Abstract
The normal development of lateral organs of the shoot requires the simultaneous repression of meristem-specific genes and the activation of organ-specific genes. ASYMMETRIC LEAVES2 (AS2) is required for the development of normal leaf shape and for the repression of KNOX genes in the leaf. AS2 is a member of the recently identified, plant-specific LATERAL ORGAN BOUNDARIES (LOB)–domain gene family. Expression of AS2 at high levels resulted in repression of the KNOX homeobox genes BREVIPEDICELLUS, KNAT2, and KNAT6 but not of the related SHOOT MERISTEMLESS gene. Overexpression of AS2 also led to a perturbation of normal adaxial-abaxial asymmetry in lateral organs, resulting in the replacement of abaxial cell types with adaxial cell types. These results indicate that AS2 is sufficient to induce adaxial cell fate and repress KNOX gene expression.
INTRODUCTION
The development of leaves and other lateral organs that arise from the shoot apical meristem (SAM) requires the coordinated activities of several distinct morphological processes. Cell division, expansion, and differentiation all contribute to the formation of mature leaves. Lateral organs contain two primary axes of asymmetry, a proximal-distal axis and an adaxial-abaxial axis. These asymmetries are established relatively early during leaf development and are defined relative to the SAM. Surgical experiments designed to examine the relationship between leaf development and the SAM demonstrated that communication between the SAM and leaf primordia is necessary for the establishment of the adaxial-abaxial pattern and for leaf outgrowth (Sussex, 1955; Snow and Snow, 1959). These experiments suggested the existence of a SAM-derived signal that is necessary for the establishment of adaxial-abaxial leaf polarity.
A number of mutations that cause alterations in leaf form have been described. Mutations in the ASYMMETRIC LEAVES1 (AS1) and AS2 genes result in the formation of rumpled, lobed leaves that curl downward and display vascular pattern defects (Tsukaya and Uchimiya, 1997; Serrano-Cartagena et al., 1999; Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001; Sun et al., 2002). AS1 and AS2 are required to maintain the repression of the class-1 KNOX genes BREVIPEDICELLUS (BP), KNAT2, and KNAT6 in the leaf. In wild-type plants, class-1 KNOX genes are expressed in the SAM and are downregulated at the P0 position before leaf initiation such that their repression predicts the position of the next primordium. In as1 and as2 mutants, expression of BP, KNAT2, and KNAT6 is expanded into the leaf blade (Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001). KNOX genes are downregulated appropriately in the P0 position in as1 and as2 mutants, however, indicating that AS1 and AS2 are not needed for the initial repression of KNOX genes in leaf primordia (Ori et al., 2000). Mutations in homologs of AS1 in Antirrhinum (PHANTASTICA [PHAN]) and maize (rough sheath2 [rs2]) also result in a loss of KNOX gene repression in the leaf (Schneeberger et al., 1998; Timmermans et al., 1999; Tsiantis et al., 1999). In addition, PHAN is required for the development of proper adaxial-abaxial polarity (Waites and Hudson, 1995; Waites et al., 1998). phan mutant leaves are radially symmetric and display abaxial characteristics. Interestingly, mutations in rs2 and AS1 do not result in adaxial-abaxial polarity defects (Schneeberger et al., 1998; Ori et al., 2000; Byrne et al., 2002), suggesting that PHAN may have a distinct function that is not shared by rs2 and AS1.
Genes that regulate adaxial-abaxial polarity in Arabidopsis include PHABULOSA (PHB) and PHAVOLUTA (PHV). Semidominant gain-of-function mutations in PHB and PHV cause the formation of radially symmetric, adaxialized lateral organs (McConnell and Barton, 1998; McConnell et al., 2001). PHB and PHV encode class-III homeodomain/Leu zipper transcription factors that are thought to specify adaxial cell fate by preventing the action of abaxial-promoting factors (McConnell et al., 2001; Bowman et al., 2002). Activation of the PHB and PHV proteins is postulated to occur via interaction with a SAM-derived ligand (McConnell et al., 2001). Members of the YABBY and KANADI gene families specify abaxial cell fate (reviewed by Bowman et al., 2002). Both of these gene families encode presumptive transcription factors that act redundantly, and ectopic expression of individual members of either family promotes abaxial identity (Bowman and Smyth, 1999; Eshed et al., 1999, 2001; Sawa et al., 1999; Siegfried et al., 1999; Kerstetter et al., 2001). Genetic data indicate that the KANADI genes act, at least in part, upstream of YABBY genes (Eshed et al., 2001), although the relationship between the two gene families is not completely clear.
AS2 encodes a member of the LATERAL ORGAN BOUNDARIES (LOB) family of plant-specific proteins (Iwakawa et al., 2002). LOB, the founding member of this gene family, is thought to play a role in boundary establishment or communication between the meristem and initiating lateral organs (Shuai et al., 2002). LOB is expressed at the base of lateral organs and is positively regulated by AS1 and AS2 (Byrne et al., 2002; Shuai et al., 2002). Here, we report the effects of the overexpression of AS2 in wild-type and as1 mutant backgrounds. We show that the expression of high levels of AS2 results in repression of the KNOX genes BP, KNAT2, and KNAT6 but not SHOOT MERISTEMLESS (STM). In addition, overexpression of AS2 results in adaxial-abaxial polarity defects, implicating AS2 in polarity establishment.
RESULTS
AS2 Is Expressed Broadly in Plant Development
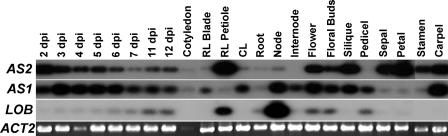
To begin to understand the role of AS2 in plant development, we first examined the expression of AS2 by reverse transcriptase–mediated (RT) PCR. AS2 transcripts were detected in most tissues tested (Figure 1). Transcripts were detected in RNA isolated from whole seedlings ranging in age from 2 to 12 days post imbibition. AS2 transcripts were detected in rosette leaves, where they were more abundant in the petiole than in the blade. RT-PCR products also were detected in cotyledons, cauline leaves, roots, inflorescence stem nodes, open flowers, young floral buds, green siliques, and all four floral organs. AS2 transcripts were not detected in internodes or pedicels. These data indicate that AS2 is expressed throughout plant development.
Figure 1.
Expression of AS2, AS1, and LOB in Wild-Type Tissues.
RT-PCR was performed on RNA isolated from seedlings at 2, 3, 4, 5, 6, 7, 11, and 12 days post imbibition (dpi), cotyledons, rosette leaf (RL) blades, rosette leaf (RL) petioles, cauline leaves (CL), roots, inflorescence stem nodes, inflorescence stem internodes, open flowers, floral buds, green siliques, flower pedicels, sepals, petals, stamens, and carpels. RT-PCR products were detected by blotting and probing with gene-specific probes after 15 cycles of amplification. ACTIN2 (ACT2) was used as a control.
as2 mutants are phenotypically similar to as1 mutants in many respects. Both mutations have similar effects on leaf morphology, cause ectopic expression of KNOX genes, and are epistatic to mutations in STM (Tsukaya and Uchimiya, 1997; Serrano-Cartagena et al., 1999; Byrne et al., 2000, 2002; Ori et al., 2000; Semiarti et al., 2001). Thus, AS1 and AS2 appear to be components of the same genetic pathway, and it has been proposed that the protein products of these genes might interact directly (Byrne et al., 2002). To compare the AS1 and AS2 expression patterns, we examined AS1 expression by RT-PCR. AS1 transcripts were detected in all tissues tested, but the overall expression profiles of AS1 and AS2 were distinct (Figure 1). For example, AS1 transcripts were detected in both petioles and blades of rosette leaves and appeared to be slightly more abundant in blade tissue, whereas AS2 transcripts were more abundant in petioles. AS1 transcripts also were detected in internode tissue, unlike AS2. Only low levels of AS1 transcripts were detected in the outer three whorls of the flower, whereas a higher level was detected in the carpel. Thus, the domains of expression of AS1 and AS2 appear to be largely overlapping but not identical.
