M., Curtiss R., 3rd In vitro and in vivo complementation of Streptococcus mutans mutants defective in adherence. Infect Immun. 1983 Nov;42(2):558–566. doi: 10.1128/iai.42.2.558-566.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawman P., Bleiweis A. S. Molecular cloning of the extracellular endodextranase of Streptococcus salivarius. J Bacteriol. 1991 Dec;173(23):7423–7428. doi: 10.1128/jb.173.23.7423-7428.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoil C., Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Moos M., Jr, Nguyen N. Y., Liu T. Y. Reproducible high yield sequencing of proteins electrophoretically separated and transferred to an inert support. J Biol Chem. 1988 May 5;263(13):6005–6008. [PubMed] [Google Scholar]
- Mooser G., Wong C. Isolation of a glucan-binding domain of glucosyltransferase (1,6-alpha-glucan synthase) from Streptococcus sobrinus. Infect Immun. 1988 Apr;56(4):880–884. doi: 10.1128/iai.56.4.880-884.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison H., Larrimore S., Curtiss R., 3rd In vitro inhibition of adherence of Streptococcus mutans strains by nonadherent mutants of S. mutans 6715. Infect Immun. 1985 Dec;50(3):826–832. doi: 10.1128/iai.50.3.826-832.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison H., Larrimore S., Curtiss R., 3rd Isolation and characterization of Streptococcus mutans mutants defective in adherence and aggregation. Infect Immun. 1981 Dec;34(3):1044–1055. doi: 10.1128/iai.34.3.1044-1055.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag D. K., Huang H. V., Berg D. E. Bidirectional chain-termination nucleotide sequencing: transposon Tn5seq1 as a mobile source of primer sites. Gene. 1988 Apr 15;64(1):135–145. doi: 10.1016/0378-1119(88)90487-8. [DOI] [PubMed] [Google Scholar]
- Oliver D. Protein secretion in Escherichia coli. Annu Rev Microbiol. 1985;39:615–648. doi: 10.1146/annurev.mi.39.100185.003151. [DOI] [PubMed] [Google Scholar]
- Poulsen S., Larson R. H., Senning R. S. Effect of dextranase on plaque formation and caries development in the rat. Scand J Dent Res. 1978 Jul;86(4):231–236. doi: 10.1111/j.1600-0722.1978.tb00623.x. [DOI] [PubMed] [Google Scholar]
- Russell R. R., Ferretti J. J. Nucleotide sequence of the dextran glucosidase (dexB) gene of Streptococcus mutans. J Gen Microbiol. 1990 May;136(5):803–810. doi: 10.1099/00221287-136-5-803. [DOI] [PubMed] [Google Scholar]
- Russell R. R. Glycosyltransferases of Streptococcus mutans strain Ingbritt. Microbios. 1978;23(93-94):136–146. [PubMed] [Google Scholar]
- Russell R. R., Shiroza T., Kuramitsu H. K., Ferretti J. J. Homology of glucosyltransferase gene and protein sequences from Streptococcus sobrinus and Streptococcus mutans. J Dent Res. 1988 Mar;67(3):543–547. doi: 10.1177/00220345880670030401. [DOI] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtele C. F., Staat R. H., Harlander S. K. Dextranases from oral bacteria: inhibition of water-insoluble glucan production and adherence to smooth surfaces by Streptococcus mutans. Infect Immun. 1975 Aug;12(2):309–317. doi: 10.1128/iai.12.2.309-317.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiroza T., Ueda S., Kuramitsu H. K. Sequence analysis of the gtfB gene from Streptococcus mutans. J Bacteriol. 1987 Sep;169(9):4263–4270. doi: 10.1128/jb.169.9.4263-4270.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Takada K., Shiota T., Curtiss R., 3rd, Michalek S. M. Inhibition of plaque and caries formation by a glucan produced by Streptococcus mutans mutant UAB108. Infect Immun. 1985 Dec;50(3):833–843. doi: 10.1128/iai.50.3.833-843.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda S., Shiroza T., Kuramitsu H. K. Sequence analysis of the gtfC gene from Streptococcus mutans GS-5. Gene. 1988 Sep 15;69(1):101–109. doi: 10.1016/0378-1119(88)90382-4. [DOI] [PubMed] [Google Scholar]
- Walker G. J., Pulkownik A., Morrey-Jones J. G. Metabolism of the polysaccharides of human dental plaque: release of dextranase in batch cultures of Streptococcus mutans. J Gen Microbiol. 1981 Nov;127(1):201–208. doi: 10.1099/00221287-127-1-201. [DOI] [PubMed] [Google Scholar]
- Wanda S. Y., Camilli A., Murchison H. M., Curtiss R., 3rd Overproduction of a dextranase inhibitor by Streptococcus sobrinus mutants. J Bacteriol. 1994 Dec;176(23):7206–7212. doi: 10.1128/jb.176.23.7206-7212.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanda S. Y., Curtiss R., 3rd Purification and characterization of Streptococcus sobrinus dextranase produced in recombinant Escherichia coli and sequence analysis of the dextranase gene. J Bacteriol. 1994 Jul;176(13):3839–3850. doi: 10.1128/jb.176.13.3839-3850.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K., Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969 Aug 25;244(16):4406–4412. [PubMed] [Google Scholar]
- Wenham D. G., Davies R. M., Cole J. A. Insoluble glucan synthesis by mutansucrase as a determinant of the cariogenicity of Streptococcus mutans. J Gen Microbiol. 1981 Dec;127(2):407–415. doi: 10.1099/00221287-127-2-407. [DOI] [PubMed] [Google Scholar]
- Wren B. W. A family of clostridial and streptococcal ligand-binding proteins with conserved C-terminal repeat sequences. Mol Microbiol. 1991 Apr;5(4):797–803. doi: 10.1111/j.1365-2958.1991.tb00752.x. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yother J., Briles D. E. Structural properties and evolutionary relationships of PspA, a surface protein of Streptococcus pneumoniae, as revealed by sequence analysis. J Bacteriol. 1992 Jan;174(2):601–609. doi: 10.1128/jb.174.2.601-609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983 Jun 1;133(1):17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]