Abstract
Aims
To analyse the correlation between MYC amplification and various clinicopathological features and outcome in a cohort of 245 patients with invasive breast carcinoma treated with surgery followed by anthracycline‐based chemotherapy. Given the high prevalence of MYC amplification in tumours of BRCA1 mutation carriers and the similarities between these and sporadic “basal‐like” carcinomas, the prevalence of MYC amplification in “basal‐like” breast carcinomas was investigated.
Methods
MYC gene copy number was assessed on tissue microarrays containing duplicate cores of 245 invasive breast carcinomas by means of chromogenic in situ hybridisation using SpotLight C‐MYC amplification probe and chromosome 8 centromeric probe (CEP8). Signals were evaluated at 400× magnification; 30 morphologically unequivocal neoplastic cells in each core were counted for the presence of the gene and CEP8 probes.
Results
Amplification was defined as a MYC:CEP8 ratio >2. Signals for both MYC and CEP8 were assessable in 196/245 (80%) tumours. MYC amplification was found in 19/196 cases (9.7%) and was not associated with tumour size, histological grade, positivity for oestrogen receptor, progesterone receptor, HER2, epidermal growth factor, cytokeratins 14, 5/6 and 17, MIB1 or p53. Only 4% of basal‐like carcinomas showed MYC amplification, compared to 8.75% and 10.7% of luminal and HER2 tumours respectively. On univariate analysis, MYC amplification displayed a significant association with shorter metastasis‐free and overall survival and proved to be an independent prognostic factor on multivariate survival analysis.
Conclusion
MYC amplification is not associated with “basal‐like” phenotype and proved to be an independent prognostic factor for breast cancer patients treated with anthracycline‐based chemotherapy.
Keywords: oncogene, MYC , CISH, prognosis, basal‐like breast cancer
The MYC proto‐oncogene maps to 8q24.1 and encodes at least three major transcripts, two of them containing the transactivation domain (c‐myc1 and c‐myc2).1,2,3,4 Depending on the context and on the isoform, this transcription factor can promote either cell proliferation or apoptosis.1,2,3,4 Given the plethora of biological roles played by MYC gene products and the lack of reliable antibodies for immunohistochemical analysis, the analysis of the clinical relevance of c‐myc overexpression in breast cancer has produced conflicting results.2 On the other hand, MYC gene amplification has been extensively studied in breast cancer.1,2 The prevalence of MYC amplifications ranges from 1.1% to 94.4% of cases, depending on the cohort of patients and the techniques used.1,2,5,6,7,8,9,10,11,12,13,14,15,16,17,18 In a recent meta‐analysis, largely based on studies where MYC gene copy numbers were analysed by means of Southern blot, slot blot or PCR‐based techniques,1MYC gene amplification was shown to be associated with high histological grade, presence of lymph node metastasis, lack of progesterone receptor and poor survival.
In situ methods are currently considered the “gold standard” for gene copy number assessment. Fluorescent in situ hybridisation (FISH) has been applied to the study of MYC amplification in breast cancer,5,6,7,9,10,11,12,13,15,16,17,19,20 with frequencies ranging from 5.3%12 to 86%.11 With this method, the correlations between MYC amplification and clinicopathological parameters have been surprisingly inconsistent and, although MYC amplification has been shown to be associated with poor prognosis,1 it is still unclear whether MYC amplification is an independent prognostic factor in invasive breast cancer.
Chromogenic in situ hybridisation (CISH) has been previously used to determine the prevalence of MYC amplification. In a cohort of 177 breast cancer patients, Rummukainen et al8 found that 15.25% of cases showed MYC amplification; this was correlated with aneuploidy, high histological grade, high S‐phase fraction, lack of progesterone receptor and shorter overall survival.8 However, in that study, only a MYC gene specific probe was employed. Although a good agreement between MYC as defined by dual colour FISH and single probe CISH for MYC gene amplification has been described (unweighted κ = 0.68),8 the correlations with clinicopathological features and prognostic impact of MYC amplification defined by each technique seem to differ.8 Owing to (i) the very high frequency of 8q gains, in the form of additional isochromomes 8, in high grade breast cancer, and (ii) the fact that by single probe CISH analysis, cases defined as “MYC non‐amplified but aneuploid” (4–5 copies/cell) also showed a poor prognosis when compared to cases with 1–3 copies of MYC gene,8 the use of a centromeric probe seems to be required to differentiate between polysomy of chromosome 8 and MYC gene amplification, and to reliably identify MYC low level amplifications (for example, five copies of MYC but only two copies of chromosome 8 centromere).
It has recently been shown that MYC gene amplifications are significantly more prevalent in tumours arising in BRCA1 mutation carriers.15,16 Given the phenotypic and molecular similarities between these tumours and sporadic basal‐like breast carcinomas,21,22,23,24,25,26 we hypothesised that MYC amplification could also play a role in the biology of basal‐like breast carcinomas.
In this study, utilising a centromeric probe for chromosome 8 (CEP8) and a gene specific probe for MYC, we investigated the correlation between MYC amplification and various clinicopathological parameters and outcome in a cohort of 245 women with invasive breast carcinoma treated with surgery followed by adjuvant anthracycline‐based chemotherapy. A second aim was to define the prevalence of MYC amplification in “basal‐like” breast carcinomas.
