Abstract
Background
The model halophile Halobacterium sp. NRC-1 was among the first Archaea to be completely sequenced and many post-genomic tools, including whole genome DNA microarrays are now being applied to its analysis. This extremophile displays tolerance to multiple stresses, including high salinity, extreme (non-mesophilic) temperatures, lack of oxygen, and ultraviolet and ionizing radiation.
Results
In order to study the response of Halobacterium sp. NRC-1 to two common stressors, salinity and temperature, we used whole genome DNA microarrays to assay for changes in gene expression under differential growth conditions. Cultures grown aerobically in rich medium at 42°C were compared to cultures grown at elevated or reduced temperature and high or low salinity. The results obtained were analyzed using a custom database and microarray analysis tools. Growth under salt stress conditions resulted in the modulation of genes coding for many ion transporters, including potassium, phosphate, and iron transporters, as well as some peptide transporters and stress proteins. Growth at cold temperature altered the expression of genes involved in lipid metabolism, buoyant gas vesicles, and cold shock proteins. Heat shock showed induction of several known chaperone genes. The results showed that Halobacterium sp. NRC-1 cells are highly responsive to environmental changes at the level of gene expression.
Conclusion
Transcriptional profiling showed that Halobacterium sp. NRC-1 is highly responsive to its environment and provided insights into some of the specific responses at the level of gene expression. Responses to changes in salt conditions appear to be designed to minimize the loss of essential ionic species and abate possible toxic effects of others, while exposure to temperature extremes elicit responses to promote protein folding and limit factors responsible for growth inhibition. This work lays the foundation for further bioinformatic and genetic studies which will lead to a more comprehensive understanding of the biology of a model halophilic Archaeon.
Background
Halophilic archaea (haloarchaea) flourish in extremely saline environments and are exceptionally tolerant of many environmental stresses [1,2]. Among haloarchaea, several closely related Halobacterium species are the best-studied, display the greatest halophilicity, and are widely distributed in nature. They are typified by the well-studied model organism, Halobacterium sp. NRC-1, which grows fastest aerobically in amino acid-rich environments at moderate temperatures and nearly saturated brine [3]. This strain has the ability to survive and grow phototrophically using the light driven proton pumping activity of bacteriorhodopsin in its purple membrane and anaerobically via substrate level phosphorylation using arginine and by respiration using dimethyl sulfoxide (DMSO) and trimethylamine N-oxide (TMAO) [4,5]. It can also survive at temperatures spanning the range from 10 to 56°C and NaCl concentrations from 2.5 to 5.3 M (saturation). Halobacterium sp. NRC-1 is highly tolerant of both ultraviolet light and ionizing radiation, the latter of which may be related to its relatively high desiccation resistance [6-9]. The remarkable tolerance of Halobacterium sp. NRC-1 to multiple extremes distinguishes this organism among extremophiles and Archaea [1,2].
The Halobacterium NRC-1 genome sequence was completed in 2000 and found to be 2.57 Mb in size [10-12], and is composed of three circular replicons: a large chromosome (2.0 Mb) and two minichromosomes, pNRC100 (191 kb) and pNRC200 (365 kb). Analysis of the genome sequence identified 2,682 likely genes (including 52 RNA genes), of which 1,658 coded for proteins with significant matches to the database. Of the matches, 591 were to conserved hypothetical proteins, and 1067 were to proteins with known or predicted functions. Interestingly, about 40 genes on pNRC100 and pNRC200 coded for proteins likely to be essential or important for cell viability, indicating that these replicons function as minichromosomes. Bioinformatic analysis identified 149 likely regulators and multiple general transcription factors, including six TBPs and seven TFBs, which were hypothesized to regulate gene expression in response to environmental changes [13].
After complete sequencing of the Halobacterium sp. NRC-1 genome, key post-genomic methods were developed, including a facile gene knockout system for reverse genetic analysis [14,15], and a whole genome DNA microarray for transcriptomic analysis [5,13]. We employed the DNA microarray system developed by Agilent Corporation using inkjet technology for in situ synthesis of oligonucleotides directly on glass slides [16]. This system provides high specificity by use of 60-mer oligonucleotide probes and high data quality due to limited technical noise. In the present work, we have successfully used a platform containing 2 × 8,455 features per slide representing duplicate microarrays with 2474 (97%) open reading frames (ORFs). Up to four unique probes were designed per ORF with a mean Tmof 81°C and a Tm range of 3°C. The microarray performance was tested through linearity of response and statistical significance using both biological and technical replicates [5].
We conducted three previous whole genome transcriptomic studies to examine the response of Halobacterium sp. NRC-1 to extreme conditions using DNA microarrays [5-7]. In the first study, we investigated cell growth by anaerobic respiration on either DMSO or TMAO as the sole terminal electron acceptor, and found the requirement of the dmsREABCD operon for growth under anaerobic respiration. Whole genome DNA microarray analysis showed that the dms operon is highly induced when cells are grown anaerobically with TMAO and comparison of dmsR+ and ΔdmsR strains showed that the induction of the dmsEABCD operon is dependent on a functional dmsR gene, consistent with its action as a transcriptional activator. Expression of the purple membrane protein bacterio-opsin (bop) gene as well as genes specifying buoyancy conferring gas vesicles were also induced under limiting oxygen conditions, indicating that cells respond by moving to more aerobic and illuminated zones where the alternate physiological capabilities may be utilized.
We also studied the response of Halobacterium sp. NRC-1 to high levels of UV radiation damage, an environmental stress that results from solar radiation present in its environment [6]. Cells were irradiated with 30–70 J/m2 UV-C, and transcriptional profiling showed the most strongly up-regulated gene was radA1, the archaeal homolog of rad51 in eukaryotes and recA in bacteria. Additional genes involved in homologous recombination, such as arj1 (recJ-like exonuclease), dbp (eukaryote-like DNA binding protein of the superfamily I DNA and RNA helicases), and rfa3 (replication protein A complex), as well as nrdJ, (cobalamin-dependent ribonucleotide reductase involved in DNA metabolism), were also significantly induced. Neither prokaryotic nor eukaryotic excision repair gene homologs were induced and there was no evidence of an SOS-like response. These results showed that homologous recombination plays an important role in the cellular response of Halobacterium sp. NRC-1 to UV damage.
In our most recently published study [7], we generated and examined mutants of Halobacterium sp. NRC-1 that are resistant to high energy ionizing radiation. Two independently-obtained mutants displaying LD50>11 kGy, which is higher than that of the extremely radiation-resistant bacterium Deinococcus radiodurans, were found to up-regulate an operon comprised of two single-stranded DNA binding protein (RPA) genes, rfa3, rfa8, and a third gene, ral, of unknown function. These results suggested that RPA facilitates DNA repair machinery and/or protects repair intermediates to maximize the ionizing radiation-resistance of this archaeon.
In the current report, we used whole genome DNA microarrays for Halobacterium sp. NRC-1 to assay the changes in gene expression in response to several common environmental conditions, high and low salinity and temperature. These data serve as a significant resource to expand our understanding of the physiological and transcriptional responses of Halobacterium sp. NRC-1 to the wide range of environmental stresses to which it is exposed.
Results and discussion
Low and high salinity
Halobacterium sp. NRC-1 flourishes in environments that are not only hypersaline (4.3 M NaCl optimum) but also highly variable in total salinity (2.5–5.3 M NaCl) as a result of common evaporatic and dilution processes [1]. Therefore, the cells must cope with high as well as dynamic concentrations of dissolved salts. The intracellular concentration of KCl is roughly equal (concentrations up to 5 M have been reported) to the external NaCl concentrations, and is used as the major compatible solute [17]. Genome-wide, predicted proteins have a median pI of 4.9, with a concentration of negative charges on their surface, characteristics which permit effective competition for hydration and allow function in a cytoplasm with low water activity [3,18]. High salt concentrations are also known to convert some DNA sequences, e.g. alternating (CG)-repeats, from the right-handed B to the left-handed Z form, facilitated by negative superhelical stress [19,20]. Organic compatible solutes commonly utilized in the response of bacterial and eukaryotic halophiles to osmotic stress, have also been reported in Halobacterium sp. NRC-1 [21,22].
The annotated genome of Halobacterium sp. NRC-1 provided an inventory of likely genes involved in maintaining the intracellular ionic conditions suitable for growth [11,12]. Genes coding for multiple active K+ transporters were found, including kdpABC, an ATP-driven K+ uptake system, and trkAH, low-affinity K+ transporters driven by the membrane potential. Genes coding for an active Na+ efflux system likely mediated by NhaC proteins were also present, corresponding to the unidirectional Na+-H+ antiporter activity described previously [23]. Interestingly, several of these genes, including those coding for kdpABC, trkA (three copies), and nhaC (one copy) were found on the extrachromosomal pNRC200 replicon. However, a comprehensive study of the adaptation of Halobacterium sp. NRC-1 to various salinities was not previously reported.
