Abstract
Background
Intralobar nephrogenic rests (ILNRs) are precursor lesions for Wilms tumours and are associated with WT1 gene mutations. ILNR‐associated Wilms tumours have a co‐clustering of WT1 and β‐catenin (CTNNB1) mutations and unique histological features characterised by a stromal‐predominant histology.
Aim
To determine the order in which WT1 and CTNNB1 mutations occur to understand the ILNR–Wilms tumour sequence.
Methods
Of nine Wilms tumours with WT1 and CTNNB1 mutations, three ILNRs lesions in two Wilms tumours were available for analysis of WT1 and CTNNB1 mutations using microdissection. Immunohistochemistry was also performed to investigate how the mutations in β‐catenin alter the localisation in Wilms tumour development.
Results
WT1 mutations were present in the ILNRs, however CTNNB1 mutations were absent. Immunohistochemistry for WT1 confirmed inactivation of WT1 in both ILNRs and Wilms tumours. Both the ILNRs and the associated Wilms tumours had similar immunostaining patterns for β‐catenin in the blastemal and epithelial components. Although rhabdomyoblasts were not included in ILNRs, the associated Wilms tumours showed rhabdomyogenic differentiation with a positive β‐catenin nuclear staining.
Conclusions
The results suggest that CTNNB1 mutation is a later event in Wilms tumourigenesis. CTNNB1 mutations might be associated with rhabdomyogenesis.
Keywords: nephroblastoma, beta‐catenin, Wnt sinalling pathway, microdissection
Nephrogenic rests are dysplastic tissues that occur during kidney development.1 They also have potential for occurrence of Wilms tumours.1 There are two types of nephrogenic rests according to the location of the renal lobe. Intralobar nephrogenic rests (ILNRs) are situated within the renal lobe, while perilobar nephrogenic rests are located at the periphery of the renal lobe.1 ILNRs are strongly associated with deletion of chromosome 11p13 or WT1 mutation since Wilms–aniridia–genitourinary retardation syndrome and Denys–Drash syndrome frequently have ILNRs.2 ILNR‐associated Wilms tumours have unique histological and genetic features characterised by a stromal‐predominant histology with rhabdomyogenesis1,3 and co‐clustering of WT1 and β‐catenin (CTNNB1) mutations.4 Therefore, it is also important to analyse CTNNB1 mutations to understand the sequential events and the role of the Wnt signalling pathway in ILNR‐associated tumours. Recently, it has been reported that CTNNB1 mutations play a critical role in tumour formation after loss of WT1 expression, based on the results that the expression profiling of WT1‐mutant tumours shows over‐expression of β‐catenin target genes rather than WT1 target genes.5 Determining whether CTNNB1 is mutated in nephrogenic rests should provide firm evidence for the temporal sequence of WT1 and CTNNB1 mutations, however this has not yet been reported. In this study, we therefore performed sequencing analysis for WT1 and CTNNB1 in the kidneys, ILNRs and Wilms tumours.
Patients and methods
Eleven Wilms tumours with WT1 mutations were selected from a collection of tumours obtained with informed consent from the North Health Ethics Committee, Auckland, New Zealand. Eight tumours have been reported to have the common CTNNB1 mutations.6 All CTNNB1 exons were sequenced for the remaining three tumours. In one of three WT1‐mutant tumours without a CTNNB1 exon 3 mutation, we detected a mutation in CTNNB1 exon 8 using the previously reported primers.5 Of a total nine Wilms tumours with WT1 and CTNNB1 mutations, two Wilms tumours containing three ILNRs were available for microdissection analysis. Cases 1 and 2 have been previously referred to as cases 25 and 35 respectively.6 In this study, we found that case 2 had a constitutional heterozygous WT1 mutation.
Microdissection and analysis of WT1 and CTNNB1 mutations
Sections (7 μm) were prepared from paraffin embedded blocks for microdissection. Microdissection was performed using a Leica AS LMD laser capture system (Leica Microsystems, Wetzlar, Germany) according to a method on the NIH Laser Capture Microdissection website (http://dir.nihd.nid.gov/lcm/lcm.htm) with a modification, ie, the sections were stained with 0.05% toluidine blue.
