Abstract
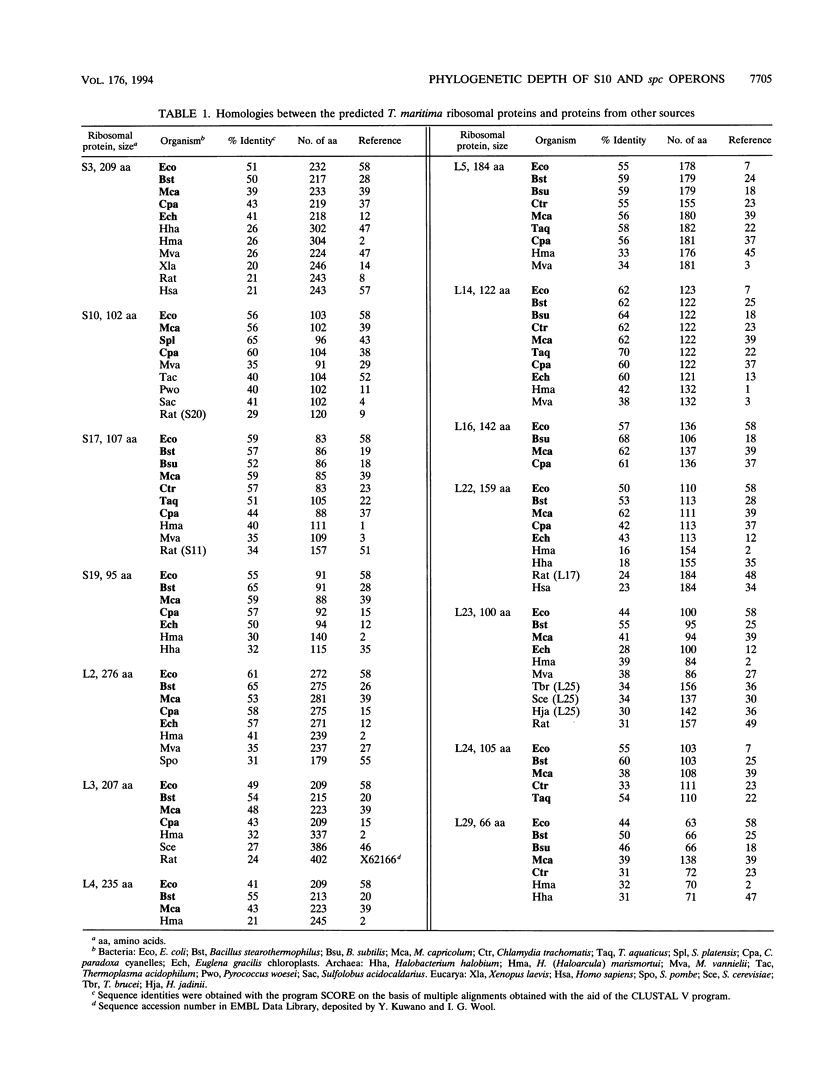
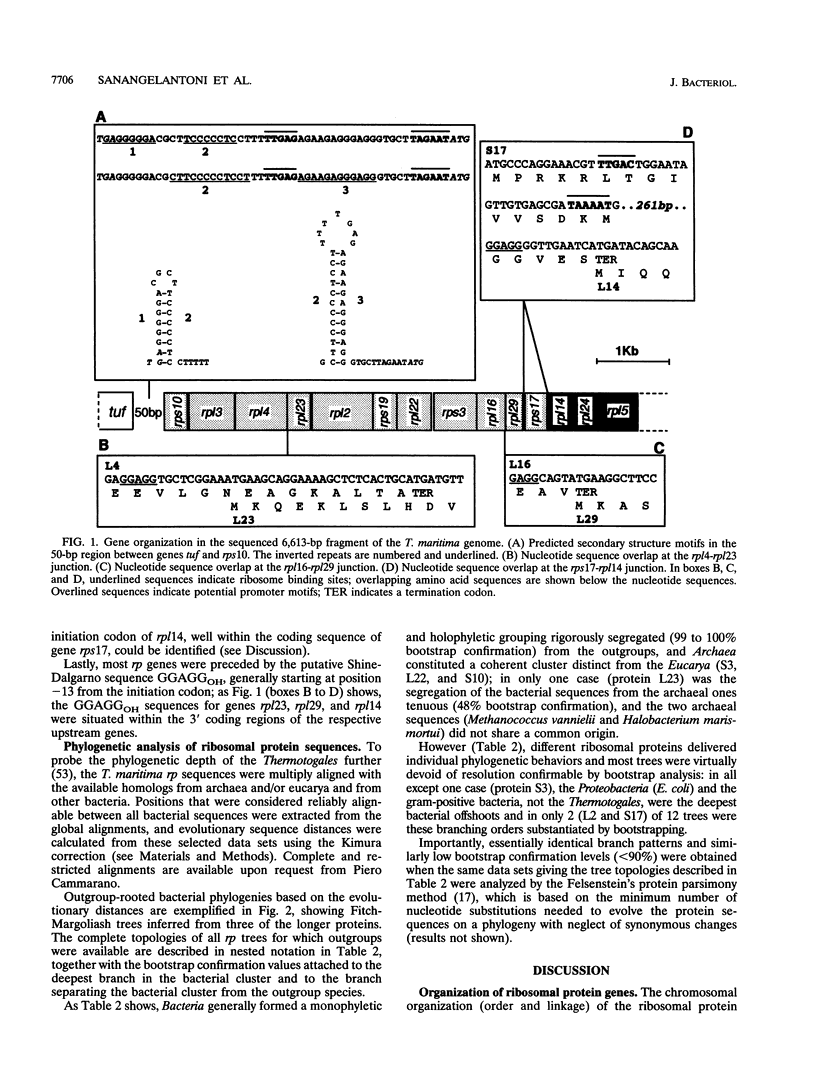
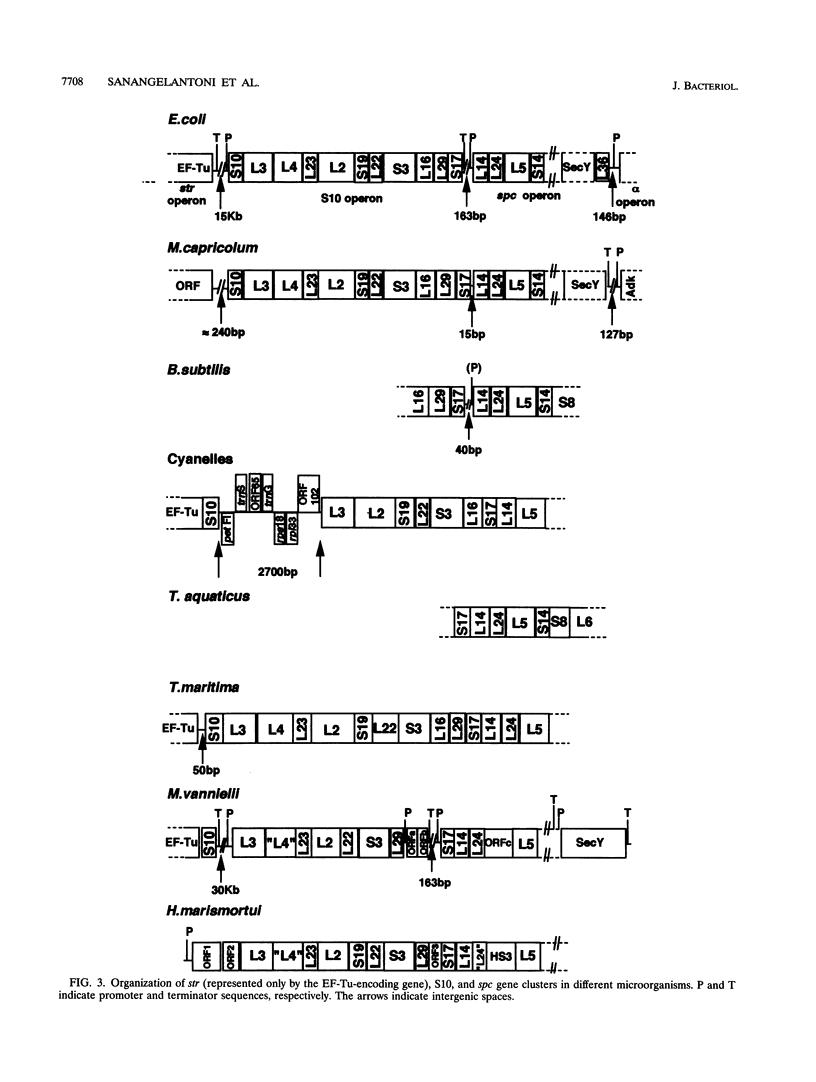
A segment of Thermotoga maritima DNA spanning 6,613 bp downstream from the gene tuf for elongation factor Tu was sequenced by use of a chromosome walking strategy. The sequenced region comprised a string of 14 tightly linked open reading frames (ORFs) starting 50 bp downstream from tuf. The first 11 ORFs were identified as homologs of ribosomal protein genes rps10, rpl3, rpl4, rpl23, rpl2, rps19, rpl22, rps3, rpl16, rpl29, and rps17 (which in Escherichia coli constitute the S10 operon, in that order); the last three ORFs were homologous to genes rpl14, rpl24, and rpl5 (which in E. coli constitute the three promoter-proximal genes of the spectinomycin operon). The 14-gene string was preceded by putative -35 and -10 promoter sequences situated 5' to gene rps10, within the 50-bp spacing between genes tuf and rps10; the same region exhibited a potential transcription termination signal for the upstream gene cluster (having tuf as the last gene) but displayed also the potential for formation of a hairpin loop hindering the terminator; this suggests that transcription of rps10 and downstream genes may start farther upstream. The similar organization of the sequenced rp genes in the deepest-branching bacterial phyla (T. maritima) and among Archaea has been interpreted as indicating that the S10-spc gene arrangement existed in the (last) common ancestor. The phylogenetic depth of the Thermotoga lineage was probed by use of r proteins as marker molecules: in all except one case (S3), Proteobacteria or the gram-positive bacteria, and not the genus Thermotoga, were the deepest-branching lineage; in only two cases, however, was the inferred branching order substantiated by bootstrap analysis.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arndt E., Krömer W., Hatakeyama T. Organization and nucleotide sequence of a gene cluster coding for eight ribosomal proteins in the archaebacterium Halobacterium marismortui. J Biol Chem. 1990 Feb 25;265(6):3034–3039. [PubMed] [Google Scholar]
- Arndt E. Nucleotide sequence of four genes encoding ribosomal proteins from the 'S10 and spectinomycin' operon equivalent region in the archaebacterium Halobacterium marismortui. FEBS Lett. 1990 Jul 16;267(2):193–198. doi: 10.1016/0014-5793(90)80923-7. [DOI] [PubMed] [Google Scholar]