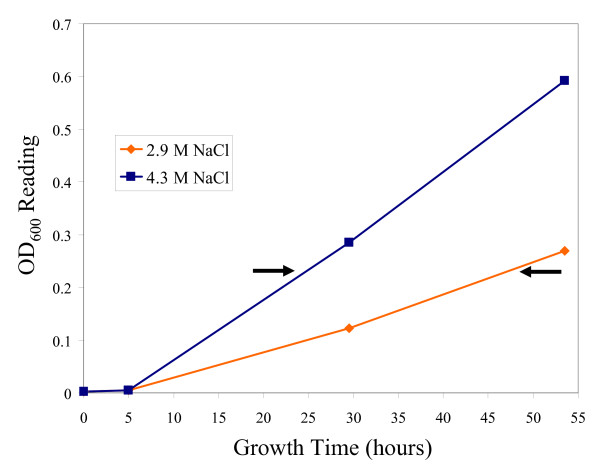
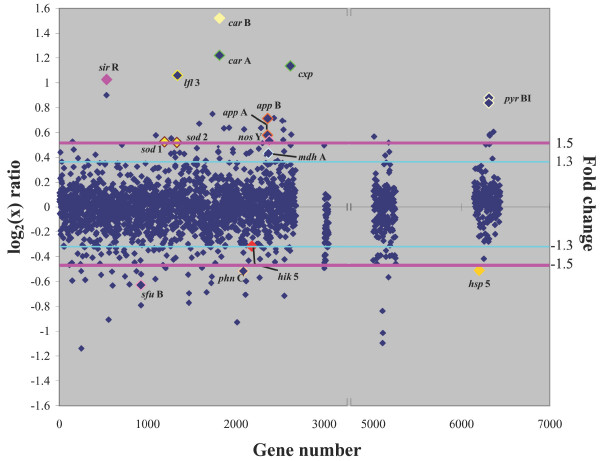
In order to determine which genes were most responsive to high and low salinity, Halobacterium sp. NRC-1 cultures were grown at three salinities, low (2.9 M NaCl), optimal (4.3 M NaCl), and high (5.0 M NaCl), in rich media at 42°C (Figs. 1 &2). The high and low salinity conditions were selected to ensure that salts did not precipitate and cells did not lyse during culture, respectively. Growth at low salinity compared to optimal salinity displayed 143 up-regulated genes and 53 down-regulated genes by 1.5-fold or greater (Fig. 3). Growth at high salinity, where the NaCl concentration (5 M) was only slightly higher than the optimal salinity (4.3 M) displayed 32 up-regulated genes and 29 down-regulated genes by 1.5-fold or greater (Fig. 4).
Figure 1.
Growth curves of Halobacterium sp. NRC-1 cultures at low salt conditions (2.9 M NaCl) and standard conditions (4.3 M NaCl). Arrows indicate the point at which cultures were harvested for microarray analysis.
Figure 2.
Growth curves of Halobacterium sp. NRC-1 cultures at high salt conditions (5.0 M NaCl) and standard conditions (4.3 M NaCl). Arrows indicate the point at which cultures were harvested for microarray analysis.
Figure 3.
Scatter plot comparing DNA microarrays hybridized with cDNA from Halobacterium sp. NRC-1 cultures grown in low (2.9 M) and standard (4.3 M) NaCl conditions. The log2 value of the Cy5/Cy3 ratio for each gene (abscissa) is plotted versus the gene number (ordinate). Gene numbers correspond to the chromosomal genes (1–2679), RNA genes (3000 to 3051), pNRC100 genes (5000 to 5256), and pNRC200 genes (6000 to 6487). Pink line represents a fold change value of 1.5 and the light blue line represents a fold change value of 1.3. Colors have been added for emphasis.
Figure 4.
Scatter plot comparing DNA microarrays hybridized with cDNA from Halobacterium sp. NRC-1 cultures grown in high (5 M) and standard (4.3 M) NaCl. The log2 value of the Cy5/Cy3 ratio for each gene (abscissa) is plotted versus the gene number (ordinate). Gene numbers correspond to the chromosomal genes (1–2679), RNA genes (3000 to 3051), pNRC100 genes (5000 to 5256), and pNRC200 genes (6000 to 6487). Pink line represents a fold change value of 1.5 and the light blue line represents a fold change value of 1.3. Colors have been added for emphasis.
Transcription of several potassium and sodium ion transporter genes was affected by salt conditions in NRC-1, mainly by low salt [24,25]. Among the putative potassium transporters, the trkA6 gene, was significantly up-regulated (2.0-fold) under low salt conditions (Table 1). This gene product may compensate for the loss of K+ under low salt conditions, via membrane leakiness, by uptake of more potassium ions. Among sodium transporters, only one (nhaC3, coded by pNRC200) gene was slightly down-regulated (1.3-fold) under low salt, while the others were unchanged. The lack of a more vigorous induction of the sodium-proton antiporter suggests that increases of Na+ concentration in cells may be tolerated during osmotic stress, possibly substituting for K+ intracellularly.
Table 1.
Selected genes significantly altered under low salinity growth
| Gene ID | Gene name | Former name | COG | Predicted function | Fold change values | log2(x) ratio | Standard deviation of log2(x) ratio |
|---|---|---|---|---|---|---|---|
| 101 | cspD1 | 1278 | cold shock protein | 1.442 | 0.502 | 0.275 | |
| 491 | dnaK | 443 | heat shock protein | -1.307 | -0.376 | 0.172 | |
| 727 | dfr | 534 | DinF-related damage-inducible protein | -2.079 | -1.019 | 0.316 | |
| 1013 | htr13 | 840 | Htr13 transducer | 1.656 | 0.649 | 0.482 | |
| 1121 | aspC2 | 436 | aspartate aminotransferase | 1.694 | 0.688 | 0.453 | |
| 1190 | sod1 | 605 | superoxide dismutase | 1.826 | 0.852 | 0.221 | |
| 1332 | sod2 | 605 | superoxide dismutase | 2.023 | 0.988 | 0.290 | |
| 1408 | ush | 737 | UDP-sugar hydrolase | -1.393 | -0.468 | 0.165 | |
| 1442 | htr12 | 840 | Htr12 transducer | 1.406 | 0.490 | 0.058 | |
| 1632 | cbiQ | 619 | cobalt transport protein | -1.265 | -0.324 | 0.214 | |
| 1634 | cbiN | 1930 | cobalt transport protein | -1.553 | -0.580 | 0.400 | |
| 1691 | rpl23p | 89 | 50S ribosomal protein L23P | -1.454 | -0.533 | 0.138 | |
| 1692 | rpl2p | 90 | 50S ribosomal protein L2P | -1.515 | -0.594 | 0.128 | |
| 1695 | rpl22p | 91 | 50S ribosomal protein L22P | -1.447 | -0.523 | 0.173 | |
| 1698 | rpl29p | 255 | 50S ribosomal protein L29P | -1.460 | -0.535 | 0.176 | |
| 1801 | hsp1 | 71 | small heat shock protein | 2.248 | 1.006 | 0.679 | |
| 1814 | carB | 458 | carbamoyl-phosphate synthase large subunit | -1.590 | -0.646 | 0.261 | |
| 1815 | carA | 505 | carbamoyl-phosphate synthase small subunit | -1.452 | -0.520 | 0.225 | |
| 1924 | trkA6 | 569 | TRK potassium uptake system protein | 2.040 | 1.006 | 0.251 | |
| 2093 | glnA | 174 | glutamine synthatase | -1.719 | -0.766 | 0.212 | |
| 2116 | xup | 2252 | xanthine/uracil permease family protein | -1.603 | -0.647 | 0.307 | |
| 2139 | atpA | 1155 | H+-transporting ATP synthase subunit A | -1.482 | -0.555 | 0.189 | |
| 2140 | atpF | 1436 | H+-transporting ATP synthase subunit F | -1.536 | -0.608 | 0.177 | |
| 2141 | atpC | 1527 | H+-transporting ATP synthase subunit C | -1.637 | -0.694 | 0.221 | |
| 2142 | atpE | 1390 | H+-transporting ATP synthase subunit E | -1.706 | -0.761 | 0.173 | |
| 2143 | atpK | 636 | H+-transporting ATP synthase subunit K | -1.658 | -0.719 | 0.173 | |
| 2144 | atpI | 1269 | H+-transporting ATP synthase subunit I | -1.816 | -0.830 | 0.295 | |
| 2281 | gst | 435 | glutathione S-transferase | 1.508 | 0.573 | 0.235 | |
| 2344 | dppD1 | oppD2 | 444 | oligopeptide ABC transporter | -1.562 | -0.611 | 0.301 |
| 2346 | dppC1 | dppC2 | 1173 | dipeptide ABC transporter permease | -1.436 | -0.488 | 0.307 |
| 2347 | dppB1 | dppB1 | 601 | dipeptide ABC transporter permease | -1.581 | -0.649 | 0.182 |
| 2358 | appA | 747 | oligopeptide binding protein | 1.893 | 0.841 | 0.485 | |
| 2359 | appB | 601 | oligopeptide ABC permease | 1.506 | 0.548 | 0.350 | |
| 2616 | cxp | 2317 | Carboxypeptidase | -2.194 | -1.113 | 0.247 | |
| 5076 | trxA1 | Thioredoxin | -1.424 | -0.491 | 0.235 | ||
| 5142 | tbpC | 2101 | transcription initiation factor IID | 1.748 | 0.743 | 0.430 | |
| 6313 | nhaC3 | 1757 | Na+/H+ antiporter | -1.280 | -0.348 | 0.146 |
Expression of many other transporter genes was altered by growth in non-optimal salinities. The sfuB iron transporter-like permease protein was down-regulated (1.6-fold) under high salt (Table 2), and the phnC gene, a component of the phosphate/phosphonate transport system was down-regulated by 1.4-fold in high salt, suggesting that Halobacterium sp. NRC-1 responds to osmotic stress by reducing uptake of these species, which may potentially be toxic [26]. The nosY gene, coding for an apparent ABC transporter component for nitrite/nitrate, was up-regulated under high salt (1.5-fold), although usage of nitrite or nitrate for respiration or as a nitrogen source has not been detected for NRC-1 [13]. Members of the two similarly organized gene clusters containing oligopeptide/dipeptide/Ni2+ transporter genes (appABCDF and dppABCDF, see Table 1), were responsive to salinity changes. Two genes in the app cluster were up-regulated between 1.5- and 1.9-fold under both high and low salt, while some genes in the dpp cluster were down-regulated 1.4- to 1.6-fold under low salt. In both cases, the permease genes, C and/or B, were more affected than the genes coding for the periplasmic and ATPase components. The increase in expression of these genes is consistent with an attempt by cells to minimize the loss of these ionic (or zwitterionic amino acids) species, whereas down-regulation may be related to growth rate or possible toxic effects. Interestingly, the proX and htr5 genes, which are proposed to code for compatible solute (possibly trimethylammonium) binding and transporter proteins (cosB and cosT, respectively, in the closely related Halobacterium salinarium [27]) were unchanged in high or low salt concentrations in NRC-1.