Materials and methods
Tissue microarrays
The tissue microarray contained replicate 0.6 mm cores of 245 invasive breast carcinomas (185 invasive ductal carcinomas, 27 invasive lobular carcinomas, 25 invasive mixed carcinomas and 8 invasive breast carcinomas of other special types). All patients were treated with therapeutic surgery (69 mastectomy and 155 wide local excision) and adjuvant anthracycline‐based chemotherapy; those with oestrogen receptor (ER) positive tumours also received endocrine therapy. Follow‐up was available for 244 patients, ranging from 0.5 to 125 months (median 67 months, mean 67 months). Full details of the characterisation of the tissue microarray and the cohort of patients are described elsewhere.27,28 Tumours were graded according to the modified Bloom–Richardson scoring system29 and size was categorised according to the TNM staging criteria. Details on the expression of ER, progesterone receptor (PR), HER2, epidermal growth factor receptor (EGFR), cytokeratin (Ck) 5/6, Ck 14 and Ck 17 are described elsewhere.27,28 Tumours were classified into basal‐like, luminal or HER2 groups according to the immunohistochemical panel proposed by Nielsen et al.30 This study was approved by the Royal Marsden Hospital Ethics Committee.
Chromogenic in situ hybridisation
CISH for MYC and chromosome 8 centromere was performed on serial tissue microarray sections as previously described,27,31 using the ready‐to‐use digoxigenin‐labelled SPoT‐Light C‐MYC amplification probe (Zymed, San Francisco, California, USA) and biotin‐labelled SPoT‐Light Chromosome 8 Centromeric Probe (Zymed, San Francisco, California, USA). Heat pretreatment of deparaffinised sections consisted of incubation for 15 min at 98°C in CISH pretreatment buffer (SPOT‐light tissue pretreatment kit, Zymed) and digested with pepsin for 6 min at room temperature according to the manufacturer's instructions. An appropriate MYC gene‐amplified breast tumour control was included in the slide run. CISH experiments were analysed by three of the authors (SMRP, SEP and JSR‐F) on a multi‐headed microscope. Only unequivocal signals were counted. Signals were evaluated at 400× and 630× magnification; 30 morphologically unequivocal neoplastic cells in each core were assessed for the presence of the gene and chromosome 8 centromere probe signals. Amplification was defined as a MYC:CEP8 ratio >2. The scoring was evaluated with observers blinded to the clinicopathological details and patients' outcome.
Statistical analysis
The StatView V.5.0 software package (SAS Institute Inc., Cary, NC, USA) was used for all calculations. Correlations between categorical variables were performed using the χ2 test, and Fisher's exact test where appropriate. Correlations between continuous and categorical variables were performed with analysis of variance. Metastasis‐free and overall survival was expressed as the number of months from diagnosis to the occurrence of an event (distant metastasis or disease‐related death, respectively). Cumulative survival probabilities were calculated using the Kaplan–Meier method. Differences between survival rates were tested with the log‐rank test. All tests were two‐tailed, with a confidence interval of 95%.
Multivariate analysis was performed using the Cox multiple hazards model. A p value of 0.05 in the univariate survival analysis was adopted as the limit for inclusion in the multivariate model, and cases with missing values were excluded from this analysis.
Results
Table 1 summarises he correlations between MYC amplification and clinicopathological features and immunohistochemical findings in 245 breast carcinomas. Briefly, 49 cores were either lost/fragmented in the CISH procedure, did not have invasive tumour or showed suboptimal signals for either MYC or CEP8. Of the 196 remaining tumours, 19 (9.7%) showed MYC:CEP8 ratios >2.0. Cases with MYC amplification were seen in the form of large clusters of MYC signals (fig 1) or multiple individual signals/nucleus (fig 1). MYC amplification was found only in grade 2 and grade 3 breast carcinomas (18 invasive ductal carcinomas and 1 mixed ductal‐lobular carcinoma/pleomorphic lobular carcinoma32). However, no significant correlation between MYC amplification and histological grade was found (only 19 grade 1 carcinomas were included in the series) and no correlation between MYC amplification and tumour size, presence of lymph node metastasis or lympho‐vascular invasion was identified. No association between MYC amplification and ER, PR, HER2, EGFR, Ck 5/6, Ck 14 or Ck 17 and p53 expression was observed. MYC amplification showed a trend for a higher proliferation rate, as defined by MIB1 expression (p = 0.1084). No correlation between basal‐phenotype, as defined by the immunohistochemical panel proposed by Nielsen et al,30 and MYC amplification was found. In fact, only 4% of basal‐like carcinomas showed MYC amplification, compared to 8.75% and 10.7% observed in luminal and HER2 tumours, respectively (p = 0.6134, NS).