AS1 and AS2 have been implicated in the regulation of LOB expression (Byrne et al., 2002), so we also examined the expression profile of LOB by RT-PCR (Figure 1). LOB was expressed in a more limited pattern than either AS1 or AS2. In contrast to that of AS1 and AS2, LOB transcript abundance increased concomitantly with seedling age. LOB transcripts were not detected in rosette leaf blades but were detected in petioles and were especially abundant in nodes. LOB expression also was detected in buds, open flowers, and pedicels but was absent from individual floral organs. These observations are largely in agreement with the previously reported LOB expression pattern, which was based on the lob::ET22 enhancer trap line (containing an enhancer trap insertion in the LOB gene) as well as LOB promoter:β-glucuronidase (GUS) fusions (Shuai et al., 2002).
Phenotypic Effects Resulting from the Overexpression of AS2
To investigate the role of AS2 in plant development, we generated transgenic plants that ubiquitously expressed AS2 under the control of the 35S promoter of Cauliflower mosaic virus. Of 76 35S:AS2 transgenic seedlings, all formed cotyledons that were narrow and curled upward, toward the adaxial side, in contrast to the flattened cotyledons of wild-type seedlings (Figures 2A to 2C). Among these seedlings, 34 formed a root system similar to that of wild-type seedlings (class I) and 42 produced a primary root that was shorter than that of wild-type seedlings (class II) (Figure 2A). The class-II seedlings could be categorized further into those that initiated lateral roots (class IIA; 12 of 42) and those that did not (class IIB; 30 of 42). Class-IIB seedlings arrested after making only a few leaves that usually failed to expand and occasionally were radial in appearance (Figure 2C). Class-I and class-IIA seedlings did not arrest, and the majority of these plants produced narrow leaves that were curled upward, twisted, and formed finger-like outgrowths from the abaxial leaf surface (Figures 2D and 2E). Upon flowering, 35S:AS2 plants usually produced abnormal, sterile flowers. Sepals occasionally exhibited protrusions on the abaxial surface, and petals often were curled inward, toward the adaxial side (Figure 2F). The inflorescence stems of 35S:AS2 plants typically did not elongate. Pedicels were shorter than those in the wild type and emerged from the inflorescence stems at abnormal angles (data not shown). Similar phenotypes resulting from the expression of AS2 from the 35S promoter have been reported, although they were not analyzed in detail (Iwakawa et al., 2002; Nakazawa et al., 2003).
Figure 2.
Phenotypes of Transgenic Plants That Overexpress AS2.
(A) Phenotypic variability in primary 35S:AS2 Columbia (Col-0) transgenic plants. From left to right: wild-type (wt), 35S:AS2 class-I, class-IIA, class-IIA, and class-IIB 12-day-old transgenic seedlings.
(B) Wild-type (Col-0) 8-day-old seedling.
(C) 35S:AS2 transgenic plant with radial leaves.
(D) 35S:AS2 transgenic plant. The arrowheads indicate outgrowth on the abaxial leaf surface.
(E) 35S:AS2 rosette leaf. The arrowhead shows a finger-like abaxial protrusion.
(F) Inflorescence of a 35S:AS2 plant. The arrowhead shows an abaxial protrusion on a sepal, and the arrow shows an inwardly curled petal.
(G) Wild-type (left) and two different 35S:AS2 plants (right).
(H) Wild-type (Col-0) inflorescence.
(I) 35S:AS2 inflorescence. The inset shows a higher magnification of 35S:AS2 pedicels.
Bars = 3 mm in (A), 1 mm in (B), (C), (F), and (I) inset, 5 mm in (D), and 2 mm in (H) for (H) and (I).
A small number of 35S:AS2 transgenic plants had relatively normal leaf morphologies, but upon flowering, they had abnormal inflorescence architecture (Figures 2G and 2I). In wild-type plants, pedicels pointed upward (Figures 2G and 2H), but 35S:AS2 plants developed pedicels that pointed downward (Figures 2G and 2I). In addition, internodes between flowers were shortened, and in the most extreme cases, there was no internode elongation between some flowers, so that multiple flowers appeared to be produced at the same node (Figure 2I). The morphological characteristics of the inflorescence resembled those of the bp mutant. BP encodes the class-1 KNOX homeodomain protein and was previously designated KNAT1 (Douglas et al., 2002; Venglat et al., 2002).
Expression of AS2 at High Levels Causes the Repression of KNOX Gene Expression
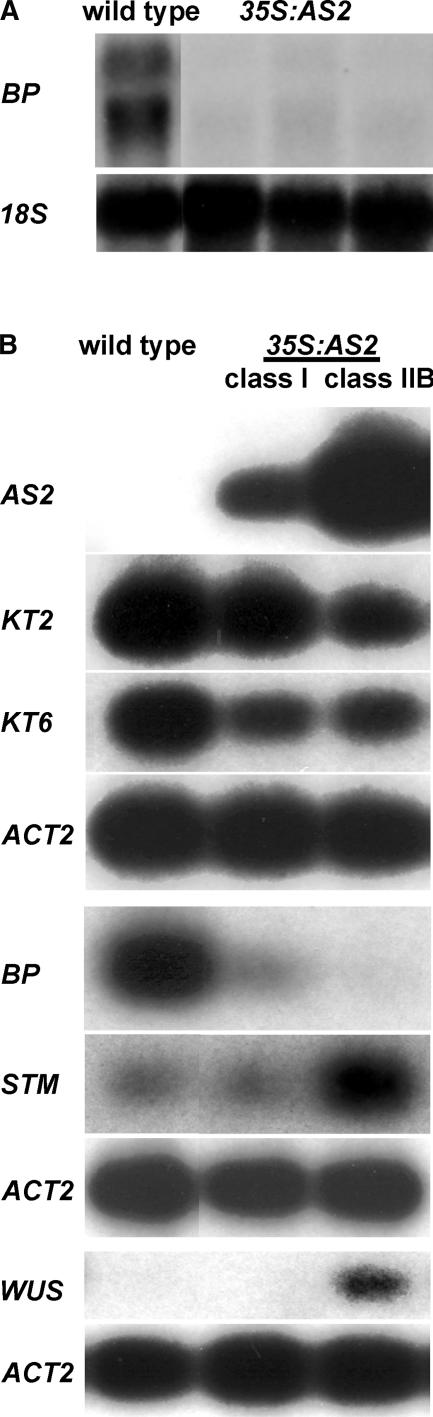
AS2, together with AS1, is required to maintain the repression of the KNOX genes BP, KNAT2, and KNAT6 in Arabidopsis leaves, because transcripts of these genes accumulate in the leaves of as1 and as2 mutants (Byrne et al., 2000, 2002; Ori et al., 2000; Semiarti et al., 2001). The observation that the inflorescences of 35S:AS2 plants resembled those of bp mutants suggested that BP repression is an outcome of AS2 overexpression. Therefore, we examined the steady state transcript levels of BP as well as the related KNOX genes STM, KNAT2, and KNAT6 in 35S:AS2 plants of varying phenotypic severity using RNA gel blot hybridization and RT-PCR. We initially examined the transcript levels of BP in 35S:AS2 plants that had relatively normal vegetative morphology, because they most closely resembled bp mutants. BP transcript levels were reduced in the inflorescences of these plants (Figure 3A).
