Abstract
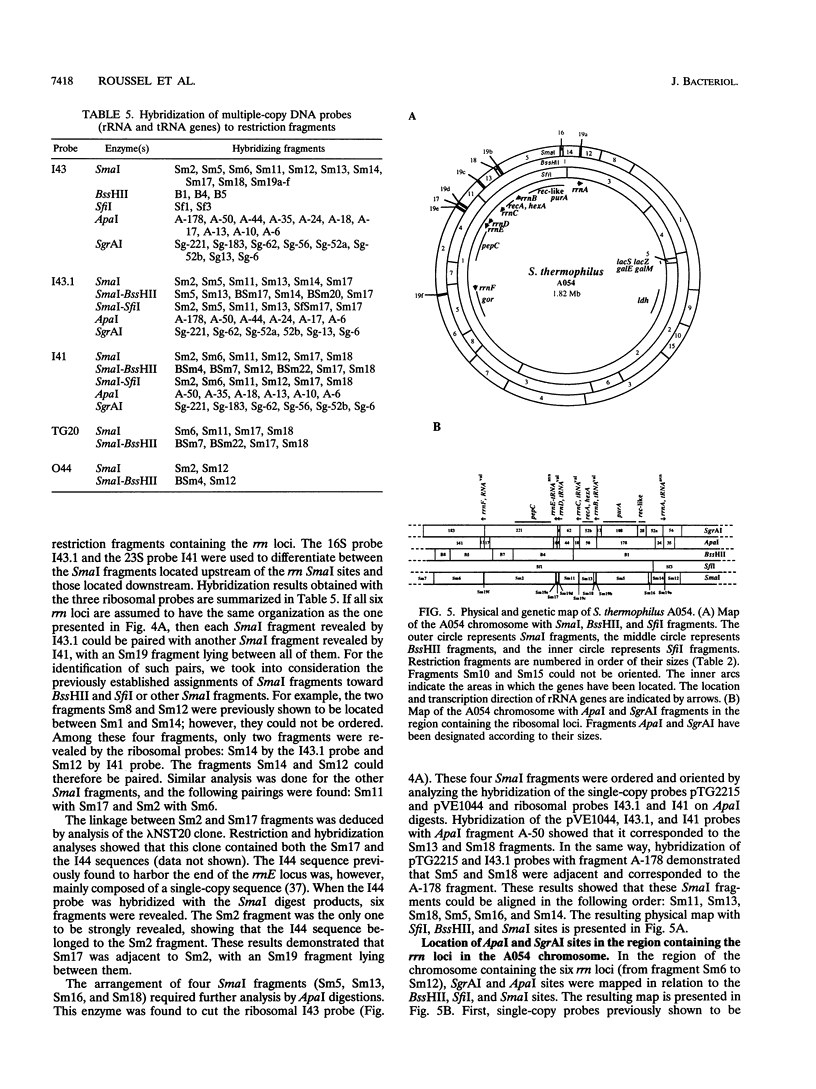
The three restriction endonucleases SfiI, BssHII, and SmaI were found to generate fragments with suitable size distributions for mapping the genome of Streptococcus thermophilus A054. A total of 5, 8, and 24 fragments were produced with SfiI, BssHII, and SmaI, respectively. An average genome size of 1,824 kb was determined by summing the total fragment sizes obtained by digestions with these three enzymes. Partial and multiple digestions of genomic DNA in conjunction with Southern hybridization were used to map SfiI, BssHII, and SmaI fragments. All restriction fragments were arranged in a unique circular chromosome. Southern hybridization analysis with specific probes allowed 23 genetic markers to be located on the restriction map. Among them, six rrn loci were precisely located. The area of the chromosome containing the ribosomal operons was further detailed by mapping some of the ApaI and SgrAI sites. Comparison of macrorestriction patterns from three clones derived from strain A054 revealed two variable regions in the chromosome. One was associated with the tandem rrnD and rrnE loci, and the other was mapped in the region of the lactose operon.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft I., Wolk C. P., Oren E. V. Physical and genetic maps of the genome of the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1989 Nov;171(11):5940–5948. doi: 10.1128/jb.171.11.5940-5948.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellis M., Pagès M., Roizès G. A simple and rapid method for preparing yeast chromosomes for pulsed field gel electrophoresis. Nucleic Acids Res. 1987 Aug 25;15(16):6749–6749. doi: 10.1093/nar/15.16.6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley R. W., Leigh J. A., Collins M. D. Intrageneric structure of Streptococcus based on comparative analysis of small-subunit rRNA sequences. Int J Syst Bacteriol. 1991 Oct;41(4):487–494. doi: 10.1099/00207713-41-4-487. [DOI] [PubMed] [Google Scholar]
- Brewer B. J. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell. 1988 Jun 3;53(5):679–686. doi: 10.1016/0092-8674(88)90086-4. [DOI] [PubMed] [Google Scholar]
- Canard B., Cole S. T. Genome organization of the anaerobic pathogen Clostridium perfringens. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6676–6680. doi: 10.1073/pnas.86.17.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canard B., Saint-Joanis B., Cole S. T. Genomic diversity and organization of virulence genes in the pathogenic anaerobe Clostridium perfringens. Mol Microbiol. 1992 Jun;6(11):1421–1429. doi: 10.1111/j.1365-2958.1992.tb00862.x. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Olson M. V. Separation of chromosomal DNA molecules from yeast by orthogonal-field-alternation gel electrophoresis. Nucleic Acids Res. 1984 Jul 25;12(14):5647–5664. doi: 10.1093/nar/12.14.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Keseler I. M., Shimkets L. J. Genome size of Myxococcus xanthus determined by pulsed-field gel electrophoresis. J Bacteriol. 