Table 1 Correlations between MYC amplification, clinicopathological parameters and immunohistochemical markers in 245 invasive breast carcinomas.
| Parameter | n | NA | Not amplified | Amplified | p Value |
|---|---|---|---|---|---|
| Size | 194 | 51 | 0.3101* | ||
| T1 | 89 | 12 | |||
| T2 | 76 | 5 | |||
| T3 | 2 | 2 | |||
| Type | 196 | 49 | 0.2266* | ||
| Ductal | 131 | 18 | |||
| Lobular | 19 | 0 | |||
| Mixed | 20 | 1 | |||
| Other | 7 | 0 | |||
| Grade | 193 | 52 | 0.1942* | ||
| 1 | 19 | 0 | |||
| 2 | 49 | 4 | |||
| 3 | 106 | 15 | |||
| LVI | 195 | 50 | 0.6219† | ||
| + | 116 | 14 | |||
| − | 60 | 5 | |||
| LN metastasis | 189 | 56 | 0.4530† | ||
| + | 105 | 14 | |||
| − | 65 | 5 | |||
| ER | 196 | 49 | >0.9999† | ||
| + | 146 | 16 | |||
| − | 31 | 3 | |||
| PR | 196 | 45 | 0.1635† | ||
| + | 132 | 17 | |||
| − | 46 | 2 | |||
| HER2 | 196 | 49 | 0.7390† | ||
| + | 25 | 3 | |||
| − | 152 | 16 | |||
| EGFR | 196 | 45 | 0.6995† | ||
| + | 20 | 1 | |||
| − | 157 | 18 | |||
| Ck 14 | 195 | 50 | >0.9999† | ||
| + | 17 | 1 | |||
| − | 159 | 18 | |||
| Ck 5/6 | 187 | 58 | >0.9999† | ||
| + | 20 | 2 | |||
| − | 148 | 17 | |||
| Ck 17 | 193 | 52 | >0.9999† | ||
| + | 20 | 3 | |||
| − | 155 | 15 | |||
| Basal markers | 195 | 50 | >0.9999† | ||
| + | 30 | 3 | |||
| − | 146 | 16 | |||
| Nielsen groups | 193 | 52 | 0.6134* | ||
| Basal | 24 | 1 | |||
| Luminal | 126 | 14 | |||
| HER2 | 25 | 3 | |||
| p53 | 193 | 52 | 0.5987† | ||
| + | 51 | 7 | |||
| − | 123 | 12 | |||
| MIB1 | 192 | 53 | 0.1084* | ||
| <10% | 73 | 4 | |||
| 10–30% | 75 | 13 | |||
| >30% | 25 | 2 |
*χ2 test; †Fisher's exact test.
Ck, cytokeratins; EGFR, epidermal growth factor receptor; ER, oestrogen receptor; LN, lymph node; LVI, lympho‐vascular invasion; NA, not assessable (lost or uninterpretable cores); PR, progesterone receptor.
Nielsen groups30: HER2: HER2 positive = ER+/−, Ck 5/6 and/or EGFR+/−; luminal = HER2 negative, ER positive, Ck 5/6 and/or EGFR+/−; basal: HER2 negative, ER negative, Ck 5/6 and/or EGFR+.
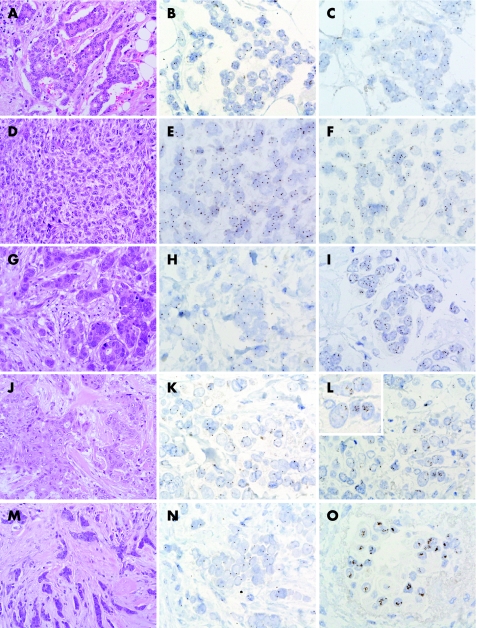
Figure 1 MYC gene copy numbers in breast cancer: grade 1 invasive ductal carcinoma (A) with 1–3 chromosome 8 centromere (CEP8, B) and MYC (C) gene signals. Metaplastic spindle cell breast carcinoma (D) with chromosome 8 polysomy (E) and an average of 4.5 copies of MYC (F); the MYC:CEP8 ratio was 1.59. Grade 2 invasive ductal carcinoma (G) harbouring “low level” MYC amplification: note the presence of 1–3 copies of CEP8 (H) but increased copy numbers of MYC (I); the MYC:CEP8 ratio was 2.24. Grade 3 invasive ductal carcinoma (J) with 1–3 copies of chromosome 8 centromere (K) and clusters of MYC gene signals (L). Inset: MYC gene signal clusters. Mixed ductal‐lobular carcinoma (M) with 1–3 CEP8 copies (N) and MYC large signal clusters (O). (Original magnification ×200—A, D, G, J and M; ×630—B, C, E, F, H, I, K, L, N and O; ×1000—inset L.)