Figure 3.
KNOX Gene Expression in 35S:AS2 Plants.
(A) RNA gel blots showing BP expression in inflorescence tissue of wild-type (left) and three different 35S:AS2 plants. Ten micrograms of total RNA was loaded in each lane. The filter was probed with BP cDNA (top gel) or 18S rDNA as a loading control (bottom gel).
(B) RT-PCR analyses of AS2, KNAT2 (KT2), KNAT6 (KT6), BP, STM, and WUS transcripts in wild-type (left), 35S:AS2 class-I (middle), and 35S:AS2 class-IIB (right) seedlings. RT-PCR products were detected by blotting and probing with gene-specific probes after 10 cycles of amplification for AS2 and 15 cycles of amplification for the other genes. ACT2 was used as a control with 10 cycles of amplification, and the appropriate control is shown below each experimental set.
We also examined KNOX gene expression in the shoots of 35S:AS2 transformants that showed vegetative abnormalities. We examined both class-I seedlings, which formed relatively normal roots and produced narrow leaves with abaxial protrusions, and class-IIB seedlings, which produced very short roots and few leaves. Class-IIB seedlings were predicted to arrest at a later stage. Examination of AS2 expression in these transgenic plants showed that AS2 transcript levels correlated with phenotypic severity. Both classes had increased AS2 expression relative to the wild type, and class-IIB seedlings had higher levels of AS2 transcripts than class-I seedlings (Figure 3B). Compared with the wild type, a reduction in KNAT2 and KNAT6 transcript levels was observed in both classes of transgenic seedlings (Figure 3B). KNAT2 transcript levels were reduced more dramatically in class-IIB seedlings, whereas the level of KNAT6 transcripts was similar in both classes. BP transcript levels were much more affected, showing a strong reduction in both classes (Figure 3B). STM transcript levels were unchanged in class-I seedlings but apparently were increased in class-IIB seedlings (Figure 3B). This apparent upregulation of STM likely was attributable to a higher ratio of meristem to leaf tissue in these seedlings. To test this possibility, transcript levels of WUSCHEL (WUS), a homeodomain gene that is expressed in the central zone of the meristem (Mayer et al., 1998), were examined. Higher levels of WUS transcripts also were detected in class-IIB seedlings (Figure 3B), consistent with the presence of more meristem tissue in these seedlings.
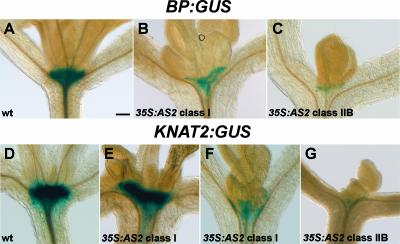
To further investigate the decrease in KNOX transcript levels in 35S:AS2 transgenic plants, the 35S:AS2 transgene was introduced into plants containing a GUS reporter gene driven by either the BP promoter (Ori et al., 2000) or the KNAT2 promoter (Dockx et al., 1995). 35S:AS2 transgenic plants were identified by selection for glufosinate ammonium resistance and stained for GUS activity. In the BP:GUS background, all 102 seedlings carrying the 35S:AS2 transgene showed a reduction in staining intensity relative to the wild type. The reduction in GUS staining correlated with phenotypic severity, being reduced most strongly in seedlings with the strongest phenotype (Figures 4A to 4C). By contrast, the majority of class-I 35S:AS2 plants retained high levels of KNAT2:GUS expression, similar to the parental line (Figures 4D and 4E). GUS staining intensity was reduced in some class-I seedlings and all class-II individuals (Figures 4F and 4G).
Figure 4.
Activity of BP and KNAT2 Promoters in 35S:AS2 Transgenic Plants.
(A) Wild-type (wt) Col-0 seedling carrying a BP:GUS construct.
(B) and (C) Class-I (B) and Class-IIB (C) 35S:AS2 transgenic plants carrying a BP:GUS construct and showing reduced GUS expression.
(D) Wild-type C-24 seedling carrying a KNAT2:GUS construct.
(E) Class-I 35S:AS2 transgenic seedling carrying a KNAT2:GUS construct and showing similar GUS activity to the wild type.
(F) Class-I 35S:AS2 transgenic seedling carrying a KNAT2:GUS construct and showing reduced GUS expression.
(G) Class-IIB 35S:AS2 transgenic seeding carrying a KNAT2:GUS construct and showing strongly reduced GUS activity.
Bar in (A) = 1 mm for (A) to (G).
35S:AS2 Plants Exhibit Adaxial-Abaxial Polarity Defects
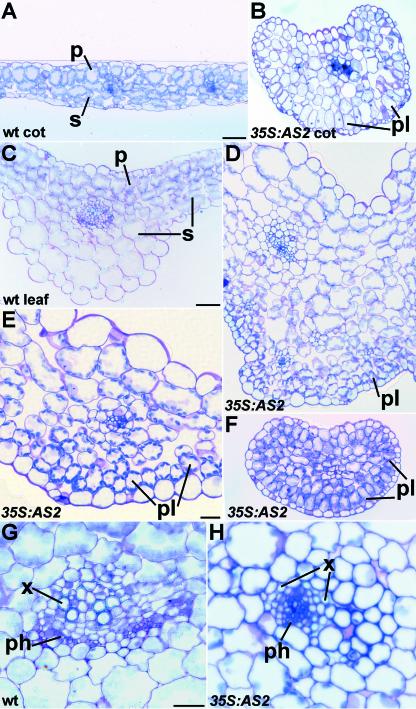
To further examine the morphological abnormalities of 35S:AS2 plants, they were embedded and sectioned. Transverse sections through cotyledons and leaves of 35S:AS2 plants revealed abnormalities in the organization of internal tissues. A distinct adaxial-abaxial polarity was visible in wild-type cotyledons and leaves. Tightly packed, elongated palisade mesophyll cells were present on the adaxial side, and loosely packed, rounded spongy mesophyll cells were present on the abaxial side (Figures 5A and 5C). This polarity was disrupted in 35S:AS2 cotyledons and leaves. Elongated, densely packed cells that resembled palisade mesophyll were present beneath the abaxial epidermis, and cells on the adaxial side of the leaf resembled spongy mesophyll (Figures 5B, 5D, and 5E). In those leaves that were radial in appearance, a subepidermal ring of palisade cells was observed around the entire leaf (Figure 5F).
Figure 5.
Overexpression of AS2 Results in Adaxial-Abaxial Polarity Defects.
Cross-section through a wild-type (wt) cotyledon (A), a 35S:AS2 cotyledon (B), a wild-type rosette leaf ([C] and [G]), and a 35S:AS2 rosette leaf ([D] to [F] and [H]). In each panel, the adaxial leaf surface is at the top of the image. cot, cotyledon; p, palisade mesophyll; ph, phloem; pl, palisade-like mesophyll; s, spongy mesophyll; x, xylem. Bar in (A) = 100 μm for (A) and (B); bar in (C) = 50 μm for (C), (D), and (F); bar in (E) = 20 μm; and bar in (G) = 20 μm for (G) and (H).
