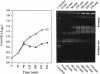
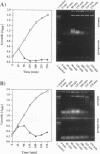
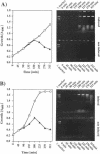
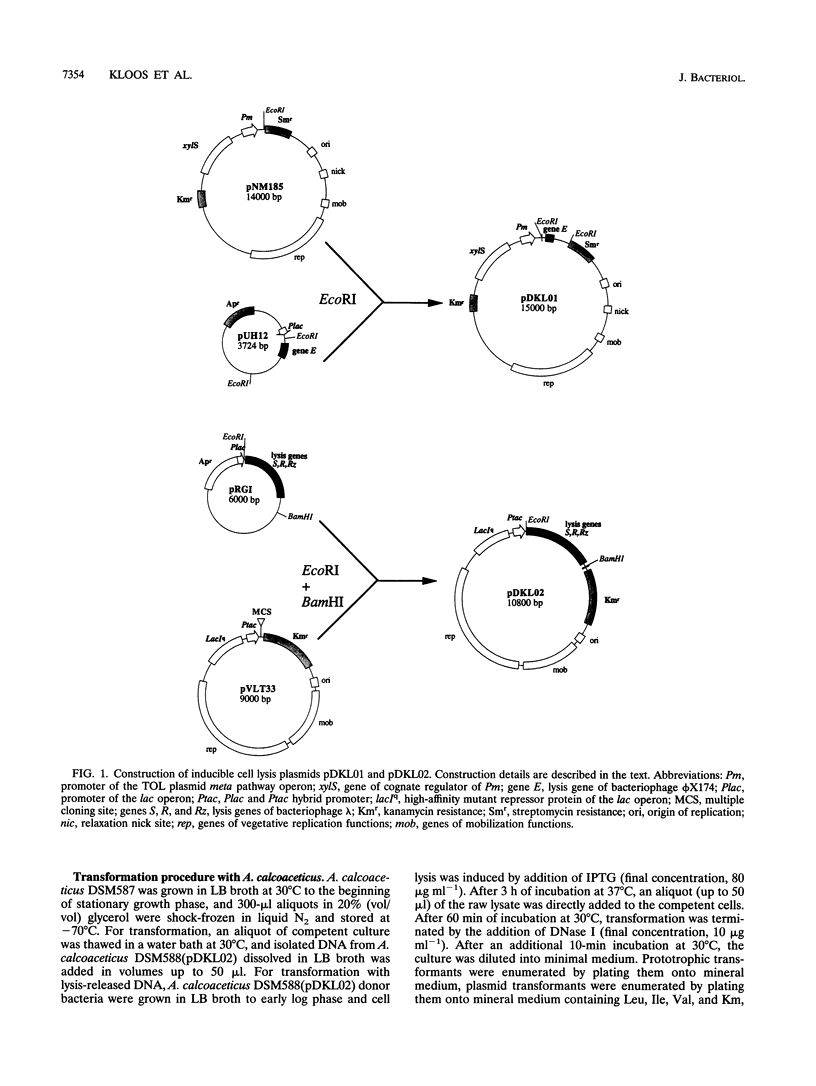
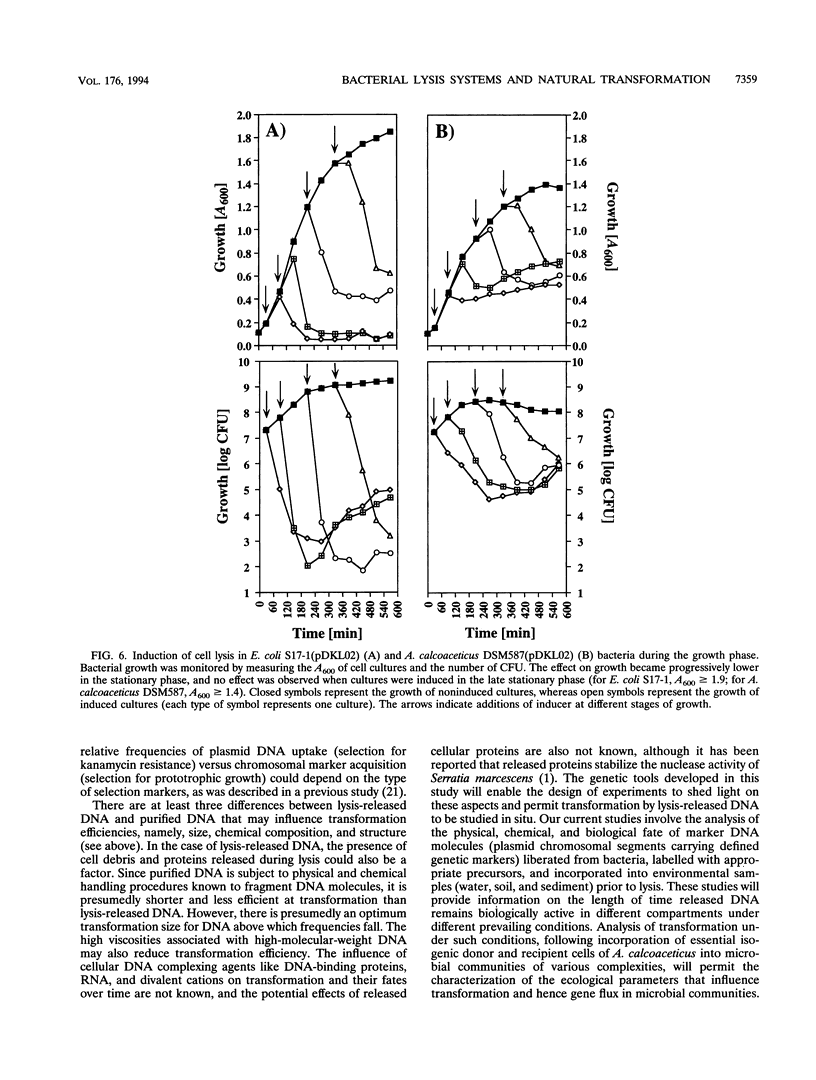
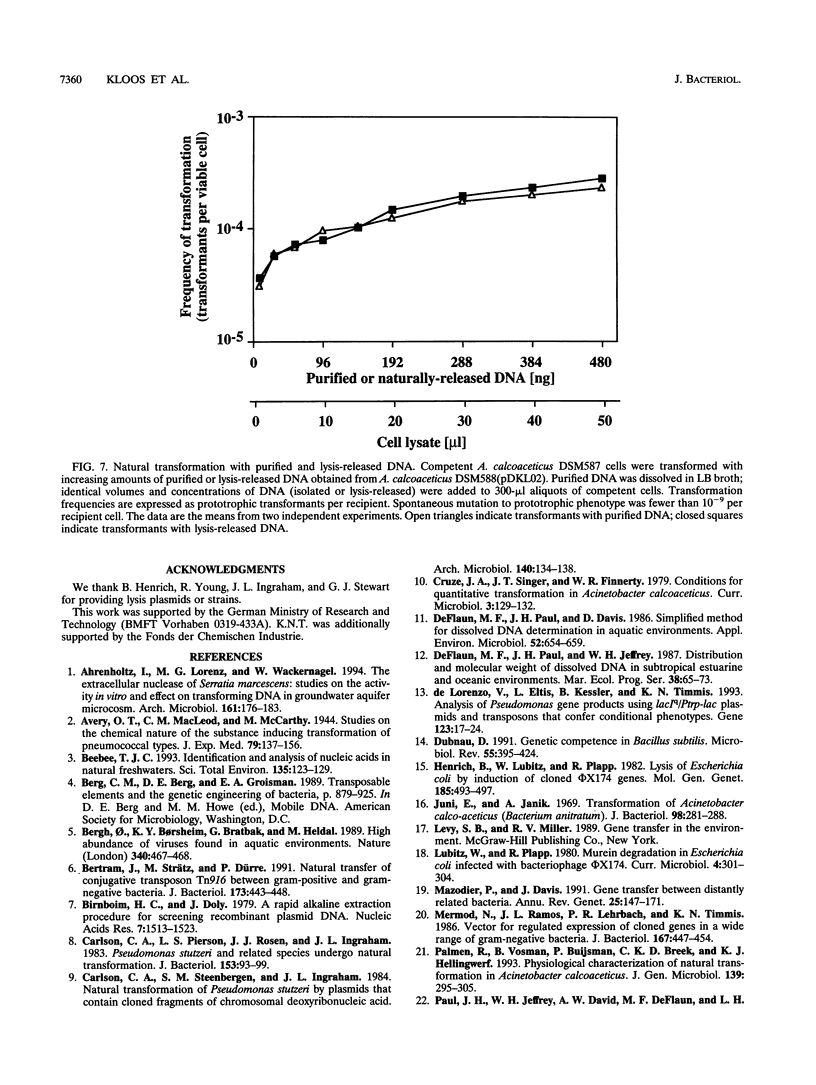
Abstract
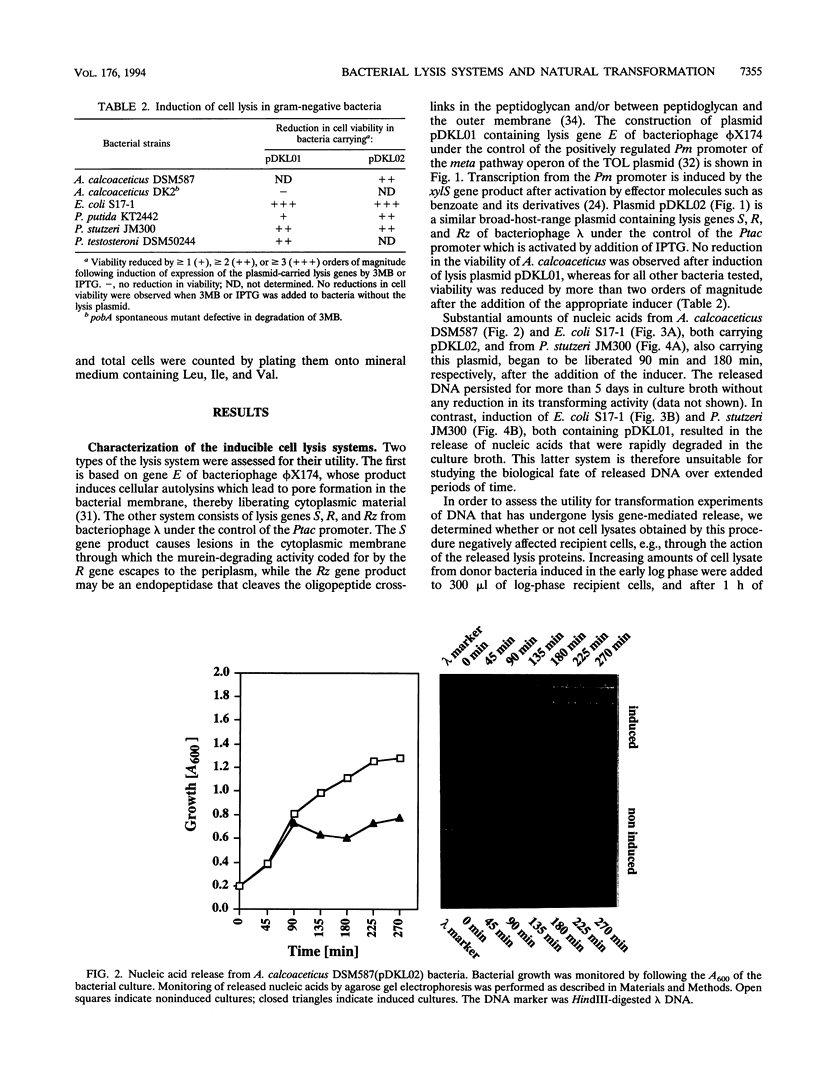
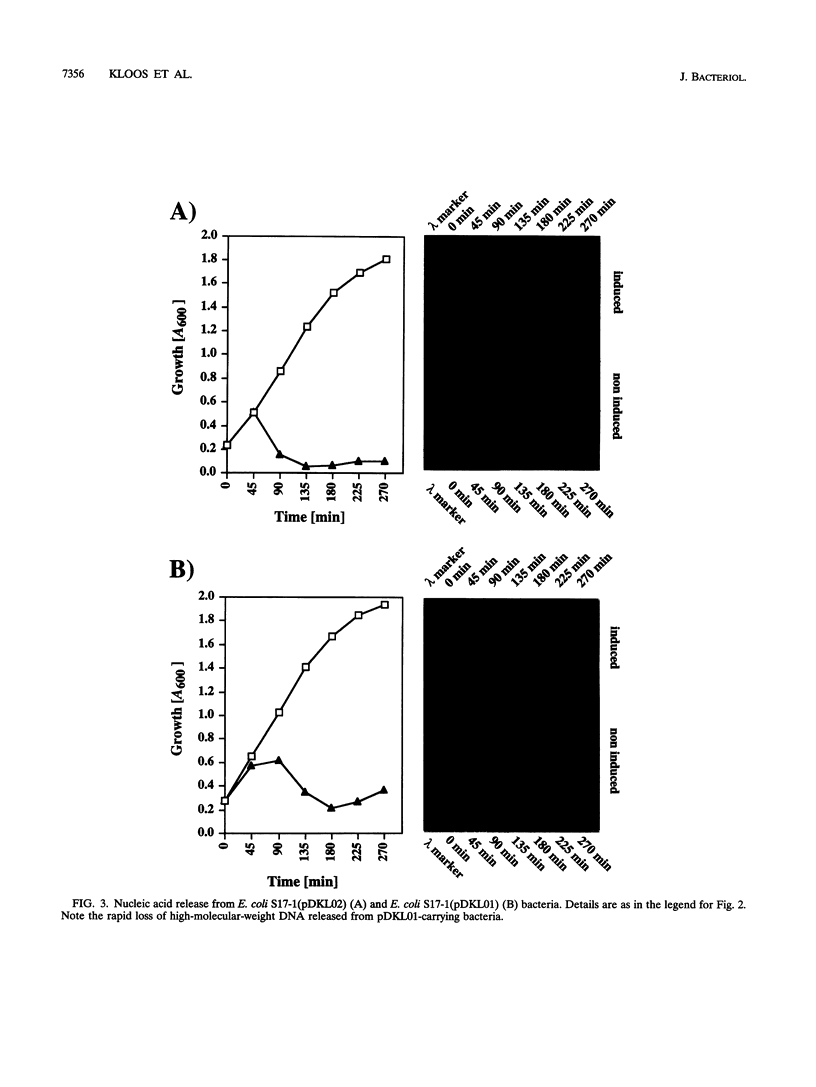
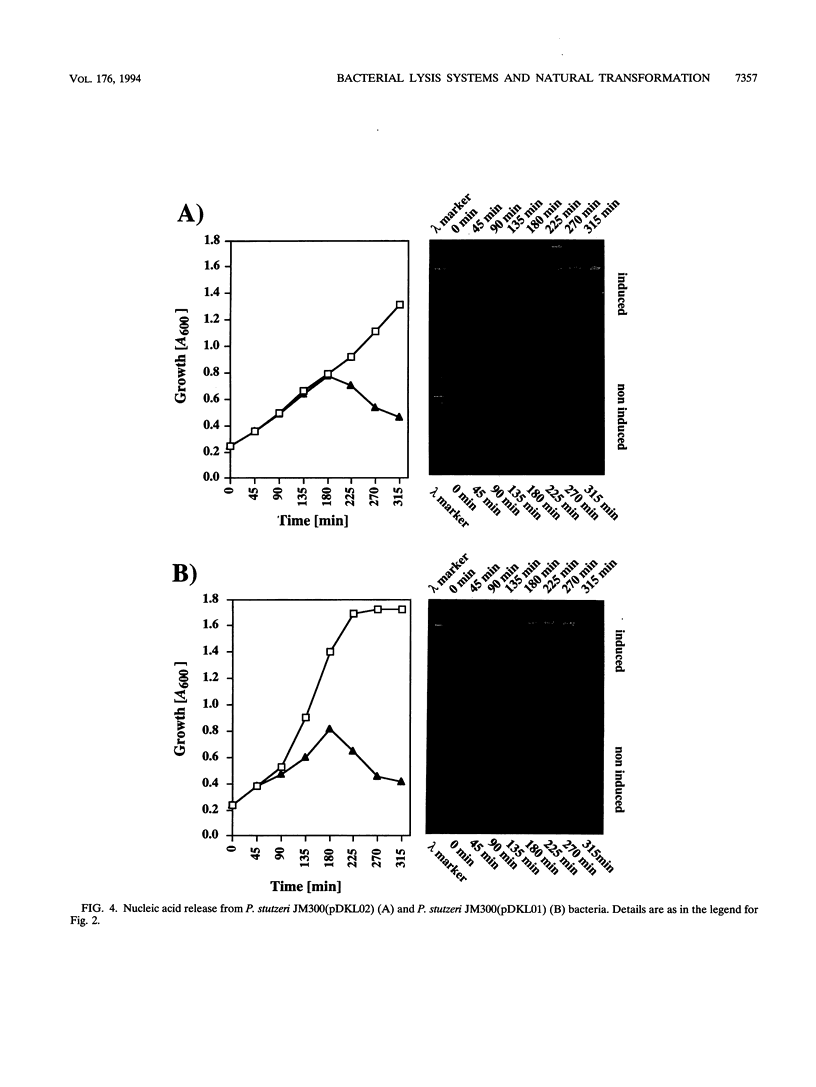
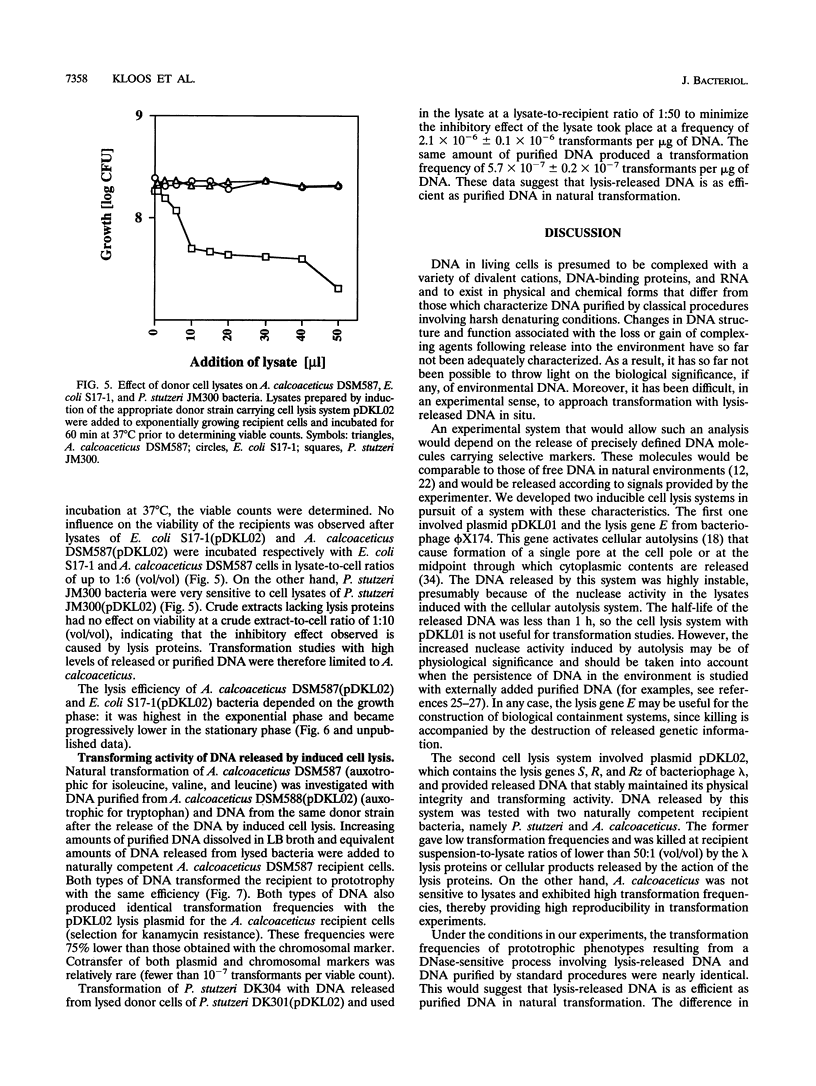
Two novel conditional broad-host-range cell lysis systems have been developed for the study of natural transformation in bacteria and the environmental fate of DNA released by cell death. Plasmid pDKL02 consists of lysis genes S, R, and Rz from bacteriophage lambda under the control of the Ptac promoter. The addition of inducer to Escherichia coli, Acinetobacter calcoaceticus, or Pseudomonas stutzeri containing plasmid pDKL02 resulted in cell lysis coincident with the release of high amounts of nucleic acids into the surrounding medium. The utility of this lysis system for the study of natural