Abstract
Aim
To investigate the complex interplay between human papilloma virus (HPV) infection and p53 gene alteration in 92 head and neck squamous cell carcinoma (HNSCC) and 28 leukoplakia samples from eastern India.
Methods
DNA isolated from the patient samples was subjected to HPV detection, loss of heterozygosity (LOH) analysis of the chromosome 17p region harbouring p53, genotyping at the p53 codon 72 locus and sequencing of the entire p53 gene to identify somatic mutations. Codon 72 heterozygotes carrying the p53 mutation were further cloned and resequenced to identify the allele harbouring the mutation.
Results
HPV positivity in the HNSCC samples was 69%; 21% of the HNSCC were found to harbour p53 mutations in the coding region of the gene. The absence of the p53 mutation in HPV positive tumours was statistically significant compared to the HPV negative tumours (p = 0.01), but the same did not hold true for p53 LOH (p = 1.0). Among the germline p53 codon 72 heterozygotes, the Pro allele was preferentially lost (p = 0.02) while the Arg allele was mutated in the majority of cases. The risk of HPV mediated tumourigenesis increased with the increase in number of Arg alleles at the codon 72 locus.
Conclusion
It is proposed that genetic and epigenetic alteration of p53 follow distinct pathways during the development of HNSCC from normal epithelium via dysplasia. The p53 mutation and HPV mediated p53 inactivation possibly constitute two independent pathways of tumourigenesis.
Keywords: HNSCC, HPV, p53 mutation, p53 LOH, p53 polymorphism
Head and neck squamous cell carcinoma (HNSCC) constitutes 5.5% of all malignancies and is the sixth most common cancer worldwide, with about 616 000 cases being diagnosed in 2000.1 It accounts for 30–40% of all cancer types in India.2 Several aetiological factors, particularly tobacco/alcohol consumption and human papilloma virus (HPV) infection, are shown to be linked with the disease.2 It is becoming increasingly apparent that infection with HPV is an important determinant of HNSCC development. HPV exerts its oncogenic property by inhibiting the function of the tumour suppressor proteins p53 and RB1.3 E6 protein of HPV can bind to the p53 protein and promote its degradation, thereby inhibiting p53 mediated responses.4 Another mechanism of E6 mediated inactivation of p53 function via inhibition of p300 mediated p53 acetylation has recently been established.5
In addition to HPV mediated inactivation/degradation, p53 function can also be impaired by mutations and/or loss of the p53 gene. Mutations of the p53 gene are common in HNSCC and can be detected early in the pathogenesis of these tumours.6,7 Two regions on chromosome 17, one at the short arm (17p13.1‐pter) harbouring the p53 gene and the other at the long arm (17q21‐24), have been identified as frequent targets for allelic loss.6,8 Loss of heterozygosity (LOH) at the p53 locus is common in HNSCC.9 Although there are reports which confirm that alteration of one or more putative tumour suppressor loci at 17p13.3 is associated with different cancer types other than HNSCC,10 a recent genome‐wide analysis on LOH in HNSCC confirmed the region proximal to the p53 gene (D17S1353) at 17p13 as the hotspot for allelic loss.11
Few earlier findings raised the possibility that the presence of high‐risk HPV types might obviate the requirement for p53 gene mutations, and initial studies on cervical carcinoma did indeed support an inverse correlation between the presence of HPV and p53 mutations.12 Further studies, however, have suggested that this apparently negative association may reflect an overall low prevalence of p53 mutations in tumours from this site, irrespective of HPV infection.13,14 The relationship between HPV infection and p53 gene alterations in HNSCC is also currently under investigation. Few recent reports found wild type p53 more frequently in HPV infected oral cancer specimens compared to those devoid of HPV infection.15,16 One of these reports also suggests that HNSCC specimens that showed frequent chromosomal loss were devoid of any HPV infection.15
Apart from inactivation of p53 by mutation and allelic loss, individuals carrying certain germline p53 polymorphisms were found to be more susceptible to HPV/tobacco mediated carcinogenesis.17,18,19 Although inconclusive, it has been reported that individuals with p53 codon 72 arginine homozygous genotype were at higher risk compared to proline homozygous genotype in developing cervical cancer on HPV infection.17,18 Recently, we have reported that p53 codon 72 arginine homozygous genotype confers high risk of developing oral leukoplakia in a tobacco dependent manner.19 It has also been reported that the arginine containing allele was preferentially mutated and retained in squamous cell carcinoma arising in Arg/Pro germline heterozygotes.20,21 Recently, it was shown that preferential loss of p53 codon 72 proline allele in squamous cell carcinomas of the vulva occurs in an HPV independent manner.22
Although there are a few reports on the relationship of p53 mutation and HPV infection status in HNSCC, the number of samples studied was low and p53 polymorphic status was not considered. In the present study, we have tried to decipher the complex interplay among p53 polymorphisms, mutations, LOH and HPV infection in a large number of HNSCC tumours from eastern India.
Materials and methods
Tumour samples
All tissue samples were collected from the patients admitted to the Cancer Centre and Welfare Home, Thakurpukur, Kolkata and Chittaranjan National Cancer Institute, Kolkata prior to their treatment. Samples were immediately frozen and stored at −80°C. All tumours were histopathologically diagnosed as squamous cell carcinoma, graded and staged according to the UICC TNM classification. All samples were collected from participants with informed consent, and the Institutional Review Board for ethical use of human samples approved the study.