Vasculature in 35S:AS2 plants did not develop normally. In wild-type leaves, vascular tissue exhibited adaxial-abaxial polarity: xylem developed on the adaxial side of the leaf, and phloem developed on the abaxial side (Figure 5G). In 35S:AS2 leaves, veins were twisted such that this polarity was variable along the proximal-distal leaf axis. Polarity often was disrupted, resulting in vascular bundles that formed at an abnormal angle relative to the plane of the leaf surface (data not shown). In the most extreme cases, phloem was encircled by xylem (Figure 5H). Occasionally, a strand of xylem developed without any associated phloem (data not shown). The venation pattern of 35S:AS2 seedlings also was different from that of the wild type. The cotyledon midvein frequently failed to develop to the tip, and lateral veins usually did not form. Occasionally, the cotyledon midvein split to form two or more veins (data not shown).
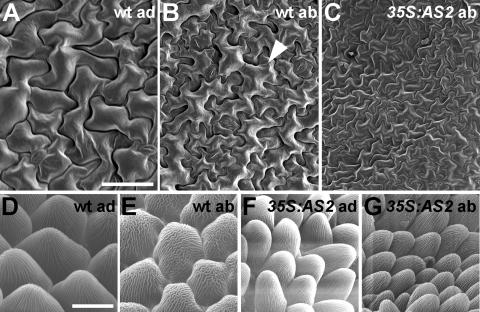
Unlike the internal tissues, polarity defects were apparent only occasionally in the epidermal cells of 35S:AS2 plants. One distinct difference between abaxial and adaxial epidermal surfaces of wild-type plants is that trichomes are present only on the adaxial leaf surface of the first four to five leaves (Telfer et al., 1997). Consequently, one obvious hallmark of leaf adaxialization is the appearance of trichomes on the abaxial side of early leaves (Kerstetter et al., 2001; McConnell et al., 2001). 35S:AS2 plants were never observed to form trichomes on the abaxial side of early leaves, so by this criterion, the epidermal surface was not adaxialized. In wild-type leaves, there also are differences in the appearance of the puzzle-shaped pavement cells on the abaxial and adaxial surfaces. Pavement cells on the adaxial blade are larger and more uniform in size than those on the abaxial surface (Figures 6A and 6B) (Bowman, 1993). We examined the shape of epidermal cells of 35S:AS2 leaves by scanning electron microscopy. The adaxial surface of 35S:AS2 leaves was not visibly different from that of wild-type leaves (data not shown). The abaxial epidermis of 35S:AS2 leaves appeared to be a mixture of adaxial and abaxial cell types. Patches of epidermis on this side of the leaf resembled the wild-type adaxial epidermis, containing cells that are uniform in size, with an absence of the characteristic large, irregular cells that typify the wild-type abaxial epidermis (Figure 6C). We also observed rare flowers that produced petals with conical cells, characteristic of the adaxial epidermis, on both epidermal surfaces (Figures 6D to 6G).
Figure 6.
Epidermal Surface of 35S:AS2 Transgenic Plants.
(A) Adaxial epidermal cells of a wild-type (wt) rosette leaf showing uniform cell size.
(B) Abaxial epidermis of a wild-type rosette leaf showing irregular cell size. The arrowhead points to a large, irregular cell.
(C) Abaxial epidermis of a 35S:AS2 rosette leaf resembling a wild-type adaxial epidermis.
(D) Adaxial epidermis of a wild-type petal.
(E) Abaxial epidermis of a wild-type petal.
(F) Adaxial epidermis of a 35S:AS2 petal.
(G) Abaxial epidermis of a 35S:AS2 petal.
Bar in (A) = 50 μm for (A) to (C); bar in (D) = 10 μm for (D) to (G).
Relationship Between AS2 and Polarity Genes
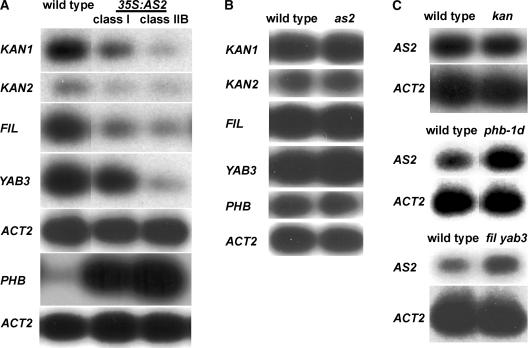
Many of the phenotypes exhibited by 35S:AS2 plants were similar to those caused by mutations in the redundant KANADI1 (KAN1) and KAN2 genes, which specify abaxial cell identity (Eshed et al., 2001; Kerstetter et al., 2001). To examine possible interactions between AS2 and the KANADI genes, we examined transcript levels of KAN1 and KAN2 in 35S:AS2 shoots. KAN1 and KAN2 transcripts were detectable in 35S:AS2 shoots, but at reduced levels compared with those in the wild type, and seemed to be reduced more dramatically in the most severe class-IIB seedlings (Figure 7A). Members of the YABBY gene family act redundantly to specify abaxial identity, so we also examined transcript levels of the YABBY genes FILAMENTOUS FLOWER (FIL) and YABBY3 (YAB3) in 35S:AS2 shoots. Transcript levels of FIL and YAB3 also were reduced in 35S:AS2 shoots compared with those in the wild type (Figure 7A). By contrast, transcript levels of PHB, a gene that regulates adaxial identity, were increased in 35S:AS2 shoots of both classes, with the more severe class-IIB seedlings showing higher levels than class-I seedlings (Figure 7A).
Figure 7.
Expression Analyses of Polarity Genes in 35S:AS2 Transgenic Plants and as2 Mutants and AS2 in Polarity Mutants.
(A) RT-PCR analyses of KAN1, KAN2, FIL, YAB3, and PHB transcript levels in 12-day-old shoots of wild-type C-24 and 35S:AS2 transgenic plants.
(B) RT-PCR analyses of KAN1, KAN2, FIL, YAB3, and PHB expression in 11-day-old shoots of wild-type Landsberg erecta and as2-1 mutant plants.
(C) RT-PCR analyses of AS2 expression in wild-type Ler and kan1 kan2 (kan), phb-1d, and fil-8 yab3-2 (fil yab3) mutant seedlings.
RT-PCR products were detected by blotting and probing with gene-specific probes after 15 cycles of amplification. Exposure times varied between experiments, and expression levels cannot be compared between experiments. ACT2 was used as a control with 10 cycles of amplification, and the appropriate control is shown below each experimental set.
To further investigate the relationship between AS2 and polarity genes, we examined the expression of KAN1, KAN2, FIL, YAB3, and PHB in as2-1 mutants. No significant differences in the transcript levels of these genes were observed in as2 mutants compared with those in the wild type (Figure 7B). We also examined transcript levels of AS2 in adaxialized mutant backgrounds. In kan1 kan2 double mutants, AS2 transcripts were detected at wild-type levels (Figure 7C). By contrast, AS2 transcript levels were increased slightly in both phb-1d mutants and fil-8 yab3-2 double mutants (Figure 7C).
