Abstract
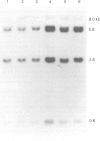
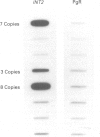
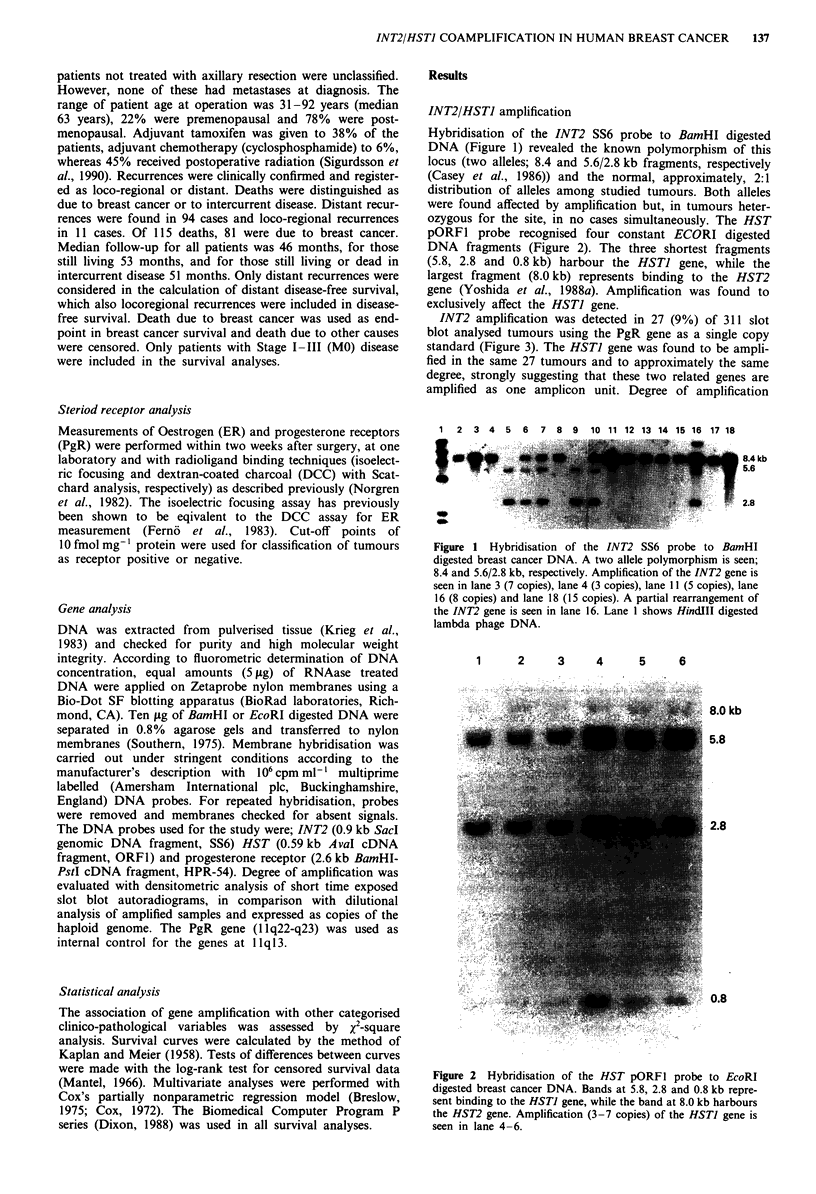
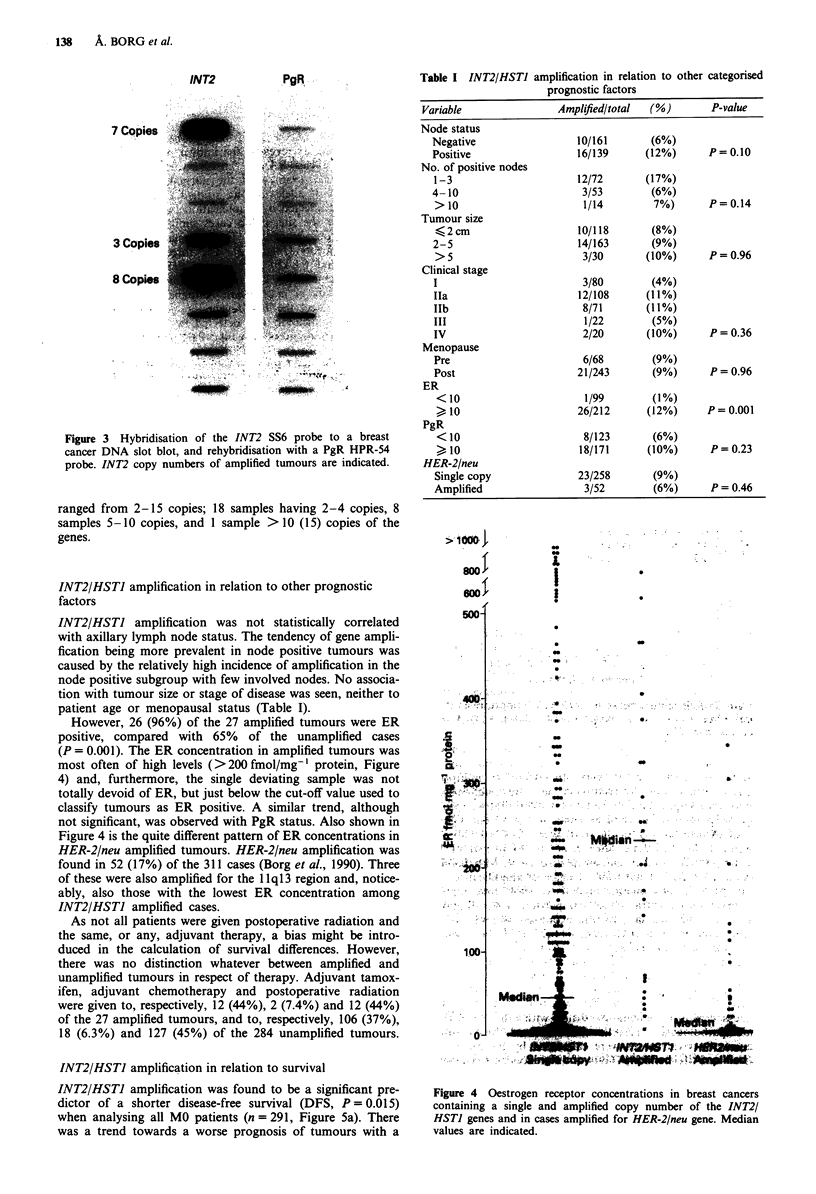
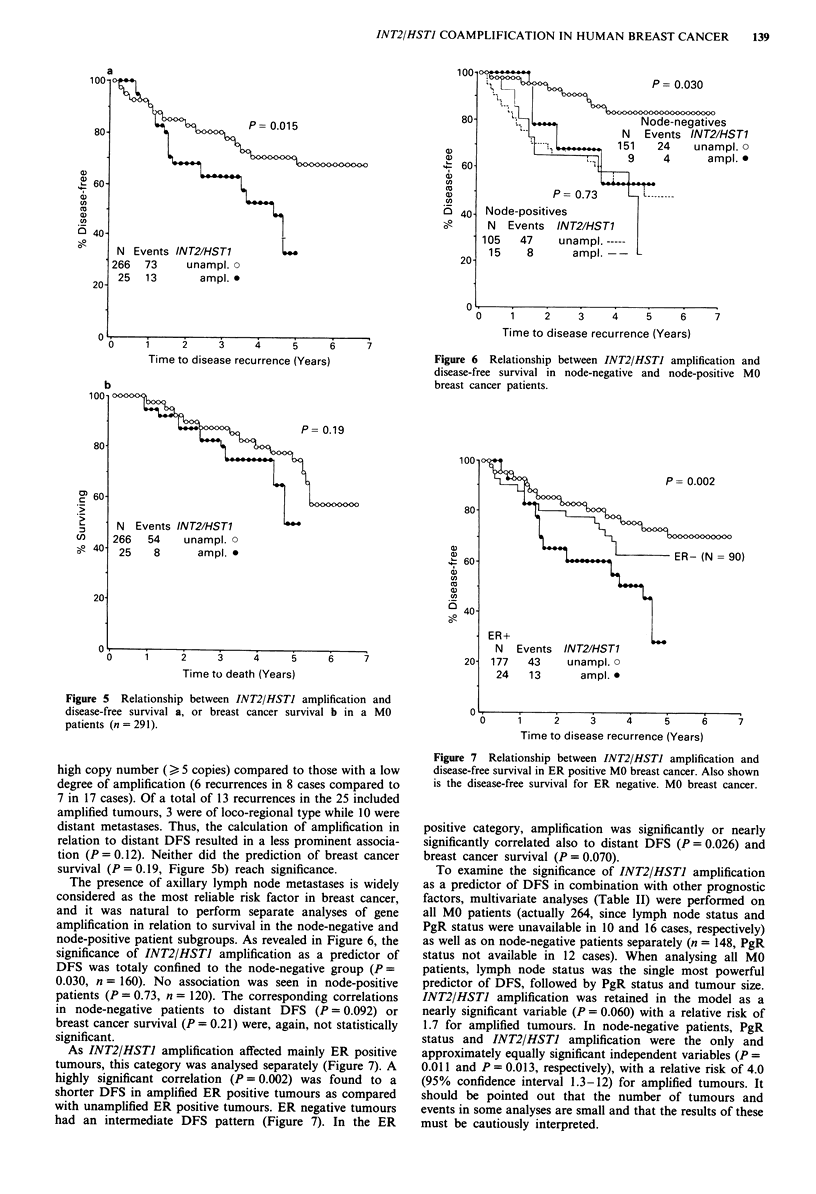
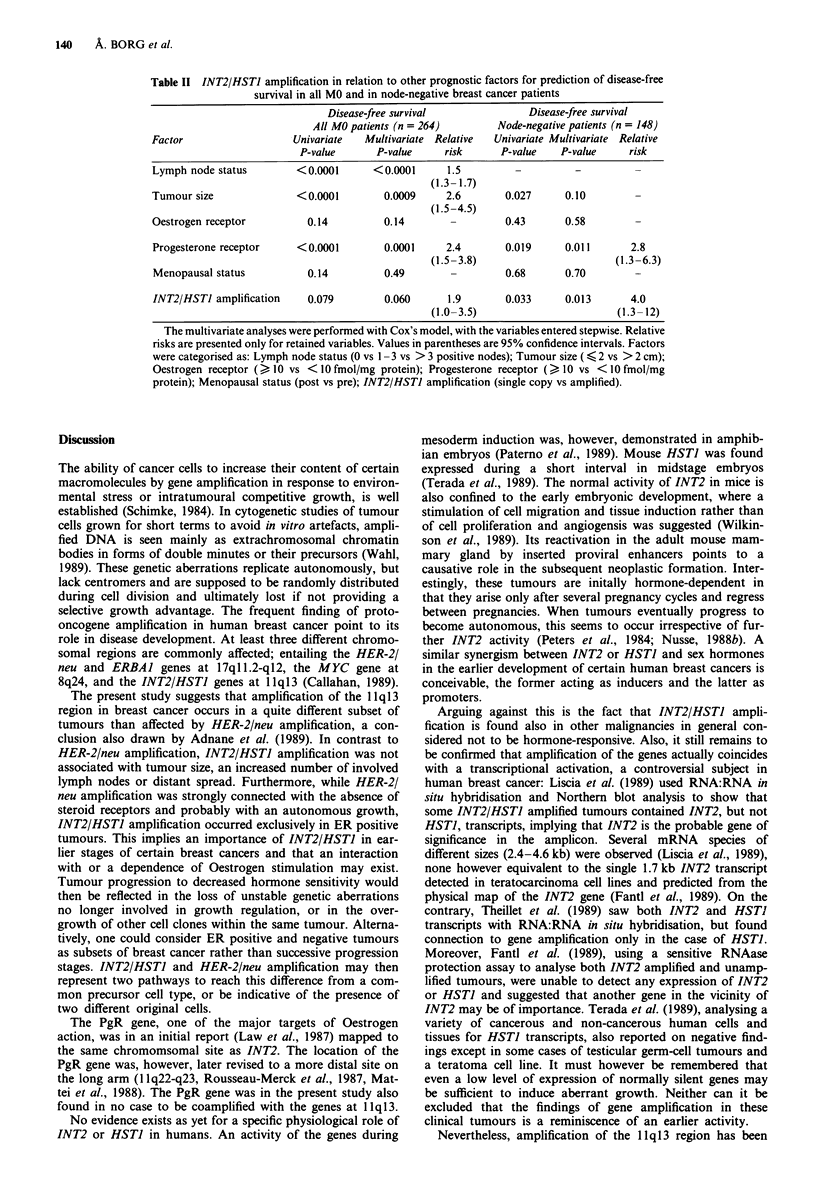
The human proto-oncogene INT2 (homologous to the mouse INT2 gene, implicated in proviral induced mammary carcinoma) has been mapped to chromosome 11q13 and found to share band localisation with, among others, the HST1 proto-oncogene. Both genes are members of the fibroblast growth factor family. In the present study, coamplification (2-15 copies) of the INT2/HST1 genes was found in 27 (9%) of 311 invasive human breast carcinomas using slot blot and Southern blot analyses. Amplification was not correlated to tumour size, axillary lymph node status or stage of disease, neither to patient age nor menopausal status. However, 26 (96%) of the 27 amplified tumours were, often strongly, Oestrogen receptor positive compared to 65% of the unamplified cases (P = 0.001). These findings are in sharp contrast to the strong correlations of HER-2/neu proto-oncogene amplification with advanced stage and steroid receptor negativity, previously observed in the same series of tumours. Patients with INT2/HST1 amplified breast cancer had a significantly shorter disease-free survival compared to those with unamplified genes (P = 0.015, median follow up 45 months). This correlation was confined to node-negative patients and persisted in multivariate analysis. No significant correlation to survival from breast cancer was found. It is concluded that amplification of the 11q13 region in breast cancer occurs in a particular subset of aggressive tumours, quite different from that identified by HER-2/neu amplification. It still remains to be shown that the selection for amplified genes at 11q13 is due to the activity of INT2, HST1 or yet another, still unidentified, neighbouring gene. However, the results are potentially of clinical value in separating a group of node-negative breast cancer for more intense treatment.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adelaide J., Mattei M. G., Marics I., Raybaud F., Planche J., De Lapeyriere O., Birnbaum D. Chromosomal localization of the hst oncogene and its co-amplification with the int.2 oncogene in a human melanoma. Oncogene. 1988 Apr;2(4):413–416. [PubMed] [Google Scholar]