Table 2.
Selected genes significantly altered under high salinity growth
| Gene ID | Gene name | Former name | COG | Predicted function | Fold change values | log2(x) ratio | Standard deviation of log2(x) ratio |
|---|---|---|---|---|---|---|---|
| 536 | sirR | 1321 | transcription repressor | 2.048 | 1.026 | 0.146 | |
| 923 | sfuB | 1178 | iron transporter-like protein | -1.598 | -0.628 | 0.384 | |
| 1190 | sod1 | 605 | superoxide dismutase | 1.441 | 0.525 | 0.073 | |
| 1332 | sod2 | 605 | superoxide dismutase | 1.435 | 0.521 | 0.045 | |
| 1339 | lfl3 | 318 | long-chain fatty-acid-CoA ligase | 2.139 | 1.060 | 0.325 | |
| 1814 | carB | 458 | carbamoyl-phosphate synthase large subunit | 2.915 | 1.521 | 0.261 | |
| 1815 | carA | 505 | carbamoyl-phosphate synthase small subunit | 2.532 | 1.222 | 0.595 | |
| 2085 | phnC | 3638 | phosphonate transport ATP-binding | -1.432 | -0.516 | 0.076 | |
| 2180 | hik5 | vng2180 | 642 | sensory histidine protein kinase (HisKA domain) | -1.246 | -0.315 | 0.072 |
| 2358 | appA | 747 | oligopeptide binding protein | 1.525 | 0.579 | 0.290 | |
| 2359 | appB | 601 | oligopeptide ABC permease | 1.687 | 0.713 | 0.339 | |
| 2367 | mdhA | 39 | L-malate dehydrogenase | 1.351 | 0.432 | 0.081 | |
| 2377 | nosY | 1277 | nitrite/nitrate ABC transporter | 1.453 | 0.534 | 0.124 | |
| 2616 | cxp | 2317 | carboxypeptidase | 2.210 | 1.137 | 0.143 | |
| 6201 | hsp5 | 71 | heat shock protein | -1.427 | -0.510 | 0.093 | |
| 6309 | pyrB | 540 | aspartate carbamoyltransferase catalytic subunit | 1.803 | 0.838 | 0.190 | |
| 6311 | pyrI | 1781 | aspartate carbamoyltransferase regulatory chain | 1.847 | 0.879 | 0.128 |
Protein kinases are believed to be involved in the salt stress response cascade in eukaryotic cells [28] as well as some prokaryotic species [29]. Among genes possibly involved in signaling in Halobacterium sp. NRC-1, the only signal transduction histidine kinase gene out of 13 in the genome that showed any significant change was hik5, for which expression under the high salt condition was slightly lowered (1.3-fold). In the large halotransducer (htr) family in NRC-1 only two of 17 htr genes, htr12 and htr13, were up-regulated under low salt (1.4- to 1.7-fold) [30]. Knowledge of the true functions of these families of genes and their precise roles in signal transduction in Halobacterium sp. NRC-1 is quite limited at present.
Several transcription factors and regulators were significantly changed under salt stress conditions. Among these, one basal transcription factor gene out of 13 in NRC-1 [12], tbpC (coded by pNRC200), was up-regulated 1.7-fold under low salt conditions. Also, the sirR gene, a putative transcriptional repressor, was significantly up-regulated under high salt (2.0-fold).
Interestingly, three stress genes were inducible under high and low salt conditions. Both sod genes, which encode superoxide dismutase, were salt-responsive. The sod2 transcript was up-regulated 1.4-fold under high salt and 2.0-fold under low salt, and sod1 was up-regulated 1.4-fold under high salt and 1.8-fold under low salt. Superoxide dismutase is responsible for limiting damage from reactive oxygen species, which are induced by extreme conditions, such as oxidative stress and excess irradiance [31] in many species. The gst gene, coding for glutathione S-transferase, another detoxification enzyme involved in cell protection from reactive oxygen species [32], was up-regulated (1.5-fold) under low salt conditions. These findings suggest that the removal of reactive oxygen species is of increased importance in osmotically stressed haloarchaea.
An earlier study [33] suggested that the small heat shock protein family coded by hsp genes may be involved in salt stress responses in the related haloarchaeon, Haloferax volcanii. We observed that two hsp genes in Halobacterium sp. NRC-1 were affected by salinity changes: hsp1 was up-regulated under low salt conditions (2.3-fold) and hsp5, coded by pNRC200, was down-regulated 1.4-fold under high salt conditions (as well as in the cold, see below). Two predicted stress genes, the cold shock gene cspD1 (1.4-fold up-regulated), and the heat shock gene dnaK (1.3-fold down-regulated), were regulated under low salt conditions, which suggests a wider role than temperature adaptation for these chaperones. Interestingly, a carboxypeptidase gene, cxp, which may function in protein turnover, was highly down-regulated in low salt (2.2-fold), and equally up-regulated in high salt.
Expression of a number of genes involved in cellular metabolism was significantly altered by salinity changes. Two highly regulated genes, carA and carB, coding for both subunits of carbamoyl-phosphate synthase, were up-regulated between 2.5- and 2.9-fold under high salt conditions and down-regulated 1.5- to 1.6-fold under low salt conditions. Any relation of these results to similar observations in Xenopus is unclear [34]. One sugar kinase, product of the ush gene, was down-regulated (1.4-fold) at low salinity, and the malate dehydrogenase gene, mdhA, was up-regulated (1.4-fold) at high salt in NRC-1. The long-chain fatty acid-CoA ligase, coded by lfl3, was up-regulated 2.1-fold in high salt. However, we did not observe a similar change in other genes involved in lipid metabolism [35]. The glutamine synthetase gene, glnA, was down-regulated (1.7-fold) under low salt conditions. The thioredoxin gene, trxA1, which is present on the inverted repeats of the pNRC replicons, was down-regulated under low salt conditions (1.4-fold). One of two aspartate aminotransferase genes, aspC2, was up-regulated under low salt (1.7-fold), and the two subunits of aspartate carbamoyltransferase, coded by pyrB and pyrI (both coded by pNRC200), were up-regulated under high salt (1.8-fold). A xanthine/uracil permease family protein, coded by xup, was down-regulated by 1.6-fold under low salt conditions, and the Na+-driven multidrug efflux pump, coded by the dfr gene, was down-regulated by 2.1-fold also under low salt. The cbiNQ genes, coding for part of the Co2+ transport system, were down-regulated 1.3- to 1.6-fold in low salt; however, this result may reflect the lower growth rate. Additional likely growth-rate dependent genes altered under low salt, included genes coding for the large ribosomal proteins, rpl2p, rpl22p, rpl23p, rpl29p (about 1.5-fold down-regulated) and the H+-transporting ATP synthase subunits, atpIKECFA (1.5 to 1.8-fold down-regulated). It is possible that the changes in gene expression of many of these salt-responsive metabolic genes in NRC-1 may be indirect, most likely as a result of growth rate changes.
