Abstract
CYP1C is the newest member of the CYP1 family of P450s; however, its physiological significance, inducers, and metabolic functions are unknown. Two full-length alleles of Fundulus heteroclitus CYP1C1 complementary DNA were cloned. The 529 amino acid protein shared the highest amino acid identity with Stenotomus chrysops CYP1C1 (81%). To investigate whether the carcinogen benzo[a ]pyrene (BaP) was a CYP1C1 inducer, we used real-time PCR to quantitatively measure tissue- and sex-specific expression of both CYP1C1 and CYP1A messenger RNAs (mRNAs) in BaP-exposed adult fish. CYP1C1 mRNA expression was constitutively higher than CYP1A in brain, spleen, eye, and gonad, while CYP1A was higher in gastrointestinal tract (GI), heart, gill, and liver. Kidney had equal but high expression of both CYP1s. There were sex differences in constitutive CYP1 expression in the GI, liver, gill, and eye. BaP exposure caused induction of CYP1C1 expression in female and male heart (31- and 17-fold), gill (seven- and four-fold), and liver (six- and five-fold), respectively. Embryo CYP1 expression was constitutively highest at 2 weeks posthatch, and whole embryos expressed 3- to 15-fold more CYP1C1 mRNA compared to CYP1A. BaP, 10 μg/l for 10 days, caused induction of both genes at 120 and 240 h postfertilization. Our results suggest that teleost CYP1C, in addition to CYP1A, is inducible by BaP, has a broad tissue distribution, and should be further investigated for its role in carcinogen bioactivation.
Keywords: CYP1C1, cytochrome P450, benzo[a]pyrene, killifish, embryos, Fundulus
Until the mid-1990s the CYP1 family of cytochrome P450 enzymes was believed to contain a single subfamily with two members, namely CYP1A1 and CYP1A2. In fish, this sub-family is generally recognized as CYP1A because mammalian CYP1A1 and CYP1A2 genes diverged less than 250 million years ago by a gene duplication event while fish diverged from land animals prior to that time (Morrison et al., 1995; Nebert and Gonzalez, 1987). However, in 1994, a second CYP1 subfamily was discovered when CYP1B1 was isolated from 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)–induced human keratinocyte cells (Sutter et al., 1994) and TCDD and polycyclic aromatic hydrocarbon (PAH)–induced mouse embryo fibroblast cells (Savas et al., 1994; Shen et al., 1994). The genes in the CYP1 family (CYP1A1, CYP1A2, and CYP1B1) are induced by environmental contaminants including PAHs and halogenated aromatic hydrocarbons (HAHs) (Murray et al., 2001) via the aryl hydrocarbon receptor (AhR) pathway. Both CYP1A1 and CYP1B1 are involved in carcinogen bioactivation of PAHs including benzo[a]pyrene (BaP) and 7,12-dimethyl-benzanthracene (Buters et al., 2003; Kim et al., 1998; Shimada et al., 1999a). In addition, the potential for CYP1A1 and CYP1B1 to be involved in estrogen genotoxicity and oxidative stress by generating the 2- and 4-hydroxyestradiol metabolites was recognized (Hayes et al., 1996).
The induction of CYP1A, particularly in fish, by PAHs has been used as a biomarker of exposure since the mid-1970s (Payne and Penrose, 1975). A number of studies have correlated CYP1A induction with biliary PAH metabolites, DNA adducts, immune suppression, and tumor formation in wild fish and following laboratory exposures (Carlson et al., 2004; Collier et al., 1992; Willett et al., 1995; Wirgin and Waldman, 1998). More recently a refractory CYP1A phenotype has been noted in fish living in highly PAH- or HAH-contaminated environments (Bello et al., 2001; Meyer et al., 2002; Nacci et al., 2002). The genetic and physiological significance of this lack of CYP1A induction in these populations is an area of ongoing research.
Another important recent finding in fish is that they also have CYP1B and now CYP1C genes. CYP1B was first cloned in plaice (Pleuronectes platessa) and was detected by Northern blot in gill tissue (Leaver and George, 2000). In both plaice and channel catfish only a single isoform of CYP1B has been identified, whereas both CYP1B1 and CYP1B2 genes have been cloned in carp (Cyprinus carpio). Carp exposed to 3-methylcholanthrene had CYP1B1 messenger RNA (mRNA) expression in liver, intestine, and gill, while CYP1B2 was only induced in the gills (El-kady et al., 2004a,b). Similarly, BaP-exposed catfish had significant CYP1B mRNA induction in blood, liver, and gonad tissues and high constitutive expression in gill (Willett et al., 2006).
The vertebrate CYP1C family was first described by Godard et al. (2005) when they identified a CYP1C1 and CYP1C2 in scup (Stenotomus chrysops) liver and head kidney. Carp express a CYP1C1 constitutively in gill tissue (Itakura et al., 2005). Analysis of sequence domains suggests that fish CYP1B and CYP1C enzymes will likely have unique catalytic functions or substrates; however, the function of these newly reported P450s is currently unknown. There has not been a CYP1C identified in mammals; however, because fish are extensively used in toxicology testing, biomonitoring, and as developmental biology and cancer models, it is important to understand the physiological roles, tissue distribution, and metabolic capacity of these CYP1 genes. It is possible that this new P450 subfamily has a role in endogenous compound metabolism, steroid metabolism, and/or xenobiotic metabolism and toxicity.
Fundulus heteroclitus (killifish or mummichog) are found in contaminated environments along the Atlantic coast of the United States with diseases including cancer and reproductive/ developmental deficits and are used as a model for marine toxicology (Leblanc et al., 1997; Meyer et al., 2002; Vogelbein et al., 1990; Zhou et al., 2000). Fundulus embryos are also sensitive to classic CYP1A inducers, which can even be measured by in ovo ethoxyresorufin-O-deethylase activity (Nacci et al., 1998; Wassenberg and Di Giulio, 2004). The genes involved in CYP1 induction, including the AhR1 and AhR2 (Karchner et al., 1999), the aryl hydrocarbon repressor (Karchner et al., 2002), the aryl hydrocarbon nuclear translocator 2 (Merson et al., 2006; Powell et al., 1999), and several other CYPs from families 1 to 3, have been cloned (Celander and Stegeman, 1997; Morrison et al., 1998; Oleksiak et al., 2000). The Fundulus antioxidant defense system has also been described (Meyer et al., 2003).
Our aim was to characterize the expression of the newest CYP1 subfamily member CYP1C1 in Fundulus adult and embryos. We found that in vivo BaP exposure induced CYP1C1 differentially compared to CYP1A which warrants additional research into the possibility that CYP1C1 may contribute to the mechanism of PAH toxicity in the heart, gill, or liver. Furthermore, embryos constitutively expressed higher levels of CYP1C1 than CYP1A suggesting a possible developmental role for this gene.
MATERIALS AND METHODS
Fish source, care and handling
A parental population of F. heteroclitus collected from an uncontaminated site at the New River inlet near Beaufort, NC was raised under the University Institutional Animal Care and Use Committee approved conditions. Sexually mature fish were bred and kept in salt water (20–25 parts per thousand [ppt]). The fish were maintained at 14:10 light-dark cycle in summer and 10:14 light-dark cycle in the winter. Adult fish were fed twice daily with tropical flake fish food (Tetramin, Tetra Werke, Germany) and live brine shrimp. First generation offspring, from wild parents, were used for the studies described here.