Table 1 lists primers and conditions for PCR for detecting WT1 and CTNNB1 mutations, and 11p13 loss of heterozygosity (LOH) (D11S1392) in microdissected ILNRs. Sequence was done with the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) and run on an ABI 3100. To confirm the sequencing results, the samples were re‐sequenced using the Thermosequenase Cycle Sequencing Kit (USB Corporation, Cleveland, OH, USA) and reactions were analysed using a LiCor 400L DNA sequencer (LiCor Inc, Lincoln, NE, USA).
Table 1 Primers and conditions for PCR.
| Primer | AT (°C) | CT | PCR product size |
|---|---|---|---|
| WT1 exon1 | |||
| F: CTG TGC CCT GCC TGT GAG | 54 | 40 | 159 bp |
| R: GGC TCC TGT TTG ATG AAG GA | |||
| WT1 exon 9 | |||
| F: TGT CCA TTT AGG TGT GAA ACC A | |||
| R: TGA AGA AAA GTT TAC GCACTT GTT | 52 | 40 | 123 bp |
| D11S1392 | |||
| F: TTG CAT CCA TAC GGA AAG TC | 58 | 40 | 200–220 bp |
| R: ACA TCT GAG ACT TGT AGT AGA AGG | |||
| CTNNB1 exon 3 | |||
| F: CGG CTG TTA GTC ACT GG | 58 | 40 | 156 bp |
| R: AAA ATC CCT GTT CCC ACT CA | |||
| CTNNB1 exon 8 | |||
| F: GAA CTT CAC CTG ACA GAT CCA | 58 | 40 | 179 bp |
| R: CTA TTC CCA TGG CAC CAG TT |
AT, annealing temperature; CT, cycle times.
Immunohistochemistry
Immunohistochemistry for WT1 was performed to confirm loss of WT1 expression in ILNRs and Wilms tumours. The adjacent tumour‐bearing normal kidney (podocytes) was used as an internal positive control. Immunohistochemistry for β‐catenin was also carried out to investigate how localisation of this protein was altered in between ILNRs and Wilms tumours. WT1 (6F‐H2, DAKO, CA, USA) and β‐catenin (E‐5, sc‐7963, Santa Cruz Biotechnology, CA, USA) were used as primary antibodies, and the DAKO Envision horseradish peroxidase system (K4001, DAKO Cytomation, CA, USA) was used to detect the primary antibodies.
Results and discussion
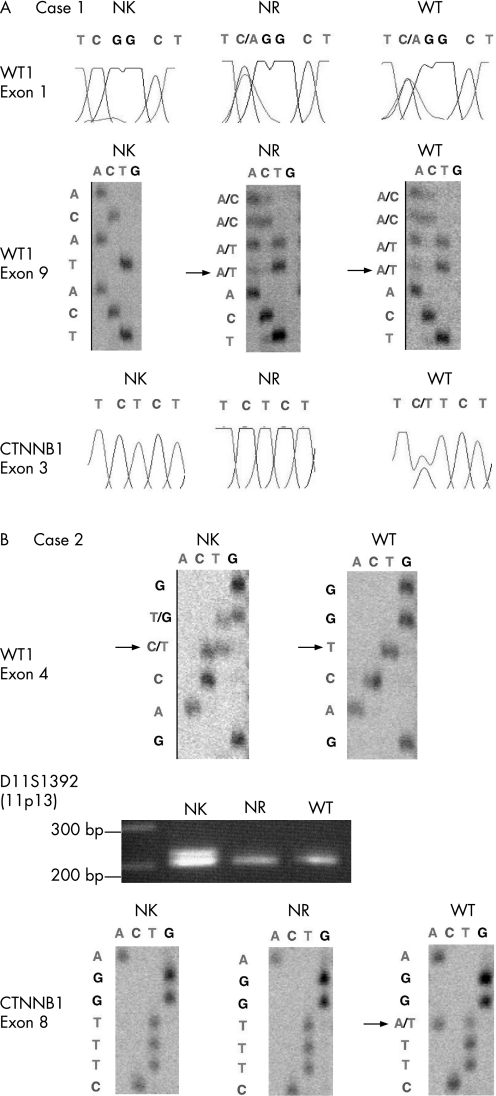