When data on the replicate cores of each tumour were treated independently, the agreement for MYC amplification, considering the categories not amplified and amplified was good, (unweighted κ = 0.7916 (0.6152–0.968)). MYC amplification was found in both cores in 11 cases and in one of the cores in 5 cases. In 3 cases, one of the cores showed amplification, whereas the other was lost during hybridisation. No statistically significant differences in tumour size, histological grade, presence of vascular invasion or lymph node metastasis, ER, PR, HER2, EGFR, Ck 14, Ck 5/6 and Ck 17 expression, and MIB1 proliferation rates were observed between cases homogeneously (ie, both cores showing MYC amplification) or heterogeneously (ie, one core showing MYC amplification and the other with normal copy numbers) amplified for MYC gene (data not shown). However, given the limited sample size of the cohort of MYC amplified cases, type 2 or β errors cannot be excluded in the present study. Interestingly, all cases with heterogeneously amplified MYC (n = 5) lacked p53 nuclear expression, whereas 6 of 11 cases with homogeneous MYC amplifications showed p53 positivity (p<0.100, χ2 test).
If MYC amplifications were defined based only on the MYC gene probe copy numbers as described by Rummukainen et al,8 12 of 19 amplified cases would have been classified as amplified, whereas 1 non‐amplified case by MYC:CEP8 ratios would have been classified as amplified (unweighted κ = 0.7286 (0.55–0.9072). MYC amplification as defined by single probe CISH showed a borderline statistical correlation with larger tumour size and higher proliferation rate (p<0.1000, χ2). No basal‐like carcinomas showed MYC amplification (0/26), whereas 7% of both luminal and HER2 tumours were MYC amplified (data not shown).
Survival analysis
In this cohort, MYC amplification, tumour size, presence of lympho‐vascular invasion, lymph node metastasis at time of diagnosis, ER, PR, Ck 14, Ck 5/6, Ck 17, p53 and proliferation index as defined by MIB1 were statistically significant prognostic factors for metastasis‐free survival on univariate analysis (table 2, fig 2A). On multivariate Cox hazard analysis, including all parameters that showed an association with metastasis‐free survival on univariate analysis, presence of MYC amplification and lymph node metastasis were shown to be independent prognostic factors (table 3).
Table 2 Univariate survival analysis of 245 patients with breast cancer treated with surgery followed by anthracycline‐based adjuvant chemotherapy.
| Parameter | n | Events | MFS Mean (SD) | p value (log rank test) | Events | OS Mean (SD) | p value (log rank test) |
|---|---|---|---|---|---|---|---|
| Size | <0.005 | >0.1000 | |||||
| T1 | 127 | 19 | 118 (3.68) | 20 | 115.4 (4.09) | ||
| T2 | 100 | 22 | 110 (4.72) | 18 | 114.9 (4.35) | ||
| T3 | 16 | 8 | 59 (7.40) | 4 | 76.4 (7.47) | ||
| Grade | <0.1000 | <0.1000 | |||||
| I | 23 | 1 | 117 (3.94) | 1 | 117 (3.64) | ||
| II | 69 | 11 | 118 (4.83) | 8 | 121 (4.93) | ||
| III | 148 | 37 | 407 (4.08) | 33 | 109 (4.00) | ||
| LN metastasis | <0.0001 | <0.0005 | |||||
| No | 83 | 5 | 129 (2.80) | 5 | 129 (2.60) | ||
| Yes | 154 | 44 | 102 (4.25) | 37 | 105 (4.32) | ||
| LVI | <0.0500 | >0.1000 | |||||
| No | 82 | 82 | 121 (4.14) | 11 | 121 (4.15) | ||
| Yes | 161 | 161 | 101 (3.66) | 31 | 104 (3.73) | ||
| ER | <0.0010 | 0.0001 | |||||
| Negative | 48 | 18 | 84 (6.78) | 17 | 86.8 (6.53) | ||
| Positive | 191 | 30 | 117 (3.11) | 24 | 119.2 (3.17) | ||
| PR | <0.0100 | <0.0005 | |||||
| Negative | 64 | 20 | 92.2 (5.96) | 20 | 92.8 (5.84) | ||
| Positive | 175 | 28 | 116.9 (3.26) | 21 | 119.7 (3.33) | ||
| HER2 | >0.1000 | >0.1000 | |||||
| Negative | 200 | 38 | 113.3 (3.25) | 32 | 115 (3.43) | ||
| Positive | 36 | 10 | 98.4 (7.84) | 9 | 102 (7.28) | ||
| EGFR | >0.1000 | <0.1000 | |||||
| Negative | 222 | 42 | 113.4 (3.08) | 35 | 115.5 (3.18) | ||
| Positive | 22 | 7 | 90.9 (9.20) | 7 | 92.3 (8.79) | ||
| Ck 14 | <0.0500 | <0.0500 | |||||