- Auer J., Spicker G., Böck A. Organization and structure of the Methanococcus transcriptional unit homologous to the Escherichia coli "spectinomycin operon". Implications for the evolutionary relationship of 70 S and 80 S ribosomes. J Mol Biol. 1989 Sep 5;209(1):21–36. doi: 10.1016/0022-2836(89)90167-8. [DOI] [PubMed] [Google Scholar]
- Burggraf S., Olsen G. J., Stetter K. O., Woese C. R. A phylogenetic analysis of Aquifex pyrophilus. Syst Appl Microbiol. 1992 Aug;15(3):352–356. doi: 10.1016/S0723-2020(11)80207-9. [DOI] [PubMed] [Google Scholar]
- Cammarano P., Palm P., Creti R., Ceccarelli E., Sanangelantoni A. M., Tiboni O. Early evolutionary relationships among known life forms inferred from elongation factor EF-2/EF-G sequences: phylogenetic coherence and structure of the archaeal domain. J Mol Evol. 1992 May;34(5):396–405. doi: 10.1007/BF00162996. [DOI] [PubMed] [Google Scholar]
- Cerretti D. P., Dean D., Davis G. R., Bedwell D. M., Nomura M. The spc ribosomal protein operon of Escherichia coli: sequence and cotranscription of the ribosomal protein genes and a protein export gene. Nucleic Acids Res. 1983 May 11;11(9):2599–2616. doi: 10.1093/nar/11.9.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y. L., Devi K. R., Olvera J., Wool I. G. The primary structure of rat ribosomal protein S3. Arch Biochem Biophys. 1990 Dec;283(2):546–550. doi: 10.1016/0003-9861(90)90682-o. [DOI] [PubMed] [Google Scholar]
- Chan Y. L., Wool I. G. The primary structure of rat ribosomal protein S20. Biochim Biophys Acta. 1990 May 24;1049(1):93–95. doi: 10.1016/0167-4781(90)90088-j. [DOI] [PubMed] [Google Scholar]
- Christopher D. A., Cushman J. C., Price C. A., Hallick R. B. Organization of ribosomal protein genes rpl23, rpl2, rps19, rpl22 and rps3 on the Euglena gracilis chloroplast genome. Curr Genet. 1988 Sep;14(3):275–285. doi: 10.1007/BF00376748. [DOI] [PubMed] [Google Scholar]
- Christopher D. A., Hallick R. B. Euglena gracilis chloroplast ribosomal protein operon: a new chloroplast gene for ribosomal protein L5 and description of a novel organelle intron category designated group III. Nucleic Acids Res. 1989 Oct 11;17(19):7591–7608. doi: 10.1093/nar/17.19.7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988 Nov 