CYP1C1 cloning
Two full-length alleles of Fundulus CYP1C1 complementary DNA (cDNA) were cloned from 5 mg/kg BaP i.p. injected Fundulus liver and gill tissues. cDNA was synthesized from total RNA using oligo (dT) primers and Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA). Because a partial Fundulus CYP1B mRNA sequence was available on GenBank (Accession #AF235140), we initially anticipated cloning a CYP1B, and primers were designed based on the CYP1B partial sequence available. The amplification of initial cDNA fragment was done with 10 μM of Primers 1 and 2 (Table 1) and an aliquot (10%) of the first-strand reaction and amplified with PCR Master mix (Roche Diagnostics Corporation, Indianapolis, IN). The 5 ′ and 3 ′ ends of the CYP1C1 cDNA were obtained by rapid amplification of cDNA ends (RACE) using the GeneRacer kit (Invitrogen) as per the supplier’s protocol. 5 ′ RACE was done with Primer 3 initially and nested PCR was then done with Primer 4.3 ′ RACE done with Primer 5 and nested with Primer 6. Primers that ultimately provided the full-length CYP1C1 were Primers 7 and 8 or 9.
TABLE 1.
Primers for Fundulus CYP1C1 Cloning and Real-Time PCR
| Oligo | Primer Sequence 5′–3′ | Location | Function |
|---|---|---|---|
| 1 | AGTTTGGTTGATGTCTTGCC | 897–916 | 1C1 forward |
| 2 | GTCAAAGGCTTGAGGGTCCT | 1459–1478 | 1C1 reverse |
| 3 | CACCTCAGGGGGTCGTGATTGACAG | 1432–1456 | 1C1 5′RACE reverse |
| 4 | GGGGGTCGTGATTGACAGACCACTG | 1425–1449 | 1C1 5′RACE reverse |
| 5 | CGCTTCACCAGCTTTGTCCCATTCA | 1323–1347 | 1C1 3′RACE forward |
| 6 | CCCAAGGACACGGTGGTCTTCATCA | 1398–1422 | 1C1 3′RACE forward |
| 7 | CTTAAGCCTTAAATCTTGATGATAGAC | 1–27 | 1C1 forward for full-length |
| 8 | CAGTATATAAAGGCTCAGTGGGACTC | 2698–2723 | 1C1 reverse for full-length |
| 9 | AACACTAAAGCAACAACTCCAGATT | 2672–2696 | 1C1 reverse for full-length |
| 10 | TCTGGACGCCTTCATCTACGA | 1295–1315 | 1C1 real-time forward |
| 11 | GTGACGTCCGATGTGGTTGA | 1357–1378 | 1C1 real-time reverse |
| 12 | CGAAGATTTTGTCCAGGTGACA | 816–837 | 1A real-time forward |
| 13 | CATTGACTTGTTGGGCAGAAACT | 874–897 | 1A real-time reverse |
| 14 | TGGTTAATTCCGATAACGAACGA | 1319–1341 | 18S real-time forward |
| 15 | CGCCACTTGTCCCTCTAAGAA | 1395–1415 | 18S real-time reverse |
For cloning, DNA bands were excised from the gel and extracted using QIAquick gel extraction kit (Qiagen, Valencia, CA). pGEM-T Easy Vector System I (Promega, Madison, WI) was used for ligation. Ligated DNA was transformed into DH5α Escherichia coli–competent cells and then plated on the Luria-Bertani broth/ampicillin/Isopropyl-β-d-1-thiogalactopyranoside (IPTG)/5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) plates. Seven white colonies from each of two separate PCR reactions were picked and miniprepped with a Qiaprep Spin miniprep kit (Qiagen). Sequencing was done by using the BigDye 3.1 sequencing kit from Applied Biosystems (Foster City, CA), except the reaction conditions were reduced to 5 μl using 0.25 μl of the BigDye mix. Sequencing reactions were analyzed on an ABI 3730Xl sequencer. Sequence homology searches were carried out using the BLAST (Basic Local Alignment Search Tool) program at http://www.ncbi.nlm.nih.gov/BLAST/, whereas sequence alignment was performed using the ClustalW program at http://workbench.sdsc.edu/ or with MegAlign in DNAStar. The final contiguous sequence of each of the 14 clones sequenced in both directions was arrived at using Lasergene sequence analysis software (DNAStar).
Adult BaP exposure
Fish were exposed in 5-l tanks containing one male and one female adult fish. Fish were kept in the tanks 7 days for acclimatization. The Fundulus (n = 20, 10 tanks) were exposed to the following treatment for 15 days (complete reproductive cycle): control (50 μl ethanol) or BaP 10 μg/l (Sigma, St Louis, MO; stock solution 1 mg/ml in 100% ethanol). Exposure conditions were 17.5–20.5°C, 14:10 light-dark period, and 20–25 ppt. Fish were fed Tetramin flakes twice daily. The tanks were checked twice every 24 h, and water was changed (85–100% static water renewal) daily at 15:00.
The fish were anaesthetized with 3-aminobenzoic acid ethyl ester (MS-222, Sigma), and their weight and length were recorded. Brain, gonad, liver, gill, gastrointestinal tract (GI), spleen, kidneys, heart, and eyes were dissected from each fish and frozen – 80°C in 1 ml RNALater (Ambion, Austin, TX). Tissues were disrupted and homogenized in 1 ml of TRIzol Reagent (Invitrogen) and extracted by the manufacturer’s protocol. The total RNA was further cleaned by using an RNeasy Kit (Qiagen) with a DNAse I digestion. The RNA concentration was determined with an Agilent 2100 Bioanalyzer. RNA was stored at – 80°C.
Embryo BaP exposure
Pooled oocytes and sperm stripped from parental population of around 50 fish were mixed together for in vitro fertilization. Fertilized eggs were randomly sorted into three treatment groups, namely untreated, dimethyl sulfoxide (DMSO) control (1 μl/ml), or BaP (10 μg/l). Each egg was placed in 1 ml of water (~21 ppt) in an autosampler vial (Fisher C4013-1) with a Teflon lined cap (Fisher B7815-8) and exposure began at approximately 4.5-h postfertilization (hpf).
Each egg was inspected daily for normal development. Every other day water was changed and eggs were redosed. Exposure lasted 10 days at which time the eggs were placed in 96-well plates on a circle of damp filter paper. After 7 days, the eggs remaining from each treatment were pooled and hatched. Simultaneously hatched fish were raised an additional 2 weeks in small finger bowls. The fry were fed brine shrimp twice daily. Water was changed in finger bowls every 48 h. Ten embryos were pooled per time point and were collected for CYP mRNA analysis at 120 and 240 hpf, immediately following hatch, and at 2 weeks posthatch (ph). Embryos were stored in RNAlater at – 80°C until they were extracted with TRIzol and RNeasy kits as described above. The exposure was repeated four times.
Analysis of water samples from exposures
Water samples (200 ml) were collected 20 min after water dosing to confirm water control and BaP concentrations at least twice during each exposure. Water samples were passed over Water Sep-Pak Vac RC C18, 500 mg (#Wat 036945, Waters Corp., Milford, MA) columns prerinsed with methanol:water ratio of 3:1 (50 ml). Columns were extracted with 15 ml methylene chloride and samples were allowed to dry under a gentle stream of nitrogen. Control samples were suspended in 500 μl of isooctane and BaP samples were suspended in 1.5 ml of iso-octane and analyzed for BaP concentrations using gas chromatography/mass spectrometry in selected ion monitoring mode. BaP standards (0.1, 0.2, 0.5, 1, and 2 ppm) were prepared in iso-octane and used to develop a calibration curve. BaP was detected as an ion with molecular weight 252 g/mol (Patel et al., 2006). Actual BaP water concentration in tanks for the adult exposure averaged 13.1 ± 2.15 μg/l. The embryo exposure average concentration was 18.1 ± 1.29 μg/l.