Case 1 was a sporadic Wilms tumour with WT1 mutations in exon 1 (137C→A, Ser 46→Term, heterozygous) and 9 (1215 ins A, frame shift‐stop (40+89), heterozygous), and a CTNNB1 mutation in exon 3 (348 TCT→TTT, Ser45Phe, heterozygous) (fig 1A). The appropriate polymorphisms were not available to determine whether two WT1 mutations occurred on different alleles or the same ones. We confirmed loss of expression of WT1 in the ILNRs and the associated Wilms tumour by immunohistochemistry (fig 2A,B), consistent with WT1 inactivation due to the mutation of both alleles. Neither WT1 nor CTNNB1 mutations were detected in the normal adjacent kidney (fig 1A). The tumour had a stromal‐predominant histology (fig 3D), which contained two ILNRs. The first ILNR was located between the renal calyx and the tumour and was thought to be a regressing rest (fig 3A). The second ILNR was a microscopic lesion (a dormant rest), which was isolated from the tumour tissue (fig 3B). Two ILNRs were microdissected and examined for WT1 and CTNNB1 mutations. Both ILNRs had the same heterozygous WT1 mutations in exons 1 and 9 as well as the tumour; however the CTNNB1 mutation was not detected (fig 1A).
Figure 1 (A) Sequencing results of WT1 and CTNNB1 in normal kidney (NK), intralobar nephrogenic rest (NR), and Wilms tumour (WT) in case 1. A heterozygous WT1 mutation in exon 1 (C to A transition) and in exon 9 mutation (insertion A) in NR and WT. The CTNNB1 mutation in exon 3 (C to T transition, heterozygous) is confined to WT. (B) Sequencing results of WT1 and CTNNB1, and 11p13 loss of heterozygosity (LOH) analysis in NK, intralobar NR, and WT in case 2. The mutation in WT1 is heterozygous (deletion C) in NK but is homozygous in WT. NR and WT have LOH at 11p13 (the middle). The CTNNB1 mutation (exon 8, T to A transition) is limited to WT.
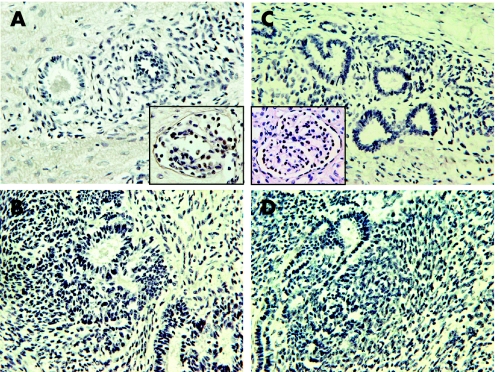
Figure 2 Immunohistochemistry for WT1 in intralobar nephrogenic rests (ILNRs) (case 1 (A), case 2 (C)), and Wilms tumours (case 1 (B), case 2 (D)). There is absence of WT1 expression in ILNRs and Wilms tumours. Insets (A) and (C) show the positive WT1 expression in podocytes in the tumour‐bearing kidney tissues as an internal control. Original magnifications, ×400.
