Abstract
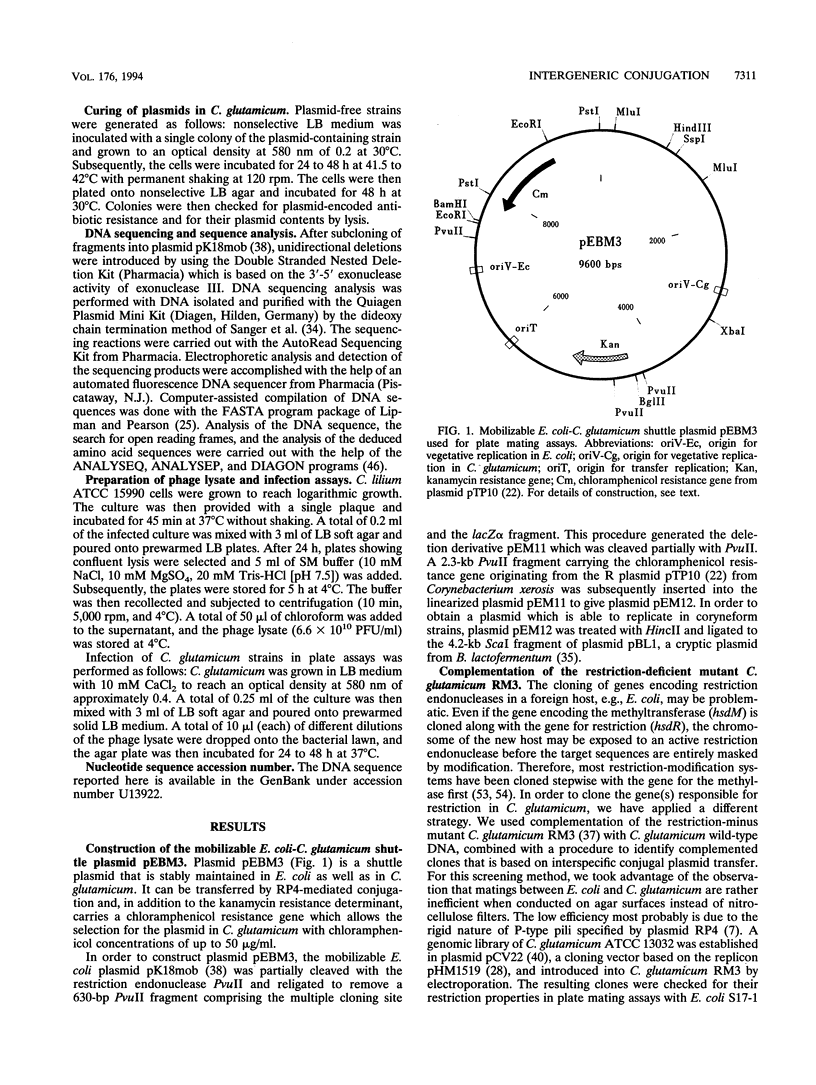
RP4-mediated transfer of mobilizable plasmids in intergeneric conjugation of Escherichia coli donors with Corynebacterium glutamicum ATCC 13032 is severely affected by a restriction system in the recipient that can be inactivated by a variety of exogenous stress factors. In this study a rapid test procedure based on intergeneric conjugal plasmid transfer that permitted the distinction between restriction-negative and restriction-positive C. glutamicum clones was developed. By using this procedure, clones of the restriction-deficient mutant strain C. glutamicum RM3 harboring a plasmid library of the wild-type chromosome were checked for their restriction properties. A complemented clone with a restriction-positive phenotype was isolated and found to contain a plasmid with a 7-kb insertion originating from the wild-type chromosome. This plasmid, termed pRES806, is able to complement the restriction-deficient phenotype of different C. glutamicum mutants. Sequence analysis revealed the presence of two open reading frames (orf1 and orf2) on the complementing DNA fragment. The region comprising orf1 and orf2 displayed a strikingly low G+C content and was present exclusively in C. glutamicum strains. Gene disruption experiments with the wild type proved that orf1 is essential for complementation, but inactivation of orf2 also resulted in a small but significant increase in fertility. These results were confirmed by infection assays with the bacteriophage CL31 from Corynebacterium lilium ATCC 15990.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- Altenbuchner J., Cullum J. DNA amplification and an unstable arginine gene in Streptomyces lividans 66. Mol Gen Genet. 1984;195(1-2):134–138. doi: 10.1007/BF00332735. [DOI] [PubMed] [Google Scholar]
- Bailey C. R., Winstanley D. J. Inhibition of restriction in Streptomyces clavuligerus by heat treatment. J Gen Microbiol. 1986 Oct;132(10):2945–2947. doi: 10.1099/00221287-132-10-2945. [DOI] [PubMed] [Google Scholar]
- Barany F., Danzitz M., Zebala J., Mayer A. Cloning and sequencing of genes encoding the TthHB8I restriction and modification enzymes: comparison with the isoschizomeric TaqI enzymes. Gene. 1992 Mar 1;112(1):3–12. doi: 10.1016/0378-1119(92)90296-2. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W. DNA restriction and modification mechanisms in bacteria. Annu Rev Microbiol. 1971;25:153–176. doi: 10.1146/annurev.mi.25.100171.001101. [DOI] [PubMed] [Google Scholar]
- Bradley D. E., Taylor D. E., Cohen D. R. Specification of surface mating systems among conjugative drug resistance plasmids in Escherichia coli K-12. J Bacteriol. 1980 Sep;143(3):1466–1470. doi: 10.1128/jb.143.3.1466-1470.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee D. K., Hammond A. W., Blakesley R. W., Adams S. M., Gerard G. F. Genetic organization of the KpnI restriction--modification system. Nucleic Acids Res. 1991 Dec 11;19(23):6505–6509. doi: 10.1093/nar/19.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N., Hedges R. W., Shaw E. J., Sykes R. B., Richmond M. H. Properties of an R factor from Pseudomonas aeruginosa. J Bacteriol. 1971 Dec;108(3):1244–1249. doi: 10.1128/jb.108.3.1244-1249.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. Interspecific gene transfer: where next? Trends Biotechnol. 1990 Aug;8(8):198–203. doi: 10.1016/0167-7799(90)90176-x. [DOI] [PubMed] [Google Scholar]
- Engel P. Plasmid transformation of Streptomyces tendae after heat attenuation of restriction. Appl Environ Microbiol. 1987 Jan;53(1):1–3. doi: 10.1128/aem.53.1.1-3.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost L. S. Bacterial conjugation: everybody's doin' it. Can J Microbiol. 1992 Nov;38(11):1091–1096. doi: 10.1139/m92-179. [DOI] [PubMed] [Google Scholar]