Quantitative real-time reverse transcription PCR
Primers for quantitative real-time PCR were designed according to the requirements of Primer Express v2.0 software. Primers for CYP1A (Primers 12 and 13), CYP1C1 (Primers 10, 11), and 18S ribosomal RNA (rRNA) (Primers 14, 15) are shown in Table 1. Amplification efficiencies of all three primer pairs were determined and were not statistically different which allows for comparisons between genes (slopes = – 3.94, – 3.55, and – 3.80 for CYP1A, CYP1C1, and 18S rRNA, respectively, p = 0.15, data not shown). RNA from tissues or 10 pooled whole embryos was isolated as described above. Synthesis of cDNA was conducted with 250 ng total RNA using Taqman Reverse Transcription (RT) Reagents (Applied Biosystems): random hexamers, multiscribe RT, RNase inhibitor, deoxynucleotide triphosphate mix, 25mM MgCl2, 10× RT buffer in a 25 μl reaction. cDNA corresponding to 20 ng for CYP1s and 10 pg for 18S of reverse-transcribed RNA served as templates for each duplicate 25 μl PCR reactions using SYBR Green PCR Master Mix (Applied Biosystems). The PCR amplifications and fluorescence detection were performed with the GeneAmp 7500 Sequence Detection System (Applied Biosystems). Universal thermal cycling conditions according to the manufacturer were used. For quality control, no template controls and no RT controls were done, as well as melt curve analyses. Samples were amplified in duplicate in separate reactions on the same plate. The amount of CYP1 mRNA, normalized to 18S, was given by the formula = 2– ΔΔCT, where CT is the threshold cycle indicating the fractional cycle number at which the amount of amplified CYP1 reached threshold. The ΔCT value is determined by subtracting the average 18S CT value from the average CYP1 CT value. Then, the calculation of ΔΔCT involves subtraction of the ΔCT value of the calibrator (in our case the calibrator was average ΔCT value of control fish response in the BaP studies or specific CYP1 form in the tissue comparisons) from the ΔCT value of each sample (ABI PRISM 7700 Sequence Detection System User’s Manual). Accordingly, CYP1 mRNA levels were reported as fold change in abundance relative to the average calibrator response. In the embryo experiments, CYP1 levels were reported relative to the untreated controls. The 18S CT values were very consistent between adult tissues and various PCR runs and averaged 16.5 ± 0.038. For embryos, the 18S CT values were 17.3 ± 0.064. Statistical differences between treatments, sexes, or genes in each tissue were determined with one-way analysis of variance (ANOVA) followed by Newman-Keuls multiple comparison test (p < 0.05) using GraphPad Prism 4.0 version.
RESULTS
Cloning of Fundulus CYP1C1
Two full-length alleles of Fundulus CYP1C1 cDNAs were cloned from 5 mg/kg BaP i.p. injected Fundulus liver and gill tissues. Allele I (GenBank Accession #DQ133570) contained a 5′ noncoding region of 182 bp, an open reading frame (ORF) of 1590 bp, and a 3′ noncoding region of 988 bp to the polyA tail. Allele II (GenBank Accession #DQ133571) contained a 5′ noncoding region of 183 bp, an ORF of 1590 bp, and a 3′ noncoding region of 992 bp to the polyA tail. The two alleles shared 99% nucleotide identity and encoded for the same deduced 529 amino acid sequence. CYP1C1 shared the highest amino acid identity with S. chrysops 1C1 (81%, GenBank Accession #AAL78297) while exhibiting only 35% identity with the Fundulus CYP1A (GenBank Accession #AAD01809). Interestingly, the CYP1C1 was only 66% similar at the amino acid level with the Fundulus CYP1B partial sequence (Gen-Bank Accession # AAL78301) which was used to design the initial cloning primers.
Despite our efforts, we were not able to successfully generate a full-length CYP1B cDNA sequence with 5′ or 3′ RACE or by PCR with degenerate primers. Similarly, we did not isolate a Fundulus CYP1C2 from any of the PCR reactions or sequenced colonies. It is possible that Fundulus do have these other CYP1 isoforms based on the fact that zebrafish, scup, carp, and fugu have both CYP1C1 and CYP1C2 genes.
Previously, Gotoh (1992) had identified six regions on CYP2 that are putative substrate recognition sites (SRS). Godard et al. (2005) have applied these same regions for analysis of the functional divergence of CYP1s. Figure 1 highlights the six SRS of Fundulus CYP1C1 aligned with scup and zebrafish CYP1C1 and CYP1C2, Fundulus CYP1A, and the Fundulus CYP1B partial sequence. As expected Fundulus CYP1C1 is most similar to the other fish CYP1C1s especially in SRS 1, 4, and 5. The least conserved SRS was 2.
FIG. 1.
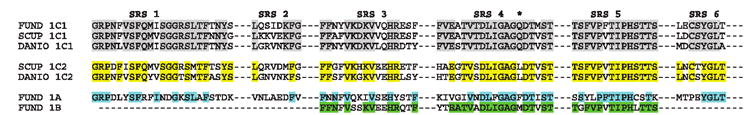
Amino acid alignment of the six substrate recognition sites (SRS) as described in (Godard et al., 2005; Gotoh, 1992) of Fundulus CYP1C1 with scup and zebrafish CYP1C1 and CYP1C2s and Fundulus CYP1A and the Fundulus CYP1B. Gray and yellow shading indicate similar sites between Fundulus CYP1C1 and other fish CYP1C1s or CYP1C2s, respectively. Blue and green shading indicate similar sites between Fundulus CYP1C1 and Fundulus CYP1A or Fundulus CYP1B, respectively. *Indicates the position of the AA that is suggested to play a role in substrate specificity.
CYP1 Expression in Adult Tissues
Constitutive expression of CYP1C1 mRNA was significantly higher compared to CYP1A in brain, eye, gonad, and spleen tissues in both sexes (ANOVA, p < 0.001). Real-time PCR results for both CYP1 genes are plotted for the four tissues that had higher constitutive CYP1C1 expression in Figure 2. The data are presented as CYP1 CT – 18S CT. The 18S rRNA signal was used as a normalization standard for each sample. Therefore, the shorter bar height represents higher mRNA expression. Relative fold induction was calculated by the equation 2 –ΔΔCT as described above. The tissue with the most CYP1C1 expression as compared to CYP1A was the testis followed by ovary where there was 108- and 65-fold more CYP1C1 compared to CYP1A, respectively (Table 2). There was 39- and 81-fold more CYP1C1 than CYP1A in male and female spleen, respectively.
FIG. 2.