- Adnane J., Gaudray P., Simon M. P., Simony-Lafontaine J., Jeanteur P., Theillet C. Proto-oncogene amplification and human breast tumor phenotype. Oncogene. 1989 Nov;4(11):1389–1395. [PubMed] [Google Scholar]
- Ali I. U., Merlo G., Callahan R., Lidereau R. The amplification unit on chromosome 11q13 in aggressive primary human breast tumors entails the bcl-1, int-2 and hst loci. Oncogene. 1989 Jan;4(1):89–92. [PubMed] [Google Scholar]
- Bale S. J., Bale A. E., Stewart K., Dachowski L., McBride O. W., Glaser T., Green J. E., 3rd, Mulvihill J. J., Brandi M. L., Sakaguchi K. Linkage analysis of multiple endocrine neoplasia type 1 with INT2 and other markers on chromosome 11. Genomics. 1989 Apr;4(3):320–322. doi: 10.1016/0888-7543(89)90336-4. [DOI] [PubMed] [Google Scholar]
- Berenson J. R., Yang J., Mickel R. A. Frequent amplification of the bcl-1 locus in head and neck squamous cell carcinomas. Oncogene. 1989 Sep;4(9):1111–1116. [PubMed] [Google Scholar]
- Borg A., Tandon A. K., Sigurdsson H., Clark G. M., Fernö M., Fuqua S. A., Killander D., McGuire W. L. HER-2/neu amplification predicts poor survival in node-positive breast cancer. Cancer Res. 1990 Jul 15;50(14):4332–4337. [PubMed] [Google Scholar]
- Brookes S., Smith R., Casey G., Dickson C., Peters G. Sequence organization of the human int-2 gene and its expression in teratocarcinoma cells. Oncogene. 1989 Apr;4(4):429–436. [PubMed] [Google Scholar]
- Burgess A. W. Int-1 and int-2: oncogenic proteins, mitogens and morphogens? Bioessays. 1988 Jan;8(1):40–42. doi: 10.1002/bies.950080111. [DOI] [PubMed] [Google Scholar]
- Callahan R. Genetic alterations in primary breast cancer. Breast Cancer Res Treat. 1989 Jul;13(3):191–203. doi: 10.1007/BF02106570. [DOI] [PubMed] [Google Scholar]