- HOLLOWAY B. W. VARIATIONS IN RESTRICTION AND MODIFICATION OF BACTERIOPHAGE FOLLOWING INCREASE OF GROWTH TEMPERATURE OF PSEUDOMONAS AERUGINOSA. Virology. 1965 Apr;25:634–642. doi: 10.1016/0042-6822(65)90091-7. [DOI] [PubMed] [Google Scholar]
- Heinemann J. A. Genetics of gene transfer between species. Trends Genet. 1991 Jun;7(6):181–185. doi: 10.1016/0168-9525(91)90433-q. [DOI] [PubMed] [Google Scholar]
- Heinemann J. A., Sprague G. F., Jr Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature. 1989 Jul 20;340(6230):205–209. doi: 10.1038/340205a0. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Hümbelin M., Suri B., Rao D. N., Hornby D. P., Eberle H., Pripfl T., Kenel S., Bickle T. A. Type III DNA restriction and modification systems EcoP1 and EcoP15. Nucleotide sequence of the EcoP1 operon, the EcoP15 mod gene and some EcoP1 mod mutants. J Mol Biol. 1988 Mar 5;200(1):23–29. doi: 10.1016/0022-2836(88)90330-0. [DOI] [PubMed] [Google Scholar]
- Jäger W., Schäfer A., Pühler A., Labes G., Wohlleben W. Expression of the Bacillus subtilis sacB gene leads to sucrose sensitivity in the gram-positive bacterium Corynebacterium glutamicum but not in Streptomyces lividans. J Bacteriol. 1992 Aug;174(16):5462–5465. doi: 10.1128/jb.174.16.5462-5465.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono M., Sasatsu M., Aoki T. R plasmids in Corynebacterium xerosis strains. Antimicrob Agents Chemother. 1983 Mar;23(3):506–508. doi: 10.1128/aac.23.3.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebl W., Bayerl A., Schein B., Stillner U., Schleifer K. H. High efficiency electroporation of intact Corynebacterium glutamicum cells. FEMS Microbiol Lett. 1989 Dec;53(3):299–303. doi: 10.1016/0378-1097(89)90234-6. [DOI] [PubMed] [Google Scholar]
- Liebl W., Ehrmann M., Ludwig W., Schleifer K. H. Transfer of Brevibacterium divaricatum DSM 20297T, "Brevibacterium flavum" DSM 20411, "Brevibacterium lactofermentum" DSM 20412 and DSM 1412, and Corynebacterium glutamicum and their distinction by rRNA gene restriction patterns. Int J Syst Bacteriol. 1991 Apr;41(2):255–260. doi: 10.1099/00207713-41-2-255. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Mazodier P., Davies J. Gene transfer between distantly related bacteria. Annu Rev Genet. 1991;25:147–171. doi: 10.1146/annurev.ge.25.120191.001051. [DOI] [PubMed] [Google Scholar]
- Mazodier P., Petter R., Thompson C. Intergeneric conjugation between Escherichia coli and Streptomyces species. J Bacteriol. 1989 Jun;171(6):3583–3585. doi: 10.1128/jb.171.6.3583-3585.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica-A T., Middleton R. B. Fertility of Salmonella typhimurium crosses with Escherichia coli. J Bacteriol. 1971 Dec;108(3):1161–1167. doi: 10.1128/jb.108.3.1161-1167.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., LeGrice S. F., Lee G., Stephens M., Sonenshein A. L., Pero J., Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186(3):339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Pátek M., Ludvík J., Benada O., Hochmannová J., Nesvera J., Krumphanzl V., Bucko M. New bacteriophage-like particles in Corynebacterium glutamicum. Virology. 1985 Jan 30;140(2):360–363. doi: 10.1016/0042-6822(85)90372-1. [DOI] [PubMed] [Google Scholar]
- Roberts R. J., Macelis D. Restriction enzymes and their isoschizomers. Nucleic Acids Res. 1992 May 11;20 (Suppl):2167–2180. doi: 10.1093/nar/20.suppl.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer A., Pühler A. Manipulation of Corynebacterium glutamicum by gene disruption and replacement. Biotechnology (N Y) 1991 Jan;9(1):84–87. doi: 10.1038/nbt0191-84. [DOI] [PubMed] [Google Scholar]