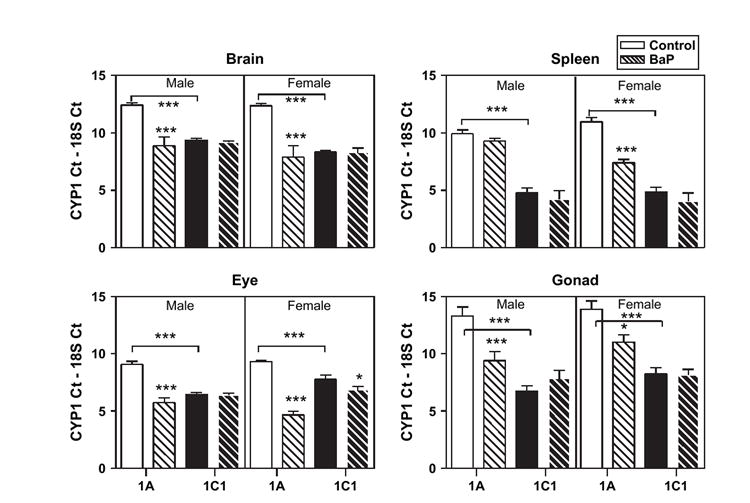
CYP1A (white bars) and CYP1C1 (black bars) mRNA expression in control (solid bars) and BaP-exposed (thatched bars) adult Fundulus. Four tissues (brain, spleen, eye, gonad) had significantly higher constitutive CYP1C1 expression in both sexes. Bar height represents the threshold cycle number for the CYP1s minus the threshold cycle number for the 18S for each sample. Note that the shorter the bar, the higher the mRNA expression. See “Materials and Methods” for details of experiments. Significant differences between treatment and control or differences between CYP1 constitutive expression are noted. Bars represent mean and standard error of the mean (*p < 0.05; ***p < 0.001; ANOVA, n = 3–5).
TABLE 2.
CYP1 mRNA Expression in Adult Fundulus Exposed 15 Days to Ethanol Control or 10 μg/l BaP
| Males | Females | |||||
|---|---|---|---|---|---|---|
| Fold BAP inductiona | Control isoformb | Fold BAP inductiona | Control isoformb | |||
| CYP1A | CYP1C | Fold abundance | CYP1A | CYP1C | Fold abundance | |
| Heart | 18.9 ± 9.70 | 16.8 ± 9.26 | 14.6 ± 2.77 more 1A versus 1C1 | 27.9 ± 10.6 | 30.5 ± 21.8 | 14.6 ± 3.64 more 1A versus 1C1 |
| Gill | 14.2 ± 2.16 | 4.47 ± 0.457 | 3.94 ± 0.478 more 1A versus 1C1 | 13.8 ± 1.47 | 6.60 ± 1.74 | 3.05 ± 0.447 more 1A versus 1C1 |
| GI | 6.75 ± 3.77 | 1.20 ± 0.14 | 8.05 ± 1.23 more 1A versus 1C1 | 1.59 ± 0.46 | 1.03 ± 0.17 | 5.12 ± 1.46 more 1A versus 1C1 |
| Liver | 1.43 ± 0.25 | 4.65 ± 1.26 | 27.4 ± 2.78 more 1A versus 1C1 | 1.70 ± 0.28 | 5.85 ± 1.73 | 33.1 ± 9.76 more 1A versus 1C1 |
| Brain | 23.6 ± 15.7 | 1.14 ± 0.08 | 8.63 ± 1.21 more 1C1 versus 1A | 37.6 ± 20.5 | 1.13 ± 0.32 | 16.7 ± 1.63 more 1C1 versus 1A |
| Eye | 12.3 ± 4.70 | 1.08 ± 0.16 | 6.27 ± 0.692 more 1C1 versus 1A | 28.2 ± 6.52 | 2.06 ± 0.422 | 3.39 ± 0.870 more 1C1 versus 1A |
| Gonad | 25.1 ± 11.2 | 0.79 ± 0.40 | 108 ± 24.6 more 1C1 versus 1A | 9.71 ± 2.96 | 1.26 ± 0.39 | 65.3 ± 20.7 more 1C1 versus 1A |
| Spleen | 1.61 ± 0.28 | 1.90 ± 0.86 | 39.2 ± 11.9 more 1C1 versus 1A | 12.4 ± 2.59 | 2.15 ± 0.75 | 80.8 ± 22.6 more 1C1 versus 1A |
| Kidney | 2.90 ± 0.653 | 1.62 ± 0.37 | 1.14 ± 0.13 more 1C1 versus 1A | 3.00 ± 0.395 | 2.30 ± 0.37 | 1.07 ± 0.20 more 1C1 versus 1A |
Calculated by ΔΔCT approach. Data normalized to 18S expression and reported as fold induction relative to controls (n = 3–5).
Calculated by ΔΔCT approach. CYP1A and CYP1C1 expression in control animals was normalized to 18S expression and is shown as relative fold abundance.
Numbers in bold are significantly different, ANOVA p < 0.05, as analyzed on 18S corrected raw data.
Gray-shaded rows represent tissues where CYP1C1 expression was constitutively higher than CYP1A expression.
There were four tissues with significantly (ANOVA, p < 0.01) higher CYP1A expression, namely, GI, heart, gill, and liver (Fig. 3). Liver had ~30-fold more CYP1A relative to CYP1C1 followed by heart (~15-fold) (Table 2). In kidney there was equal expression of CYP1A and CYP1C1 (data not shown).
FIG. 3.
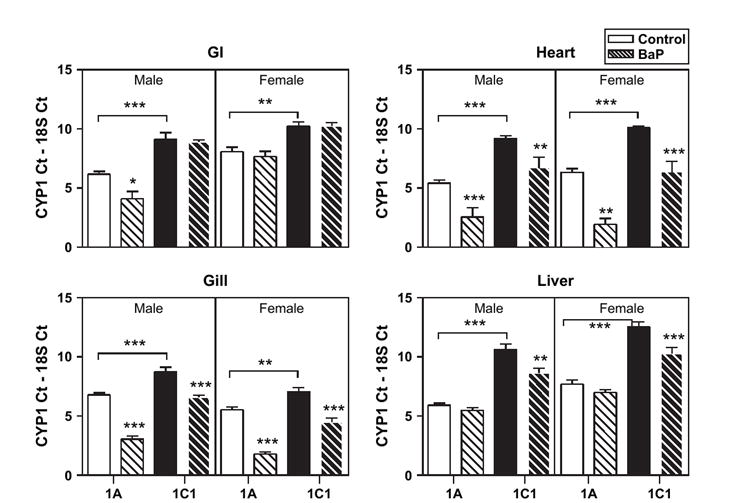
CYP1A (white bars) and CYP1C1 (black bars) mRNA expression in control (solid bars) and BaP-exposed (thatched bars) adult Fundulus. Four tissues (GI, heart, gill, liver) had significantly higher constitutive CYP1A expression in both sexes. See Figure 2 for explanation of bars. Significant differences between treatment and control or differences between CYP1 constitutive expression are noted. Bars represent mean and standard error of the mean (*p < 0.05; **p < 0.01; ***p < 0.001; ANOVA, n = 3–5).
When sex differences in constitutive gene expression were compared, there was higher CYP1A mRNA expression in male GI (four-fold) and liver (3.5-fold) and the female gill (2.5-fold) (ANOVA, p < 0.01, Fig. 4). Constitutive CYP1C1 expression was significantly higher in male eye (2.5-fold) and liver (4.5-fold, p < 0.01) and female gill (3.5-fold, p < 0.001). For all other tissues, the expression of CYP1A or CYP1C1 was not significantly different between the sexes.
FIG. 4.
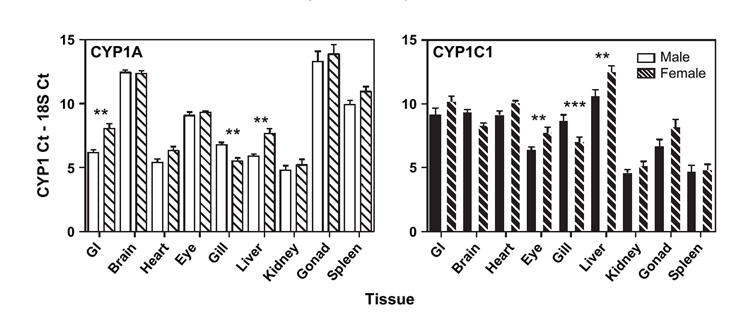
Constitutive CYP1A (white bars) and CYP1C1 (black bars) mRNA expression in adult male (solid bars) and female (thatched bars) Fundulus. See Figure 2 for explanations of bars. Significant differences between males and females within a tissue are noted (**p < 0.01; ***p < 0.001; ANOVA, n = 3–5).