| Negative | 221 | 41 | 114.0 (3.05) | 34 | 116.0 (3.13) | ||
| Positive | 22 | 8 | 84.5 (10.13) | 8 | 86.6 (9.47) | ||
| Ck 5/6 | <0.0500 | <0.0100 | |||||
| Negative | 210 | 39 | 114.0 (3.11) | 32 | 116.2 (3.19) | ||
| Positive | 25 | 9 | 84.8 (9.47) | 9 | 86.8 (8.95) | ||
| Ck 17 | <0.0005 | <0.0001 | |||||
| Negative | 213 | 35 | 116.4 (2.97) | 28 | 118.5 (3.06) | ||
| Positive | 28 | 12 | 78.1 (9.24) | 12 | 80.3 (8.72) | ||
| p53 | <0.0500 | <0.0010 | |||||
| Negative | 158 | 25 | 117 (3.41) | 18 | 120 (3.52) | ||
| Positive | 67 | 20 | 102 (6.21) | 20 | 103 (6.06) | ||
| MIB1 | <0.0100 | <0.0050 | |||||
| <10% | 96 | 14 | 118.3 (4.22) | 11 | 122.4 (3.66) | ||
| 10–30% | 97 | 20 | 111.7 (4.86) | 16 | 111.4 (5.48) | ||
| >30% | 33 | 13 | 86.7 (8.87) | 13 | 88.8 (8.44) | ||
| MYC gene | 0.0050 | <0.0050 | |||||
| Non‐amplified | 176 | 32 | 114.2 (3.43) | 27 | 117 (3.34) | ||
| Amplified | 19 | 8 | 86.1 (12.82) | 8 | 84.4 (12.17) |
Ck, cytokeratin; ER, oestrogen receptor; LN, lymph node metastasis; LVI, lympho‐vascular invasion; MFS, metastasis‐free survival; OS, overall survival; PR, progesterone receptor.
Figure 2 Univariate analysis of the prognostic impact of MYC gene amplification on metastasis‐free (A) and overall survival (B).
Table 3 Cox hazard analysis of metastasis‐free survival (n = 174 patients).
| Parameter | Coefficient (95% CI) | SE | p value | Risk ratio (95% CI) |
|---|---|---|---|---|
| Size (TNM) | 0.3898 (−0.1435 to 0.9231) | 0.2721 | 0.1520 | 1.4767 (0.8663 to 2.517) |
| Lympho‐vascular invasion | 0.3711 (−0.5735 to 1.3157) | 0.4819 | 0.4413 | 1.4493 (0.5636 to 3.7272) |
| Lymph node metastasis | 1.8411 (0.6886 to 2.9937) | 0.5880 | 0.0017* | 6.3037 (1.9909 to 19.9591) |
| Oestrogen receptor | −0.4204 (−1.6631 to 0.8223) | 0.6340 | 0.5073 | 0.6568 (0.1896 to 2.2756) |
| Progesterone receptor | −0.2974 (−1.2645 to 0.6697) | 0.4934 | 0.5467 | 0.7428 (0.2824 to 1.9537) |
| Ck 14 | 0.5152 (−1.2607 to 2.291) | 0.9060 | 0.5696 | 1.6739 (0.2835 to 9.8847) |
| Ck 5/6 | 0.2402 (−1.3223 to 1.8028) | 0.7972 | 0.7631 | 1.2716 (0.2665 to 6.0666) |
| Ck 17 | 0.201 (−0.8505 to 1.2525) | 0.5365 | 0.7079 | 1.2226 (0.4272 to 3.499) |
| p53 | 0.6324 (−0.1644 to 1.4292) | 0.4065 | 0.1198 | 1.8822 (0.8484 to 4.1754) |
| MIB1 | 0.0813 (−0.5283 to 0.691) | 0.3111 | 0.7937 | 1.0847 (0.5896 to 1.9958) |
| MYC amplification | 0.9361 (0.0445 to 1.8278) | 0.4549 | 0.0396* | 2.5501 (1.0455 to 6.2202) |
Ck, cytokeratin.
*Significant p values.
Univariate survival analysis revealed MYC amplification, lymph node metastasis at time of diagnosis, ER, PR, Ck 14, Ck 5/6, Ck 17, p53, and proliferation index as defined by MIB1 as prognostic factors for overall survival (table 2, fig 2B). On multivariate Cox hazard analysis, including all parameters that showed an association with overall survival on univariate analysis, MYC amplification, the presence of lymph node metastasis and p53 expression were shown to be independent prognostic factors for OS (table 4).
Table 4 Cox hazard analysis of overall survival (n = 174 patients).
| Parameter | Coefficient (95% CI) | SE | p value | Risk ratio (95% CI) |
|---|---|---|---|---|
| Lymph node metastasis | 1.956 (0.8375 to 3.0746) | 0.5707 | 0.0006* | 7.0712 (2.3106 to 21.6405) |
| Oestrogen receptor | −0.6014 (−1.9193 to 0.7166) | 0.6724 | 0.3711 | 0.5481 (0.1467 to 2.0474) |
| Progesterone receptor | −0.6242 (−1.6386 to 0.3902) | 0.5175 | 0.2278 | 0.5357 (0.1943 to 1.4773) |
| Ck 14 | 0.2507 (−1.5519 to 2.0532) | 0.9197 | 0.7852 | 1.2849 (0.2118 to 7.7928) |
| Ck 5/6 | 0.0155 (−1.5148 to 1.5457) | 0.7807 | 0.9842 | 1.0156 (0.2199 to 4.6914) |
| Ck 17 | 0.5896 (−0.4127 to 1.5919) | 0.5114 | 0.2489 | 1.8032 (0.6619 to 4.9129) |
| p53 | 1.0136 (0.1685 to 1.8586) | 0.4311 | 0.0187* | 2.7554 (1.1836 to 6.4147) |
| MIB1 | 0.0419 (−0.5907 to 0.6746) | 0.3228 | 0.8966 | 1.0428 (0.554 to 1.9632) |
| MYC amplification | 1.2281 (0.3018 to 2.1544) | 0.4726 | 0.0094* | 3.4147 (1.3523 to 8.6229) |
Ck: cytokeratin.