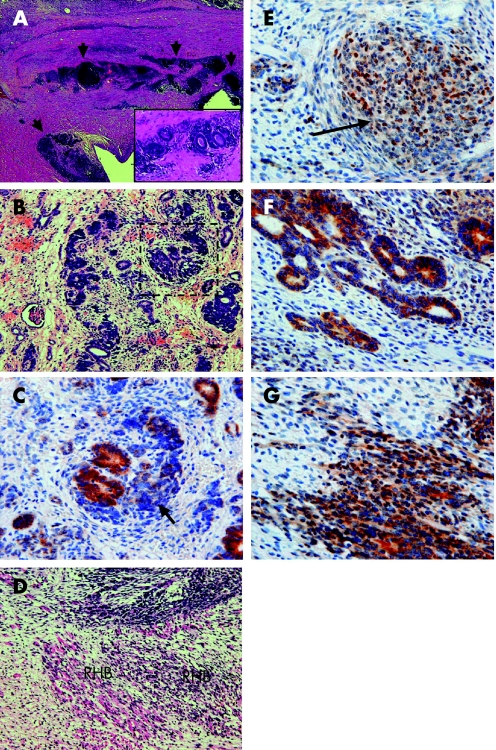
Figure 3 Histopathological and immunohistochemical features of case 1: H&E (A, inset A, B, D), β‐catenin (C, E–H). First (indicated by arrowheads) (A) and second intralobar nephrogenic rests (ILNRs) (B). Inset A shows a hyper‐power view of the blastemal cells around the epithelial structures in the sclerosing stroma. Immunohistochemistry for β‐catenin in the ILNR (a hyper‐power view of the dashed rectangle in (B)). The tumour histology shows a stromal predominant histology with rhabdomyogenesis (D). β‐Catenin is localised in the cytoplasm of the blastemal cells (indicated by arrows) and in the cytoplasm and the cell membrane of the epithelial structures in the ILNR and the associated tumour (C, F). β‐Catenin is occasionally localised in the nuclei of the blastemal cells and the rhabdomyoblasts in the tumour tissue (D, G). Original magnifications, ×40 (A); ×200 (B, inset A); ×400 (C–H).
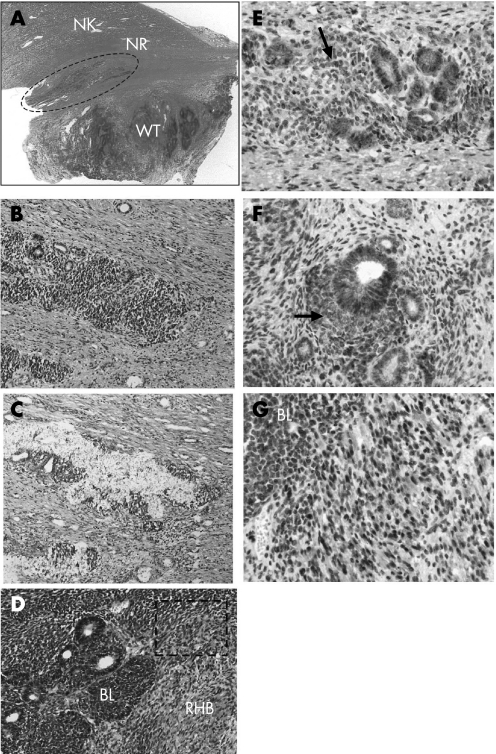
Case 2 had a constitutional heterozygous WT1 mutation in exon 4 (711 del C, frame shift‐stop (236+3)] in the tumour‐bearing kidney (fig 1B). The tumour had the homozygous WT1 mutation in exon 4 (fig 1B), and had a stromal‐predominant histology with rhabdomyogenesis (fig 4D). A macroscopically visible ILNR (a hyperplastic rest) was found between the tumour and the normal kidney (fig 4A–C). Although the CTNNB1 mutation was absent in exon 3 in the previous report,6 we detected a CTNNB1 mutation in exon 8 (1361TGG→AGG, W383R, heterozygous) in the tumour tissue (fig 1B). The mutation is not a hot spot but has recently been reported in Wilms tumour by another group.6 Microdissection was repeatedly carried out for the ILNR three times. Although the sequencing of CTNNB1 was successful, that of WT1 was not determined. Thus, instead of WT1 analysis, 11p13 LOH analysis was performed. We detected 11p13 LOH in both the ILNRs and the tumour (fig 1B). Therefore, as the normal kidney contains a heterozygous WT1 mutation, the ILNR must have the homozygous WT1 mutation as well as the tumour tissue. The homozygous WT1 mutation in the ILNRs and the Wilms tumour was confirmed by absence of WT1 expression in both tissues by immunohistochemistry (fig 2C,D). The CTNNB1 mutation was not identified in the ILNR (fig 1B). The observation that the ILNRs from both cases described here had a mutation status distinct from kidney and tumour excluded the possibility that the nephrogenic rests samples were contaminated by normal kidney or tumour tissue (table 2).