25;16(22):10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creti R., Citarella F., Tiboni O., Sanangelantoni A., Palm P., Cammarano P. Nucleotide sequence of a DNA region comprising the gene for elongation factor 1 alpha (EF-1 alpha) from the ultrathermophilic archaeote Pyrococcus woesei: phylogenetic implications. J Mol Evol. 1991 Oct;33(4):332–342. doi: 10.1007/BF02102864. [DOI] [PubMed] [Google Scholar]
- Di Cristina M., Menard R., Pierandrei-Amaldi P. Xenopus laevis ribosomal protein S1a cDNA sequence. Nucleic Acids Res. 1991 Apr 25;19(8):1943–1943. doi: 10.1093/nar/19.8.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrard J. L., Kuntz M., Weil J. H. The nucleotide sequence of five ribosomal protein genes from the cyanelles of Cyanophora paradoxa: implications concerning the phylogenetic relationship between cyanelles and chloroplasts. J Mol Evol. 1990 Jan;30(1):16–25. doi: 10.1007/BF02102449. [DOI] [PubMed] [Google Scholar]
- Henkin T. M., Moon S. H., Mattheakis L. C., Nomura M. Cloning and analysis of the spc ribosomal protein operon of Bacillus subtilis: comparison with the spc operon of Escherichia coli. Nucleic Acids Res. 1989 Sep 25;17(18):7469–7486. doi: 10.1093/nar/17.18.7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herfurth E., Hirano H., Wittmann-Liebold B. The amino-acid sequences of the Bacillus stearothermophilus ribosomal proteins S17 and S21 and their comparison to homologous proteins of other ribosomes. Biol Chem Hoppe Seyler. 1991 Oct;372(10):955–961. doi: 10.1515/bchm3.1991.372.2.955. [DOI] [PubMed] [Google Scholar]
- Herwig S., Kruft V., Wittmann-Liebold B. Primary structures of ribosomal proteins L3 and L4 from Bacillus stearothermophilus. Eur J Biochem. 1992 Aug 1;207(3):877–885. doi: 10.1111/j.1432-1033.1992.tb17119.x. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Bleasby A. J., Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992 Apr;8(2):189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- Jahn O., Hartmann R. K., Erdmann V. A. Analysis of the spc ribosomal protein operon of Thermus aquaticus. Eur J Biochem. 1991 May 8;197(3):733–740. doi: 10.1111/j.1432-1033.1991.tb15965.x. [DOI] [PubMed] [Google Scholar]