BaP Effects on CYP1 Expression in Adult Tissues
In male fish, 15 days of 10-μg/l BaP exposure caused CYP1A induction in every tissue analyzed except spleen and liver (Figs. 2 and 3, Table 2). In females, significant induction was noted in all but the GI and liver. The sex differences in CYP1A tissue expression were maintained following BaP exposure in GI, gill, and liver. There was significantly ~2- to 4-fold higher induction of CYP1A in female spleen, eye, and gill compared to male CYP1A induction. In BaP-treated fish, male CYP1A expression compared to female CYP1A expression was significantly higher in GI (19-fold) and liver (three-fold).
Compared to CYP1A, CYP1C1 was significantly induced by BaP in fewer tissues. The highest CYP1C1 induction was in male and female heart 17- and 31-fold, respectively. CYP1C1 induction ranged from four- to seven-fold in gill and liver tissues. Like CYP1A expression, the sex differences in constitutive CYP1C1 expression (i.e., higher expression in female gill and male liver) were maintained following BaP exposure with the exception of in the eye where there was no sex difference following BaP exposure.
CYP1 Expression in Embryos
RNA was isolated from embryos at 4.5, 120, 240 hpf, at hatch (17–18 days pf; 408–432 hpf), and 2 weeks ph. Constitutive CYP1 expression increased with age for both CYP1A and CYP1C1 (Fig. 5). In embryos between 120 hpf and freshly hatched, there was between 12- and 15-fold higher CYP1C1 expression compared to CYP1A. However, by 2 weeks ph embryos expressed only three-fold more CYP1C1 than CYP1A (Fig. 6).
FIG. 5.
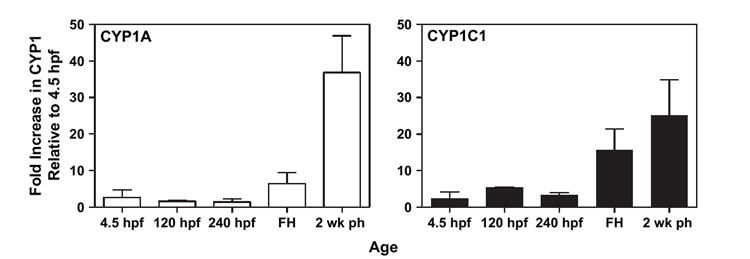
Constitutive CYP1A (white bars) and CYP1C1 (black bars) mRNA level in a pool of 10 whole embryos (n = 4). See “Materials and Methods” for details of experiments. Each bar represents the mean and standard error of the mean of the fold increase in CYP1 expression at the different ages relative to 4.5 hpf (FH = at hatch).
FIG. 6.
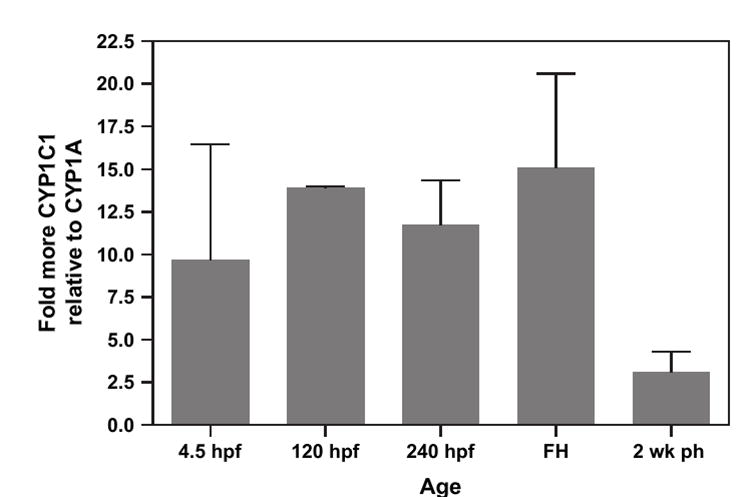
Fold-higher CYP1C1 mRNA level expression relative to CYP1A in control whole embryos. Each bar represents fold increases in CYP1C1 expression relative to CYP1A within an age.
BaP Effects on CYP1 Expression in Embryos
Both CYP1A and CYP1C1 were induced in embryos by exposure to 10 μg/l BaP for 10 days (Fig. 7). The highest induction of both genes corresponded with the end of exposure (240 hpf) and dropped off to control levels by hatch. CYP1A was much more highly induced relative to controls than CYP1C1 was (on average 330 vs. 18-fold, respectively).
FIG. 7.
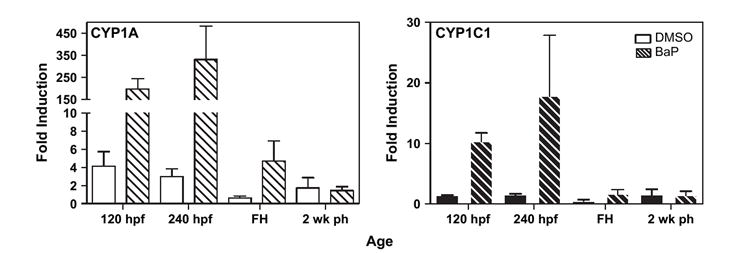
BaP effects on CYP1A (white bars) and CYP1C1 (black bars) mRNA level in a pool of 10 whole embryos (n = 4). Each bar represents mean and standard error of the mean of the fold increases in CYP1 expression in DMSO control (solid bars) or BaP treatment (thatched bars) relative to the untreated group at each age.
DISCUSSION
The results of this study demonstrate that CYP1C1 mRNA is widely distributed in adult Fundulus tissues, is present in embryos through development, and it is inducible by BaP in embryos and select tissues. Prior to this report, the distribution and inducibility of CYP1C was largely unknown. In untreated male scup liver, CYP1 expression was highest for CYP1A followed by CYP1C1 and CYP1C2 (Godard et al., 2005). We also found 27- to 33-fold higher liver CYP1A compared to CYP1C1 expression in control Fundulus. In contrast to scup, where liver expression of the CYP1s was higher than in head kidney, we found both CYP1A and CYP1C1 expressed more highly in kidney. In our studies the highest CYP1C1 constitutive expression was found in spleen, kidney, eye, gill, and gonad, respectively. In carp, CYP1C1 has been reported by Northern blot in gills but not kidney, liver, or intestine (Itakura et al., 2005).