*Significant p values.
MYC amplification defined by single probe CISH was not significantly correlated with metastasis‐free or overall survival (p>0.05, data not shown).
Discussion
Chromogenic in situ hybridisation has proven to be a useful technique to determine gene copy numbers and gene amplification on formalin‐fixed, paraffin‐embedded tissue sections.8,27,31 Unlike FISH, CISH allows a direct comparison between morphological features of neoplastic cells and the presence of gene amplification.8,27,31 Furthermore, CISH analysis is relatively quick; in the present study, the whole analysis of two probes (ie, CEP8 and MYC) in 245 replicate cores took less than a week, the neoplastic cells were easily recognisable and only 20% of the cases were not interpretable for one or the other probe.
In the present study, MYC amplification was shown to be a poor prognostic factor for distant metastasis and overall survival, in agreement with previous studies.1,2,8,13,18,33,34,35 However, we also show that the prognostic impact of MYC amplification is independent of size, histological grade, lympho‐vascular invasion, lymph node status, and expression of ER, PR, basal markers, p53 and proliferation rate. Although this study is retrospective, the differences in survival rates are not confounded by the therapeutic regimens, given that all patients were treated with surgery and adjuvant anthracycline‐based chemotherapy. Alternatively, the independent prognostic impact of MYC amplification observed in this cohort of patients could be explained by a reduced sensitivity of MYC amplified cases to anthracycline‐based chemotherapy. Although this possibility could not be completely ruled out without a non‐treated control arm, a direct mechanistic association between MYC amplification and reduced sensitivity to anthracyclines has never been described in vivo.
The distribution of MYC amplification within subclones of breast carcinomas is reported to be remarkably heterogeneous.14 In our analysis, of 16 cases harbouring MYC amplification where two cores of each tumour rendered results for both CEP8 and MYC, MYC was homogeneously amplified (ie, both cores showed amplification) in 68.75% and heterogeneously amplified in 31.25% of cases (ie, one core with MYC amplification and another with normal copy numbers). If only one core per tumour was used in the present study, 3 of 19 cases with MYC amplification would have been classified as “non‐amplified”. These results suggest that tissue microarray analysis of MYC amplification need more than one core per tumour.
In the present study, single probe CISH analysis was slightly less sensitive that CISH with two probes (MYC and CEP8). As expected, all but one case with large signal clusters and/or >5 signals/cell using one probe also displayed MYC:CEP8 ratios >2. However, the use of a centromeric probe allowed for the identification of cases with MYC low level amplification (MYC:CEP8 ratios >2, but ⩽5 MYC gene signals on average). This is not surprising given that the presence of >5 copies of a given chromosome in primary breast cancer and breast cancer cell lines is an exceedingly rare biological phenomenon36 (The Cancer Genome Anatomy Project, http://cgap.nci.nih.gov/Chromosomes). The agreement between MYC amplification as defined by CISH using one or two probes was similar to that described for CISH with one probe and dual colour FISH.8
We, and others, have recently shown that sporadic basal‐like tumours share several morphological, immunophenotypic, epigenetic and genetic characteristics with tumours arising in BRCA1 mutation carriers.22,23,24,25,26 Although MYC amplification has been reported to be remarkably frequent in tumours arising in BRCA1 mutation carriers, our results suggest that this is not one of the underlying genetic events driving the biology of sporadic basal‐like carcinomas, as MYC amplification was seen in only 1 of 25 cases (4%). This finding further corroborates the results of Adler et al,37 who showed that MYC amplification is one of the drivers of the activated serum signature, but is not related to the basal‐like phenotype. In fact, the basal‐like signature does not seem to be activated by MYC.37 However, some special types of basal‐like breast carcinomas, namely medullary carcinomas12 and metaplastic breast cancers (Reis‐Filho et al, unpublished observations), seem to have frequent MYC amplifications. Further studies analysing the prevalence of MYC copy number gains in a larger cohort of basal‐like breast carcinomas, including the special types, are warranted.