Figure 4 Histopathological and immunohistochemical features of case 2: H&E (A, B, D), toluidine blue (C), β‐catenin (E, F, G). An intralobar nephrogenic rest (ILNR) (surrounded by the dashed circle) is irregularly located between the normal kidney and the Wilms tumour (A). A hyper‐power view of the ILNR (B) and the microdissected area within the nephrogenic rest (C). The tumour histology includes an area of blastemal and epithelial components with rhabdomyogenesis (D). Expression of β‐catenin in the blastemal cells (indicated by arrows) and the epithelial component in the ILNR (E) is similar to that in the tumour (F). Nuclear staining of β‐catenin in the rhabdomyoblasts (hyper‐magnification of the dashed rectangle in (D)) (G) and weak cytoplasmic expression in the blastemal cells (G). Original magnifications, ×1 (A); ×200 (B, C, D); ×400 (E, F, G).
Table 2 Mutation status in WT1 and CTNNB1 in kidney, ILNR, and Wilms tumour.
| Kidney | ILNR | WT | |
|---|---|---|---|
| Case 1 | (2)* | ||
| WT1 | Negative | Exon 1 | Exon 1 |
| 137C→A | 137C→A | ||
| Ser 46→Term | Ser 46→Term | ||
| Heterozygous | Heterozygous | ||
| Exon 9 | Exon 9 | ||
| 1215 ins A | 1215 ins A | ||
| Frame shift‐stop (40+89) Heterozygous | Frame shift‐stop (40+89) Heterozygous | ||
| CTNNB1 | Negative | Negative | Exon 3 |
| 348 TCT→TTT | |||
| Ser45Phe | |||
| Heterozygous | |||
| Case 2 | |||
| WT1 | Exon 4 | Exon 4 | Exon 4 |
| 711 del C | 711 del C | 711 del C | |
| Frame shift‐stop (236+3) | Frame shift‐stop (236+3) | Frame shift‐stop (236+3) | |
| Heterozygous | Homozygous† | Homozygous | |
| 11p13 (D11S1392) | ROH | LOH | LOH |
| CTNNB1 | Negative | Negative | Exon 8 |
| 1361TGG→AGG | |||
| W383R | |||
| Heterozygous |
LOH, loss of heterozygosity; ROH, retention of heterozygosity.
*Two ILNR had the same results.
†The result was estimated by 11p13 LOH and WT1 mutation status in the kidney and tumour.
The localisation of β‐catenin between ILNRs and associated Wilms tumours was compared to investigate changes due to CTNNB1 mutations in Wilms tumour development. The immunohistochemical localisation of β‐catenin in the blastemal and epithelial components in the ILNRs and the associated Wilms tumours (cases 1 and 2) were similar (fig 3C,E,F and fig 4E,F): β‐catenin was localised in the cytoplasm of the blastemal cells and in the cell membrane and cytoplasm of the epithelial component in the ILNRs and the Wilms tumour, although nuclear staining of β‐catenin was focally positive in the blastemal cells in the tumours (fig 3E). The major difference was that the ILNRs did not contain rhabdomyoblasts but the associated Wilms tumours had occasional differentiation into rhabdomyoblasts with a nuclear accumulation of β‐catenin (fig 3G and fig 4G). In general, rhabdomyoblasts are rarely seen in ILNRs,1 therefore CTNNB1 mutations might be associated with rhabdomyogenesis. Another interesting result in immunohistochemistry was the mutation in exon 8 resulting in a nuclear accumulation of β‐catenin in rhabdomyoblasts in case 2. Mutations of CTNNB1 have been observed commonly in exon 3, which result in activation of the Wnt signalling pathway by stabilising β‐catenin protein, leading to its translocation to the nucleus.7 However, mutations in exon 8 might also stabilise β‐catenin protein by disrupting the APC/axin binding region of this protein.5
This study investigated the temporal sequence of WT1 and CTNNB1 mutations in the development of Wilms tumour. We have shown that mutation in the CTNNB1 occurs as a later event of Wilms tumourigenesis. It has been reported that mutation of WT1, 11p13 LOH, and 11p15 LOH are earlier events, while p53 mutation and 16q LOH are later events in Wilms tumour development.8,9,10 Our results support a sequence involving inactivation of WT1 as a very early event followed by CTNNB1 mutation and activation of the Wnt signalling pathway.5 Rhabdomyoblasts were absent in the ILNRs where WT1 was inactivated but the Wnt signalling pathway was not activated, while rhabdomyogenesis was present in the ILNR‐associated Wilms tumours where the Wnt signalling pathway is activated. Therefore, the Wnt signalling pathway might be related to myogenesis rather than loss of WT1 function.