transformation with DNA released from lysed cells was assessed with differentially marked but otherwise isogenic donor-recipient pairs of P. stutzeri JM300 and A. calcoaceticus BD4. Transformation frequencies obtained with lysis-released DNA and DNA purified by conventional methods and assessed by the use of antibiotic resistance (P. stutzeri) or amino acid prototrophy (A. calcoaceticus) for markers were comparable. A second cell lysis plasmid, pDKL01, contains the lysis gene E from bacteriophage phi X174 and causes lysis of E. coli and P. stutzeri bacteria by activating cellular autolysins. Whereas DNA released from pDKL02-containing bacteria persists in the culture broth for days, that from induced pDKL01-containing bacteria is degraded immediately after release. The lysis system involving pDKL02 is thus useful for the study of both the fate of DNA released naturally into the environment by dead cells and gene transfer by natural transformation in the environment in that biochemically unmanipulated DNA containing defined sequences and coding for selective phenotypes can be released into a selected environment at a specific time point. This will allow kinetic measurements that will answer some of the current ecological questions about the fate and biological potential of environmental DNA to be made.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahrenholtz I., Lorenz M. G., Wackernagel W. The extracellular nuclease of Serratia marcescens: studies on the activity in vitro and effect on transforming DNA in a groundwater aquifer microcosm. Arch Microbiol. 1994;161(2):176–183. doi: 10.1007/BF00276480. [DOI] [PubMed] [Google Scholar]
- Bergh O., Børsheim K. Y., Bratbak G., Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989 Aug 10;340(6233):467–468. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- Bertram J., Strätz M., Dürre P. Natural transfer of conjugative transposon Tn916 between gram-positive and gram-negative bacteria. J Bacteriol. 1991 Jan;173(2):443–448. doi: 10.1128/jb.173.2.443-448.