No previous studies, to our knowledge, have shown either CYP1B or CYP1C expression in embryonic fish. With quantitative real-time reverse transcriptase–PCR we were able to measure both CYP1A and CYP1C1 expression as early as 4.5 hpf suggesting either a maternal contribution of these RNAs to the embryo or early synthesis of these mRNAs. Constitutive expression levels of both CYP1A and CYP1C1 were low and not increasing from 4.5 to 240 hpf (Fig. 5). In Fundulus development, 120 hpf (~stages 27 and 28) is after the onset of circulation and corresponds to when the ventricle of heart forms a definite chamber, the otoliths are dense concrete bodies and body movement increases. The heart chambers are differentiated and the liver and gut become apparent at about 168 hpf (stage 31). At 240 hpf (stages 34 and 35), the lower jaw forms and the mouth may begin to open (Armstrong and Child, 1965). In our experiments at 240 hpf, the eggs are removed from the exposure and placed in a well on filter paper to dry out prior to hatch. Hatch occurs on day 17 or 18, and by 2 weeks ph embryos are approximately 11 mm long. CYP1A and CYP1C1 expression increased dramatically at hatch (6- to 16-fold over the 4.5 hpf levels) and continued to increase by 2 weeks ph (25-to 37-fold). When the relative amounts of CYP1 were compared in the untreated embryos, we found that CYP1C1 expression was higher than CYP1A at all time points and the highest relative expression was between 120 hpf and hatch (Fig. 6). These studies suggest that CYP1C1 may be more important than CYP1A during development. We have ongoing studies using in situ hybridization to investigate the specific location of CYP1C1 expression because in the studies described here whole embryos were pooled and homogenized.
In fish and mammals, BaP exposure is associated with suite of toxicities including immunosuppression, oxidative stress, mutagenicity, and stable DNA adduct formation. Bay region diol epoxide metabolites are largely believed to be formed by the action of CYP1 enzymes and are associated with stable DNA adduct formation and cancer initiation. In most species and tissues, there are relatively low levels of CYP1 constitutively expressed, however, they can be highly induced by exposure to AhR-ligands including BaP. Fundulus exposed to BaP show dose-related increases in biliary PAH metabolites, CYP1A mRNA, EROD activity, CYP1A protein, and DNA adduct formation (Patel et al., 2006; Willett et al., 1995, 2001). We found CYP1A mRNA was significantly induced by BaP exposure in all tissues investigated except female GI, male spleen and livers. While the lack of the induction in the liver was unexpected, it may be because the long exposure time (15 days) may have lead to high CYP1A protein and thus rapid biotransformation of BaP in the liver lowering the concentrations in this tissue. The highest fold induction of CYP1A was in the heart, brain, and testis. Similar results were reported in zebrafish exposed to 1.5 and 3 μg/l waterborne BaP for 56 days. In zebrafish, there was a dose-related increase in CYP1A mRNA expression in heads but not in liver (Hoffman and Oris, 2006).
Understanding the inducers and target tissues of the newer fish CYP1s will be important for further understanding of the potential contribution of these CYP1s in bioactivating or inactivating environmental contaminants. In fish, there are somewhat mixed results concerning PAH induction of the more recently discovered CYP1B and CYP1C genes. In plaice, constitutive CYP1B expression was noted in gill, but it was not induced by 50 mg/kg i.p. injection of β-naphthoflavone (Leaver and George, 2000). In contrast, 20 mg/kg BaP caused induction of CYP1B mRNA in channel catfish blood, gonad, and liver. Induction of CYP1B mRNA was also detected in primary cultured catfish gill cells when treated with BaP, TCDD, and polychlorinated biphenyls (PCBs) 77, 126, and 169 but not by treatment with PCB153 or 4,4′-dichlorodiphenyltrichloroethane (Willett et al., 2006). In carp, 100 mg/kg 3-methylcholanthrene caused induced expression of CYP1B1 in liver, intestine, and gills, but CYP1B2 expression was only induced in gills (El-kady et al., 2004a,b). The carp CYP1C1 was not induced in liver, intestine, gill, or kidney 24 h following a 100 mg/kg i.p. injection of β-naphthoflavone (Itakura et al., 2005). Our results represent the first evidence that CYP1C1 is inducible by PAH exposure. The difference between carp and Fundulus may be species related or may be due to less sensitivity in the Northern blot methods used in the carp study compared to the real-time PCR used here. CYP1C1 was significantly induced in adult Fundulus gill, liver, and heart following a 15-day waterborne 10-μg/l exposure. Highest induction was found in the heart, a known target organ for PAH toxicities in the early life stages.
To date, nothing is known about the protein expression or the metabolic substrates of the fish CYP1B and CYP1Cs. Hypotheses can be generated by comparing the amino acid sequences particularly in the substrate recognition sites. In their analysis of fish CYP1Cs, Godard et al. (2005) proposed that SRS1, SRS3, and SRS4 likely contribute the most to differences in substrate recognition between CYP1C1s and CYP1C2s. Particularly in SRS4, where the I-helix interacts with the heme-oxygen, there is a conserved threonine that is preceded by two amino acids (*Fig. 1) that are suggested to play a role in substrate specificity. In Fundulus CYP1C1, this residue is a glutamine, in scup and zebrafish CYP1C2s it is a leucine, in Fundulus CYP1A it is a phenylalanine, and in CYP1B it is a methionine. Differences in SRS2 and SRS6 have been suggested to be involved in differentiating substrate specificities between the CYP1Cs and the CYP1Bs (Godard et al., 2005). To better understand the differences in metabolic capacity of these CYP1s, the enzymes should be recombinantly expressed. In mammals, for example, ethoxyresorufin is a better substrate for CYP1A1 compared to CYP1B1. CYP1B1 is an estradiol hydroxylase primarily at the C-4 position compared to the C-2 position for CYP1A1 (Hayes et al., 1996). In some species, CYP1B1 is more active than CYP1A1 in metabolizing BaP to the proximate toxicant BaP-7,8-diol (Shimada et al., 1999b).
PAHs cause embryo toxicity with symptoms including cranial-facial malformations, yolk sac and pericardial edema, subcutaneous hemorrhaging, and reduced growth (Billiard et al., 1999; Incardona et al., 2004; Wassenberg and Di Giulio, 2004). Neither a BaP exposure 10 days at 10 μg/l (this study) nor BaP 7 days at 100 μg/l caused significant deformities in Fundulus embryos (Wassenberg and Di Giulio, 2004). However, when fish were cotreated with PAHs and classic CYP1A inhibitors including α-naphthoflavone and piperonyl butoxide, embryonic EROD activity was significantly decreased while the deformity index was dramatically increased (Wassenberg and Di Giulio, 2004). This study suggested that inhibiting CYP1A increased the toxicity of PAHs and could possibly implicate the involvement of another CYP in the toxicity. More recently, work in zebrafish using AhR2 and CYP1A morpholinos showed that knocking down AhR2 reduced cardiac toxicity of β-naphthoflavone and α-naphthoflavone cotreatments. However, when CYP1A was knocked down, it enhanced the toxicity of both β-naphthoflavone and the β-naphthoflavone and α-naphthoflavone cotreatment (Billiard et al., 2006). Therefore, the PAH-mediated toxicity appears to be AhR mediated, but CYP1A seems to be providing a protective role. In our embryo study, both CYP1A and CYP1C1 were induced by BaP. However, CYP1A was maximally induced relative to controls ~300-fold, whereas CYP1C1 was only induced about 18-fold. The extensive induction of CYP1A may be providing protection against BaP toxicity and further research is necessary to understand the significance of constitutively higher CYP1C1 expression in embryos and its induction by BaP. It is also unknown whether the classic CYP1A inhibitors will also inhibit CYP1C1 expression.
In summary, we have described a new CYP1C1 gene in Fundulus an important model species in marine environmental toxicology. CYP1C1 mRNA had a wide tissue distribution and was inducible in three toxicologically relevant tissues (heart, gill, and liver). Additional research may find that this enzyme is involved both in PAH bioactivation to carcinogenic intermediates and/or in the mechanisms associated with PAH embryo toxicity.
