Abstract
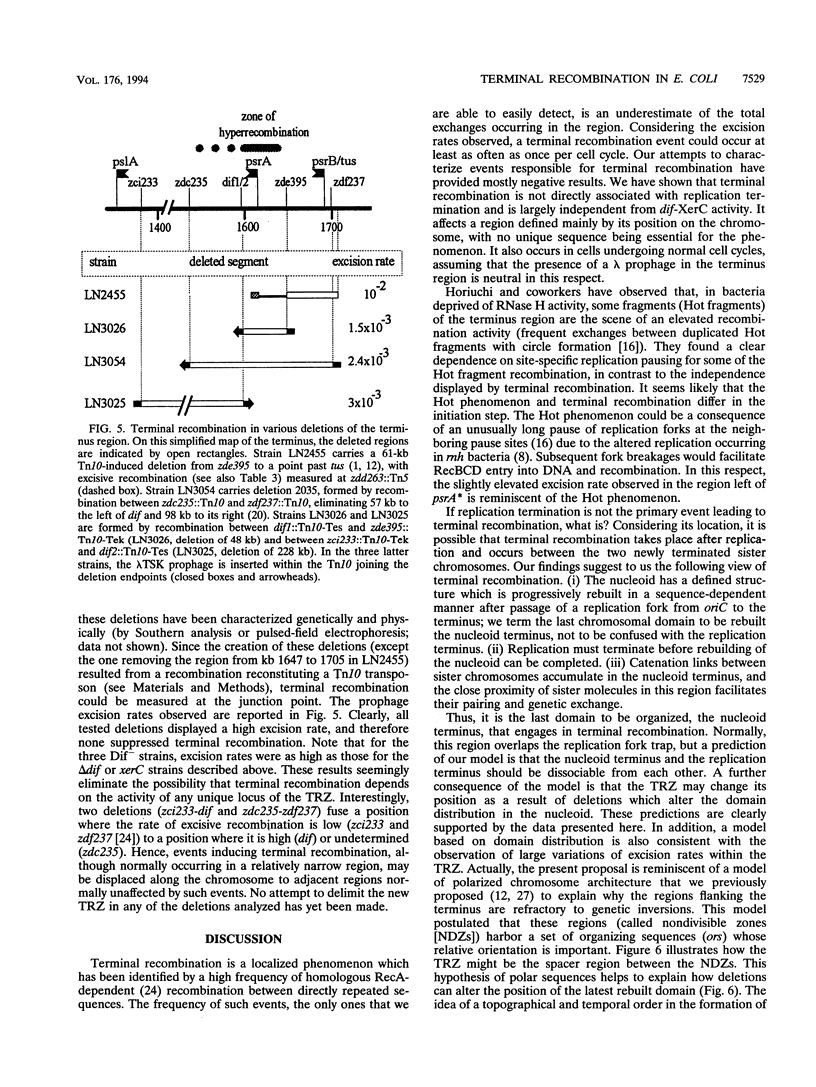
The terminus region of the Escherichia coli chromosome is the scene of frequent homologous recombination. This can be demonstrated by formation of deletions between directly repeated sequences which flank a genetic marker whose loss can be easily detected. We report here that terminal recombination events are restricted to a relatively large terminal recombination zone (TRZ). On one side of the TRZ, the transition from the region with a high excision rate to the normal (low) excision rates of the rest of the chromosome occurs along a DNA stretch of less than 1 min. No specific border of this domain has been defined. To identify factors inducing terminal recombination, we examined its relation to two other phenomena affecting the same region, site-specific recombination at the dif locus and site-specific replication pausing. Both the location and the efficiency of terminal recombination remained unchanged after inactivation of the dif-specific recombination system. Similarly, inactivation of site-specific replication pausing or displacement of the replication fork trap so that termination occurs about 200 kb away from the normal region had no clear effect on this phenomenon. Therefore, terminal recombination is not a direct consequence of either dif-specific recombination or replication termination. Furthermore, deletions encompassing the wild-type TRZ do not eliminate hyperrecombination. Terminal recombination therefore cannot be attributed to the activity of some unique sequence of the region. A possible explanation of terminal hyperrecombination involves nucleoid organization and its remodeling after replication: we propose that post replicative reconstruction of the nucleoid organization results in a displacement of the catenation links between sister chromosomes to the last chromosomal domain to be rebuilt. Unrelated to replication termination, this process would facilitate interactions between the catenated molecules and would make the domain highly susceptible to recombination between sister chromosomes.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bird R. E., Louarn J., Martuscelli J., Caro L. Origin and sequence of chromosome replication in Escherichia coli. J Mol Biol. 1972 Oct 14;70(3):549–566. doi: 10.1016/0022-2836(72)90559-1. [DOI] [PubMed] [Google Scholar]
- Blakely G., Colloms S., May G., Burke M., Sherratt D. Escherichia coli XerC recombinase is required for chromosomal segregation at cell division. New Biol. 1991 Aug;3(8):789–798. [PubMed] [Google Scholar]
- Blakely G., May G., McCulloch R., Arciszewska L. K., Burke M., Lovett S. T., Sherratt D. J. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell. 1993 Oct 22;75(2):351–361. doi: 10.1016/0092-8674(93)80076-q. [DOI] [PubMed] [Google Scholar]
- Béjar S., Bouché J. P. A new dispensable genetic locus of the terminus region involved in control of cell division in Escherichia coli. Mol Gen Genet. 1985;201(2):146–150. doi: 10.1007/BF00425651. [DOI] [PubMed] [Google Scholar]
- Clerget M. Site-specific recombination promoted by a short DNA segment of plasmid R1 and by a homologous segment in the terminus region of the Escherichia coli chromosome. New Biol. 1991 Aug;3(8):780–788. [PubMed] [Google Scholar]
- Cornet F., Mortier I., Patte J., Louarn J. M. Plasmid pSC101 harbors a recombination site, psi, which is able to resolve plasmid multimers and to substitute for the analogous chromosomal Escherichia coli site dif. J Bacteriol. 1994 Jun;176(11):3188–3195. doi: 10.1128/jb.176.11.3188-3195.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellay R., Frey J., Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene. 1987;52(2-3):147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- François V., Louarn J., Louarn J. M. The terminus of the Escherichia coli chromosome is flanked by several polar replication pause sites. Mol Microbiol. 1989 Aug;3(8):995–1002. doi: 10.1111/j.1365-2958.1989.tb00250.x. [DOI] [PubMed] [Google Scholar]
- François V., Louarn J., Patte J., Louaran J. M. A system for in vivo selection of genomic rearrangements with predetermined endpoints in Escherichia coli using modified Tn10 transposons. Gene. 1987;56(1):99–108. doi: 10.1016/0378-1119(87)90162-4. [DOI] [PubMed] [Google Scholar]