AS2 Overexpression in the as1 Mutant Background
Existing genetic and molecular data indicate that AS1 and AS2 function together (Serrano-Cartagena et al., 1999; Ori et al., 2000; Semiarti et al., 2001; Byrne et al., 2002). To determine if the effects of AS2 overexpression require a functional AS1 gene, we introduced the 35S:AS2 transgene into an as1 mutant background. Of 35 as1 seedlings carrying the 35S:AS2 transgene, 7 resembled as1 single mutant plants during vegetative growth, whereas the remaining 28 showed additional phenotypes caused by the AS2 transgene. Those transgenic plants that did not show additional phenotypes were assumed to express low levels of the transgene and were not examined further. Two as1 35S:AS2 seedlings formed narrow cotyledons that curled upward, similar to the effect of 35S:AS2 in a wild-type background (data not shown). The remaining as1 35S:AS2 seedlings produced leaves that curled upward at the edges (Figure 8B), in contrast to as1 mutant leaves, which curled downward (Figure 8A). The upward leaf curling was similar to the effect of the 35S:AS2 transgene in a wild-type background (Figure 2D), although less pronounced. Abaxial protrusions, which were observed commonly in 35S:AS2 transgenic plants in a wild-type background (Figures 2D and 2E), were not formed in the as1 mutant background (Figure 8B). Upon flowering, most as1 35S:AS2 plants produced flowers that had short pedicels that emerged from the stem at abnormal angles (Figure 8C). Internodes between flowers were shorter than those in as1 single mutants, inflorescence stems were thickened, and fertility was reduced in these plants. We detected no change in BP transcript levels in these plants (data not shown).
Figure 8.
Morphology of as1 35S:AS2 Transgenic Plants.
(A) 25-day-old as1 mutant plant.
(B) 25-day-old as1 35S:AS2 plant. Arrowheads show leaves curling upward at the edges.
(C) as1 35S:AS2 plant after flowering.
Bars in (A) and (B) = 2 mm; bar in (C) = 3 mm.
DISCUSSION
AS2 Functions in Adaxial-Abaxial Polarity
Overexpression revealed a role for AS2 in the establishment of adaxial-abaxial polarity, something that was not apparent from the loss-of-function phenotype. Several aspects of the 35S:AS2 phenotype indicate that AS2 can promote adaxial identity in lateral organs. The cotyledons and leaves of 35S:AS2 plants were curled upward, toward the adaxial side of the leaf, and had limited blade expansion. Mutations that affect adaxial-abaxial polarity typically show reduced lamina expansion (reviewed by Bowman et al., 2002), consistent with the model that juxtaposition of adaxial and abaxial domains is required for leaf blade outgrowth (Waites and Hudson, 1995). Examination of the internal tissues of 35S:AS2 cotyledons and leaves revealed the presence of cells that resembled palisade mesophyll on the abaxial side. Leaves showing the most extreme phenotype were radial in appearance, and palisade cells ringed the leaf circumference. The abaxial epidermis of leaves and floral organs also was affected in 35S:AS2 plants, often appearing to be a mixture of abaxial and adaxial identities.
The organization of vascular tissues frequently was altered in 35S:AS2 leaves. The orientation of phloem and xylem poles relative to the plane of the leaf surface was abnormal, and some veins appeared to twist as they traveled through the leaf. In some cases, xylem developed on both sides of phloem or developed in the absence of identifiable phloem (data not shown). Vascular patterning also was abnormal in as1 and as2 mutant leaves, which may indicate a role for these genes in the establishment of the vascular network (Semiarti et al., 2001; Sun et al., 2002). Vascular-pattern defects also are associated with the loss of adaxial-abaxial polarity in phb-1d mutants and transgenic plants that ectopically express KAN1 (McConnell and Barton, 1998; Eshed et al., 2001; Kerstetter et al., 2001). These observations may indicate a relationship between vascular patterning and adaxial-abaxial polarity. One possibility is that the formation of a vascular network requires the presence of distinct adaxial and abaxial domains. In this case, overexpression of AS2 might affect vascular patterning indirectly.
In many respects, 35S:AS2 leaves resembled those of kan1 kan2 double mutants (Eshed et al., 2001). KANADI genes act redundantly to promote abaxial cell fate, and in plants that have lost both KAN1 and KAN2 function, adaxial cell types develop in place of abaxial cell types (Eshed et al., 2001). Both 35S:AS2 leaves and kan1 kan2 mutant leaves develop outgrowths on the abaxial leaf surface. In these leaves, there appears to be an incomplete conversion of abaxial cell types to adaxial cell types, which would result in the development of patches of cells with adaxial identity adjacent to cells with abaxial identity. The outgrowths may represent ectopic margins, which are formed when adaxial and abaxial domains are adjacent to each other (Waites and Hudson, 1995).
The phenotypes caused by overexpression implicate AS2 in the development of normal adaxial-abaxial polarity in lateral organs. However, as2 mutants do not show a conspicuous polarity defect, although it has been suggested that the downward curling of as2 mutant leaves, as well as other aspects of the phenotype, are consistent with abaxialization (Ori et al., 2000). Likewise, as1 mutants, which exhibit a phenotype nearly identical to that of as2, do not display obvious polarity defects, nor do as1 as2 double mutants (Serrano-Cartagena et al., 1999; Byrne et al., 2000; Ori et al., 2000). This is in contrast to mutations in the Antirrhinum PHAN gene, which result in the formation of radially symmetric, abaxialized leaves (Waites and Hudson, 1995; Waites et al., 1998). PHAN and AS1 are related genes, both encoding MYB-domain proteins required for KNOX gene repression in the leaf (Waites et al., 1998; Byrne et al., 2000). Therefore, the lack of functional conservation with respect to leaf polarity was initially surprising (Byrne et al., 2000). Our observations implicating AS2 in the establishment of adaxial-abaxial polarity may help to clarify this issue. AS2 appears to act redundantly with respect to polarity establishment but not with respect to other aspects of leaf morphogenesis. Functional redundancy is a common theme in genetic pathways that regulate polarity (reviewed by Bowman et al., 2002). The identities of factors that act redundantly with AS2 are not known, but the closely related LOB-DOMAIN (LBD) genes are likely candidates. LBD36, also known as ASL1 (Iwakawa et al., 2002), encodes a LOB-domain protein that is closely related to AS2, and overexpression of LBD36 results in phenotypes similar to those of 35S:AS2 plants, including the repression of BP expression (Nakazawa et al., 2003; W.-c. Lin and P.S. Springer, unpublished data).
We do not yet understand the relationship between AS2 and other polarity genes. Transcript levels of the abaxializing genes KAN1, KAN2, FIL, and YAB3 were reduced in 35S:AS2 plants, especially in those with the most extreme phenotypes, whereas transcript levels of the adaxializing gene PHB were increased in 35S:AS2 plants. The change in expression of these polarity genes might indicate that AS2 acts upstream of them to positively regulate PHB and negatively regulate KANADI and YABBY genes. However, transcript levels of PHB, KANADI, and YABBY genes were not altered in as2 mutants, indicating that AS2 is not required for the regulation of these genes. This finding suggests that the effect of AS2 overexpression on polarity-gene expression may be indirect, but it also is consistent with AS2 acting redundantly to control the expression of these genes. KANADI and YABBY genes are expressed on the abaxial side of lateral organs (Siegfried et al., 1999; Kerstetter et al., 2001), whereas PHB is expressed on the adaxial side (McConnell et al., 2001), so a change in the expression of these genes in 35S:AS2 seedlings might simply reflect the presence of adaxialized cell types on the abaxial side of leaves. In this case, AS2 might define a separate pathway for polarity determination. AS2 transcripts were upregulated in adaxialized phb-1d mutants and fil yab3 double mutants but appeared to be unaffected in kan1 kan2 double mutants. These data suggest that AS2 is positively regulated by PHB and negatively regulated by YABBY genes, but whether this regulation is direct or indirect is not clear. The relationship between the abaxializing factors KAN1, KAN2, and the YABBY genes also is not clear (reviewed by Bowman et al., 2002).