In the present study, MYC amplification was an independent predictor of distant metastasis. Interestingly, the activated serum/“wound response” gene expression signature,38 which has been shown to be induced by MYC amplification,37,39 is strongly associated with increased risk of distant metastasis. Taken together, these results suggest that MYC amplification may play an important biological role in the later stages of tumour progression,13 in particular in the activation of a transcriptional programme that promotes the development of breast cancer metastasis.38,39,40
In conclusion, this study shows that although MYC amplification is heterogeneous in breast carcinomas, it can be reliably and rapidly assessed on tissue microarrays by means of CISH using a combination of a gene specific probe for MYC and a centromeric probe for chromosome 8. However, more than one core of each tumour should be included. Further studies analysing the concordance of results on MYC amplification obtained with CISH performed on whole tissue sections and tissue microarrays are warranted. MYC amplification has proven to be an independent prognostic factor for metastasis‐free and overall survival. Prospective studies assessing the additional prognostic information provided by MYC amplification analysis in addition to that offered by traditional clinicopathological and immunohistochemical parameters are required to further confirm these findings.
Take‐home messages
Chromogenic in situ hybridisation is a useful technique to determine gene amplification in high throughput tissue microarray based studies.
MYC is heterogeneously amplified in breast cancer.
MYC amplification is an independent prognostic factor for metastasis‐free and overall survival for patients treated with anthracycline‐based chemotherapy.
Abbreviations
CEP8 - chromosome 8 centromeric probe
CISH - chromogenic in situ hybridisation
Ck - cytokeratin
EGFR - epidermal growth factor receptor
ER - oestrogen receptor
FISH - fluorescent in situ hybridisation
PR - progesterone receptor
Footnotes
Funding: This study was supported by Breakthrough Breast Cancer.
Competing interests: None declared.
References
- 1.Deming S L, Nass S J, Dickson R B.et al C‐myc amplification in breast cancer: a meta‐analysis of its occurrence and prognostic relevance. Br J Cancer 2000831688–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liao D J, Dickson R B. c‐Myc in breast cancer. Endocr Relat Cancer 20007143–164. [DOI] [PubMed] [Google Scholar]
- 3.Patel J H, Loboda A P, Showe M K.et al Analysis of genomic targets reveals complex functions of MYC. Nat Rev Cancer 20044562–568. [DOI] [PubMed] [Google Scholar]
- 4.Pelengaris S, Khan M, Evan G. c‐MYC: more than just a matter of life and death. Nat Rev Cancer 20022764–776. [DOI] [PubMed] [Google Scholar]
- 5.Blancato J, Singh B, Liu A.et al Correlation of amplification and overexpression of the c‐myc oncogene in high‐grade breast cancer: FISH, in situ hybridisation and immunohistochemical analyses. Br J Cancer 2004901612–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Persons D L, Borelli K A, Hsu P H. Quantitation of HER‐2/neu and c‐myc gene amplification in breast carcinoma using fluorescence in situ hybridization. Mod Pathol 199710720–727. [PubMed] [Google Scholar]
- 7.Robanus‐Maandag E C, Bosch C A, Kristel P M.et al Association of C‐MYC amplification with progression from the in situ to the invasive stage in C‐MYC‐amplified breast carcinomas. J Pathol 200320175–82. [DOI] [PubMed] [Google Scholar]
- 8.Rummukainen J K, Salminen T, Lundin J.et al Amplification of c‐myc oncogene by chromogenic and fluorescence in situ hybridization in archival breast cancer tissue array samples. Lab Invest 2001811545–1551. [DOI] [PubMed] [Google Scholar]
- 9.Rummukainen J K, Salminen T, Lundin J.et al Amplification of c‐myc by fluorescence in situ hybridization in a population‐based breast cancer tissue array. Mod Pathol 2001141030–1035. [DOI] [PubMed] [Google Scholar]
- 10.Shimada M, Imura J, Kozaki T.et al Detection of Her2/neu, c‐MYC and ZNF217 gene amplification during breast cancer progression using fluorescence in situ hybridization. Oncol Rep 200513633–641. [PubMed] [Google Scholar]
- 11.Visscher D W, Wallis T, Awussah S.et al Evaluation of MYC and chromosome 8 copy number in breast carcinoma by interphase cytogenetics. Genes Chromosomes Cancer 1997181–7. [PubMed] [Google Scholar]
- 12.Al‐Kuraya K, Schraml P, Torhorst J.et al Prognostic relevance of gene amplifications and coamplifications in breast cancer. Cancer Res 2004648534–8540. [DOI] [PubMed] [Google Scholar]
- 13.Aulmann S, Adler N, Rom J.et al c‐myc amplifications in primary breast carcinomas and their local recurrences. J Clin Pathol 200659424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glockner S, Buurman H, Kleeberger W.et al Marked intratumoral heterogeneity of c‐myc and cyclinD1 but not of c‐erbB2 amplification in breast cancer. Lab Invest 2002821419–1426. [DOI] [PubMed] [Google Scholar]