- Casey G., Smith R., McGillivray D., Peters G., Dickson C. Characterization and chromosome assignment of the human homolog of int-2, a potential proto-oncogene. Mol Cell Biol. 1986 Feb;6(2):502–510. doi: 10.1128/mcb.6.2.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delli Bovi P., Curatola A. M., Kern F. G., Greco A., Ittmann M., Basilico C. An oncogene isolated by transfection of Kaposi's sarcoma DNA encodes a growth factor that is a member of the FGF family. Cell. 1987 Aug 28;50(5):729–737. doi: 10.1016/0092-8674(87)90331-x. [DOI] [PubMed] [Google Scholar]
- Dickson C., Peters G. Potential oncogene product related to growth factors. 1987 Apr 30-May 6Nature. 326(6116):833–833. doi: 10.1038/326833a0. [DOI] [PubMed] [Google Scholar]
- Fernö M., Borg A., Norgren A. A comparison of two steroid receptor assays in breast cancer: dextran coated charcoal and isoelectric focusing. Anticancer Res. 1983 Jul-Aug;3(4):243–246. [PubMed] [Google Scholar]
- Hatada I., Tokino T., Ochiya T., Matsubara K. Co-amplification of integrated hepatitis B virus DNA and transforming gene hst-1 in a hepatocellular carcinoma. Oncogene. 1988 Nov;3(5):537–540. [PubMed] [Google Scholar]
- Krieg P., Amtmann E., Sauer G. The simultaneous extraction of high-molecular-weight DNA and of RNA from solid tumors. Anal Biochem. 1983 Oct 15;134(2):288–294. doi: 10.1016/0003-2697(83)90299-3. [DOI] [PubMed] [Google Scholar]
- Law M. L., Kao F. T., Wei Q., Hartz J. A., Greene G. L., Zarucki-Schulz T., Conneely O. M., Jones C., Puck T. T., O'Malley B. W. The progesterone receptor gene maps to human chromosome band 11q13, the site of the mammary oncogene int-2. Proc Natl Acad Sci U S A. 1987 May;84(9):2877–2881. doi: 10.1073/pnas.84.9.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidereau R., Callahan R., Dickson C., Peters G., Escot C., Ali I. U. Amplification of the int-2 gene in primary human breast tumors. Oncogene Res. 1988 Feb;2(3):285–291. [PubMed] [Google Scholar]
- Liscia D. S., Merlo G. R., Garrett C., French D., Mariani-Costantini R., Callahan R. Expression of int-2 mRNA in human tumors amplified at the int-2 locus. Oncogene. 1989 Oct;4(10):1219–1224. [PubMed] [Google Scholar]
- Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep. 1966 Mar;50(3):163–170. [PubMed] [Google Scholar]
- Mattei M. G., Krust A., Stropp U., Mattei J. F., Chambon P. Assignment of the human progesterone receptor to the q22 band of chromosome 11. Hum Genet. 1988 Jan;78(1):96–97. doi: 10.1007/BF00291245. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Larsson C., Julier C., Byström C., Skogseid B., Wells S., Oberg K., Carlson M., Taggart T., O'Connell P. Localization of the genetic defect in multiple endocrine neoplasia type 1 within a small region of chromosome 11. Am J Hum Genet. 1989 May;44(5):751–755. [PMC free article] [PubMed] [Google Scholar]
- Norgren A., Borg A., Fernö M., Johansson U., Lindahl B., Tsiobanelis K. Improved method for assay of estradiol and progesterone receptors with special reference to breast cancer. Anticancer Res. 1982 Sep-Oct;2(5):315–320. [PubMed] [Google Scholar]
- Nusse R. The int genes in mammary tumorigenesis and in normal development. Trends Genet. 1988 Oct;4(10):291–295. doi: 10.1016/0168-9525(88)90172-2. [DOI] [PubMed] [Google Scholar]
- Paterno G. D., Gillespie L. L., Dixon M. S., Slack J. M., Heath J. K. Mesoderm-inducing properties of INT-2 and kFGF: two oncogene-encoded growth factors related to FGF. Development. 1989 May;106(1):79–83. doi: 10.1242/dev.106.1.79. [DOI] [PubMed] [Google Scholar]
- Peters G., Brookes S., Smith R., Placzek M., Dickson C. The mouse homolog of the hst/k-FGF gene is adjacent to int-2 and is activated by proviral insertion in some virally induced mammary tumors. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5678–5682. doi: 10.1073/pnas.86.15.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters G., Lee A. E., Dickson C. Activation of cellular gene by mouse mammary tumour virus may occur early in mammary tumour development. Nature. 1984 May 17;309(5965):273–275. doi: 10.1038/309273a0. [DOI] [PubMed] [Google Scholar]