Interactions between AS2 and KNOX Genes
The as2 loss-of-function phenotype demonstrates that AS2 plays an important role, together with AS1, in the development of normal leaf morphology. One aspect of this role involves the maintenance of KNOX gene repression in the leaf. The KNOX genes BP, KNAT2, and KNAT6 are misexpressed in as1 and as2 mutant leaves (Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001). STM also appears to be misexpressed late in development in as1 leaves (Semiarti et al., 2001). However, it is not clear if all aspects of the as1 and as2 mutant phenotypes are caused by ectopic KNOX gene expression. Individual mutations in BP, KNAT2, or STM do not suppress as1 or as2 mutant phenotypes (Byrne et al., 2002), indicating that misexpression of individual KNOX genes is not required for the as1 and as2 mutant phenotypes.
Several lines of evidence indicate that AS2 overexpression results in the repression of KNOX gene expression. 35S:AS2 plants produced an inflorescence that resembled that of the bp mutant, and steady state BP transcript levels were decreased in all 35S:AS2 plants examined. Steady state KNAT2 and KNAT6 transcript levels also were reduced to a lesser extent in 35S:AS2 transgenic plants, with the largest reduction seen in plants that showed the highest levels of AS2 expression. These transgenic plants (class IIB) also had extremely short roots and arrested after producing a small number of leaf primordia. This aspect of the phenotype may be attributable to the combined reduction of BP, KNAT2, and KNAT6 transcripts.
KNOX genes are downregulated normally in the P0 of as1 and as2 mutants (Ori et al., 2000), but this repression is not maintained. Thus, the initial repression of KNOX genes must be independent of AS1 and AS2. When AS2 is overexpressed, it is able to induce the repression of the KNOX genes BP, KNAT2, and KNAT6 in the meristem. It appears that the transcriptional repression of KNAT2 and KNAT6 requires a higher level of AS2 expression than does the repression of BP. In agreement with this observation, BP is expressed at higher levels than KNAT2 and KNAT6 in as2 mutant leaves (Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001). Together, these data suggest that the mechanism by which BP expression is restricted from lateral organs may be distinct from the mechanism of KNAT2 and KNAT6 restriction.
AS2 encodes a nuclear protein (Iwakawa et al., 2002) and therefore might directly repress KNOX gene transcription. Alternatively, the repression of KNOX genes by AS2 might be indirect. In certain contexts, KNOX genes appear to be coexpressed with AS1 and/or AS2, which suggests that the mechanism of repression is indirect. In fil yab3 double mutants, KNOX genes are misexpressed in the leaf, whereas AS1 expression is unaffected (Kumaran et al., 2002) and AS2 expression is upregulated (Figure 7C). These findings indicate that AS1 and AS2 are not sufficient to repress KNOX gene expression in this mutant background. In addition, in tomato, the domain of expression of Le-PHAN, an AS1/PHAN/rs2 ortholog, overlaps with the expression domain of a class-I KNOX gene in the meristem (Koltai and Bird, 2000).
Relationship between AS2 and AS1
Genetic data indicate that AS1 and AS2 function together to regulate leaf morphogenesis (Semiarti et al., 2001; Byrne et al., 2002; Iwakawa et al., 2002). Consistent with this idea, the majority of the phenotypes associated with AS2 overexpression, including leaf polarity defects and BP repression, are suppressed in an as1 mutant background. The expression patterns of AS1 and AS2 were not identical in wild-type tissues, because AS2 was more abundant than AS1 in some tissues and vice versa. Furthermore, AS1 is expressed throughout the leaf blade (Byrne et al., 2000), whereas AS2 expression may be adaxially restricted (Iwakawa et al., 2002). In 35S:AS2 plants, AS1 and AS2 transcripts may be colocalized in cells that do not normally express both genes. The discrete expression patterns of AS1 and AS2 may indicate that, in addition to shared functions, each gene plays a distinct role in plant development. In this regard, it is interesting that transgenic plants overexpressing AS1 do not resemble 35S:AS2 plants (Theodoris et al., 2003). Overexpression of AS1 resulted in the formation of leaves that were narrower than wild-type leaves and that expressed a BP:GUS reporter at wild-type levels (Theodoris et al., 2003). Thus, AS1 appears to function somewhat differently than AS2.
AS2 transcripts were detected on the adaxial face of embryonic cotyledons (Iwakawa et al., 2002), consistent with a role in adaxial specification. AS2 transcripts were more abundant in the leaf petiole than in the blade, suggesting that AS2 might be expressed predominantly at the base of the leaf. The related LOB-domain gene LOB also is expressed at the adaxial leaf base, which suggests a similar role for LOB in leaf polarity (Shuai et al., 2002). AS2 positively regulates LOB expression (Byrne et al., 2002), which further implicates LOB in the regulation of polarity. LOB transcript abundance, however, was not affected in 35S:AS2 seedlings (data not shown), indicating that AS2 is not sufficient to induce LOB expression. Ectopic expression of LOB led to the formation of small leaves that were curled upward but showed no obvious polarity defects (Shuai et al., 2002). Therefore, if LOB plays a role in the establishment of adaxial-abaxial polarity, additional factors must be required.
AS2 encodes a LOB-domain protein of unknown function. The LOB domain contains a predicted coiled coil that may function in protein–protein interactions (Shuai et al., 2002). The presence of a coiled-coil domain suggests that AS2 might function via interactions with one or more additional proteins. One candidate interacting protein is AS1, although the nonidentical expression patterns of AS1 and AS2 suggest that their functions may overlap only partially.
AS2 and Lateral Organ Development
Overexpression has revealed a role for AS2 in the regulation of adaxial-abaxial polarity in lateral organs and has provided confirmation for the role of AS2 in KNOX gene repression. The relationship between adaxial-abaxial polarity control and KNOX gene repression is not clear, although there does not appear to be a direct connection. KNOX genes also are repressed by members of the YABBY gene family (Kumaran et al., 2002). YABBY genes are expressed early in leaf development and are important regulators of abaxial identity. In fil yab3 double mutants, which are adaxialized, KNOX genes are derepressed (Kumaran et al., 2002). Conversely, in 35S:AS2 seedlings, which also are adaxialized, KNOX genes are repressed. The opposite effects on KNOX gene expression in these different backgrounds suggest that there is no direct relationship between adaxial-abaxial polarity control and KNOX gene regulation. Thus, KNOX gene repression and adaxial promotion may be two distinct and unrelated functions of AS2. Genes that are important for polarity determination also are expressed in the early leaf, and many early leaf genes may function to repress KNOX genes. KNOX genes negatively regulate gibberellin biosynthesis (Sakamoto et al., 2001; Hay et al., 2002), so an important outcome of KNOX gene repression in the leaf is the upregulation of gibberellin signaling pathways, leading to differentiation. KNOX genes also are misexpressed in the blade-on-petiole1 (bop1) mutant (Ha et al., 2003). The molecular identity of the BOP1 gene has not been reported, but it should shed additional light on the important process of KNOX gene repression in the leaf.