- Kaul R., Gray G. J., Koehncke N. R., Gu L. J. Cloning and sequence analysis of the Chlamydia trachomatis spc ribosomal protein gene cluster. J Bacteriol. 1992 Feb;174(4):1205–1212. doi: 10.1128/jb.174.4.1205-1212.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura J., Kimura M. The complete amino acid sequences of the 5 S rRNA binding proteins L5 and L18 from the moderate thermophile Bacillus stearothermophilus ribosome. FEBS Lett. 1987 Jan 1;210(1):85–90. doi: 10.1016/0014-5793(87)81303-0. [DOI] [PubMed] [Google Scholar]
- Kimura M., Kimura J., Ashman K. The complete primary structure of ribosomal proteins L1, L14, L15, L23, L24 and L29 from Bacillus stearothermophilus. Eur J Biochem. 1985 Aug 1;150(3):491–497. doi: 10.1111/j.1432-1033.1985.tb09049.x. [DOI] [PubMed] [Google Scholar]
- Krömer W. J., Hatakeyama T., Kimura M. Nucleotide sequences of Bacillus stearothermophilus ribosomal protein genes: part of the ribosomal S10 operon. Biol Chem Hoppe Seyler. 1990 Jul;371(7):631–636. doi: 10.1515/bchm3.1990.371.2.631. [DOI] [PubMed] [Google Scholar]
- Köpke A. K., Wittmann-Liebold B. Comparative studies of ribosomal proteins and their genes from Methanococcus vannielii and other organisms. Can J Microbiol. 1989 Jan;35(1):11–20. doi: 10.1139/m89-003. [DOI] [PubMed] [Google Scholar]
- Lechner K., Heller G., Böck A. Organization and nucleotide sequence of a transcriptional unit of Methanococcus vannielii comprising genes for protein synthesis elongation factors and ribosomal proteins. J Mol Evol. 1989 Jul;29(1):20–27. doi: 10.1007/BF02106178. [DOI] [PubMed] [Google Scholar]
- Leer R. J., van Raamsdonk-Duin M. M., Hagendoorn M. J., Mager W. H., Planta R. J. Structural comparison of yeast ribosomal protein genes. Nucleic Acids Res. 1984 Sep 11;12(17):6685–6700. doi: 10.1093/nar/12.17.6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao D., Dennis P. P. Molecular phylogenies based on ribosomal protein L11, L1, L10, and L12 sequences. J Mol Evol. 1994 Apr;38(4):405–419. doi: 10.1007/BF00163157. [DOI] [PubMed] [Google Scholar]
- Liao D., Dennis P. P. The organization and expression of essential transcription translation component genes in the extremely thermophilic eubacterium Thermotoga maritima. J Biol Chem. 1992 Nov 15;267(32):22787–22797. [PubMed] [Google Scholar]
- Lindahl L., Sor F., Archer R. H., Nomura M., Zengel J. M. Transcriptional organization of the S10, spc and alpha operons of Escherichia coli. Biochim Biophys Acta. 1990 Aug 27;1050(1-3):337–342. doi: 10.1016/0167-4781(90)90191-4. [DOI] [PubMed] [Google Scholar]