- Sakamoto H., Mori M., Taira M., Yoshida T., Matsukawa S., Shimizu K., Sekiguchi M., Terada M., Sugimura T. Transforming gene from human stomach cancers and a noncancerous portion of stomach mucosa. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3997–4001. doi: 10.1073/pnas.83.11.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimke R. T. Gene amplification, drug resistance, and cancer. Cancer Res. 1984 May;44(5):1735–1742. [PubMed] [Google Scholar]
- Sigurdsson H., Baldetorp B., Borg A., Dalberg M., Fernö M., Killander D., Olsson H. Indicators of prognosis in node-negative breast cancer. N Engl J Med. 1990 Apr 12;322(15):1045–1053. doi: 10.1056/NEJM199004123221505. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Theillet C., Le Roy X., De Lapeyrière O., Grosgeorges J., Adnane J., Raynaud S. D., Simony-Lafontaine J., Goldfarb M., Escot C., Birnbaum D. Amplification of FGF-related genes in human tumors: possible involvement of HST in breast carcinomas. Oncogene. 1989 Jul;4(7):915–922. [PubMed] [Google Scholar]
- Thomas K. A. Transforming potential of fibroblast growth factor genes. Trends Biochem Sci. 1988 Sep;13(9):327–328. doi: 10.1016/0968-0004(88)90098-9. [DOI] [PubMed] [Google Scholar]
- Tsuda H., Hirohashi S., Shimosato Y., Hirota T., Tsugane S., Yamamoto H., Miyajima N., Toyoshima K., Yamamoto T., Yokota J. Correlation between long-term survival in breast cancer patients and amplification of two putative oncogene-coamplification units: hst-1/int-2 and c-erbB-2/ear-1. Cancer Res. 1989 Jun 1;49(11):3104–3108. [PubMed] [Google Scholar]
- Tsuda T., Nakatani H., Matsumura T., Yoshida K., Tahara E., Nishihira T., Sakamoto H., Yoshida T., Terada M., Sugimura T. Amplification of the hst-1 gene in human esophageal carcinomas. Jpn J Cancer Res. 1988 May;79(5):584–588. doi: 10.1111/j.1349-7006.1988.tb00026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimoto Y., Yunis J., Onorato-Showe L., Erikson J., Nowell P. C., Croce C. M. Molecular cloning of the chromosomal breakpoint of B-cell lymphomas and leukemias with the t(11;14) chromosome translocation. Science. 1984 Jun 29;224(4656):1403–1406. doi: 10.1126/science.6610211. [DOI] [PubMed] [Google Scholar]