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson C. A., Pierson L. S., Rosen J. J., Ingraham J. L. Pseudomonas stutzeri and related species undergo natural transformation. J Bacteriol. 1983 Jan;153(1):93–99. doi: 10.1128/jb.153.1.93-99.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deflaun M. F., Paul J. H., Davis D. Simplified method for dissolved DNA determination in aquatic environments. Appl Environ Microbiol. 1986 Oct;52(4):654–659. doi: 10.1128/aem.52.4.654-659.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D. Genetic competence in Bacillus subtilis. Microbiol Rev. 1991 Sep;55(3):395–424. doi: 10.1128/mr.55.3.395-424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrich B., Lubitz W., Plapp R. Lysis of Escherichia coli by induction of cloned phi X174 genes. Mol Gen Genet. 1982;185(3):493–497. doi: 10.1007/BF00334146. [DOI] [PubMed] [Google Scholar]
- Juni E., Janik A. Transformation of Acinetobacter calco-aceticus (Bacterium anitratum). J Bacteriol. 1969 Apr;98(1):281–288. doi: 10.1128/jb.98.1.281-288.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazodier P., Davies J. Gene transfer between distantly related bacteria. Annu Rev Genet. 1991;25:147–171. doi: 10.1146/annurev.ge.25.120191.001051. [DOI] [PubMed] [Google Scholar]
- Mermod N., Ramos J. L., Lehrbach P. R., Timmis K. N. Vector for regulated expression of cloned genes in a wide range of gram-negative bacteria. J Bacteriol. 1986 Aug;167(2):447–454. doi: 10.1128/jb.167.2.447-454.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmen R., Vosman B., Buijsman P., Breek C. K., Hellingwerf K. J. Physiological characterization of natural transformation in Acinetobacter calcoaceticus. J Gen Microbiol. 1993 Feb;139(2):295–305. doi: 10.1099/00221287-139-2-295. [DOI] [PubMed] [Google Scholar]
- Paul J. H., Jeffrey W. H., David A. W., Deflaun M. F., Cazares L. H. Turnover of extracellular DNA in eutrophic and oligotrophic freshwater environments of southwest Florida. Appl Environ Microbiol. 