- 15.Adem C, Soderberg C L, Hafner K.et al ERBB2, TBX2, RPS6KB1, and MYC alterations in breast tissues of BRCA1 and BRCA2 mutation carriers. Genes Chromosomes Cancer 2004411–11. [DOI] [PubMed] [Google Scholar]
- 16.Grushko T A, Dignam J J, Das S.et al MYC is amplified in BRCA1‐associated breast cancers. Clin Cancer Res 200410499–507. [DOI] [PubMed] [Google Scholar]
- 17.Rodríguez‐Pinilla S M, Rodríguez‐Gil Y, Moreno‐ Bueno G.et al Sporadic invasive breast carcinomas with medullary features display a basal‐like phenotype. An immunohistochemical and gene amplification study. Am J Surg Pathol 200731501–508. [DOI] [PubMed] [Google Scholar]
- 18.Schmitt F C, Reis‐Filho J S. c‐myc, not her‐2/neu, can predict the prognosis of breast cancer patients: how novel, how accurate, and how significant? Breast Cancer Res 20035188–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janocko L E, Brown K A, Smith C A.et al Distinctive patterns of Her‐2/neu, c‐myc, and cyclin D1 gene amplification by fluorescence in situ hybridization in primary human breast cancers. Cytometry 200146136–149. [DOI] [PubMed] [Google Scholar]
- 20.Palacios J, Honrado E, Osorio A.et al Phenotypic characterization of BRCA1 and BRCA2 tumors based in a tissue microarray study with 37 immunohistochemical markers. Breast Cancer Res Treat 2005905–14. [DOI] [PubMed] [Google Scholar]
- 21.Sorlie T, Tibshirani R, Parker J.et al Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 20031008418–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turner N, Tutt A, Ashworth A. Hallmarks of “BRCAness” in sporadic cancers. Nat Rev Cancer 20044814–819. [DOI] [PubMed] [Google Scholar]
- 23.Turner N, Reis‐Filho J S. Basal‐like breast cancer and the BRCA1 phenotype. Oncogene 2006255846–5853. [DOI] [PubMed] [Google Scholar]
- 24.Turner N, Reis‐Filho J S, Russel A M.et al BRCA1 dysfunction in sporadic basal‐like breast cancer. Oncogene 2007262126–2132. [DOI] [PubMed] [Google Scholar]
- 25.Foulkes W D, Stefansson I M, Chappuis P O.et al Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst 2003951482–1485. [DOI] [PubMed] [Google Scholar]
- 26.Lakhani S R, Reis‐Filho J S, Fulford L.et al Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin Cancer Res 2005115175–5180. [DOI] [PubMed] [Google Scholar]
- 27.Reis‐Filho J S, Savage K, Lambros M B.et al Cyclin D1 protein overexpression and CCND1 amplification in breast carcinomas: an immunohistochemical and chromogenic in situ hybridisation analysis. Mod Pathol 200619999–1009. [DOI] [PubMed] [Google Scholar]
- 28.Reis‐Filho J S, Steele D, Di Palma S.et al Distribution and significance of nerve growth factor receptor (NGFR/p75NTR) in normal, benign and malignant breast tissue. Mod Pathol 200619307–319. [DOI] [PubMed] [Google Scholar]
- 29.Elston C W, Ellis I O. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long‐term follow‐up, Histopathology 199119403–410. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen T O, Hsu F D, Jensen K.et al Immunohistochemical and clinical characterization of the basal‐like subtype of invasive breast carcinoma. Clin Cancer Res 2004105367–5374. [DOI] [PubMed] [Google Scholar]
- 31.Lambros M B, Simpson P T, Jones C.et al Unlocking pathology archives for molecular genetic studies: a reliable method to generate probes for chromogenic and fluorescent in situ hybridization. Lab Invest 200686398–408. [DOI] [PubMed] [Google Scholar]
- 32.Reis‐Filho J S, Simpson P T, Jones C.et al Pleomorphic lobular carcinoma of the breast: role of comprehensive molecular pathology in characterization of an entity. J Pathol 20052071–13. [DOI] [PubMed] [Google Scholar]
- 33.Schlotter C M, Vogt U, Bosse U.et al C‐myc, not HER‐2/neu, can predict recurrence and mortality of patients with node‐negative breast cancer. Breast Cancer Res 20035R30–R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim C, Bryant J, Horne Z.et al Trastuzumab sensitivity of breast cancer with co‐amplification of HER2 and cMYC suggests pro‐apoptotic function of dysregulated cMYC in vivo (abstract). Breast Cancer Res Treat 20059446 [Google Scholar]
- 35.Borg A, Baldetorp B, Ferno M.et al c‐myc amplification is an independent prognostic factor in postmenopausal breast cancer. Int J Cancer 199251687–691. [DOI] [PubMed] [Google Scholar]
- 36.Mertens F, Johansson B, Hoglund M.et al Chromosomal imbalance maps of malignant solid tumors: a cytogenetic survey of 3185 neoplasms. Cancer Res 1997572765–2780. [PubMed] [Google Scholar]
- 37.Adler A S, Lin M, Horlings H.et al Genetic regulators of large‐scale transcriptional signatures in cancer. Nat Genet 200638421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang H Y, Sneddon J B, Alizadeh A A.et al Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol 20042E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adler A S, Chang H Y. From description to causality: mechanisms of gene expression signatures in cancer. Cell Cycle 200651148–1151. [DOI] [PubMed] [Google Scholar]
- 40.Chang H Y, Nuyten D S, Sneddon J B.et al Robustness, scalability, and integration of a wound‐response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci USA 20051023738–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]