Growth in the cold
Optimum growth for Halobacterium sp. NRC-1 occurs at 42°C, but measurable growth is observed down to a temperature of 15°C, with very slow growth at temperatures as low as 10°C [11,36]. The growth temperature optimum and minimum for NRC-1 are typical of most haloarchaea, but significantly higher than for some cold-adapted species, e.g. Halorubrum lacusprofundi, where the temperature minimum for growth has been reported down to -2°C [36,37].
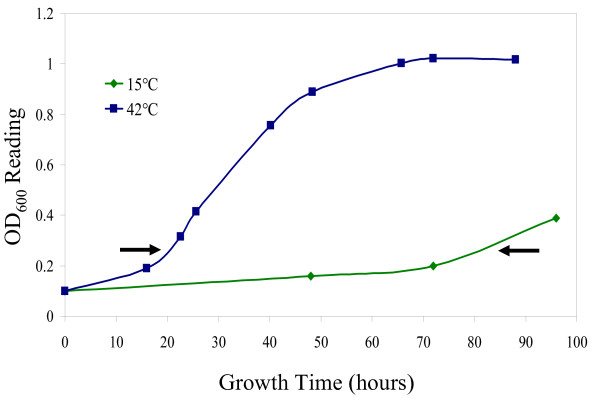
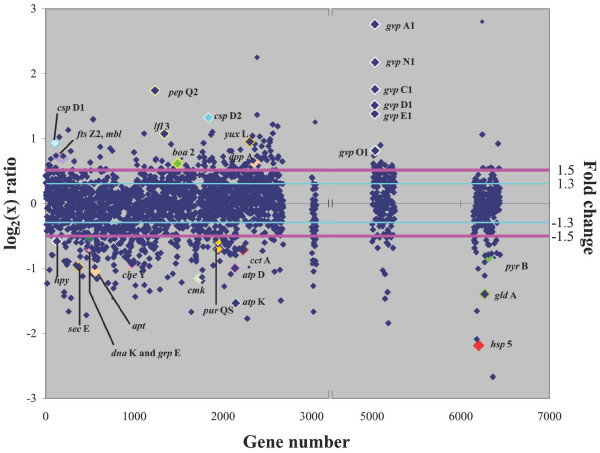
In order to determine which genes in Halobacterium sp. NRC-1 are responsive to reduced temperatures, cultures were grown at low (15°C) and optimal (42°C) temperatures (Fig. 5). Growth at 15°C resulted in up-regulation of 151 genes and down-regulation of 287 genes by 1.5-fold or greater (Fig. 6). Interestingly, 29% of the up-regulated and 37% of the down-regulated genes are of unknown function, suggesting the involvement of novel genes in adaptation of Halobacterium sp. NRC-1 to cold temperature.
Figure 5.
Growth curves of Halobacterium sp. NRC-1 cultures grown in cold (15°C) and standard (42°C) temperatures. Arrows indicate the point where cultures were harvested for microarray analysis.
Figure 6.
Scatter plot comparing DNA microarrays hybridized with cDNA from Halobacterium sp. NRC-1 cultures grown in cold (15°C) and standard (42°C) temperatures. The log2 value of the Cy5/Cy3 ratio for each gene (abscissa) is plotted versus the gene number (ordinate). Gene numbers correspond to the chromosomal genes (1–2679), RNA genes (3000 to 3051), pNRC100 genes (5000 to 5256), and pNRC200 genes (6000 to 6487). Pink line represents a fold change value of 1.5 and the light blue line represents a fold change value of 1.3. Colors have been added for emphasis.
An interesting class of responsive genes was those involved in membrane lipid metabolism. The gene coding for the first step in synthesis of polar lipids, sn-1-glycerol phosphate dehydrogenase (gldA), which is coded by pNRC200, was down regulated 2.7-fold (Table 3), suggesting that polar lipid biosynthesis is altered in Halobacterium sp. NRC-1 cells growing at low temperature. There is also an up-regulation of genes encoding dehydrogenases, such as acd4 (1.5-fold) coding for acyl-CoA dehydrogenase, and genes coding for increased turnover of polar lipids, e.g. lfl3 (2.2-fold), coding for a long-chain fatty acid-CoA ligase, and aca (1.4-fold), coding an acetoacetyl-CoA thiolase. These results are consistent with Halobacterium sp. NRC-1 altering the composition of its lipids in the cold, as has been previously reported in other microorganisms [38-42].
Table 3.
Selected genes significantly altered under low temperature growth
| Gene ID | Gene name | Former name | COG | Predicted function | Fold change values | log2(x) ratio | Standard deviation of log2(x) ratio |
|---|---|---|---|---|---|---|---|
| 101 | cspD1 | 1278 | cold shock protein | 2.307 | 0.935 | 0.912 | |
| 134 | hpyA | 2036 | archaeal histone | -1.525 | -0.584 | 0.269 | |
| 153 | mbl | vng153 | mreB-like protein | 1.685 | 0.708 | 0.365 | |
| 192 | ftsZ2 | 206 | cell division protein | 1.635 | 0.696 | 0.203 | |
| 254 | tfbG | 1405 | transcription initiation factor IIB | 2.259 | 1.132 | 0.356 | |
| 375 | secE | 2443 | protein translocase | -2.004 | -0.982 | 0.244 | |
| 491 | dnaK | 443 | heat shock protein | -1.654 | -0.695 | 0.303 | |
| 494 | grpE | 576 | heat shock protein | -1.477 | -0.522 | 0.353 | |
| 559 | apt | 503 | adenine phosphoribosyltransferase | -2.143 | -1.041 | 0.410 | |
| 679 | acd4 | 1960 | acyl-CoA dehydrogenase | 1.516 | 0.531 | 0.477 | |
| 683 | fbaA | 1830 | possible 1,6-fructose biphosphate aldolase | -1.606 | -0.673 | 0.171 | |
| 823 | gspE2 | 0630 | type II secretion system protein | -1.624 | -0.623 | 0.472 | |
| 887 | gyrB | 187 | DNA gyrase subunit B | -1.422 | -0.458 | 0.397 | |
| 905 | pmu2 | 1109 | phosphomannomutase | -1.800 | -0.833 | 0.206 | |
| 974 | cheY | 784 | chemotaxis protein | -1.956 | -0.923 | 0.349 | |
| 1233 | pepQ2 | 6 | X-pro aminopeptidase homolog | 3.430 | 1.741 | 0.330 | |
| 1339 | lfl3 | 318 | long-chain fatty-acid-CoA ligase | 2.174 | 1.075 | 0.366 | |
| 1488 | boa2 | 3413 | bacterio-opsin activator-like protein | 1.551 | 0.612 | 0.231 | |
| 1690 | rpl4e | 0088 | 50S ribosomal protein L4E | -1.685 | -0.675 | 0.474 | |
| 1703 | rps4e | 1471 | 30S ribosomal protein S4E | -1.598 | -0.599 | 0.477 | |
| 1727 | cmk | 1102 | cytidydylate kinase | -2.254 | -1.154 | 0.231 | |
| 1836 | cspD2 | 1278 | cold shock protein | 3.112 | 1.322 | 0.990 | |
| 1944 | purS | vng1944 | 1828 | phosphoribosylformylglycinamidine synthase, PurS component | -1.523 | -0.606 | 0.032 |
| 1945 | purQ | purL2 | 47 | phosphoribosylformylglycinamide synthase I, glutamine amidotransferase domain | -1.509 | -0.590 | 0.108 |
| 2063 | aca | 183 | probable acetyl-CoA acetyltransferase | 1.421 | 0.483 | 0.263 | |
| 2135 | atpD | 1394 | H+-transporting ATP synthase subunit D | -2.012 | -0.998 | 0.173 | |
| 2138 | atpB | 1156 | H+-transporting ATP synthase subunit B | -1.381 | -0.449 | 0.221 | |
| 2142 | atpE | 1390 | H+-transporting ATP synthase subunit E | -1.716 | -0.728 | 0.389 | |
| 2143 | atpK | 636 | H+-transporting ATP synthase subunit K | -2.926 | -1.535 | 0.203 | |
| 2144 | atpI | 1269 | H+-transporting ATP synthase subunit I | -1.802 | -0.798 | 0.388 | |
| 2226 | cctA | 459 | Thermosome subunit alpha | -1.735 | -0.713 | 0.489 | |
| 2302 | yuxL | 1506 | acylaminoacyl-peptidase | 2.096 | 0.951 | 0.601 | |
| 2349 | dppA | 747 | dipeptide ABC transporter dipeptide-binding | 1.609 | 0.634 | 0.486 | |
| 5028 | gvpE1 | GvpE protein | 3.097 | 1.381 | 0.883 | ||
| 5029 | gvpD1 | GvpD protein | 3.396 | 1.513 | 0.875 | ||
| 5030 | gvpA1 | GvpA protein | 8.018 | 2.761 | 0.860 | ||
| 5032 | gvpC1 | GvpC protein | 3.985 | 1.761 | 0.844 | ||
| 5033 | gvpN1 | GvpN protein | 5.161 | 2.174 | 0.764 | ||
| 5034 | gvpO1 | GvpO protein | 1.824 | 0.814 | 0.395 | ||
| 6201 | hsp5 | 71 | heat shock protein | -4.784 | -2.187 | 0.446 | |
| 6270 | gldA | 371 | sn-glycerol-1-phosphate dehydrogenase | -2.676 | -1.396 | 0.266 | |
| 6309 | pyrB | 540 | aspartate carbamoyltransferase catalytic subunit | -1.767 | -0.817 | 0.121 | |
| 6311 | pyrI | 1781 | aspartate carbamoyltransferase regulatory chain | -1.835 | -0.855 | 0.244 |
The genomes of several haloarchaea have shown the presence of multiple cold shock genes, the products of which may bind to single-stranded DNA and RNA, functioning as RNA chaperones, and facilitating the initiation of translation under low temperatures [36,43,44]. As expected, expression of both cold shock genes, cspD1 and cspD2, was altered in NRC-1 cells during growth at 15°C, with the former up-regulated 2.3-fold and the latter up-regulated 3.1-fold. In addition, three heat shock genes, dnaK, grpE and hsp5, were down-regulated from 1.5- to 4.8-fold in the cold. Most of these changes, some of which were among the highest fold-changes observed in our investigations, were reversed when cells were heat shocked (see below).