Microdissection and DNA isolation
The normal cells present as contaminants in the primary tumour tissues were removed by a microdissection procedure.23 More than 50–60 serial tumour sections (10–20 μm) were dissected and placed on glass slides using a cryostat (model CM 1800, Leica, Heidelberg, Germany). The representative 5 μm tumour sections from different regions of the tumour were stained with H&E for diagnosis as well as for marking the tumour rich regions. The normal cells present in the tumour sections were removed by microdissection. The adjacent normal tissues of the primary tumour were similarly dissected for the presence of tumour cell infiltration. For those cases in which the “normal” tissues were contaminated with tumour cells, peripheral blood leucocytes of the corresponding patient were taken as normal. The microdissected tumour samples containing <60% tumour cells were not taken for further analysis. Genomic DNA was isolated from tissues by standard proteinase K digestion followed by phenol–chloroform extraction.24
HPV detection and typing
The presence of HPV in the HNSCC samples was detected by performing PCR using primers (MY09 and MY11) from the consensus L1 region.2 Typing of HPV‐16/18 in the L1 positive samples was done by means of PCR using specific primers from the E6 region of HPV‐16 and the E7 region of HPV‐18. The PCR products were electrophoresed in 2% agarose gel, stained with ethidium bromide, visualised under ultraviolet light, and photographed. For final confirmation of the HPV types, after gel electrophoresis the PCR products were transferred on to nylon membranes for Southern hybridisation with [32P]‐labelled HPV type specific probes.23 DNA from the SiHa (for HPV‐16) and HeLa (for HPV‐18) cell lines and the HPV type specific plasmids were used as positive controls.
LOH analysis
We have used five highly polymorphic (heterozygosity ⩾85% except D17S261 which had ∼68% heterozygosity) CA repeat markers in the present study. Two markers were from chromosome 17p12 (D17S261 and D17S1803) and three from chromosome 17p13.1 (TP53, D17S 520 and D17S 1852). These microsatellite loci were amplified by PCR in a 10 μl reaction volume containing 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 0.2 mM of each dNTP, 2 pmol of each primer, an optimal concentration of MgCl2 (1.5–2.0 mM) determined separately for each primer pair and 50–100 ng of template genomic DNA. Reaction mixtures were heated to 94°C for 3 min, cooled to 72°C to add 0.2 units Taq DNA polymerase (Life Technologies, Carlsbad, California, USA) and cycled for 30 times in a GeneAmp 9700 (Applied Biosystems, Foster City, California, USA) PCR machine. Each cycle consists of 30 s at 94°C, 30 s at appropriate annealing temperature (50–60°C) and 30 s at 72°C; final extension was carried at 72°C for 7 min. Samples without DNA for each primer pair was used as negative controls. Prior to PCR, one of the paired primers was end labelled with (γ32P) dATP (sp. act. 3000 Ci/mM, Amersham, UK) using T4 polynucleotide kinase (GIBCO BRL, Gaithersburg, Maryland, USA). Amplified DNA (1 μl) was mixed with formamide dye, denatured and then electrophoresed on a denaturing 6% polyacrylamide gel at 2300V for 3–4 hours. After electrophoresis gels were dried and exposed to x ray films (KODAK X‐Omat) for autoradiography followed by densitometric scanning (CS‐900; Simadju, Osaka, Japan) of the autoradiographs. LOH was determined by estimating the ratios of the band intensities of paired tumour and normal DNA. For informative cases, allelic loss was scored if there was complete loss of one allele or if the relative band intensity of one allele was decreased by at least 50% in the tumours, compared with the same allele in the normal control. LOH indices of >1.5 and <0.67 were considered to be loss of smaller and larger alleles, respectively.25
Screening of p53 gene mutation
Analysis of p53 exons 4–9 by direct sequencing
The genomic DNA from all the tissue samples was subjected to PCR amplification using the primers F53 (CCT GAA AAC AAC GTT CTG GTA A) and 9R53 (TAG ACT GGA AAC TTT CCA CTT G) to generate a 2.96 kb amplicon spanning exons 4–9 of the p53 gene [NCBI accession number for the reference sequence is X54156]. PCR was performed in a reaction volume of 25 μl, in 2.5 μl PCR buffer (Life Technologies) with 1.5 μM MgCl2, 10 pmol of each primer, 200 μM of each dNTP and 1.5 U platinum Taq‐DNA‐polymerase high fidelity (Invitrogen, Carlsbad, California, USA). Cycling conditions were 94°C/5 min (initial denaturation), 94°C/30 s, 52°C–58°C/30 s, 72°C/1–3 min (35 cycles) and 72°C/7 min on a GeneAmp 9700 (Applied Biosystems) thermal cycler. This amplicon was subjected to sequencing using exon specific sequencing primers.26 Sequencing was performed with a Dye Deoxy Terminator Cycle sequencing Kit (ABI‐Perkin Elmer, Weiterstadt, Germany) in an ABI‐Prism 3100 Avant Genetic Analyser (ABI‐Perkin Elmer). Exons 4–9 were completely sequenced in all cases. Mutations were confirmed by sequencing the opposite strand, and quality of the base calling was checked by phred score analysis using ABI sequence analysis software V.5.1.1.
Analysis of p53 exons 2, 3,10 and 11 by single strand conformational polymorphism followed by sequencing
PCR was performed for exons 2–3, 10 and 11 separately to determine the mutation in these exons using primers described elsewhere26 in a reaction volume of 25 μl, in 2.5 μl PCR buffer (Life Technologies) with 1.5 μM MgCl2, 10 pmol of each primer, 200 μM of each dNTP and 1.5 U Taq‐DNA‐polymerase (Life Technologies). Cycling conditions were 94°C/5 min (initial denaturation), 94°C/30 s, 52°C–62°C/30 s, 72°C/30 s (35 cycles) and 72°C/7 min on a GeneAmp 9700 (Applied Biosystems) thermal cycler. The amplified products of sizes 196–300 bp were subjected to single strand conformational polymorphism analysis as follows. Aliquots (20 μl) of the samples were mixed with equal amounts of denaturing dye and run in a 12% native polyacrylamide gel in 1× TBE (89 mm Tris Base, 89 mm borate, 2 mm EDTA) at 160 volts (constant) for 18–22 hours depending on the fragment size. The gels were then silver stained according to the manufacturer's protocol (Amersham Biosciences, Uppsala, Sweden). Any sample showing alteration in band mobility was subjected to further analysis by sequencing.
Genotyping of p53 codon 72 Arg/Pro polymorphism
p53 codon 72 Arg/Pro polymorphism in the tumour samples was analysed by PCR followed by BstUI restriction enzyme digestion. PCR was performed with primers p53+ (TCC CCC TTG CCG TCC CAA) and p53− (CGT GCA AGT CAC AGA CTT) that flanked the codon 72 Arg/Pro polymorphic site. The resulting 279 bp PCR product was subjected to restriction digestion with BstUI (New England Biolabs Inc., Beverly, Massachusetts, USA). The digest contained either 279 bp DNA fragment (in the absence of the polymorphic BstUI site) or a combination of 160 bp and 119 bp DNA fragments (in the presence of the polymorphic BstUI restriction site).