1990 Aug;172(8):4206–4213. doi: 10.1128/jb.172.8.4206-4213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu G., Vollrath D., Davis R. W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986 Dec 19;234(4783):1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- Colmin C., Pebay M., Simonet J. M., Decaris B. A species-specific DNA probe obtained from Streptococcus salivarius subsp. thermophilus detects strain restriction polymorphism. FEMS Microbiol Lett. 1991 Jun 15;65(2):123–128. doi: 10.1016/0378-1097(91)90290-q. [DOI] [PubMed] [Google Scholar]
- Duwat P., Ehrlich S. D., Gruss A. A general method for cloning recA genes of gram-positive bacteria by polymerase chain reaction. J Bacteriol. 1992 Aug;174(15):5171–5175. doi: 10.1128/jb.174.15.5171-5175.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellwood M., Nomura M. Chromosomal locations of the genes for rRNA in Escherichia coli K-12. J Bacteriol. 1982 Feb;149(2):458–468. doi: 10.1128/jb.149.2.458-468.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow J. A., Collins M. D. DNA base composition, DNA-DNA homology and long-chain fatty acid studies on streptococcus thermophilus and Streptococcus salivarius. J Gen Microbiol. 1984 Feb;130(2):357–362. doi: 10.1099/00221287-130-2-357. [DOI] [PubMed] [Google Scholar]
- Gasc A. M., Kauc L., Barraillé P., Sicard M., Goodgal S. Gene localization, size, and physical map of the chromosome of Streptococcus pneumoniae. J Bacteriol. 1991 Nov;173(22):7361–7367. doi: 10.1128/jb.173.22.7361-7367.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giffard P. M., Rathsam C., Kwan E., Kwan D. W., Bunny K. L., Koo S. P., Jacques N. A. The ftf gene encoding the cell-bound fructosyltransferase of Streptococcus salivarius ATCC 25975 is preceded by an insertion sequence and followed by FUR1 and clpP homologues. J Gen Microbiol. 1993 May;139(5):913–920. doi: 10.1099/00221287-139-5-913. [DOI] [PubMed] [Google Scholar]
- Giffard P. M., Simpson C. L., Milward C. P., Jacques N. A. Molecular characterization of a cluster of at least two glucosyltransferase genes in Streptococcus salivarius ATCC 25975. J Gen Microbiol. 1991 Nov;137(11):2577–2593. doi: 10.1099/00221287-137-11-2577. [DOI] [PubMed] [Google Scholar]
- Green C. J., Vold B. S. Sequence analysis of a cluster of twenty-one tRNA genes in Bacillus subtilis. Nucleic Acids Res. 1983 Aug 25;11(16):5763–5774. doi: 10.1093/nar/11.16.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C. J., Vold B. S. Staphylococcus aureus has clustered tRNA genes. J Bacteriol. 1993 Aug;175(16):5091–5096. doi: 10.1128/jb.175.16.5091-5096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guédon G., Pébay M., Colmin C., Simonet J. M., Decaris B. The 23S-5S spacer of two rRNA loci of Streptococcus salivarius subsp thermophilus includes a promoter. Biochimie. 1992 Jun;74(6):585–588. doi: 10.1016/0300-9084(92)90159-c. [DOI] [PubMed] [Google Scholar]
- Hantman M. J., Sun S., Piggot P. J., Daneo-Moore L. Chromosome organization of Streptococcus mutans GS-5. J Gen Microbiol. 1993 Jan;139(1):67–77. doi: 10.1099/00221287-139-1-67. [DOI] [PubMed] [Google Scholar]