A striking observation was that gas vesicle gene expression and content were increased when Halobacterium sp. NRC-1 was grown in the cold. In particular, the rightward transcribed genes of the buoyant gas vesicle gene cluster (gvpACNO coded on both pNRC replicons), were significantly up-regulated, showing increases from 1.8- to 8.0-fold in the cold. The first two genes of the leftward operon, gvpDE, which encode putative regulators for gas vesicle biosynthesis, were also up-regulated (3- and 3.4-fold). Microscopic examination of cells yielded results that were consistent with the microarray data, i.e. an increase in gas vesicle content observable in cells grown at lower temperatures (data not shown).
Previously, increases in the supercoiling of DNA have been reported upon cold shock in E. coli [45,46]; congruently, the levels of certain DNA topoisomerases change during cold shock [47,48]. Surprisingly, the Halobacterium sp. NRC-1 gyrB gene was found to be down-regulated 1.4-fold in the cold, and we did not observe significant changes in gyrA, top6AB, or topA. Interestingly, the archaeal histone gene (hpyA) was down-regulated (1.5-fold), while the actin (mbl) and tubulin-like (ftsZ2) genes were up-regulated (1.7- and 1.6-fold respectively) in the cold, suggesting reduced need for genomic compaction but increased requirement for intracellular organization in the cold.
At reduced temperatures, the solubility of gases and the stability of toxic reactive oxygen species may increase [49,50]. However, we did not observe a corresponding increase in either superoxide dismutase (sod1 and sod2) gene or the glutathione S-transferase (gst) gene. Further, the peptidyl-prolyl cis-trans isomerase (slyD) and the peptidyl-prolyl isomerase (ppiA) genes were not up regulated. These results suggest that at low temperatures, NRC-1 does not enhance the use of these gene products to remove oxygen radicals or interact with hydrophobic patches and aid in the folding of proteins [51].
A variety of important genes necessary for metabolism and cellular functions were altered during growth at 15°C, likely as a result of decreased growth rate. Genes coding for the H+-transporting ATP synthase (atpIKEBD), several of the 30 and 50S ribosomal proteins (rps and rpl genes), transport (secE, gspE2), purine/pyrimidine metabolism (purSQ, apt, cmk, pyrBI), and chemotaxis (cheY) were all down-regulated (1.4 to 3.0-fold). For genes of carbohydrate metabolism, fbaA, a sugar aldolase, and pmu2, a sugar kinase, were about 1.7-fold down-regulated. Among genes related to peptide metabolism, e.g. the dipeptide transporter coded by the dpp gene, dppA, was up-regulated (1.6-fold), as were two aminopeptidase genes, pepQ2 (3.4-fold) and yuxL (2.1-fold). These findings suggest an effort to increase the amino acid pool available to cells in the cold; however, decreased growth rate would result from inhibition of ATP production, protein synthesis and export, DNA metabolism, and taxis.
Heat shock
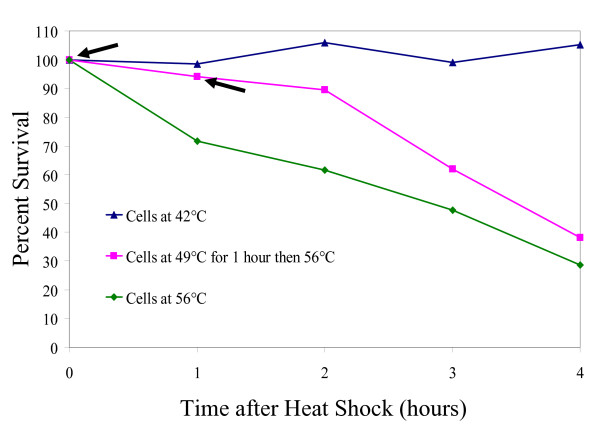
Halobacterium sp. NRC-1 is known to be slightly thermotolerant, exhibiting relatively normal growth up to 48°C. As the temperature is increased from the growth optimum of 42°C, the growth rate declines and temperatures above 50°C prevent growth and provoke photobleaching. After treatment of cells at 49°C, normal growth resumes after shifting back to more moderate temperatures. Pretreatment of NRC-1 cells at 49°C for 1 hour, and then shifting to 56°C, increased survival 2.5-fold compared to cells directly shifted from 42 to 56°C, indicating a classic heat shock effect (Fig. 7) [52].
Figure 7.
Percent survival of Halobacterium sp. NRC-1 after heat shock at 56°C with or without incubation at an intermediate temperature (49°C). Arrows indicate the point where cultures were harvested for microarray analysis.
In order to determine which genes are responsive to heat shock, Halobacterium sp. NRC-1 cultures which were shifted for one hour from 42 to 49°C were compared to cultures remaining at 42°C. Heat shock at 49°C resulted in up-regulation of 64 genes and down-regulation of 43 genes by 1.5-fold or greater (Fig. 8). Genes of unknown function constituted 34% of the up-regulated and 28% of the down-regulated genes.
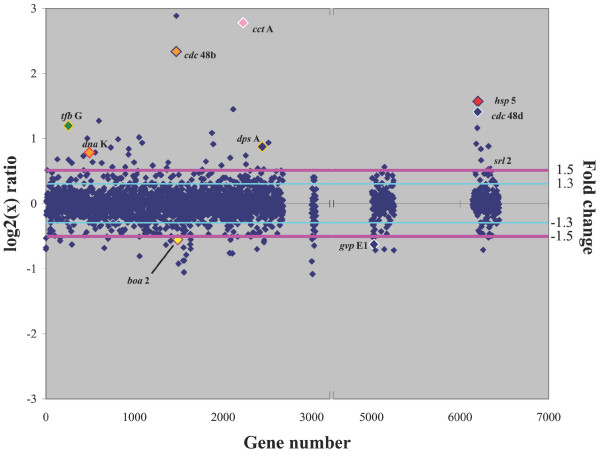
Figure 8.
Scatter plot comparing DNA microarrays hybridized with cDNA from Halobacterium sp. NRC-1 cultures with or without heat shock (at 49°C). The log2 value of the Cy5/Cy3 ratio for each gene (abscissa) is plotted versus the gene number (ordinate). Gene numbers correspond to the chromosomal genes (1–2679), RNA genes (3000 to 3051), pNRC100 genes (5000 to 5256), and pNRC200 genes (6000 to 6487). Pink line represents a fold change value of 1.5 and the light blue line represents a fold change value of 1.3. Colors have been added for emphasis.
Of the four genes coding small heat shock proteins belonging to the Hsp20/α-crystallin family in Halobacterium sp. NRC-1, which are known to protect against irreversible aggregation of cellular proteins and assist in protein refolding during stress conditions [53], hsp5 was the most highly affected, being up-regulated 3.0-fold after heat shock (Table 4). Among genes encoding the dnaK/dnaJ/grpE Hsp70 family chaperones involved in disaggregation and reactivation of proteins [54], only the dnaK gene was found to be highly up-regulated, 1.8-fold after heat shock. The cctA gene, coding the chaperonin-containing t-complex polypeptide (Cct) thermosome family was up-regulated 8.7-fold [53]. This family has been hypothesized to be involved in general environmental stress [55-57]; however, in this transcriptomic study, we only observed an increase in transcript under heat shock in NRC-1. In addition, members of the AAA+ ATPase class have been found to be up-regulated during heat shock and aid in the refolding of proteins [58,59]. Two members of this class of genes, cdc48b (5.6-fold) and cdc48d (3.4-fold), the latter of which is coded by pNRC200, were significantly up-regulated during heat shock.