Analysis of the codon 72 status of the mutant p53 allele in Arg/Pro germline heterozygous samples
A 2.96 kb PCR fragment was amplified with primers spanning exons 4−9 (as described above, except that 1.5 U platinum Taq‐DNA‐polymerase (Invitrogen, USA) was used) from each of these codon 72 heterozygous tumour genomic DNA carrying p53 mutations. PCR fragments were gel purified using a Qiagen gel purification kit (Hilden, Germany) and subjected to TA cloning according to the manufacturer's protocol (Qiagen TA Cloning Kit). Ten recombinant white colonies from each library were then subjected to colony PCR and subsequent digestion with BstUI to select the Arg or Pro containing cloned fragments. Two such representative clones for each of the Arg and Pro variant were resequenced to determine which of these two alleles harbours the mutation.
Identifying the lost allele in the samples showing LOH
Identification of the lost allele in tumour tissues of the patients who are germline heterozygous at the codon 72 locus was done by sequencing or restriction fragment length polymorphism analysis of both the normal and tumour genomic DNAs. Absence of heterozygosity at the codon 72 locus in the tumour DNA of the paired normal heterozygous individuals clearly indicates the nature of the lost allele.
Statistical analysis
Pearson's χ2 test or Fisher's exact test (when sample number was <5 in any cell) was used for comparison of HPV+ and HPV− tumours with respect to sex, age at diagnosis, tobacco habits, site affected, tumour stage, histopathology and p53 mutation and LOH status. A trend test was conducted to determine any increase in HPV mediated tumourigenesis risk with the increase in number of risk alleles.27 The χ2 test for goodness of fit was done to compare between observed and expected values using web‐based freely available software (http://www.graphpad.com/quickcalcs/index.cfm).
Results
Description of Study Samples
A pool of 120 tissue samples, comprising 92 operated primary HNSCC tumour and 28 dysplastic leukoplakia together with their corresponding normal tissues or peripheral blood leucocytes were analysed in this study. Of the 120 patients, 101 were male and 19 were female; mean age was 46.94 (8.42) years. Tumours were divided into three categories on the basis of affected primary site: oral cavity and oropharyngeal; laryngeal; and orofacial.
Fifty‐five tumours were from the oral cavity and oropharyngeal region (23 buccal mucosa, 19 tongue, 6 cheek, 3 lip, 2 oropharynx, 1 tonsil, 1 palate), 12 were from the laryngeal region (3 neck and 9 larynx) and 20 were from the orofacial region (13 maxilla, 7 mandible); for 5 samples no site was identified. All 28 leukoplakia samples were from the buccal mucosa Histopathologically, the HNSCC tumours were classified as stage I (5 tumours), stage II (25 tumours), stage III (34 tumours) and stage IV (22 tumours). Similarly, 28 leukoplakia tissues were classified histologically into three groups: mild (n = 3), moderate (n = 12), and severe dysplasia (n = 13). Among these 120 tumours, 46 were lymph node positive and another 60 were diagnosed as free from any lymph node involvement. No information regarding lymph node involvement was available for 14 patients. Eighty‐two patients were habitual users of tobacco by different means (bidi, pan, cigarette, etc) and 22 did not have any tobacco habit. No information regarding tobacco consumption was available for 14 patients. Table 1 summarises the age, sex, tobacco habit, nodal involvement, and tumour site and stage of the study subjects.
Table 1 Study subjects.
| No (%) | |
|---|---|
| HNSCC and leukoplakia samples (n = 120) | |
| Male | 101 (84.2) |
| Female | 19 (15.8) |
| Mean age (SD) (y) | 46.94 (8.42) |
| Lymph node positive | 46 ( 38.3) |
| Lymph node negative | 60 ( 50) |
| Unknown | 14 (11.6) |
| Tobacco user | 82 ( 68.3) |
| Tobacco free | 22 ( 18.3) |
| Unknown | 16 (13.3) |
| HNSCC (n = 92) | |
| Oral cavity and oropharyngeal tumours (n = 55) | |
| Buccal mucosa | 23 (25.0) |
| Tongue | 19 (20.7) |
| Cheek | 6 (6.5) |
| Lip | 3 (3.3) |
| Oropharynx | 2 (2.2) |
| Tonsil | 1 (1.1) |
| Palate | 1 (1.1) |
| Laryngeal tumours (n = 12) | |
| Larynx | 9 (9.8) |
| Neck | 3 (3.3) |
| Orofacial tumours (n = 20) | |
| Maxilla | 13 (14.1) |
| Mandible | 7 (7.6) |
| Unknown (n = 5) | 5 (5.4) |
| Histopathology (n = 92) | |
| Stage I | 5 (5.4) |
| Stage II | 25 (27.2) |
| Stage III | 34 (37.0) |
| Stage IV | 22 (23.9) |
| Unknown | 6 (6.5) |
| Leukoplakia (n = 28) | |
| Leukoplakia site (n = 28 ) | |
| Buccal mucosa | 28 (100.0) |
| Histopathological stage (n = 28) | |
| Mild dysplasia | 3 (10.71) |
| Moderate dysplasia | 12 (42.85) |
| Severe dysplasia | 13 (46.43) |
HNSCC, head and neck squamous cell carcinoma.
HPV status of HNSCC tumours used in the study
Among the 92 HNSCC samples, HPV status was identified for 86 samples. Fifty‐nine of the 86 tumours were HPV positive (69%) and 27 were HPV negative (31%). For six samples HPV status was not determined because of the unavailability of the DNA samples. The distribution of HPV in the tumours was biased, with more tumours carrying HPV DNA than expected (p = 0.0006). HPV subtype 16 was found to be present in 56 tumours (95%), and only three tumours were infected with subtype 18 (5%) among HPV positive samples. Similar frequencies of HPV positive HNSCC tumours in the Indian population have previously been reported.28 HPV positive and negative tumours were compared with respect to various clinicopathological characteristics (table 2). The two groups of tumours did not differ significantly with regard to patient age at diagnosis (p = 0.74), tobacco use (p = 0.3), tumour site (p = 0.2), histology (p = 0.2) and stages I/II and III/IV (p = 0.6) (table 2). There was a difference between the distribution of men and women with regard to HPV+ and HPV− tumours, the male patients being significantly more HPV positive (p = 0.036, table 2). Interestingly, we also observed that about 43% (12/28) of the leukoplakia samples were HPV positive. HPV positivity was 60.7% (8/13) in the severe dysplasia and 26.7% (4/15) in the mild/moderate dysplasia samples. However, due to small sample size we did not analyse this data any further.