- Herman R. E., McKay L. L. Cloning and expression of the beta-D-galactosidase gene from Streptococcus thermophilus in Escherichia coli. Appl Environ Microbiol. 1986 Jul;52(1):45–50. doi: 10.1128/aem.52.1.45-50.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis E. D., Widom R. L., LaFauci G., Setoguchi Y., Richter I. R., Rudner R. Chromosomal organization of rRNA operons in Bacillus subtilis. Genetics. 1988 Nov;120(3):625–635. doi: 10.1093/genetics/120.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoureux M., Prévost H., Cavin J. F., Diviès C. Recognition of Leuconostoc oenos strains by the use of DNA restriction profiles. Appl Microbiol Biotechnol. 1993 Jul;39(4-5):547–552. doi: 10.1007/BF00205049. [DOI] [PubMed] [Google Scholar]
- Le Bourgeois P., Lautier M., Mata M., Ritzenthaler P. Physical and genetic map of the chromosome of Lactococcus lactis subsp. lactis IL1403. J Bacteriol. 1992 Nov;174(21):6752–6762. doi: 10.1128/jb.174.21.6752-6762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblond P., Francou F. X., Simonet J. M., Decaris B. Pulsed-field gel electrophoresis analysis of the genome of Streptomyces ambofaciens strains. FEMS Microbiol Lett. 1990 Oct;60(1-2):79–88. doi: 10.1016/0378-1097(90)90349-u. [DOI] [PubMed] [Google Scholar]
- Lee J. J., Smith H. O., Redfield R. J. Organization of the Haemophilus influenzae Rd genome. J Bacteriol. 1989 Jun;171(6):3016–3024. doi: 10.1128/jb.171.6.3016-3024.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llanos R. M., Hillier A. J., Davidson B. E. Cloning, nucleotide sequence, expression, and chromosomal location of ldh, the gene encoding L-(+)-lactate dehydrogenase, from Lactococcus lactis. J Bacteriol. 1992 Nov;174(21):6956–6964. doi: 10.1128/jb.174.21.6956-6964.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okahashi N., Sasakawa C., Okada N., Yamada M., Yoshikawa M., Tokuda M., Takahashi I., Koga T. Construction of NotI restriction map of the Streptococcus mutans genome. J Gen Microbiol. 1990 Nov;136(11):2217–2223. doi: 10.1099/00221287-136-11-2217. [DOI] [PubMed] [Google Scholar]
- Pittet A. C., Hottinger H. Sequence of a hexameric tRNA gene cluster associated with rRNA genes in Lactobacillus bulgaricus. Nucleic Acids Res. 1989 Jun 26;17(12):4873–4873. [PMC free article] [PubMed] [Google Scholar]
- Poolman B., Royer T. J., Mainzer S. E., Schmidt B. F. Carbohydrate utilization in Streptococcus thermophilus: characterization of the genes for aldose 1-epimerase (mutarotase) and UDPglucose 4-epimerase. J Bacteriol. 1990 Jul;172(7):4037–4047. doi: 10.1128/jb.172.7.4037-4047.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poolman B., Royer T. J., Mainzer S. E., Schmidt B. F. Lactose transport system of Streptococcus thermophilus: a hybrid protein with homology to the melibiose carrier and enzyme III of phosphoenolpyruvate-dependent phosphotransferase systems. J Bacteriol. 1989 Jan;171(1):244–253. doi: 10.1128/jb.171.1.244-253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudhomme M., Méjean V., Martin B., Claverys J. P. Mismatch repair genes of Streptococcus pneumoniae: HexA confers a mutator phenotype in Escherichia coli by negative complementation. J Bacteriol. 