Table 4.
Selected genes significantly altered under heat shock
| Gene ID | Gene name | Former name | COG | Predicted function | Fold change values | log2(x) ratio | Standard deviation of log2(x) ratio |
|---|---|---|---|---|---|---|---|
| 254 | tfbG | 1405 | transcription initiation factor IIB | 2.412 | 1.199 | 0.450 | |
| 491 | dnaK | 443 | heat shock protein | 1.747 | 0.786 | 0.233 | |
| 1472 | cdc48b | 464 | cell division cycle protein | 5.631 | 2.340 | 0.637 | |
| 1488 | boa2 | 3413 | bacterio-opsin activator-like protein | -1.471 | -0.553 | 0.107 | |
| 2226 | cctA | 459 | thermosome subunit alpha | 8.712 | 2.777 | 1.038 | |
| 2443 | dpsA | 783 | DNA protecting protein under starved conditions | 1.854 | 0.876 | 0.205 | |
| 5028 | gvpE1 | GvpE protein | -1.553 | -0.626 | 0.162 | ||
| 6199 | cdc48d | 464 | cell division cycle protein | 3.371 | 1.413 | 1.080 | |
| 6201 | hsp5 | 71 | heat shock protein | 3.050 | 1.573 | 0.313 | |
| 6320 | srl2 | vng6320 | Smc and Rad50 like ATPase | 1.471 | 0.493 | 0.420 |
Some researchers have reported an increase in DNA repair proteins during heat shock responses [60,61]. In our experiments, about 5% of the up-regulated genes were involved with DNA metabolism; however, only one DNA repair gene, srl2, a SMC/Rad50-like ATPase found on pNRC200 was slightly up-regulated (1.5-fold). Among other genes coding DNA binding proteins, we also observed an increase in dpsA (1.9-fold), a stress inducible DNA binding protein, consistent with an earlier proteomic study [52].
Many genes seem to be coordinately regulated when Halobacterium sp. NRC-1 is exposed to the cold and heat, including twenty-five genes that were inversely regulated under the two conditions. Although many of these genes are of unknown function, several genes known to be involved in heat shock (hsp5, dnaK, cctA), as well as two putative regulators (gvpE1 and boa2) were identified. In addition, ten genes were regulated in the same direction under both conditions, including tfbG (2.4-fold up-regulated), one of seven TFB genes in NRC-1.
Conclusion
Using whole genome microarrays, we have identified genes likely to be involved in adaptation of the model halophilic Archaeon Halobacterium sp. NRC-1 to environmental stresses. Under all environmental stresses examined thus far, well-characterized stress response genes were observed; however, expression changes in unidentified or novel genes were also common. Heat shock showed induction of several chaperone genes, likely to protect cellular proteins from denaturation and breakdown. Growth in the cold suggested the alteration of lipid metabolism, an increased potential for flotation (to escape to a warmer zone) and a slowdown of protein production (possibly in preparation for dormancy). Growth in high salinity resulted in down-regulation of selected ion transporters, presumably to reduce the entry of toxic species. Responses to growth in low salinity also pointed to the need to maintain proper intracellular ionic conditions via the modulation of many transporter genes. Further bioinformatic and genetic analysis of the genes responsive to the many stressors that halophilic archaea respond to in their environment will lead to a fuller understanding of the biology of these interesting microbes.
Methods
Culturing and DNA microarray analysis
Halobacterium sp. NRC-1 cultures were grown in standard CM+ medium with shaking at 220 rpm on a New Brunswick Scientific platform shaker. For growth in the cold, cultures were aerobically grown at 15°C. For heat shock experiments, cells were grown at 42°C followed by incubation at 49°C for one hour and 56°C for three or four hours. For salt experiments, cells were grown at 42°C with either 2.9, 4.3, or 5.0 M NaCl. Pre-cultures grown under the same conditions used in the growth curve were used as inocula. For the heat killing profile, Halobacterium sp. NRC-1 cultures used were grown under standard laboratory conditions (aerobically at 42°C in CM+ medium). Each growth curve and heat killing profile was based on the average of three cultures. For each microarray experiment, three cultures were grown in the same manner as those used for the growth curves and RNA was pooled from all three cultures for cDNA synthesis and hybridization. Cultures grown at high and low salinities and 15°C were harvested at early exponential phase (OD600 = 0.19 to 0.23) for microarray analysis. Cultures used for the heat shock microarrays were grown to early exponential phase and then incubated at 49°C for 1 hour. Before harvesting the cells to collect RNA, cultures were swirled briefly in an ethanol-dry ice bath to rapidly cool the cultures and "freeze" the RNA profile. Total RNA was isolated using the Agilent total RNA isolation mini kit (Agilent Technologies, Palo Alto, CA) and then treated with RNase-free DNase I. cDNA was prepared with equal amounts of RNA from control and experimental samples and then fluorescently labeled with Cy3-dCTP and Cy5-dCTP. Concentrations of RNA and cDNA were measured using a Nanodrop (ND-1000) spectrophotometer (NanoDrop Technologies, Wilmington, DE). Incorporation of the Cy3 and Cy5 labels was checked via gel electrophoresis and scanning on a GE Typhoon fluorescence scanner (GE Healthcare, Piscataway, NJ). Washing and hybridization of the arrays was performed as recommended by Agilent and as previously described [5]. Slides were scanned for Cy-3 and Cy-5 signals with an Agilent DNA-microarray scanner. Probe signals were extracted with the Agilent Feature Extraction Software and analyzed using the statistical methods described below. Oligonucleotide arrays, in situ synthesized using ink-jet technology, were used for transcriptome analysis of Halobacterium sp. NRC-1. Oligomer (60-mer) probes were previously designed for 2474 ORFs utilizing the program OligoPicker [16]. Two replicate microarrays were performed for the high and low salinity and 15°C growth experiments, and three replicate microarrays were performed for the heat shock experiment. Data shown are based on the analysis of all arrays performed for each of the given conditions.
Microarray data processing
The Agilent Feature Extraction program was used for image analysis and processing of the microarray image file. Background signals were subtracted from raw signals using the area either on or around the features. Dye biases created by differences in the red and green channel signals caused by different efficiencies in labeling, emission and detection were also estimated and corrected. Signals from each channel were normalized using the LOWESS algorithm to remove intensity-dependent effects within the calculated values. Further statistical analyses were performed as described below.
Statistical analysis
The illuminant intensity, log2(x) value, and standard deviation of the log2(x) value were calculated for the normalized red and green probe values for each gene in each microarray. The illuminant intensity was calculated through the logarithm of the geometric mean of Cy5 and Cy3 processed signal intensities by first calculating:. Then for a finite-sized sample of size N, we calculated the intensity using the arithmetic mean: . Biased estimators for sample means of log2(x) ratios were calculated by the arithmetic mean of log2(x) ratios for a set of N probes for a gene; where ri is the processed Cy5 illuminant intensity level and gi is the processed Cy3 illuminate intensity value for the ith probe: [62]. Standard deviations for sample means of log2(x) ratios were then calculated: . Changes in transcript levels were considered significant if they were changed more than 1.3- to 1.5-fold using a linear transform function, with the upper figure used to calculate total numbers of altered genes under specific conditions. See Tables 1, 2, 3, 4 for a listing of notable regulated genes, their fold change, log2(x) value, and standard deviation under the high and low salinity and temperature conditions tested.
Database and cluster analysis
All microarray data were stored in our HaloArray database [63] mirrored on two Linux servers. The systems use Apache server software, MySQL databases, and custom Perl script-based webtools designed for access and data analysis. This database is a part of our comprehensive HaloWeb database [64], which includes our genome sequence and annotation data, and is indexed in the Thompson ISI Web of Knowledge and Current Web Contents.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
SD conceived the study, and JAC and PD conducted the experimental analysis, and together with SD, analyzed the data and wrote the manuscript. JAC and JK did the statistical analysis and JK developed the HaloArray database and microarray analysis tool, and JAM designed the microarrays and initiated the DNA microarrays experiments. All authors read and approved the final manuscript.
Contributor Information
James A Coker, Email: coker@umbi.umd.edu.
Priya DasSarma, Email: dassarmp@umbi.umd.edu.
Jeffrey Kumar, Email: kumar@umbi.umd.edu.
Jochen A Müller, Email: jmueller@jewel.morgan.edu.
Shiladitya DasSarma, Email: dassarma@umbi.umd.edu.
Acknowledgements
We would like to thank the reviewers for helping to improve the manuscript. This work was funded by NSF grants MCB-029617 and 0450695.