1989 Jul;55(7):1823–1828. doi: 10.1128/aem.55.7.1823-1828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab R., Neal G., Garrett J., Grimaila R., Fusselman R., Young R. Mutational analysis of bacteriophage lambda lysis gene S. J Bacteriol. 1986 Sep;167(3):1035–1042. doi: 10.1128/jb.167.3.1035-1042.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J. L., Stolz A., Reineke W., Timmis K. N. Altered effector specificities in regulators of gene expression: TOL plasmid xylS mutants and their use to engineer expansion of the range of aromatics degraded by bacteria. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8467–8471. doi: 10.1073/pnas.83.22.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski G., Lorenz M. G., Sayler G., Wackernagel W. Persistence of free plasmid DNA in soil monitored by various methods, including a transformation assay. Appl Environ Microbiol. 1992 Sep;58(9):3012–3019. doi: 10.1128/aem.58.9.3012-3019.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski G., Lorenz M. G., Wackernagel W. Adsorption of plasmid DNA to mineral surfaces and protection against DNase I. Appl Environ Microbiol. 1991 Apr;57(4):1057–1061. doi: 10.1128/aem.57.4.1057-1061.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski G., Lorenz M. G., Wackernagel W. Use of polymerase chain reaction and electroporation of Escherichia coli to monitor the persistence of extracellular plasmid DNA introduced into natural soils. Appl Environ Microbiol. 1993 Oct;59(10):3438–3446. doi: 10.1128/aem.59.10.3438-3446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Franklin F. C., Williams P. A. Regulation of the degradative pathway enzymes coded for by the TOL plasmid (pWWO) from Pseudomonas putida mt-2. J Bacteriol. 1978 Jun;134(3):757–764. doi: 10.1128/jb.134.3.757-764.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992 Sep;56(3):430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V., Eltis L., Kessler B., Timmis K. N. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene. 1993 Jan 15;123(1):17–24. doi: 10.1016/0378-1119(93)90533-9. [DOI] [PubMed] [Google Scholar]