1991 Nov;173(22):7196–7203. doi: 10.1128/jb.173.22.7196-7203.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pébay M., Colmin C., Guédon G., De Gaspéri C., Decaris B., Simonet J. M. Detection of intraspecific DNA polymorphism in Streptococcus salivarius subsp. thermophilus by a homologous rDNA probe. Res Microbiol. 1992 Jan;143(1):37–46. doi: 10.1016/0923-2508(92)90032-j. [DOI] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel Y., Colmin C., Simonet J. M., Decaris B. Strain characterization, genome size and plasmid content in the Lactobacillus acidophilus group (Hansen and Mocquot). J Appl Bacteriol. 1993 May;74(5):549–556. [PubMed] [Google Scholar]
- Salzano G., Moschetti G., Villani F., Coppola S. Biotyping of Streptococcus thermophilus strains by DNA fingerprinting. Res Microbiol. 1993 Jun;144(5):381–387. doi: 10.1016/0923-2508(93)90195-8. [DOI] [PubMed] [Google Scholar]
- Schroeder C. J., Robert C., Lenzen G., McKay L. L., Mercenier A. Analysis of the lacZ sequences from two Streptococcus thermophilus strains: comparison with the Escherichia coli and Lactobacillus bulgaricus beta-galactosidase sequences. J Gen Microbiol. 1991 Feb;137(2):369–380. doi: 10.1099/00221287-137-2-369. [DOI] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Slos P., Bourquin J. C., Lemoine Y., Mercenier A. Isolation and characterization of chromosomal promoters of Streptococcus salivarius subsp. thermophilus. Appl Environ Microbiol. 1991 May;57(5):1333–1339. doi: 10.1128/aem.57.5.1333-1339.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R., Andachi Y., Muto A. Evolution of tRNAs and tRNA genes in Acholeplasma laidlawii. Nucleic Acids Res. 1991 Dec 25;19(24):6787–6792. doi: 10.1093/nar/19.24.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanskanen E. I., Tulloch D. L., Hillier A. J., Davidson B. E. Pulsed-Field Gel Electrophoresis of SmaI Digests of Lactococcal Genomic DNA, a Novel Method of Strain Identification. Appl Environ Microbiol. 1990 Oct;56(10):3105–3111. doi: 10.1128/aem.56.10.3105-3111.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulloch D. L., Finch L. R., Hillier A. J., Davidson B. E. Physical map of the chromosome of Lactococcus lactis subsp. lactis DL11 and localization of six putative rRNA operons. J Bacteriol. 1991 May;173(9):2768–2775. doi: 10.1128/jb.173.9.2768-2775.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vold B. S. Structure and organization of genes for transfer ribonucleic acid in Bacillus subtilis. Microbiol Rev. 1985 Mar;49(1):71–80. doi: 10.1128/mr.49.1.71-80.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widom R. L., Jarvis E. D., LaFauci G., Rudner R. Instability of rRNA operons in Bacillus subtilis. J Bacteriol. 1988 Feb;170(2):605–610. doi: 10.1128/jb.170.2.605-610.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohda M., Okada H., Kumagai H. Molecular cloning and nucleotide sequencing of the aspartate racemase gene from lactic acid bacteria Streptococcus thermophilus. Biochim Biophys Acta. 1991 Jun 13;1089(2):234–240. doi: 10.1016/0167-4781(91)90013-c. [DOI] [PubMed] [Google Scholar]