Table 2 Comparison of clinicopathological parameters between human papilloma virus (HPV) positive and negative head and neck squamous cell carcinoma (HNSCC) samples.
| Clinicopathological parameters | HPV+ | HPV− | p Value |
|---|---|---|---|
| Male | 52 | 19 | 0.036 |
| Female | 7 | 9 | |
| Age at diagnosis (y) | |||
| ⩽40 | 9 | 5 | 0.74 |
| 41–50 | 21 | 7 | |
| 51–60 | 20 | 8 | |
| ⩾61 | 9 | 6 | |
| Tobacco + | 35 | 22 | 0.3 |
| Tobacco − | 16 | 5 | |
| Oral cavity | 34 | 20 | 0.2 |
| Orofacial | 14 | 6 | |
| Laryngeal | 10 | 1 | |
| WDSCC | 23 | 12 | 0.2 |
| MDSCC | 21 | 5 | |
| PDSCC | 7 | 0 | |
| Stage I/II | 22 | 7 | 0.6 |
| Stage III/IV | 37 | 17 |
Only HNSCC samples are summarised. Any difference in numbers from Table 1 is due to the unavailability of HPV status data for six HNSCC samples.
MDSCC, moderately differentiated squamous cell carcinoma; PDSCC, poorly differentiated squamous cell carcinoma; WDSCC, well differentiated squamous cell carcinoma.
p53 mutation profile of HPV positive and negative HNSCC tumours
High prevalence of HPV in HNSCC samples offered an opportunity to examine whether p53 inactivation by point mutation and HPV E6 mediated inactivation occurred in a mutually exclusive manner or not in the process of tumourigenesis. Sixty‐eight HNSCC samples from the 86 tumours analysed for HPV were randomly selected and screened for mutations in the coding region of the gene. An entire coding sequence (exons 2–11) of the p53 gene was examined for these samples by single strand conformational polymorphism followed by sequencing or direct sequencing exclusively. Fourteen HNSCC tumours (21%) were found to harbour p53 mutations in the coding region of the gene (1 novel 31 bp deletion,29 1 frameshift and 12 missense mutations; table 3).
Table 3 p53 mutation profile of head and neck squamous cell carcinoma samples.
| Tumour | Stage | HPV status | Mutation | Region of protein mutated | Mutation type | R/P normal | R/P tumour |
|---|---|---|---|---|---|---|---|
| D116 | III | No | E8, nt 14456 (codon 263) (−1,FS) | Non‐conserved (before S10) | Structural | R | R |
| 222 | II | No | E5, nt 13203 (codon 175) g>a, Arg>His | Conserved (L2 Zn region) | Contact | P | P |
| 292 | III | 16 | E8 31 bp (nt 14442–14472) deletion | Conserved (S10 +splice site) | Structural | P | P |
| 308 | II | 16 | E5, nt 13203 (codon 175) g>a, Arg>His | Conserved (L2 Zn region) | Contact | R | R |
| 1332 | III | No | E4, nt 12271 (codon 116) c>g, Ser>Cys | Non‐conserved (before L1) | Contact | R | R |
| 2326 | IV | No | E6, nt 13428 (codon 223) c>t, Pro>Leu | Non‐conserved (within S7–S8) | Structural | R | R |
| 2592 | III | No | E7, nt 14074 (codon 249) g>t, Arg>Ser | Conserved (L3 DNA binding) | Contact | R/P | R |
| 4248 | III | 16 | E8, nt 14487 (codon 273) g>a, Arg>His | Conserved (S10 DNA contact) | Contact | P | P |
| 5919 | III | No | E5, nt 13107 (codon 143) t>c, Val>Ala | Non‐conserved (S3 β sandwich) | Structural | R | R |
| 5943 | II | No | E5, nt 13216 (codon 179) t>g, His>Gln | Conserved (L2) | Contact | R/P | R/P |
| 5959 | II | 16 | E8, nt 14487 (codon 273) g>a, Arg>His | Conserved (S10 DNA contact) | Contact | R/P | R/P |
| 5966 | III | No | E8, nt 14505 (codon 279) g>a, Gly>Glu | Conserved (H2) | Contact | P | P |
| 7059 | IV | No | E5, nt 13196 (codon 173) g>t, Arg>His | Conserved (L2) | Contact | R/P | R/P |
| 7999 | IV | 18 | E7, nt 14069 (codon 248) c>t, Arg>Trp | Conserved (DNA contact) | Contact | R | R |
E, exon; FS, frame shift; R, arginine allele; P, proline allele; nt, nucleotide number in the corresponding p53 sequence [NCBI accession number X54156].
All mutations were present within the hotspot region (exons 4–8). Table 3 depicts the location of the mutation (exon/intron), nucleotide number [NCBI accession number for the reference sequence is X54156], codon affected, exact base change and the resulting amino acid change in the tumour samples. p53 protein has five evolutionary conserved regions, which are essential in maintaining the structural and functional integrity of the protein. It was observed that 10/14 changes in the coding region (71%) were in the conserved residues, thereby severely impairing normal p53 function.
Any possible association between the presence of a mutation in the p53 gene and HPV status of the HNSCC tumours was examined. As shown in table 4, absence of p53 mutation in HPV positive tumours was statistically significant compared to the HPV negative tumours (p = 0.01). In other words, HPV positive tumours are more likely to contain wild type p53, whereas HPV negative tumours are likely to suffer p53 mutation.