References
- DasSarma S. Extreme microbes. American Scientist. 2007;95:224–231. [Google Scholar]
- DasSarma S. Extreme halophiles are models for astrobiology. Microbe. 2006;1:120–126. [Google Scholar]
- DasSarma S, Berquist BR, Coker JA, DasSarma P, Müller JA. Post-genomics of the model haloarchaeon Halobacterium sp. NRC-1. Saline Systems. 2006;2:3. doi: 10.1186/1746-1448-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliga NS, Kennedy SP, Ng WV, Hood L, DasSarma S. Genomic and genetic dissection of an archaeal regulon. Proc Natl Acad Sci U S A. 2001;98:2521–2525. doi: 10.1073/pnas.051632498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller JA, DasSarma S. Genomic analysis of anaerobic respiration in the archaeon Halobacterium sp. strain NRC-1: dimethyl sulfoxide and trimethylamine N-oxide as terminal electron acceptors. J Bacteriol. 2005;187:1659–1667. doi: 10.1128/JB.187.5.1659-1667.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCready S, Müller JA, Boubriak I, Berquist BR, Ng WL, DasSarma S. UV irradiation induces homologous recombination genes in the model archaeon, Halobacterium sp. NRC-1. Saline Systems. 2005;1:3. doi: 10.1186/1746-1448-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVeaux LC, Müller JA, Smith J, Petrisko J, Wells DP, DasSarma S. Extremely radiation-resistant mutants of a halophilic archaeon with increased single-strand DNA binding protein (RPA) gene expression. Radiation Res. 2007;(in press) doi: 10.1667/RR0935.1. [DOI] [PubMed] [Google Scholar]
- Baliga NS, Bjork SJ, Bonneau R, Pan M, Iloanusi C, Kottemann MC, Hood L, DiRuggiero J. Systems level insights into the stress response to UV radiation in the halophilic archaeon Halobacterium NRC-1. Genome Res. 2004;14:1025–1035. doi: 10.1101/gr.1993504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead K, Kish A, Pan M, Kaur A, Reiss DJ, King N, Hohmann L, DiRuggiero J, Baliga NS. An integrated systems approach for understanding cellular responses to gamma radiation. Mol Syst Biol. 2006;2:47. doi: 10.1038/msb4100091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WV, Ciufo SA, Smith TM, Bumgarner RE, Baskin D, Faust J, Hall B, Loretz C, Seto J, Slagel J, Hood L, DasSarma S. Snapshot of a large dynamic replicon in a halophilic archaeon: megaplasmid or minichromosome? Genome Res. 1998;8:1131–1141. doi: 10.1101/gr.8.11.1131. [DOI] [PubMed] [Google Scholar]
- Ng WV, Kennedy SP, Mahairas GG, Berquist B, Pan M, Shukla HD, Lasky SR, Baliga NS, Thorsson V, Sbrogna J, Swartzell S, Weir D, Hall J, Dahl TA, Welti R, Goo YA, Leithauser B, Keller K, Cruz R, Danson MJ, Hough DW, Maddocks DG, Jablonski PE, Krebs MP, Angevine CM, Dale H, Isenbarger TA, Peck RF, Pohlschroder M, Spudich JL, Jung KW, Alam M, Freitas T, Hou S, Daniels CJ, Dennis PP, Omer AD, Ebhardt H, Lowe TM, Liang P, Riley M, Hood L, DasSarma S. Genome sequence of Halobacterium species NRC-1. Proc Natl Acad Sci U S A. 2000;97:12176–12181. doi: 10.1073/pnas.190337797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasSarma S. In: Microbial Genomes. Fraser CM, Read TD and Nelson KE, editor. Totowa, NJ, Humana Press, Inc.; 2004. Genome sequence of an extremely halophilic archaeon; pp. 383–399. [Google Scholar]
- Berquist BR, Müller JA, DasSarma S. In: Extremophiles. Oren A and Rainey F, editor. Vol. 35 , Elsevier/Academic Press; 2006. Genetic systems for halophilic archaea; pp. 649–680. (Methods in Microbiology). [Google Scholar]
- Peck RF, Echavarri-Erasun C, Johnson EA, Ng WV, Kennedy SP, Hood L, DasSarma S, Krebs MP. brp and blh are required for synthesis of the retinal cofactor of bacteriorhodopsin in Halobacterium salinarum. J Biol Chem. 2001;276:5739–5744. doi: 10.1074/jbc.M009492200. [DOI] [PubMed] [Google Scholar]
- Wang G, Kennedy SP, Fasiludeen S, Rensing C, DasSarma S. Arsenic resistance in Halobacterium sp. strain NRC-1 examined by using an improved gene knockout system. J Bacteriol. 2004;186:3187–3194. doi: 10.1128/JB.186.10.3187-3194.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, Lefkowitz SM, Ziman M, Schelter JM, Meyer MR, Kobayashi S, Davis C, Dai H, He YD, Stephaniants SB, Cavet G, Walker WL, West A, Coffey E, Shoemaker DD, Stoughton R, Blanchard AP, Friend SH, Linsley PS. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- Roberts MF. Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Systems. 2005;1:5. doi: 10.1186/1746-1448-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy SP, Ng WV, Salzberg SL, Hood L, DasSarma S. Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence. Genome Res. 2001;11:1641–1650. doi: 10.1101/gr.190201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, DasSarma S. Isolation and chromosomal distribution of natural Z-DNA-forming sequences in Halobacterium halobium. J Biol Chem. 1996;271:19724–19731. doi: 10.1074/jbc.271.33.19724. [DOI] [PubMed] [Google Scholar]
- Kim J, Yang C, DasSarma S. Analysis of left-handed Z-DNA formation in short d(CG)n sequences in Escherichia coli and Halobacterium halobium plasmids. Stabilization by increasing repeat length and DNA supercoiling but not salinity. J Biol Chem. 1996;271:9340–9346. doi: 10.1074/jbc.271.16.9340. [DOI] [PubMed] [Google Scholar]
- DasSarma S, DasSarma P. Encyclopedia of Life Sciences. , Wiley Press; 2006. Halophiles. [Google Scholar]
- Robert H, Le Marrec C, Blanco C, Jebbar M. Glycine betaine, carnitine, and choline enhance salinity tolerance and prevent the accumulation of sodium to a level inhibiting growth of Tetragenococcus halophila. Appl Environ Microbiol. 2000;66:509–517. doi: 10.1128/AEM.66.2.509-517.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanyi JK. Salt-dependent properties of proteins from extremely halophilic bacteria. Bacteriol Rev. 1974;38:272–290. doi: 10.1128/br.38.3.272-290.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner DJ. Lysis and dissolution of cells and envelopes of an extremely halophilic bacterium. J Bacteriol. 1964;87:1147–1156. doi: 10.1128/jb.87.5.1147-1156.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenchov B, Vescio EM, Sprott GD, Zeidel ML, Mathai JC. Salt tolerance of archaeal extremely halophilic lipid membranes. J Biol Chem. 2006;281:10016–10023. doi: 10.1074/jbc.M600369200. [DOI] [PubMed] [Google Scholar]
- Kaur A, Pan M, Meislin M, Facciotti MT, El-Gewely R, Baliga NS. A systems view of haloarchaeal strategies to withstand stress from transition metals. Genome Res. 2006;16:841–854. doi: 10.1101/gr.5189606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoeva MV, Storch KF, Klein C, Oesterhelt D. A novel mode of sensory transduction in archaea: binding protein-mediated chemotaxis towards osmoprotectants and amino acids. EMBO J. 2002;21:2312–2322. doi: 10.1093/emboj/21.10.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao HL, Pih KT, Lim JH, Kang SG, Jin JB, Kim SH, Hwang I. An Arabidopsis GSK3/shaggy-like gene that complements yeast salt stress-sensitive mutants is induced by NaCl and abscisic acid. Plant Physiol. 1999;119:1527–1534. doi: 10.1104/pp.119.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoumskaya MA, Paithoonrangsarid K, Kanesaki Y, Los DM, Zinchenko VV, Tanticharoen M, Suzuki I, Murata N. Identical Hik-Rre systems are involved in perception and transduction of salt signals and hyperosmotic signals but regulate the expression of individual genes to different extents in Synechocystis. J Biol Chem. 2005;280:21531–21538. doi: 10.1074/jbc.M412174200. [DOI] [PubMed] [Google Scholar]
- Koch MK, Oesterhelt D. MpcT is the transducer for membrane potential changes in Halobacterium salinarum. Mol Microbiol. 2005;55:1681–1694. doi: 10.1111/j.1365-2958.2005.04516.x. [DOI] [PubMed] [Google Scholar]
- Cheeseman JM, Herendeen LB, Cheeseman AT, Clough BF. Photosynthesis and photoprotection in mangrove under field conditions. Plant Cell Environ. 1997;20:579–588. doi: 10.1111/j.1365-3040.1997.00096.x. [DOI] [Google Scholar]