- Schäfer A., Kalinowski J., Pühler A. Increased fertility of Corynebacterium glutamicum recipients in intergeneric matings with Escherichia coli after stress exposure. Appl Environ Microbiol. 1994 Feb;60(2):756–759. doi: 10.1128/aem.60.2.756-759.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A., Kalinowski J., Simon R., Seep-Feldhaus A. H., Pühler A. High-frequency conjugal plasmid transfer from gram-negative Escherichia coli to various gram-positive coryneform bacteria. J Bacteriol. 1990 Mar;172(3):1663–1666. doi: 10.1128/jb.172.3.1663-1666.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A., Tauch A., Jäger W., Kalinowski J., Thierbach G., Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994 Jul 22;145(1):69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- Seep-Feldhaus A. H., Kalinowski J., Pühler A. Molecular analysis of the Corynebacterium glutamicum lysl gene involved in lysine uptake. Mol Microbiol. 1991 Dec;5(12):2995–3005. doi: 10.1111/j.1365-2958.1991.tb01859.x. [DOI] [PubMed] [Google Scholar]
- Seiler H. Identification key for coryneform bacteria derived by numerical taxonomic studies. J Gen Microbiol. 1983 May;129(5):1433–1471. doi: 10.1099/00221287-129-5-1433. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Michaud W., Levin H. L., Boeke J. D., Hieter P. Trans-kingdom promiscuity. Nature. 1990 Jun 14;345(6276):581–582. doi: 10.1038/345581b0. [DOI] [PubMed] [Google Scholar]
- Sonnen H., Schneider J., Kutzner H. J. Characterization of phi GA1, an inducible phage particle from Brevibacterium flavum. J Gen Microbiol. 1990 Mar;136(3):567–571. doi: 10.1099/00221287-136-3-567. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Staden R. The current status and portability of our sequence handling software. Nucleic Acids Res. 1986 Jan 10;14(1):217–231. doi: 10.1093/nar/14.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Carlier C., Courvalin P. Conjugative plasmid transfer from Enterococcus faecalis to Escherichia coli. J Bacteriol. 1988 Sep;170(9):4388–4391. doi: 10.1128/jb.170.9.4388-4391.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Carlier C., Poyart-Salmeron C., Courvalin P. Shuttle vectors containing a multiple cloning site and a lacZ alpha gene for conjugal transfer of DNA from Escherichia coli to gram-positive bacteria. Gene. 1991 Jun 15;102(1):99–104. doi: 10.1016/0378-1119(91)90546-n. [DOI] [PubMed] [Google Scholar]
- Trieu-Cuot P., Gerbaud G., Lambert T., Courvalin P. In vivo transfer of genetic information between gram-positive and gram-negative bacteria. EMBO J. 1985 Dec 16;4(13A):3583–3587. doi: 10.1002/j.1460-2075.1985.tb04120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertès A. A., Inui M., Kobayashi M., Kurusu Y., Yukawa H. Presence of mrr- and mcr-like restriction systems in coryneform bacteria. Res Microbiol. 1993 Mar-Apr;144(3):181–185. doi: 10.1016/0923-2508(93)90043-2. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. G., Murray N. E. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- Wilson G. G. Organization of restriction-modification systems. Nucleic Acids Res. 1991 May 25;19(10):2539–2566. doi: 10.1093/nar/19.10.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh P., Oreglia J., Sicard A. M. Transfection of Corynebacterium lilium protoplasts. J Gen Microbiol. 1985 Dec;131(12):3179–3183. doi: 10.1099/00221287-131-12-3179. [DOI] [PubMed] [Google Scholar]
- Yeh P., Sicard A. M., Sinskey A. J. General organization of the genes specifically involved in the diaminopimelate-lysine biosynthetic pathway of Corynebacterium glutamicum. Mol Gen Genet. 1988 Apr;212(1):105–111. doi: 10.1007/BF00322451. [DOI] [PubMed] [Google Scholar]