Many of the factors known to play roles in determining adaxial-abaxial polarity are putative transcription factors that are unique to plants (Siegfried et al., 1999; Eshed et al., 2001; Kerstetter et al., 2001; McConnell et al., 2001). The process of polarity establishment appears to be highly redundant, because most if not all genes that function in this process act redundantly (reviewed by Bowman et al., 2002). AS2 also appears to act redundantly to regulate organ polarity, a function that was not revealed by loss-of-function mutations. This finding suggests the importance of ectopic expression and overexpression experiments in functional studies. It will be important to investigate the related LBD genes for possible overlapping functions with AS2.
METHODS
Plant Material, Growth Conditions, and Transformation
Arabidopsis thaliana plants were grown as described previously (Shuai et al., 2002). Binary T-DNA vectors were introduced into Agrobacterium tumefaciens strain GV3101 (Koncz and Schell, 1986). Arabidopsis plants were transformed by floral dip (Clough and Bent, 1998). The 35S:AS2 transgene was introduced into the wild-type Columbia ecotype or into as1-1 mutant plants in a Landsberg erecta background. The BP:GUS reporter was in a Columbia background (Ori et al., 2000), and the KNAT2:GUS reporter was in the C-24 ecotype (Dockx et al., 1995). Transformants were selected on Murashige and Skoog (1962) medium containing 50 μM glufosinate ammonium (Crescent Chemical, Islandia, NY) or on soil by spraying with Finale (1000-fold dilution; AgrEvo Environmental Health, Montvale, NJ). Spraying was initiated at 9 days after germination and was performed every 2 days for 6 days.
Constructs
For the overexpression of AS2, an EcoRI-SacI fragment containing the 35S promoter of Cauliflower mosaic virus and a XbaI-HindIII fragment containing the OCTOPINE SYNTHASE transcription terminator were excised from SLJ4D4 (Jones et al., 1992) and cloned into binary vector pCAMBIA3300 (www.cambia.org) to generate plasmid pCL0011. The AS2 coding region was amplified from EST clone VBVYB03 with primers LBD6-F (5′-ATTTCCCCTCTGAGCAACAG-3′) and LBD6-R (5′-AAGACGGATCAACAGTACGG-3′) under the following conditions: denaturation at 94°C for 3 min, followed by 24 cycles of 94°C for 45 s, 56°C for 45 s, and 72°C for 1 min using Ex-Taq Polymerase (Takara, Shiga, Japan) under conditions specified by the manufacturer. The amplified fragment was cloned into the pGEM-T Easy vector (Promega, Madison, WI), sequenced to verify its integrity, and subcloned subsequently into pCL0011 to create the 35S:AS2 construct.
Expression Analyses
Total RNA was isolated using TRIzol (Invitrogen, Carlsbad, CA). RNA gel blot hybridizations were performed as described previously (Martienssen et al., 1989). For reverse transcriptase–mediated PCR studies, cDNA was synthesized from 2 μg of total RNA using an oligo(dT) primer and Superscript Reverse Transcriptase (Invitrogen). One-twentieth volume of each cDNA was used as a template for PCR amplification. The gene-specific primers and PCR conditions were as described below. PCR conditions for BP amplification were as follows: denaturation at 94°C for 3 min, followed by 15 cycles of 94°C for 45 s, 64°C for 45 s, and 72°C for 1 min, using primers KNAT1-F (5′-GCTCCACCTGATGTGGTTGA-3′) and KNAT1-R (5′-TGTTGAGGATGTGAATGGGA-3′). PCR conditions for KNAT2, KNAT6, STM, and WUS amplification were as follows: denaturation at 94°C for 3 min, followed by 15 cycles of 94°C for 45 s, 57°C for 45 s, and 72°C for 1 min. Gene-specific primers were KNAT2-F (5′-ACC- ACCGGAGACAATCAAAG-3′) and KNAT2-R (5′-TCCGCTGCTATGTCATCATC-3′) (Byrne et al., 2000); KNAT6-F (5′-TGGCAGACTCGACACCAGTA-3′) and KNAT6-R (5′-CCGGTGAAAATCGTGTCTCT-3′); STM-F (5′-TTAGGGAGCCTCAAGCAAGA-3′) and STM-R (5′-TAC- AAACTGCATGTCCTCCG-3′); and WUS-5 (5′-GAATCAAACACACATGGAGC-3′) and WUS-3 (5′-AGAGGAAGCGTACGTCGATG-3′).
Primers and amplification conditions for LOB and ACT2 were as described previously (Shuai et al., 2002). KAN1, KAN2, FIL, YAB3, and PHB cDNAs were amplified with the gene-specific primers KANADI1-5 (5′-ACAACAACGCTTACCGATCA-3′) and KANADI1-R (5′-ATTTCTCGTGCCAATCTGGT-3′); KANADI2-5 (5′-TCATGCCAAGATTCCCAG-3′) and KANADI2-R (5′-TTAGTGAGATCGACCCAGAG-3′); FIL-3 (5′-GCTATG- TCCAATGCAACTTT-3′) and FIL-4 (5′-TTCTTGGCAGCAGCACTAAA-3′); YAB3-5 (5′-ACTTCTCATCTACGGACCAG-3′) and YAB3-R (5′-TCAGCC- ATGAGTCCAAAGTG-3′); and PHB-5 (5′-TGATGGTCCATTCGATGA- GC-3′) and PHB-3 (5′-TCTAAACTCACGAGGCCGCA-3′). PCR conditions for these genes were as follows: 94°C for 3 min, followed by 15 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 1 min. One-half of each PCR sample was blotted and hybridized as described previously (Springer et al., 1995). Exposure times varied depending on the experiment, and for this reason, expression levels cannot be compared directly between experiments.
Histology and Microscopy
Tissue samples were fixed and processed for scanning electron microscopy as described previously (Shuai et al., 2002). For cross-sections, leaves were fixed in 2.5% (v/v) glutaraldehyde (Electron Microscopy Sciences, Fort Washington, PA) in 0.1 M phosphate buffer, pH 6.8. Tissue was placed under vacuum in fixation solution until it sank and then fixed overnight at 4°C. Leaves were rinsed with phosphate buffer several times and dehydrated through an ethanol series up to 95% ethanol. The ethanol was replaced gradually by historesin (Leica Microsystems, Wetzlar, Germany) through a series of ethanol/historesin mixtures up to 100% historesin. Tissue remained in 100% historesin overnight. All steps before polymerization were performed at 4°C. Resin was polymerized at room temperature. Three-micrometer sections were cut, mounted on glass slides, and stained with 0.5% toluidine blue. 35S:AS2 BP:GUS and 35S:AS2 KNAT2:GUS plants were selected, stained for GUS activity, and imaged as described previously (Shuai et al., 2002).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact P. Springer, patricia.springer@ucr.edu.
Acknowledgments
We thank Kathleen Eckard for help with sample embedding and sectioning and Mary Byrne for sharing primer sequences and other unpublished data, as1 and as2 mutant seeds, and for helpful discussions. We thank Benjamin Aronson and Elizabeth Bell for critical reading of the manuscript. We thank Sarah Hake for providing BP:GUS seeds, Jan Traas for providing KNAT2:GUS seeds, John Bowman for providing kan1 kan2 mutant seeds, and Venkatesan Sundaresan for providing fil-8 yab3-2 double mutant seeds. phb-1d mutant seeds were provided by the ABRC (Columbus, OH). This work was supported National Science Foundation Grant IBN-9875371 to P.S.S.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.014969.
References
- Bowman, J. (1993). Arabidopsis: An Atlas of Morphology and Development. (New York: Springer).