- Townsend DM, Tew KD. The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene. 2003;22:7369–7375. doi: 10.1038/sj.onc.1206940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels CJ, McKee AH, Doolittle WF. Archaebacterial heat-shock proteins. EMBO J. 1984;3:745–749. doi: 10.1002/j.1460-2075.1984.tb01878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindley TE, Laberge T, Hall A, Hewett-Emmett D, Walsh PJ, Anderson PM. Sequence, expression and evolutionary relationships of carbamoyl phosphate synthetase I in the toad Xenopus laevis. J Exp Zool Part A Ecol Genet Physiol. 2007;307:163–175. doi: 10.1002/jez.a.364. [DOI] [PubMed] [Google Scholar]
- Bidle KA, Hanson TE, Howell K, Nannen J. HMG-CoA reductase is regulated by salinity at the level of transcription in Haloferax volcanii. Extremophiles. 2007;11:49–55. doi: 10.1007/s00792-006-0008-3. [DOI] [PubMed] [Google Scholar]
- Reid IN, Sparks WB, Lubow S, McGrath M, Livio M, Valenti J, Sowers KR, Shukla HD, MacAuley S, Miller T, Suvanasuthi R, Belas R, Colman A, Robb FT, DasSarma P, Müller JA, Coker JA, Cavicchioli R, Chen F, DasSarma S. Terrestrial models for extraterrestrial life: methanogens and halophiles at Martian temperatures. International Journal of Astrobiology. 2006;5:89–97. doi: 10.1017/S1473550406002916. [DOI] [Google Scholar]
- Franzmann PD, Stackebrandt E, Sanderson K, Volkman JK, Cameron DE, Stevenson PL, McMeekin TA, Burton HR. Halobacterium lacusprofundi sp. nov., a halophilic bacterium isolated from Deep Lake, Antarctica. Syst Appl Microbiol. 1988;11:20–27. [Google Scholar]
- Russell NJ, Hamamoto T. In: Extremophiles: microbial life in extreme environments. Horikoshi K and Grant D, editor. New York, Wiley-Liss; 1998. Psychrophiles; pp. 25–45. [Google Scholar]
- Koga Y, Morii H. Biosynthesis of ether-type polar lipids in archaea and evolutionary considerations. Microbiol Mol Biol Rev. 2007;71:97–120. doi: 10.1128/MMBR.00033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols DS, Miller MR, Davies NW, Goodchild A, Raftery M, Cavicchioli R. Cold adaptation in the Antarctic archaeon Methanococcoides burtonii involves membrane lipid unsaturation. J Bacteriol. 2004;186:8508–8515. doi: 10.1128/JB.186.24.8508-8515.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tindall BJ. Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol Lett. 1990;66:199–202. doi: 10.1111/j.1574-6968.1990.tb03996.x. [DOI] [Google Scholar]
- Kamekura M, Kates M. Structural diversity of membrane lipids in members of Halobacteriaceae. Biosci Biotechnol Biochem. 1999;63:969–972. doi: 10.1271/bbb.63.969. [DOI] [PubMed] [Google Scholar]
- Cavicchioli R. Cold-adapted archaea. Nat Rev Microbiol. 2006;4:331–343. doi: 10.1038/nrmicro1390. [DOI] [PubMed] [Google Scholar]
- Giaquinto L, Curmi PM, Siddiqui KS, Poljak A, DeLong E, DasSarma S, Cavicchioli R. The structure and function of cold shock proteins in archaea. J Bacteriol. 2007;189:5738–5748. doi: 10.1128/JB.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima T, Kataoka K, Ogata Y, Inoue R, Sekimizu K. Increase in negative supercoiling of plasmid DNA in Escherichia coli exposed to cold shock. Mol Microbiol. 1997;23:381–386. doi: 10.1046/j.1365-2958.1997.2181582.x. [DOI] [PubMed] [Google Scholar]
- Higgins CF, Dorman CJ, Ní Bhriain N. In: The Bacterial Chromosome. Drlica K and Riley M, editor. Washington D.C., American Society for Microbiology Press; 1990. Environmental influences on DNA supercoiling: a novel mechanism for the regulation of gene expression; pp. 421–432. [Google Scholar]
- Lopez-Garcia P, Forterre P. Control of DNA topology during thermal stress in hyperthermophilic archaea: DNA topoisomerase levels, activities and induced thermotolerance during heat and cold shock in Sulfolobus. Mol Microbiol. 1999;33:766–777. doi: 10.1046/j.1365-2958.1999.01524.x. [DOI] [PubMed] [Google Scholar]
- Weinberg MV, Schut GJ, Brehm S, Datta S, Adams MWW. Cold shock of a hyperthermophile archaeon: Pyrococcus furiosus exhibits multiple responses to a suboptimal growth temperature with a key role for membrane-bound glycoproteins. J Bacteriol. 2005;187:336–348. doi: 10.1128/JB.187.1.336-348.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medigue C, Krin E, Pascal G, Barbe V, Bernsel A, Bertin PN, Cheung F, Cruveiller S, D'Amico S, Duilio A, Fang G, Feller G, Ho C, Mangenot S, Marino G, Nilsson J, Parrilli E, Rocha EP, Rouy Z, Sekowska A, Tutino ML, Vallenet D, von Heijne G, Danchin A. Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 2005;15:1325–1335. doi: 10.1101/gr.4126905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson TA, Karapetian A, Miller AF, Brunold TC. Probing the geometric and electronic structures of the low-temperature azide adduct and the product-inhibited form of oxidized manganese superoxide dismutase. Biochemistry. 2005;44:1504–1520. doi: 10.1021/bi048639t. [DOI] [PubMed] [Google Scholar]
- Ideno A, Yoshida T, Iida T, Furutani M, Maruyama T. FK506-binding protein of the hyperthermophilic archaeon, Thermococcus sp. KS-1, a cold-shock-inducible peptidyl-prolyl cis-trans isomerase with activities to trap and refold denatured proteins. Biochem J. 2001;357:465–471. doi: 10.1042/0264-6021:3570465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla HD. Proteomic analysis of acidic chaperones, and stress proteins in extreme halophile Halobacterium NRC-1: a comparative proteomic approach to study heat shock response. Proteome Sci. 2006;4:6. doi: 10.1186/1477-5956-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narberhaus F. Alpha-crystallin-type heat shock proteins: socializing minichaperones in the context of a multichaperone network. Microbiol Mol Biol Rev. 2002;66:64–93. doi: 10.1128/MMBR.66.1.64-93.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/S0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Yokota SI, Yanagi H, Yura T, Kubota H. Upregulation of cytosolic chaperonin CCT subunits during recovery from chemical stress that causes accumulation of unfolded proteins. Eur J Biochem. 2000;267:1658–1564. doi: 10.1046/j.1432-1327.2000.01157.x. [DOI] [PubMed] [Google Scholar]
- Yamada A, Sekiguchi M, Mimura T, Ozeki Y. The role of plant CCTalpha in salt- and osmotic-stress tolerance. Plant Cell Physiol. 2002;43:1043–1048. doi: 10.1093/pcp/pcf120. [DOI] [PubMed] [Google Scholar]
- Kuo YP, Thompson DK, St. Jean A, Charlebois RL, Daniels CJ. Characterization of two heat shock genes from Haloferax volcanii: a model system for transcription regulation in the Archaea. J Bacteriol. 1997;179:6318–6324. doi: 10.1128/jb.179.20.6318-6324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy M, Akoev V, Zolkiewski M. Domain stability in the AAA+ ATPase ClpB from Escherichia coli. Arch Biochem Biophys. 2006;453:63–69. doi: 10.1016/j.abb.2006.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K, Wantanabe Y, Yohda M, Yoshida M. Heat-inactivated proteins are rescued by the DnaK.J-GrpE set and ClpB chaperones. Proc Natl Acad Sci U S A. 1999;96:7184–7189. doi: 10.1073/pnas.96.13.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockley KR, Ward DE, Chhabra SR, Conners SB, Montero CI, Kelly RM. Heat shock response by the hyperthermophilic archaeon Pyrococcus furiosus. Appl Environ Microbiol. 2003;69:2365–2371. doi: 10.1128/AEM.69.4.2365-2371.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KL, Roberts C, Disz T, Vonstein V, Hwang K, Overbeek R, Olson PD, Projan SJ, Dunman PM. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J Bacteriol. 2006;188:6739–6756. doi: 10.1128/JB.00609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goryachev AB, Macgregor PF, Edwards AM. Unfolding of microarray data. J Comput Biol. 2001;8:443–461. doi: 10.1089/106652701752236232. [DOI] [PubMed] [Google Scholar]
- The HaloArray databases. http://halo2.umbi.umd.edu/cgi-bin/scripts/form2.pl. http://halo2.umbi.umd.edu/cgi-bin/scripts/form2.pl http://halo3.umbi.umd.edu/cgi-bin/scripts/form2.pl.
- The HaloWeb database. http://halo.umbi.umd.edu http://halo.umbi.umd.edu