Table 4 p53 mutation in relation to human papilloma virus (HPV) status of head and neck squamous cell carcinoma.
| Status of p53 mutation | HPV+ | HPV− | Fisher's p |
|---|---|---|---|
| p53 mutation present (14) | 5 | 9 | 0.01 |
| p53 mutation absent (54) | 40 | 14 |
The presence of the p53 mutation was also checked for any correlation with molecular and clinicopathological parameters like p53 LOH (p = 1.0), tobacco consumption (p = 0.15), lymphogenic tumour spread (p = 1.0) and tumour stage (p = 0.1), but none were found to have any relation to p53 mutation status of the tumour (data not shown). None of the leukoplakia samples showed any mutation within the hotspot region of the p53 gene.
LOH profile of HPV positive and negative HNSCC tumours
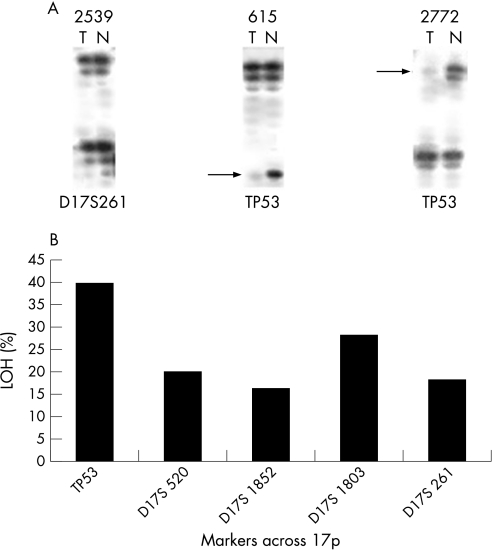
The prevalence of HPV in HNSCC samples was also correlated with the p53 inactivation by alleleic loss in order to determine any relationship between the two events in the process of tumourigenesis. Seventy‐three HNSCC samples, randomly selected from the 92 tumours, at different stages of development, were genotyped for LOH analysis at five different loci on chromosome 17 using highly polymorphic microsatellite markers; two from 17p12 (D17S261 and D17S1803) and three from the 17p13.1 region (TP53, D17S520 and D17S1852). These markers allowed us to examine the pattern of chromosomal loss around the p53 locus in the short arm of chromosome 17. The choice of markers was based on high heterozygosity index and vicinity to the p53 locus. It was of interest to examine whether there is any difference between the nature of chromosomal loss at the TP53 marker as compared to the other markers in HPV positive and negative tumours. Figure 1A shows some representative autoradiograms in a few chromosome 17p markers. Sample 2539 is an example of retention of both alleles in the tumour tissue. The other two pairs in the same panel (615 and 2772) are examples of loss of lower and upper alleles respectively in the tumour tissue. Among the markers analysed in the short arm of chromosome 17, TP53 at the 17p13.1 displayed the highest frequency of LOH (40%; fig 1B). The TP53 microsatellite marker is ∼20 kb upstream of the p53 gene. Therefore, the high rate of chromosomal alterations at the TP53 locus in these samples suggests the loss of p53 gene to be the major deciding event in HNSCC. Another marker, D17S1803 at chromosome 17p12, also exhibited a high frequency of LOH (28%), suggesting the presence of a putative tumour suppressor gene near this locus. The presence of LOH at the TP53 marker was correlated with molecular and clinicopathological parameters like p53 mutation, tobacco consumption, lymphogenic tumour spread and tumour stage (data not shown). Only tobacco consumption was found to bear a positive relation with TP53 LOH status of the tumour (p = 0.01)—that is, chances of chromosomal alterations in the TP53 marker near the p53 gene increased with tobacco intake.
Figure 1 Loss of heterozygosity (LOH) analysis of head and neck squamous cell carcinoma (HNSCC) samples with chromosome 17p markers. (A) Representative autoradiographs showing LOH at different markers on chromosome 17. The markers (shown at the bottom of each panel) were amplified from paired tumour (T) and normal (N) tissues taken from selected patients (shown at the top of each panel), analysed by denaturing 6% polyacrylamide gel electrophoresis and autoradiographed. The first pair is an example of retention of both alleles. The other two pairs are examples of loss of lower and upper alleles respectively. (B) Frequency of LOH at different markers on chromosome 17 in primary HNSCC. Names of the markers and their cytogenetic positions are shown at the bottom of each histogram.
Among the 73 tumours analysed for chromosome 17p LOH, HPV infection status was determined for 67 tumours. Table 5 shows the distribution of LOH of 549 alleles analysed in HPV positive and negative HNSCC tumours obtained from genotyping 67 tumours with five microsatellite markers from chromosome 17p. Of these 67 tumours, TP53 status was known for 52 samples. So, for the TP53 marker, LOH of 104 alleles (from 52 tumours) was distributed between HPV positive and negative HNSCC tumours. The results showed that the difference in LOH at chromosome 17p as a whole and TP53 locus in particular between HPV positive and negative tumours was not statistically significant (table 5).
Table 5 Chromosome 17p loss of heterozygosity in relation to human papilloma virus (HPV) status of head and neck squamous cell carcinoma.
| Markers | No of markers | No of tumours | HPV+ | HPV− | p Value | |
|---|---|---|---|---|---|---|
| All chromosome 17 markers | 5 | 67 | Alleles lost | 37 | 22 | 0.2 |
| Alleles retained | 352 | 138 | ||||
| TP53 marker | 1 | 52 | Alleles lost | 14 | 6 | 1.0 |
| Alleles retained | 60 | 24 |
Association between HPV status of HNSCC tumours and codon 72 polymorphism in the p53 gene
Since it is known that HPV E6 oncoprotein degrades the Arg form of p53 at a faster rate than the Pro form, comparisons were made to find any association between HPV positivity of HNSCC tumour and its codon 72 polymorphic status. The codon 72 Arg (R) or Pro (P) status was examined in 86 tumours for which HPV status was also known. A Ptrend analysis was done in order to check for any increase in risk of HPV mediated tumourigenesis with increase in Arg (R) or Pro (P) allele. Arg allele was found to be the risk allele because the risk of HPV mediated tumourigenesis increased with the increase in number of Arg alleles (Ptrend = 0.033, table 6).