- François V., Louarn J., Patte J., Rebollo J. E., Louarn J. M. Constraints in chromosomal inversions in Escherichia coli are not explained by replication pausing at inverted terminator-like sequences. Mol Microbiol. 1990 Apr;4(4):537–542. doi: 10.1111/j.1365-2958.1990.tb00621.x. [DOI] [PubMed] [Google Scholar]
- Hidaka M., Akiyama M., Horiuchi T. A consensus sequence of three DNA replication terminus sites on the E. coli chromosome is highly homologous to the terR sites of the R6K plasmid. Cell. 1988 Nov 4;55(3):467–475. doi: 10.1016/0092-8674(88)90033-5. [DOI] [PubMed] [Google Scholar]
- Hill T. M., Kopp B. J., Kuempel P. L. Termination of DNA replication in Escherichia coli requires a trans-acting factor. J Bacteriol. 1988 Feb;170(2):662–668. doi: 10.1128/jb.170.2.662-668.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill T. M., Pelletier A. J., Tecklenburg M. L., Kuempel P. L. Identification of the DNA sequence from the E. coli terminus region that halts replication forks. Cell. 1988 Nov 4;55(3):459–466. doi: 10.1016/0092-8674(88)90032-3. [DOI] [PubMed] [Google Scholar]
- Horiuchi T., Fujimura Y., Nishitani H., Kobayashi T., Hidaka M. The DNA replication fork blocked at the Ter site may be an entrance for the RecBCD enzyme into duplex DNA. J Bacteriol. 1994 Aug;176(15):4656–4663. doi: 10.1128/jb.176.15.4656-4663.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri G. S., MacAllister T., Sista P. R., Bastia D. The replication terminator protein of E. coli is a DNA sequence-specific contra-helicase. Cell. 1989 Nov 17;59(4):667–674. doi: 10.1016/0092-8674(89)90012-3. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kuempel P. L., Henson J. M., Dircks L., Tecklenburg M., Lim D. F. dif, a recA-independent recombination site in the terminus region of the chromosome of Escherichia coli. New Biol. 1991 Aug;3(8):799–811. [PubMed] [Google Scholar]
- Lee E. H., Kornberg A., Hidaka M., Kobayashi T., Horiuchi T. Escherichia coli replication termination protein impedes the action of helicases. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9104–9108. doi: 10.1073/pnas.86.23.9104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louarn J. M., Bird R. E. Size distribution and molecular polarity of newly replicated DNA in Escherichia coli. Proc Natl Acad Sci U S A. 1974 Feb;71(2):329–333. doi: 10.1073/pnas.71.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louarn J. M., Louarn J., François V., Patte J. Analysis and possible role of hyperrecombination in the termination region of the Escherichia coli chromosome. J Bacteriol. 1991 Aug;173(16):5097–5104. doi: 10.1128/jb.173.16.5097-5104.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louarn J., Patte J., Louarn J. M. Evidence for a fixed termination site of chromosome replication in Escherichia coli K12. J Mol Biol. 1977 Sep 25;115(3):295–314. doi: 10.1016/0022-2836(77)90156-5. [DOI] [PubMed] [Google Scholar]
- Perkins J. D., Heath J. D., Sharma B. R., Weinstock G. M. XbaI and BlnI genomic cleavage maps of Escherichia coli K-12 strain MG1655 and comparative analysis of other strains. J Mol Biol. 1993 Jul 20;232(2):419–445. doi: 10.1006/jmbi.1993.1401. [DOI] [PubMed] [Google Scholar]
- Rebollo J. E., François V., Louarn J. M. Detection and possible role of two large nondivisible zones on the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9391–9395. doi: 10.1073/pnas.85.24.9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A. S., Wake R. G. A unique DNA intermediate associated with termination of chromosome replication in Bacillus subtilis. Cell. 1984 Dec;39(3 Pt 2):683–689. doi: 10.1016/0092-8674(84)90475-6. [DOI] [PubMed] [Google Scholar]
- de Massy B., Béjar S., Louarn J., Louarn J. M., Bouché J. P. Inhibition of replication forks exiting the terminus region of the Escherichia coli chromosome occurs at two loci separated by 5 min. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1759–1763. doi: 10.1073/pnas.84.7.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B., Fayet O., Kogoma T. Multiple origin usage for DNA replication in sdrA(rnh) mutants of Escherichia coli K-12. Initiation in the absence of oriC. J Mol Biol. 1984 Sep 15;178(2):227–236. doi: 10.1016/0022-2836(84)90141-4. [DOI] [PubMed] [Google Scholar]



