Abstract
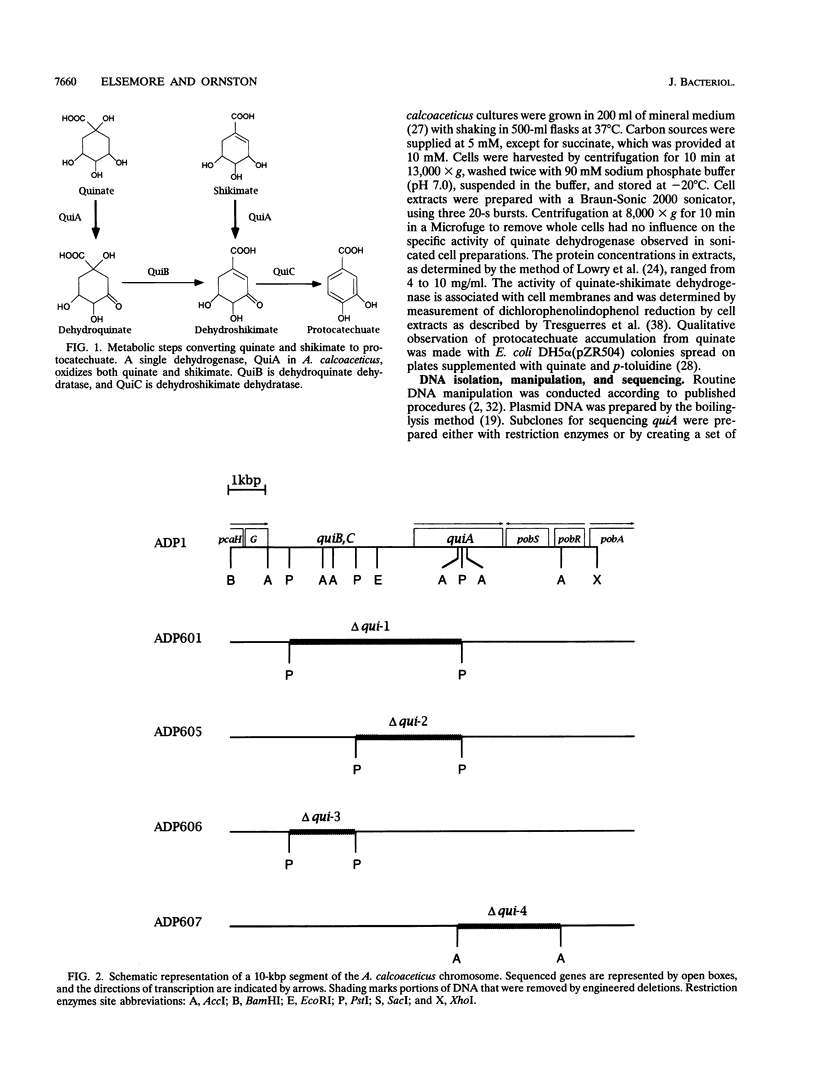
An 18-kbp Acinetobacter calcoaceticus chromosomal segment contains the pcaIJFBDKCHG operon, which is required for catabolism of protocatechuate, and pobSRA, genes associated with conversion of p-hydroxybenzoate to protocatechuate. The genetic function of the 6.5 kbp of DNA between pcaG and pobS was unknown. Deletions in this DNA were designed by removal of fragments between restriction sites, and the deletion mutations were introduced into A. calcoaceticus by natural transformation. The mutations prevented growth with either quinate or shikimate, growth substrates that depend upon qui gene function for their catabolism to protocatechuate. The location of quiA, a gene encoding quinate-shikimate dehydrogenase, was indicated by its expression in one of the deletion mutants, and the position of the gene was confirmed by determination of its 2,427-bp nucleotide sequence. The deduced amino acid sequence of QuiA confirmed that it is a member of a family of membrane-associated, pyrrolo-quinoline quinone-dependent dehydrogenases, as had been suggested by earlier biochemical investigations. Catabolism of quinate and skikimate is initiated by NAD(+)-dependent dehydrogenases in other microorganisms, so it is evident that different gene pools were called upon to provide the ancestral enzyme for this metabolic step.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmed S. I., Giles N. H. Organization of enzymes in the common aromatic synthetic pathway: evidence for aggregation in fungi. J Bacteriol. 1969 Jul;99(1):231–237. doi: 10.1128/jb.99.1.231-237.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averhoff B., Gregg-Jolly L., Elsemore D., Ornston L. N. Genetic analysis of supraoperonic clustering by use of natural transformation in Acinetobacter calcoaceticus. J Bacteriol. 1992 Jan;174(1):200–204. doi: 10.1128/jb.174.1.200-204.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun C., Zumft W. G. The structural genes of the nitric oxide reductase complex from Pseudomonas stutzeri are part of a 30-kilobase gene cluster for denitrification. J Bacteriol. 1992 Apr;174(7):2394–2397. doi: 10.1128/jb.174.7.2394-2397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain R. B. The identity of shikimate dehydrogenase and quinate dehydrogenase in Aspergillus niger. Biochem J. 1972 Apr;127(2):15P–15P. [PMC free article] [PubMed] [Google Scholar]
- Cánovas J. L., Wheelis M. L., Stanier R. Y. Regulation of the enzymes of the beta-ketoadipate pathway in Moraxella calcoacetica. 2. The role of protocatechuate as inducer. Eur J Biochem. 1968 Jan;3(3):293–304. doi: 10.1111/j.1432-1033.1968.tb19529.x. [DOI] [PubMed] [Google Scholar]
- DiMarco A. A., Averhoff B. A., Kim E. E., Ornston L. N. Evolutionary divergence of pobA, the structural gene encoding p-hydroxybenzoate hydroxylase in an Acinetobacter calcoaceticus strain well-suited for genetic analysis. Gene. 1993 Mar 15;125(1):25–33. doi: 10.1016/0378-1119(93)90741-k. [DOI] [PubMed] [Google Scholar]