Table 6 Distribution of p53 codon 72 genotypes in human papilloma virus (HPV) positive and negative head and neck squamous cell carcinoma samples.
| p53 codon 72 status | HPV+ | HPV− | OR (95% CI) |
|---|---|---|---|
| P/P tumour tissue | 6 | 9 | Referent |
| R/P tumour tissue | 23 | 8 | 4.3 (1.2 to 15.6)* |
| R/R tumour tissue | 30 | 10 | 4.5 (1.3 to 15.4)† |
P, Pro; R, Arg.
*p = 0.048, †p = 0.02, Ptrend = 0.033.
It was clearly observed that the chance of HPV mediated tumourigenesis was significantly higher in people with codon 72 Arg/Arg and Arg/Pro status in comparison to Pro/Pro homozygotes taken as referent genotype (OR = 4.5, 95% CI 1.3 to 15.4, p = 0.02; and OR = 4.3, 95% CI 1.2 to 15.6, p = 0.048).
p53 LOH and mutation in relation to R/P background
According to recent reports, some cancer types exhibit non‐random allele loss with respect to codon 72 polymorphism in exon 4 of the p53 gene.20,21 In the present study, HNSCC samples with germline codon 72 Arg/Pro heterozygous background were used to test this phenomenon. A germline heterozygous background allows determination of the differential fate of the two alleles (Arg or Pro) during a somatic event—mutation or allelic loss. For the 18 germline heterozygous samples present, the status of the two alleles in the corresponding tumour samples was determined by sequencing or restriction fragment length polymorphism analysis of this particular locus. Fourteen HNSCC samples (78%) have lost the Pro allele, while in four tumours the R allele was lost (χ2 = 5.6, p = 0.02, table 7).
Table 7 p53 loss of heterozygosity (LOH) in relation to codon 72 Arg/Pro status of head and neck squamous cell carcinoma.
| Status | Allelic changes | No | χ2 | p Value |
|---|---|---|---|---|
| p53 LOH (18) | Loss of Pro allele | 14 | 5.6 | 0.02 |
| Loss of Arg allele | 4 |
Next, any bias in the occurrence of p53 mutation in the background of Arg or Pro at the codon 72 position of the p53 gene was investigated. Among the 14 exonic mutants, four occurred in the germline heterozygous background (Arg/Pro); the remaining 10 were in either the Arg/Arg homozygous background (n = 6) or the Pro/Pro homozygous background (n = 4). For the four mutated heterozygotic HNSCC samples in this study, the mutated allele was Arg in 3 cases and Pro in 1 case. To sum up, the p53 mutations revealed that the mutant allele was Arg at codon 72 in most cases (9/14). However, this was not statistically significant (p = 0.3), probably due to the small number of samples.
Discussion
HPV is a very important determinant of HNSCC development. In contrast to the western population, the prevalence of HPV in HNSCC patients is very high in India. In this particular study, HPV positivity was detected in 69% of the HNSCC tumours, which corroborates with previous reports on Indian HNSCC patients.28 Similarly, the presence of p53 mutation was detected in ∼21% of the HNSCC analysed, which is also comparable with earlier reports on p53 point mutations in Indian HNSCC tissues which showed mutations in 17–21% of patients.30,31,32 The majority of the p53 mutations identified were found to be in exons 5 and 8 of the gene. In comparison to the low rate of p53 point mutations in India, the p53 point mutations reported in HNSCC from the USA, Europe and Japan ranged from 39% to 69%.30 Exon 7 of p53 has been reported as the most frequently mutated exon in HNSCC from the USA,33 and exons 5 and 8 as the most affected in Japanese HNSCC patients.34 Another frequent way of p53 inactivation is the loss of the chromosome 17p region in several tumours. The TP53 microsatellite marker in chromosome 17p exhibited the highest frequency of LOH (40%) among five different microsatellite markers in chromosome 17p analysed in this study. A recent genome‐wide analysis of LOH in HNSCC confirmed the region proximal to the p53 gene (D17S1353) at 17p13 as the hot spot for allelic loss.11 Incidentally, D17S1353 and TP53 microsatellite markers used in this study are both from the same chromosomal region; this further strengthens our observation.
The presence of the p53 mutation and LOH in HNSCC was subsequently correlated with other clinicopathological parameters such as HPV status, tobacco consumption, lymphogenic tumour spread and tumour stage. A positive correlation between p53 LOH and tobacco consumption was observed (p = 0.01). However, there was no significant correlation between tobacco consumption and p53 mutation. Interestingly, an inverse correlation between the presence of p53 mutation and HPV infection was observed (p = 0.01), but no such correlation was observed with p53 LOH. Thus, this observation suggests that HPV infection and p53 mutation in HNSCC tumour rarely coexist. This is possibly because they can independently lead to p53 inactivation. Two recent reports also suggested that HPV positive HNSCC tumours harbour infrequent mutation in the p53 gene.15,16
In tumours, where both copies of the p53 gene are inactivated by point mutation and allelic loss, it would be interesting to test whether there is any preference between these two events towards the specific germline alleles at the codon 72 position. Indeed, in the present study, it was observed that in germline codon 72 heterozygotes, the Proline allele of the p53 gene was preferentially lost while the Arginine allele was preferentially mutated and retained. It may be argued that the mutated Arginine allele perhaps gains some additional function, thereby providing a selective growth advantage in the process of tumour development. Evidence supporting this argument comes from the fact that the Arginine form of some mutated p53 protein binds more tightly to p73 protein compared to the Proline form of the same mutants.21 Comparable results were obtained in a few other studies with different cancer types.20,22,35,36,37,38 However, another report was contradictory to the above findings, which stated that ovarian tumours homozygous or hemizygous for the proline allele had a significantly higher frequency of p53 mutations.39 Thus, more studies on mutant p53 function under different polymorphic background are required to resolve these issues.
Take‐home messages
HPV positivity and p53 mutation were detected in 69% and 21% of HNSCC tumours respectively.
The majority of the p53 mutations identified were found to be in exons 5 and 8 of the gene.
An inverse correlation between the presence of the p53 mutation and HPV infection was observed (p = 0.01), but no such correlation was observed with p53 LOH.
In germline p53 codon 72 heterozygotes, Proline allele was preferentially lost while the Arginine allele was preferentially mutated and retained.