Take‐home messages
Intralobar nephrogenic rests (ILNRs) are precursor lesions for Wilms tumours and are associated with WT1 mutations.
WT1 and β‐catenin (CTNNB1) mutations are frequently co‐clustered in Wilms tumour.
Mutations of WT1 occur as a first event in ILNRs followed secondarily by CTNNB1 mutations in Wilms tumour.
CTNNB1 mutations might be associated with rhabdomyogenesis in Wilms tumour.
Acknowledgements
We thank Dr Benjamin Tycko for the sequence information of the CTNNB1 primers.
Abbreviations
ILNR - intralobar nephrogenic rest
LOH - loss of heterozygosity
Footnotes
Funding: This work was supported by the Health Research Council of New Zealand, the Cancer Society of New Zealand, and the NZ Lottery Health Grants Board.
Competing interests: None.
Ethics approval for this study was obtained from North Health Ethics Committee, Auckland, New Zealand.
References
- 1.Beckwith J B, Kiviat N B, Bonadio J F. Nephrogenic rests, nephroblastomatosis, and the pathogenesis of Wilms tumor. Pediatr Pathol 1990101–36. [DOI] [PubMed] [Google Scholar]
- 2.Beckwith J B. Nephrogenic rests and the pathogenesis of Wilms tumor: developmental and clinical considerations. Am J Med Genet 199879268–273. [DOI] [PubMed] [Google Scholar]
- 3.Beckwith J B. Precursor lesions of Wilms tumor: clinical and biological implications. Med Pediatr Oncol 199321158–168. [DOI] [PubMed] [Google Scholar]
- 4.Maiti S, Alam R, Amos C I.et al Frequent association of beta‐catenin and WT1 mutations in Wilms tumors. Cancer Res 2000606288–6292. [PubMed] [Google Scholar]
- 5.Li C M, Kim C E, Margolin A A.et al CTNNB1 mutations and overexpression of Wnt/beta‐catenin target genes in WT1‐mutant Wilms tumors. Am J Pathol 20041651943–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukuzawa R, Heathcott R W, Sano M.et al Myogenesis in Wilms tumors is associated with mutations of the WT1 gene and activation of Bcl‐2 and the Wnt signaling pathway. Pediatr Dev Pathol 20047125–137. [DOI] [PubMed] [Google Scholar]
- 7.Eberhart C G, Argani P. Wnt signaling in human development: beta‐catenin nuclear translocation in fetal lung, kidney, placenta, capillaries, adrenal, and cartilage. Pediatr Dev Pathol 20014351–357. [DOI] [PubMed] [Google Scholar]
- 8.Park S, Bernard A, Bove K E.et al Inactivation of WT1 in nephrogenic rests, genetic precursors to Wilms tumour. Nat Genet 19935363–367. [DOI] [PubMed] [Google Scholar]
- 9.Charles A K, Brown K W, Berry P J. Microdissecting the genetic events in nephrogenic rests and Wilms tumor development. Am J Pathol 1998153991–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maitra A, Salahuddin S, Timmons C F.et al Allelotyping nephrogenic rests: putative precursor lesions of Wilms tumors. Pediatr Dev Pathol 19992488–489. [DOI] [PubMed] [Google Scholar]