- Bowman, J.L., Eshed, Y., and Baum, S.F. (2002). Establishment of polarity in angiosperm lateral organs. Trends Genet. 18, 134–141. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., and Smyth, D.R. (1999). CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126, 2387–2396. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Barley, R., Curtis, M., Arroyo, J.M., Dunham, M., Hudson, A., and Martienssen, R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408, 967–971. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Simorowski, J., and Martienssen, R.A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129, 1957–1965. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.J. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Dockx, J., Quaedvlieg, N., Keultjes, G., Kock, P., Weisbeek, P., and Smeekens, S. (1995). The homeobox gene ATK1 of Arabidopsis thaliana is expressed in the shoot apex of the seedling and in flowers and inflorescence stems of mature plants. Plant Mol. Biol. 28, 723–737. [DOI] [PubMed] [Google Scholar]
- Douglas, S.J., Chuck, G., Dengler, R.E., Pelecanda, L., and Riggs, C.D. (2002). KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell 14, 547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., and Bowman, J.L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99, 199–209. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., Perea, J.V., and Bowman, J.L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11, 1251–1260. [DOI] [PubMed] [Google Scholar]
- Ha, C.M., Kim, G.-T., Kim, B.C., Jun, J.H., Soh, M.S., Ueno, Y., Machida, Y., Tsukaya, H., and Nam, H.G. (2003). The BLADE-ON-PETIOLE 1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis. Development 130, 161–172. [DOI] [PubMed] [Google Scholar]
- Hay, A., Kaur, H., Phillips, A., Hedden, P., Hake, S., and Tsiantis, M. (2002). The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr. Biol. 12, 1557–1565. [DOI] [PubMed] [Google Scholar]
- Iwakawa, H., Ueno, Y., Semiarti, E., Onouchi, H., Kojima, S., Tsukaya, H., Hasebe, M., Soma, T., Ikezaki, M., Machida, C., and Machida, Y. (2002). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43, 467–478. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G., Shlumukov, L., Carland, F., English, J., Scofield, S.R., Bishop, G.J., and Harrison, K. (1992). Effective vectors for transformation, expression of heterologous genes, and assaying transposon excision in transgenic plants. Transgenic Res. 1, 285–297. [DOI] [PubMed] [Google Scholar]
- Kerstetter, R.A., Bollman, K., Taylor, R.A., Bomblies, K., and Poethig, R.S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411, 706–709. [DOI] [PubMed] [Google Scholar]
- Koltai, H., and Bird, D.M. (2000). Epistatic repression of PHANTASTICA and class 1 KNOTTED genes is uncoupled in tomato. Plant J. 22, 455–459. [DOI] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of the TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204, 383–396. [Google Scholar]
- Kumaran, M.K., Bowman, J.L., and Sundaresan, V. (2002). YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell 14, 2761–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen, R.A., Barkan, A., Freeling, M., and Taylor, W.C. (1989). Molecular cloning of a maize gene involved in photosynthetic membrane organization that is regulated by Robertson's Mutator. EMBO J. 8, 1633–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, K.F., Schoof, H., Haecker, A., Lenhard, M., Jürgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., and Barton, M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125, 2935–2942. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Nakazawa, M., Ichikawa, T., Ishikawa, A., Kobayashi, H., Tsuhara, Y., Kawashima, M., Suzuki, K., Muto, S., and Matsui, M. (2003). Activation tagging, a novel tool to dissect the functions of a gene family. Plant J. 34, 741–750. [DOI] [PubMed] [Google Scholar]
- Ori, N., Eshed, Y., Chuck, G., Bowman, J.L., and Hake, S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127, 5523–5532. [DOI] [PubMed] [Google Scholar]
- Sakamoto, T., Kamiya, N., Ueguchi-Tanaka, M., Iwahori, S., and Matsuoka, M. (2001). KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 15, 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, S., Watanabe, K., Goto, K., Kanaya, E., Morita, E.H., and Okada, K. (1999). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13, 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger, R., Tsiantis, M., Freeling, M., and Langdale, J.A. (1998). The rough sheath2 gene negatively regulates homeobox gene expression during maize leaf development. Development 125, 2857–2865. [DOI] [PubMed] [Google Scholar]
- Semiarti, E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., and Machida, Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128, 1771–1783. [DOI] [PubMed] [Google Scholar]
- Serrano-Cartagena, J., Robles, P., Ponce, M.R., and Micol, J.L. (1999). Genetic analysis of leaf form mutants from the Arabidopsis Information Service collection. Mol. Gen. Genet. 261, 725–739. [DOI] [PubMed] [Google Scholar]
- Shuai, B., Reynaga-Peña, C.G., and Springer, P.S. (2002). The LATERAL ORGAN BOUNDARIES gene defines a novel, plant-specific gene family. Plant Physiol. 129, 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried, K.R., Eshed, Y., Baum, S.F., Otsuga, D., Drews, G.N., and Bowman, J.L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126, 4117–4128. [DOI] [PubMed] [Google Scholar]
- Snow, M., and Snow, R. (1959). The dorsiventrality of leaf primordia. New Phytol. 58, 188–207. [Google Scholar]
- Springer, P.S., McCombie, W.R., Sundaresan, V., and Martienssen, R.A. (1995). Gene trap tagging of PROLIFERA, an essential MCM2-3-5-like gene in Arabidopsis. Science 268, 877–880. [DOI] [PubMed] [Google Scholar]
- Sun, Y., Zhou, Q., Zhang, W., Fu, Y., and Huang, H. (2002). ASYMMETRIC LEAVES1, an Arabidopsis gene that is involved in the control of cell differentiation in leaves. Planta 214, 694–702. [DOI] [PubMed] [Google Scholar]
- Sussex, I.M. (1955). Morphogenesis in Solanum tuberosum L.: Experimental investigation of leaf dorsiventrality and orientation in the juvenile shoot. Phytomorphology 5, 286–300. [Google Scholar]
- Telfer, A., Bollman, K.M., and Poethig, R.S. (1997). Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124, 645–654. [DOI] [PubMed] [Google Scholar]
- Theodoris, G., Inada, N., and Freeling, M. (2003). Conservation and molecular dissection of ROUGH SHEATH2 and ASYMMETRIC LEAVES1 function in leaf development. Proc. Natl. Acad. Sci. USA 100, 6837–6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans, M.C.P., Hudson, A., Becraft, P.W., and Nelson, T. (1999). ROUGH SHEATH2: A Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284, 151–153. [DOI] [PubMed] [Google Scholar]
- Tsiantis, M., Schneeberger, R., Golz, J.F., Freeling, M., and Langdale, J.A. (1999). The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science 284, 154–156. [DOI] [PubMed] [Google Scholar]
- Tsukaya, H., and Uchimiya, H. (1997). Genetic analyses of the formation of the serrated margin of leaf blades in Arabidopsis: Combination of a mutational analysis of leaf morphogenesis with the characterization of a specific marker gene expressed in hydathodes and stipules. Mol. Gen. Genet. 256, 231–238. [DOI] [PubMed] [Google Scholar]
- Venglat, S.P., Dumonceaux, T., Rozwadowski, K., Parnell, L., Babic, V., Keller, W., Martienssen, R., Selvaraj, G., and Datla, R. (2002). The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites, R., and Hudson, A. (1995). phantastica: A gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121, 2143–2154. [Google Scholar]
- Waites, R., Selvadurai, H.R.N., Oliver, I.R., and Hudson, A. (1998). The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93, 779–789. [DOI] [PubMed] [Google Scholar]