- DiMarco A. A., Averhoff B., Ornston L. N. Identification of the transcriptional activator pobR and characterization of its role in the expression of pobA, the structural gene for p-hydroxybenzoate hydroxylase in Acinetobacter calcoaceticus. J Bacteriol. 1993 Jul;175(14):4499–4506. doi: 10.1128/jb.175.14.4499-4506.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doten R. C., Ngai K. L., Mitchell D. J., Ornston L. N. Cloning and genetic organization of the pca gene cluster from Acinetobacter calcoaceticus. J Bacteriol. 1987 Jul;169(7):3168–3174. doi: 10.1128/jb.169.7.3168-3174.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg-Jolly L. A., Ornston L. N. Properties of Acinetobacter calcoaceticus recA and its contribution to intracellular gene conversion. Mol Microbiol. 1994 Jun;12(6):985–992. doi: 10.1111/j.1365-2958.1994.tb01086.x. [DOI] [PubMed] [Google Scholar]
- Gregg-Jolly L. A., Ornston L. N. Recovery of DNA from the Acinetobacter calcoaceticus chromosome by gap repair. J Bacteriol. 1990 Oct;172(10):6169–6172. doi: 10.1128/jb.172.10.6169-6172.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartnett C., Neidle E. L., Ngai K. L., Ornston L. N. DNA sequences of genes encoding Acinetobacter calcoaceticus protocatechuate 3,4-dioxygenase: evidence indicating shuffling of genes and of DNA sequences within genes during their evolutionary divergence. J Bacteriol. 1990 Feb;172(2):956–966. doi: 10.1128/jb.172.2.956-966.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins A. R., Giles N. H., Kinghorn J. R. Genetical and biochemical aspects of quinate breakdown in the filamentous fungus Aspergillus nidulans. Biochem Genet. 1982 Apr;20(3-4):271–286. doi: 10.1007/BF00484424. [DOI] [PubMed] [Google Scholar]
- Hawkins A. R., Lamb H. K., Moore J. D., Charles I. G., Roberts C. F. The pre-chorismate (shikimate) and quinate pathways in filamentous fungi: theoretical and practical aspects. J Gen Microbiol. 1993 Dec;139(12):2891–2899. doi: 10.1099/00221287-139-12-2891. [DOI] [PubMed] [Google Scholar]
- Holloway B. W., Morgan A. F. Genome organization in Pseudomonas. Annu Rev Microbiol. 1986;40:79–105. doi: 10.1146/annurev.mi.40.100186.000455. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Juni E. Genetics and physiology of Acinetobacter. Annu Rev Microbiol. 1978;32:349–371. doi: 10.1146/annurev.mi.32.100178.002025. [DOI] [PubMed] [Google Scholar]
- Juni E., Janik A. Transformation of Acinetobacter calco-aceticus (Bacterium anitratum). J Bacteriol. 1969 Apr;98(1):281–288. doi: 10.1128/jb.98.1.281-288.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen N. T., Tamaki S., Kobayashi D., Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988 Oct 15;70(1):191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Ornston L. N. Regulation of catabolic pathways in Pseudomonas. Bacteriol Rev. 1971 Jun;35(2):87–116. doi: 10.1128/br.35.2.87-116.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornston L. N., Stanier R. Y. The conversion of catechol and protocatechuate to beta-ketoadipate by Pseudomonas putida. J Biol Chem. 1966 Aug 25;241(16):3776–3786. [PubMed] [Google Scholar]
- Parke D. Application of p-Toluidine in Chromogenic Detection of Catechol and Protocatechuate, Diphenolic Intermediates in Catabolism of Aromatic Compounds. Appl Environ Microbiol. 1992 Aug;58(8):2694–2697. doi: 10.1128/aem.58.8.2694-2697.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsko G. A., Kenyon G. L., Gerlt J. A., Ringe D., Kozarich J. W. On the origin of enzymatic species. Trends Biochem Sci. 1993 Oct;18(10):372–376. doi: 10.1016/0968-0004(93)90091-z. [DOI] [PubMed] [Google Scholar]