It is suggested that both leukoplakia and HNSCC development can take place in HPV dependent and independent pathways, and p53 mutation is a comparatively later event in the process of tumourigenesis.
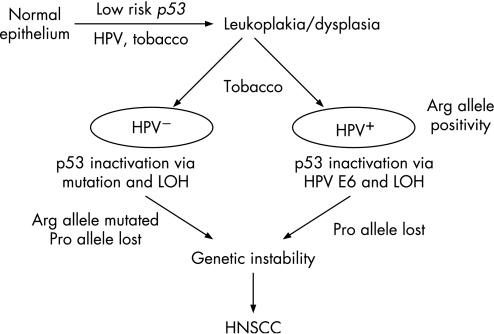
Based on the findings of this study, a hypothetical model for HNSCC predisposition mediated by genetic alterations in p53 and environmental exposures can be proposed (fig 2). From prospective studies, it was reported that the majority of HNSCC (∼85%) developed from normal squamous epithelium via dysplasia.40 Considering this three stage model of HNSCC development (from normal epithelium to dysplasia and finally to HNSCC), it may be proposed that the development of dysplasia from normal epithelium was modulated by the low risk inherited polymorphisms in the p53 gene in a tobacco dependent manner as evident from our recent study.19 Since HPV infection but not p53 mutation was observed in the leukoplakia samples in this study, it might be suggested that both leukoplakia/dysplasia and HNSCC development can take place in HPV dependent and independent pathways, and p53 mutation is a comparatively later event in the process of tumourigenesis. In the case of HNSCC, individuals with the Arg allele were at significant risk of developing HPV dependent tumours, perhaps because the Arg form of p53 is degraded at a faster rate than the Pro form,17 and is thus selected out for tumour development. The presence of high‐risk HPV types might obviate a requirement for p53 gene point mutations in the HPV positive HNSCC tumours, whereas this was frequently observed in the HPV negative tumours. p53 inactivation via allelic loss in both HPV positive and negative tumours can occur randomly. The overall effect of all these genetic insults on p53 renders it inactive, or in some cases oncogenic, giving rise to genetic instability, which eventually drives the cells to the cancer state.
Figure 2 Hypothetical pathways for head and neck squamous cell carcinoma (HNSCC) development. Genetic and epigenetic alteration of p53 follows distinct pathways during the development of HNSCC from normal epithelium via leukoplakia/dysplasia. Transition from normal to leukoplakia/dysplasia is associated with the germline p53 codon 72 polymorphisms,19 whereas p53 mutation and human papilloma virus (HPV) mediated p53 inactivation are later events (transition from leukoplakia/dysplasia to HNSCC) and possibly constitute two independent pathways of tumourigenesis.
Acknowledgements
We are grateful to the Director, Chittaranjan National Cancer Institute, Kolkata, India and Cancer Centre and Welfare Home, Thakurpukur, Kolkata, India for providing access to patients; and all members of the Human Genetics and Genomics Division of IICB for their kind cooperation and encouragement during the study. SM is grateful to the Council of Scientific and Industrial Research, New Delhi, India for predoctoral fellowships.
Abbreviations
HNSCC - head and neck squamous cell carcinoma
HPV - human papilloma virus
LOH - loss of heterozygosity
Footnotes
Funding: This work was supported by research grants from the Department of Biotechnology (grant number BT/MB/05/002/94) and the Department of Science and Technology (grant number SP/SO/D‐75/96) of the Government of India.
Competing interests: None.
References
- 1.Parkin D M, Bray F, Ferlay J.et al Estimating the world cancer burden: Globocan 2000. Int J Cancer 200194153–156. [DOI] [PubMed] [Google Scholar]
- 2.D'Costa J, Saranath D, Dedhia P.et al Detection of HPV‐16 genome in human oral cancers and potentially malignant lesions from India. Oral Oncol 199834413–420. [DOI] [PubMed] [Google Scholar]
- 3.Filippova M, Parkhurst L, Duerksen‐Hughes P J. The human papillomavirus 16 E6 protein binds to Fas‐associated death domain and protects cells from Fas‐triggered apoptosis. J Biol Chem 200427925729–25744. [DOI] [PubMed] [Google Scholar]
- 4.Kessis T D, Slebos R J, Nelson W G.et al Human papillomavirus 16 E6 expression disrupts the p53‐mediated cellular response to DNA damage. Proc Natl Acad Sci USA 1993903988–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas M C, Chiang C M. E6 oncoprotein represses p53‐dependent gene activation via inhibition of protein acetylation independently of inducing p53 degradation. Mol Cell 200517251–264. [DOI] [PubMed] [Google Scholar]
- 6.Boyle J O, Hakim J, Koch W.et al The incidence of p53 mutations increases with progression of head and neck cancer. Cancer Res 1993534477–4480. [PubMed] [Google Scholar]
- 7.Greenblatt M S, Bennett W P, Hollstein M.et al Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res 1994544855–4878. [PubMed] [Google Scholar]
- 8.Kiaris H, Spanakis N, Ergazaki M.et al Loss of heterozygosity at 9p and 17q in human laryngeal tumors. Cancer Lett 199597129–134. [DOI] [PubMed] [Google Scholar]
- 9.Adamson R, Jones A S, Field J K. Loss of heterozygosity studies on chromosome 17 in head and neck cancer using microsatellite markers. Oncogene 199492077–2082. [PubMed] [Google Scholar]
- 10.Sarkar C, Chattopadhyay P, Ralte A M.et al Loss of heterozygosity of a locus in the chromosomal region 17p13.3 is associated with increased cell proliferation in astrocytic tumors. Cancer Genet Cytogenet 2003144156–164. [DOI] [PubMed] [Google Scholar]
- 11.Beder L B, Gunduz M, Ouchida M.et al Genome‐wide analyses on loss of heterozygosity in head and neck squamous cell carcinomas. Lab Invest 20038399–105. [DOI] [PubMed] [Google Scholar]
- 12.Crook T, Wrede D, Tidy J A.et al Clonal p53 mutation in primary cervical cancer: association with human‐papillomavirus‐negative tumours. Lancet 19923391070–1073. [DOI] [PubMed] [Google Scholar]