- Mager D. L., Freeman J. D. A human gene related to the ribosomal protein L23 gene of Halobacterium marismortui. Nucleic Acids Res. 1990 Sep 11;18(17):5301–5301. doi: 10.1093/nar/18.17.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankin A. S. The nucleotide sequence of the genes coding for the S19 and L22 equivalent ribosomal proteins from Halobacterium halobium. FEBS Lett. 1989 Mar 27;246(1-2):13–16. doi: 10.1016/0014-5793(89)80243-1. [DOI] [PubMed] [Google Scholar]
- Metzenberg S., Joblet C., Verspieren P., Agabian N. Ribosomal protein L25 from Trypanosoma brucei: phylogeny and molecular co-evolution of an rRNA-binding protein and its rRNA binding site. Nucleic Acids Res. 1993 Oct 25;21(21):4936–4940. doi: 10.1093/nar/21.21.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalowski C. B., Pfanzagl B., Löffelhardt W., Bohnert H. J. The cyanelle S10 spc ribosomal protein gene operon from Cyanophora paradoxa. Mol Gen Genet. 1990 Nov;224(2):222–231. doi: 10.1007/BF00271555. [DOI] [PubMed] [Google Scholar]
- Neumann-Spallart C., Jakowitsch J., Kraus M., Brandtner M., Bohnert H. J., Löffelhardt W. rps10, unreported for plastid DNAs, is located on the cyanelle genome of Cyanophora paradoxa and is cotranscribed with the str operon genes. Curr Genet. 1991 Apr;19(4):313–315. doi: 10.1007/BF00355061. [DOI] [PubMed] [Google Scholar]
- Ohkubo S., Muto A., Kawauchi Y., Yamao F., Osawa S. The ribosomal protein gene cluster of Mycoplasma capricolum. Mol Gen Genet. 1987 Dec;210(2):314–322. doi: 10.1007/BF00325700. [DOI] [PubMed] [Google Scholar]
- Olsen G. J., Woese C. R., Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994 Jan;176(1):1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanangelantoni A. M., Tiboni O. The chromosomal location of genes for elongation factor Tu and ribosomal protein S10 in the cyanobacterium Spirulina platensis provides clues to the ancestral organization of the str and S10 operons in prokaryotes. J Gen Microbiol. 1993 Nov;139(11):2579–2584. doi: 10.1099/00221287-139-11-2579. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholzen T., Arndt E. Organization and nucleotide sequence of ten ribosomal protein genes from the region equivalent to the spectinomycin operon in the archaebacterium Halobacterium marismortui. Mol Gen Genet. 1991 Aug;228(1-2):70–80. doi: 10.1007/BF00282450. [DOI] [PubMed] [Google Scholar]