Acknowledgments
This work was supported by National Institute of Environmental Health Sciences grant R01ES012710 and instruments provided by National Center for Research Resources grant RR016476 from the MFGN INBRE program. We thank Annette Ford and Kate Argote for technical assistance with exposures and fish care.
References
- Armstrong PB, Child JS. Stages in the normal development of Fundulus heteroclitus. Biol Bull. 1965;128:143–169. [Google Scholar]
- Bello SM, Franks DG, Stegeman JJ, Hahn ME. Acquired resistance to Ah receptor agonists in a population of Atlantic killifish (Fundulus heteroclitus) inhabiting a marine superfund site: In vivo and in vitro studies on the inducibility of xenobiotic metabolizing enzymes. Toxicol Sci. 2001;60:77–91. doi: 10.1093/toxsci/60.1.77. [DOI] [PubMed] [Google Scholar]
- Billiard SM, Querbach K, Hodson PV. Toxicity of retene to early life stages of two freshwater fish species. Environ Toxicol Chem. 1999;18:2070–2077. [Google Scholar]
- Billiard SM, Timme-Laragy AR, Wassenberg DM, Cockman C, DiGiulio RT. The role of the aryl hydrocarbon receptor pathway in mediating synergistic developmental toxicity of polycyclic aromatic hydrocarbons in zebrafish. Toxicol Sci. 2006;92(2):526–536. doi: 10.1093/toxsci/kfl011. [DOI] [PubMed] [Google Scholar]
- Buters JT, Quintanilla-Martinez L, Schober W, Soballa VJ, Hintermair J, Wolff T, Gonzalez FJ, Greim H. CYP1B1 determines susceptibility to low doses of 7,12-dimethylbenz(a)anthracene-induced ovarian cancers in mice: Correlation of CYP1B1-mediated DNA adducts with carcinogenicity. Carcinogenesis. 2003;24(2):327–334. doi: 10.1093/carcin/24.2.327. [DOI] [PubMed] [Google Scholar]
- Carlson EA, Li Y, Zelikoff JT. Benzo[a]pyrene-induced immunotoxicity in Japanese medaka (Oryzias latipes): Relationship between lymphoid CYP1A activity and humoral immune suppression. Toxicol Appl Pharmacol. 2004;201:40–52. doi: 10.1016/j.taap.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Celander M, Stegeman JJ. Isolation of a cytochrome P450 3A cDNA sequence (CYP3A30) from the marine teleost Fundulus heteroclitus and phylogenetic analyses of CYP3A genes. Biochem Biophys Res Commun. 1997;236:306–312. doi: 10.1006/bbrc.1997.6956. [DOI] [PubMed] [Google Scholar]
- Collier TK, Singh SV, Awasthi YC, Varanasi U. Hepatic xenobiotic metabolizing enzymes in two species of benthic fish showing different prevalences of contaminant-associated liver neoplasms. Toxicol Appl Pharmacol. 1992;113:319–324. doi: 10.1016/0041-008x(92)90131-b. [DOI] [PubMed] [Google Scholar]
- El-kady MAH, Mitsuo R, Kaminishi Y, Itakura T. cDNA cloning, sequence analysis and expression of 3-methylcholanthrene-inducible cytochrome P450 1b1 in carp (Cyprinus carpio) Environ Sci. 2004a;11(4):231–240. [PubMed] [Google Scholar]
- El-kady MAH, Mitsuo R, Kaminishi Y, Itakura T. Isolation of cDNA of novel cytochrome P450 1B gene, CYP1B2, from carp (Cyprinus carpio) and its induced expression in gills. Environ Sci. 2004b;11(6):345–354. [PubMed] [Google Scholar]
- Godard CA, Goldsone JV, Said MR, Dickerson RL, Woodin BR, Stegeman JJ. The new vertebrate CYP1C family: Cloning of new subfamily members and phylogenetic analysis. Biochem Biophys Res Commun. 2005;331:1016–1024. doi: 10.1016/j.bbrc.2005.03.231. [DOI] [PubMed] [Google Scholar]
- Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992;267(1):83–90. [PubMed] [Google Scholar]
- Hayes CL, Spink DC, Spink BC, Cao JQ, Walker NJ, Sutter TR. 17β-estradiol hydroxylation catalyzed by human cytochrome P450 1B1. Proc Natl Acad Sci USA. 1996;93:9776–9781. doi: 10.1073/pnas.93.18.9776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman JL, Oris JT. Altered gene expression: A mechanism for reproductive toxicity in zebrafish exposed to benzo[a]pyrene. Aquat Toxicol. 2006;78:332–340. doi: 10.1016/j.aquatox.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Incardona JP, Collier TK, Scholz NL. Defects in cardiac function precede morphological abnormalities in fish embryos exposed to polycyclic aromatic hydrocarbons. Toxicol Appl Pharmacol. 2004;196:191–205. doi: 10.1016/j.taap.2003.11.026. [DOI] [PubMed] [Google Scholar]
- Itakura T, El-kady MAH, Mitsuo R, Kaminishi Y. Complementary DNA cloning and constitutive expression of cytochrome P450 1C1 in the gills of carp (Cyprinus carpio) Environ Sci. 2005;12(2):111–120. [PubMed] [Google Scholar]
- Karchner SI, Franks DG, Powell WH, Hahn ME. Regulatory interactions among three members of the vertebrate aryl hydrocarbon receptor family: AhR repressor, AhR1 and AhR2. J Biol Chem. 2002;277(9):6949–6959. doi: 10.1074/jbc.M110779200. [DOI] [PubMed] [Google Scholar]
- Karchner SI, Powell WH, Hahn ME. Identification and functional characterization of two highly divergent aryl hydrocarbon receptors (AhR1 and AhR2) in the teleost Fundulus heteroclitus. J Biol Chem. 1999;274(47):33814–33824. doi: 10.1074/jbc.274.47.33814. [DOI] [PubMed] [Google Scholar]
- Kim JH, Stansbury KH, Walker NJ, Trush MA, Strickland PT, Sutter TR. Metabolism of benzo(a)pyrene and benzo(a)pyrene-7,8-diol by human cytochrome P450 1B1. Carcinogenesis. 1998;19(10):1847–1853. doi: 10.1093/carcin/19.10.1847. [DOI] [PubMed] [Google Scholar]
- Leaver MJ, George SG. A cytochrome P4501B gene from a fish, Pleuronectes platessa. Gene. 2000;256:83–91. doi: 10.1016/s0378-1119(00)00373-5. [DOI] [PubMed] [Google Scholar]
- Leblanc J, Couillard CM, Brethes J-CF. Modification of the reproduction period in mummichog (Fundulus heteroclitus) living downstream from a bleached kraft pulp mill in the Miramichi Estuary, New Brunswick, Canada. Can J Fish Aquat Sci. 1997;54:2564–2573. [Google Scholar]
- Merson RR, Franks DG, Karchner SI, Hahn ME. Development and characterization of polyclonal antibodies against the aryl hydrocarbon receptor protein family (AHR1, AHR2, and AHR repressor) of Atlantic killifish Fundulus heteroclitus. Comp Biochem Physiol. 2006;142(Part C):85–94. doi: 10.1016/j.cbpc.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Meyer JN, Nacci D, Di Giulio RT. Cytochrome P4501A (CYP1A) in killifish (Fundulus heteroclitus): Heritability of altered expression and relationship to survival in contaminated sediments. Toxicol Sci. 2002;68:69–81. doi: 10.1093/toxsci/68.1.69. [DOI] [PubMed] [Google Scholar]