- 13.Helland A, Holm R, Kristensen G.et al Genetic alterations of the TP53 gene, p53 protein expression and HPV infection in primary cervical carcinomas. J Pathol 1993171105–114. [DOI] [PubMed] [Google Scholar]
- 14.Fujita M, Inoue M, Tanizawa O.et al Alterations of the p53 gene in human primary cervical carcinoma with and without human papillomavirus infection. Cancer Res 1992525323–5328. [PubMed] [Google Scholar]
- 15.Braakhuis B J, Snijders P J, Keune W J.et al Genetic patterns in head and neck cancers that contain or lack transcriptionally active human papillomavirus. J Natl Cancer Inst 200496998–1006. [DOI] [PubMed] [Google Scholar]
- 16.Dai M, Clifford G M, le Calvez F.et al Human papillomavirus type 16 and TP53 mutation in oral cancer: matched analysis of the IARC multicenter study. Cancer Res 200464468–471. [DOI] [PubMed] [Google Scholar]
- 17.Storey A, Thomas M, Kalita A.et al Role of a p53 polymorphism in the development of human papillomavirus‐associated cancer. Nature 1998393229–234. [DOI] [PubMed] [Google Scholar]
- 18.Mitra S, Misra C, Singh R K.et al Association of specific genotype and haplotype of p53 gene with cervical cancer in India. J Clin Pathol 20055826–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitra S, Sikdar N, Misra C.et al Risk assessment of p53 genotypes and haplotypes in tobacco‐associated leukoplakia and oral cancer patients from eastern India. Int J Cancer 2005117786–793. [DOI] [PubMed] [Google Scholar]
- 20.Schneider‐Stock R, Boltze C, Peters B.et al Selective loss of codon 72 proline p53 and frequent mutational inactivation of the retained arginine allele in colorectal cancer. Neoplasia 20046529–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marin M C, Jost C A, Brooks L A.et al A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat Genet 20002547–54. [DOI] [PubMed] [Google Scholar]
- 22.Brooks L A, Tidy J A, Gusterson B.et al Preferential retention of codon 72 arginine p53 in squamous cell carcinomas of the vulva occurs in cancers positive and negative for human papillomavirus. Cancer Res 2000606875–6877. [PubMed] [Google Scholar]
- 23.Dasgupta S, Chakraborty S B, Roy A.et al Differential deletions of chromosome 3p are associated with the development of uterine cervical carcinoma in Indian patients. Mol Pathol 200356263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch EFManiatis T.Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory, 1989
- 25.MacGrogan D, Levy A, Bostwick D.et al Loss of chromosome arm 8p loci in prostate cancer: mapping by quantitative allelic imbalance. Genes Chromosomes Cancer 199410151–159. [DOI] [PubMed] [Google Scholar]
- 26.Varley J M, McGown G, Thorncroft M.et al Are there low‐penetrance TP53 alleles? Evidence from childhood adrenocortical tumors. Am J Hum Genet 199965995–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schoonjans F. Medcalc software, 7.4 ed. Mariakerke 1993
- 28.Balaram P, Nalinakumari K R, Abraham E.et al Human papillomaviruses in 91 oral cancers from Indian betel quid chewers—high prevalence and multiplicity of infections. Int J Cancer 199561450–454. [DOI] [PubMed] [Google Scholar]
- 29.Roychoudhury S, Mitra S. Gene symbol: TP53. Disease: squamous cell carcinoma of head and neck, Hum Genet 2005116233. [PubMed] [Google Scholar]
- 30.Saranath D, Tandle A T, Teni T R.et al p53 inactivation in chewing tobacco‐induced oral cancers and leukoplakias from India. Oral Oncol 199935242–250. [PubMed] [Google Scholar]
- 31.Kannan K, Munirajan A K, Krishnamurthy J.et al Low incidence of p53 mutations in betel quid and tobacco chewing‐associated oral squamous carcinoma from India. Int J Oncol 1999151133–1136. [DOI] [PubMed] [Google Scholar]
- 32.Heinzel P A, Balaram P, Bernard H U. Mutations and polymorphisms in the p53, p21 and p16 genes in oral carcinomas of Indian betel quid chewers. Int J Cancer 199668420–423. [DOI] [PubMed] [Google Scholar]
- 33.Somers K D, Merrick M A, Lopez M E.et al Frequent p53 mutations in head and neck cancer. Cancer Res 1992525997–6000. [PubMed] [Google Scholar]
- 34.Sakai E, Tsuchida N. Most human squamous cell carcinomas in the oral cavity contain mutated p53 tumor‐suppressor genes. Oncogene 19927927–933. [PubMed] [Google Scholar]
- 35.Schneider‐Stock R, Mawrin C, Motsch C.et al Retention of the arginine allele in codon 72 of the p53 gene correlates with poor apoptosis in head and neck cancer. Am J Pathol 20041641233–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bergamaschi D, Gasco M, Hiller L.et al p53 polymorphism influences response in cancer chemotherapy via modulation of p73‐dependent apoptosis. Cancer Cell 20033387–402. [DOI] [PubMed] [Google Scholar]
- 37.Furihata M, Takeuchi T, Matsumoto M.et al p53 mutation arising in Arg72 allele in the tumorigenesis and development of carcinoma of the urinary tract. Clin Cancer Res 200281192–1195. [PubMed] [Google Scholar]
- 38.Tada M, Furuuchi K, Kaneda M.et al Inactivate the remaining p53 allele or the alternate p73? Preferential selection of the Arg72 polymorphism in cancers with recessive p53 mutants but not transdominant mutants. Carcinogenesis 200122515–517. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Kringen P, Kristensen G B.et al Effect of the codon 72 polymorphism (c.215G>C, p.Arg72Pro) in combination with somatic sequence variants in the TP53 gene on survival in patients with advanced ovarian carcinoma. Hum Mutat 20042421–34. [DOI] [PubMed] [Google Scholar]
- 40.Shiu M N, Chen T H. Impact of betel quid, tobacco and alcohol on three‐stage disease natural history of oral leukoplakia and cancer: implication for prevention of oral cancer. Eur J Cancer Prev 20041339–45. [DOI] [PubMed] [Google Scholar]