- Schultz L. D., Friesen J. D. Nucleotide sequence of the tcml gene (ribosomal protein L3) of Saccharomyces cerevisiae. J Bacteriol. 1983 Jul;155(1):8–14. doi: 10.1128/jb.155.1.8-14.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiridonova V. A., Akhmanova A. S., Kagramanova V. K., Köpke A. K., Mankin A. S. Ribosomal protein gene cluster of Halobacterium halobium: nucleotide sequence of the genes coding for S3 and L29 equivalent ribosomal proteins. Can J Microbiol. 1989 Jan;35(1):153–159. doi: 10.1139/m89-023. [DOI] [PubMed] [Google Scholar]
- Suzuki K., Wool I. G. The primary structure of rat ribosomal protein L17. Biochem Biophys Res Commun. 1991 Jul 15;178(1):322–328. doi: 10.1016/0006-291x(91)91817-v. [DOI] [PubMed] [Google Scholar]
- Suzuki K., Wool I. G. The primary structure of rat ribosomal protein L23a. The application of homology search to the identification of genes for mammalian and yeast ribosomal proteins and a correlation of rat and yeast ribosomal proteins. J Biol Chem. 1993 Feb 5;268(4):2755–2761. [PubMed] [Google Scholar]
- Tanaka T., Kuwano Y., Ishikawa K., Ogata K. Nucleotide sequence of cloned cDNA specific for rat ribosomal protein S11. J Biol Chem. 1985 May 25;260(10):6329–6333. [PubMed] [Google Scholar]
- Tesch A., Klink F. Nucleotide sequence of the gene coding for ribosomal protein S10 from the archaeum Thermoplasma acidophilum. Nucleic Acids Res. 1992 Aug 11;20(15):4090–4090. doi: 10.1093/nar/20.15.4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiboni O., Cantoni R., Creti R., Cammarano P., Sanangelantoni A. M. Phylogenetic depth of Thermotoga maritima inferred from analysis of the fus gene: amino acid sequence of elongation factor G and organization of the Thermotoga str operon. J Mol Evol. 1991 Aug;33(2):142–151. doi: 10.1007/BF02193628. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. T., Tan Y. M., Tan Y. H. Isolation of a cDNA encoding human 40S ribosomal protein s3. Nucleic Acids Res. 1990 Nov 25;18(22):6689–6689. doi: 10.1093/nar/18.22.6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski G., Zurawski S. M. Structure of the Escherichia coli S10 ribosomal protein operon. Nucleic Acids Res. 1985 Jun 25;13(12):4521–4526. doi: 10.1093/nar/13.12.4521. [DOI] [PMC free article] [PubMed] [Google Scholar]