- Meyer JN, Smith JD, Winston GW, Di Giulio RT. Antioxidant defenses in killifish (Fundulus heteroclitus) exposed to contaminated sediments and model prooxidants: Short-term and heritable responses. Aquat Toxicol. 2003;65:377–395. doi: 10.1016/j.aquatox.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Morrison HG, Oleksiak MF, Cornell NW, Sogin ML, Stegeman JJ. Identification of cytochrome P-450 1A (CYP1A) genes from two teleost fish, toadfish (Opsanus tau) and scup (Stenotomus chrysops), and phylogenetic analysis of CYP1A genes. Biochem J. 1995;308:97–104. doi: 10.1042/bj3080097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison HG, Weil EJ, Karchner SI, Sogin ML, Stegeman JJ. Molecular cloning of CYP1A from the estuarine fish Fundulus heteroclitus and phylogenetic analysis of CYP1 genes: Update with new species. Comp Biochem Physiol. 1998;121C:231–240. doi: 10.1016/s0742-8413(98)10044-0. [DOI] [PubMed] [Google Scholar]
- Murray GI, Melvin WT, Greenlee WF, Burke MD. Regulation, function and tissue-specific expression of cytochrome P450 CYP1B1. Annu Rev Pharmacol Toxicol. 2001;41:297–316. doi: 10.1146/annurev.pharmtox.41.1.297. [DOI] [PubMed] [Google Scholar]
- Nacci D, Coiro L, Kuhn A, Champlin D, Munns W, Specker J, Cooper K. Nondestructive indicator of ethoxyresorufin-O-deethylase activity in embryonic fish. Environ Toxicol Chem. 1998;17(12):2481–2486. [Google Scholar]
- Nacci DE, Kohan M, Pelletier M, George E. Effects of benzo(a)pyrene exposure on a fish population resistant to the toxic effects of dioxin-like compounds. Aquat Toxicol. 2002;57:203–215. doi: 10.1016/s0166-445x(01)00196-5. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Gonzalez FJ. P450 genes: Structure, evolution, and regulation. Annu Rev Biochem. 1987;56:945–993. doi: 10.1146/annurev.bi.56.070187.004501. [DOI] [PubMed] [Google Scholar]
- Oleksiak MF, Wu S, Parker C, Karchner SI, Stegeman JJ. Identification, functional characterization, and regulation of a new cytochrome P450 subfamily, the CYP2Ns. J Biol Chem. 2000;275:2312–2321. doi: 10.1074/jbc.275.4.2312. [DOI] [PubMed] [Google Scholar]
- Patel MR, Scheffler BE, Wang L, Willett KL. Effects of benzo(a)pyrene exposure on killifish (Fundulus heteroclitus) aromatase activities and mRNA. Aquat Toxicol. 2006;77:267–278. doi: 10.1016/j.aquatox.2005.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne JF, Penrose WR. Induction of aryl hydrocarbon (benzo(a)pyrene) hydroxylase in fish by petroleum. Bull Environ Contam Toxicol. 1975;14:112–116. doi: 10.1007/BF01685608. [DOI] [PubMed] [Google Scholar]
- Powell WH, Karchner SI, Bright R, Hahn ME. Functional diversity of vertebrate ARNT proteins: Identification of ARNT2 as the predominant form of ARNT in the marine teleost, Fundulus heteroclitus. Arch Biochem Biophys. 1999;361(1):156–163. doi: 10.1006/abbi.1998.0992. [DOI] [PubMed] [Google Scholar]
- Savas U, Bhattacharyya KK, Christou M, Alexander DL, Jefcoate CR. Mouse cytochrome P-450EF, representative of a new 1B subfamily of cytochrome P-450s. Cloning, sequence determination, and tissue expression. J Biol Chem. 1994;269:14905–14911. [PubMed] [Google Scholar]
- Shen Z, Liu J, Wells RL, Elkind MM. cDNA cloning, sequence analysis, and induction by aryl hydrocarbons of a murine cytochrome P450 gene, Cyp1b1. DNA Cell Biol. 1994;13(7):763–769. doi: 10.1089/dna.1994.13.763. [DOI] [PubMed] [Google Scholar]
- Shimada T, Gillam EM, Oda Y, Tsumura F, Sutter TR, Guengerich FP, Inoue K. Metabolism of benzo(a)pyrene to trans-7,8-dihydroxy-7,8-dihydrobenzo(a)pyrene by recombinant human cytochrome P450 1B1 and purified liver epoxide hydrolase. Chem Res Toxicol. 1999a;12:623–629. doi: 10.1021/tx990028s. [DOI] [PubMed] [Google Scholar]
- Shimada T, Watanabe J, Kawajiri K, Sutter TR, Guengerich FP, Gillam EMJ, Inoue K. Catalytic properties of polymorphic human cytochrome P450 1B1 variants. Carcinogenesis. 1999b;20(8):1607–1613. doi: 10.1093/carcin/20.8.1607. [DOI] [PubMed] [Google Scholar]
- Sutter TR, Tang YM, Hayes CL, Wo Y-Y, Jabs EW, Li X, Yin H, Cody CW, Greenlee WF. Complete cDNA sequence of a human dioxin-inducible mRNA identifies a new gene subfamily of cytochrome P450 that maps to chromosome 2. J Biol Chem. 1994;269(18):13092–13099. [PubMed] [Google Scholar]
- Vogelbein WK, Fournie JW, Van Veld PA, Huggett RJ. Hepatic neoplasms in the mummichog Fundulus heteroclitus from a creosote-contaminated site. Cancer Res. 1990;50:5978–5986. [PubMed] [Google Scholar]
- Wassenberg DM, Di Giulio RT. Synergistic embryotoxicity of polycyclic aromatic hydrocarbon aryl hydrocarbon receptor agonists with cytochrome P4501A inhibitors in Fundulus heteroclitus. Environ Health Perspect. 2004;112:1658–1664. doi: 10.1289/ehp.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett K, Steinberg MA, Thomsen J, Narasimhan TK, Safe SH, McDonald SJ, Beatty KB, Kennicutt MC. Exposure of killifish to benzo(a)pyrene: Comparative metabolism, DNA adduct formation and aryl hydrocarbon (Ah) receptor agonist activities. Comp Biochem Physiol. 1995;112B(1):93–103. [Google Scholar]
- Willett KL, Ganesan S, Patel M, Metzger C, Quiniou S, Waldbieser G, Scheffler B. In vivo and in vitro CYP1B mRNA expression in channel catfish. Mar Environ Res. 2006;62:S332–S336. doi: 10.1016/j.marenvres.2006.04.015. [DOI] [PubMed] [Google Scholar]
- Willett KL, Wassenberg D, Lienesch L, Di Giulio RT, Reichert WL. In vivo and in vitro inhibition of CYP1A-dependent activity in Fundulus heteroclitus by the polynuclear aromatic hydrocarbon (PAH) fluoranthene. Toxicol Appl Pharmacol. 2001;177:264–271. doi: 10.1006/taap.2001.9296. [DOI] [PubMed] [Google Scholar]
- Wirgin I, Waldman JR. Altered gene expression and genetic damage in North American fish populations. Mutat Res. 1998;399:193–219. doi: 10.1016/s0027-5107(97)00256-x. [DOI] [PubMed] [Google Scholar]
- Zhou T, John-Alder HB, Weis JS, Weis P. Endocrine disruption: Thyroid dysfunction in mummichogs (Fundulus heteroclitus) from a polluted habitat. Mar Environ Res. 2000;50(1–5):393–397. doi: 10.1016/s0141-1136(00)00042-8. [DOI] [PubMed] [Google Scholar]