- Tsutsumi M., Sakamoto H., Yoshida T., Kakizoe T., Koiso K., Sugimura T., Terada M. Coamplification of the hst-1 and int-2 genes in human cancers. Jpn J Cancer Res. 1988 Apr;79(4):428–432. doi: 10.1111/j.1349-7006.1988.tb01609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada A., Sakamoto H., Katoh O., Yoshida T., Yokota J., Little P. F., Sugimura T., Terada M. Two homologous oncogenes, HST1 and INT2, are closely located in human genome. Biochem Biophys Res Commun. 1988 Dec 15;157(2):828–835. doi: 10.1016/s0006-291x(88)80324-3. [DOI] [PubMed] [Google Scholar]
- Wahl G. M. The importance of circular DNA in mammalian gene amplification. Cancer Res. 1989 Mar 15;49(6):1333–1340. [PubMed] [Google Scholar]
- Wilkinson D. G., Peters G., Dickson C., McMahon A. P. Expression of the FGF-related proto-oncogene int-2 during gastrulation and neurulation in the mouse. EMBO J. 1988 Mar;7(3):691–695. doi: 10.1002/j.1460-2075.1988.tb02864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M. C., Wada M., Satoh H., Yoshida T., Sakamoto H., Miyagawa K., Yokota J., Koda T., Kakinuma M., Sugimura T. Human HST1 (HSTF1) gene maps to chromosome band 11q13 and coamplifies with the INT2 gene in human cancer. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4861–4864. doi: 10.1073/pnas.85.13.4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Miyagawa K., Odagiri H., Sakamoto H., Little P. F., Terada M., Sugimura T. Genomic sequence of hst, a transforming gene encoding a protein homologous to fibroblast growth factors and the int-2-encoded protein. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7305–7309. doi: 10.1073/pnas.84.20.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Muramatsu H., Muramatsu T., Sakamoto H., Katoh O., Sugimura T., Terada M. Differential expression of two homologous and clustered oncogenes, Hst1 and Int-2, during differentiation of F9 cells. Biochem Biophys Res Commun. 1988 Dec 15;157(2):618–625. doi: 10.1016/s0006-291x(88)80295-x. [DOI] [PubMed] [Google Scholar]
- Zhou D. J., Casey G., Cline M. J. Amplification of human int-2 in breast cancers and squamous carcinomas. Oncogene. 1988 Mar;2(3):279–282. [PubMed] [Google Scholar]