- Rosenberg S. L., Hegeman G. D. Clustering of functionally related genes in Pseudomonas aeruginosa. J Bacteriol. 1969 Jul;99(1):353–355. doi: 10.1128/jb.99.1.353-355.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanley M. S., Harrison A., Parales R. E., Kowalchuk G., Mitchell D. J., Ornston L. N. Unusual G + C content and codon usage in catIJF, a segment of the ben-cat supra-operonic cluster in the Acinetobacter calcoaceticus chromosome. Gene. 1994 Jan 28;138(1-2):59–65. doi: 10.1016/0378-1119(94)90783-8. [DOI] [PubMed] [Google Scholar]
- Simon R., O'Connell M., Labes M., Pühler A. Plasmid vectors for the genetic analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol. 1986;118:640–659. doi: 10.1016/0076-6879(86)18106-7. [DOI] [PubMed] [Google Scholar]
- Stanier R. Y., Ornston L. N. The beta-ketoadipate pathway. Adv Microb Physiol. 1973;9(0):89–151. [PubMed] [Google Scholar]
- Stanier R. Y., Palleroni N. J., Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966 May;43(2):159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- Tamaki T., Horinouchi S., Fukaya M., Okumura H., Kawamura Y., Beppu T. Nucleotide sequence of the membrane-bound aldehyde dehydrogenase gene from Acetobacter polyoxogenes. J Biochem. 1989 Oct;106(4):541–544. doi: 10.1093/oxfordjournals.jbchem.a122889. [DOI] [PubMed] [Google Scholar]
- Tresguerres M. E., De Torrontegui G., Cánovas J. L. The metabolism of quinate by Acinetobacter calco-aceticus. Arch Mikrobiol. 1970;70(2):110–118. doi: 10.1007/BF00412202. [DOI] [PubMed] [Google Scholar]
- Tresguerres M. E., de Torrontegui G., Ingledew W. M., Cánovas J. L. Regulation of the enzymes of the beta-ketoadipate pathway in Moraxella. Control of quinate oxidation by protocatechuate. Eur J Biochem. 1970 Jul;14(3):445–450. doi: 10.1111/j.1432-1033.1970.tb00309.x. [DOI] [PubMed] [Google Scholar]
- Tsou A. Y., Ransom S. C., Gerlt J. A., Buechter D. D., Babbitt P. C., Kenyon G. L. Mandelate pathway of Pseudomonas putida: sequence relationships involving mandelate racemase, (S)-mandelate dehydrogenase, and benzoylformate decarboxylase and expression of benzoylformate decarboxylase in Escherichia coli. Biochemistry. 1990 Oct 23;29(42):9856–9862. doi: 10.1021/bi00494a015. [DOI] [PubMed] [Google Scholar]
- Wellington C. L., Beatty J. T. Overlapping mRNA transcripts of photosynthesis gene operons in Rhodobacter capsulatus. J Bacteriol. 1991 Feb;173(4):1432–1443. doi: 10.1128/jb.173.4.1432-1443.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelis M. L., Stanier R. Y. The genetic control of dissimilatory pathways in Pseudomonas putida. Genetics. 1970 Oct;66(2):245–266. doi: 10.1093/genetics/66.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Young D. A., Bauer C. E., Williams J. C., Marrs B. L. Genetic evidence for superoperonal organization of genes for photosynthetic pigments and pigment-binding proteins in Rhodobacter capsulatus. Mol Gen Genet. 1989 Jul;218(1):1–12. doi: 10.1007/BF00330558. [DOI] [PubMed] [Google Scholar]
- van Kleef M. A., Duine J. A. Bacterial NAD(P)-independent quinate dehydrogenase is a quinoprotein. Arch Microbiol. 1988 May;150(1):32–36. doi: 10.1007/BF00409714. [DOI] [PubMed] [Google Scholar]