Abstract
Hydrolysis of common membrane phospholipids occurs in response to various environmental stresses, but the control and cellular function of this hydrolysis are not fully understood. Hydrogen peroxide (H2O2) is a pivotal signaling molecule involved in various stress responses. Here, we show that the plasma membrane–bound phospholipase D, PLDδ, is activated in response to H2O2 and that the resulting phosphatidic acid (PA) functions to decrease H2O2-promoted programmed cell death. The Arabidopsis genome has 12 PLD genes, and knockout of PLDδ abolishes specifically the oleate-stimulated PLD activity. H2O2 treatment of Arabidopsis cells activates PLD enzyme activity, and ablation of PLDδ abolishes that activation. PLDδ-null cells display increased sensitivity to H2O2-induced cell death. The addition of PA to PLDδ-null cells mitigates the H2O2 effect, whereas suppression of the H2O2-induced PA formation in wild-type cells increases the effect. PLDδ-ablated plants exhibit increased susceptibility to stress. These results demonstrate that activation of oleate-stimulated PLDδ constitutes an important step in the plant response to H2O2 and increasing plant stress tolerance.
INTRODUCTION
Plants generate and accumulate hydrogen peroxide (H2O2) in response to a wide variety of biotic and abiotic stresses, including UV irradiation, excess light, wounding, heat, chilling, water deficits, and pathogens (Prasad et al., 1994; Sharma et al., 1996; Bowler and Fluhr, 2000; Breusegem et al., 2001; Mittler, 2002; Neill et al., 2002a; Kwak et al., 2003; Park et al., 2003; Yoshioka et al., 2003). Increasing evidence shows that H2O2 triggers various cellular changes and that the responses are dose dependent. Low dosages of H2O2 enhance plant tolerance to multiple stresses and pathogens, whereas high dosages promote programmed cell death (Greenberg, 1996; Alvarez et al., 1998; Delledonne et al., 2001). Cell death is a basic biological process that functions in many aspects of animal and plant development and in their responses to hormones and environmental stresses (Greenberg, 1996; Neill et al., 2002a). Although the mechanisms and regulation of plant programmed cell death remain to be defined, H2O2 has emerged as an important signal in the promotion of plant cell death (Bethke and Jones, 2001; Delledonne et al., 2001; Ren et al., 2002; Dat et al., 2003). It has been suggested that H2O2 induces cell death by initiating a signal transduction pathway rather than by direct killing as a result of phytotoxicity (Levine et al., 1994; Neill et al., 2002a). However, the cellular components involved in the plant response to H2O2 are not fully understood (Bowler and Fluhr, 2000; Breusegem et al., 2001; Mittler, 2002; Neill et al., 2002b).
Hydrolysis of membrane phospholipids occurs in plants under various stress conditions, including wounding, freezing, drought, salt, and pathogen elicitation (Chapman, 1998; Laxalt and Munnik, 2002; Wang, 2002). The activation of phospholipase D (PLD) has been thought to constitute an important and early step in stress-induced phospholipid hydrolysis (Ryu and Wang, 1996; Lee et al., 1997; Wang et al., 2000; Austin-Brown and Chapman, 2002; Meijer et al., 2002). PLD, which hydrolyzes phospholipids to phosphatidic acid (PA) and a free head group, forms a predominant family of phospholipases in plants. Arabidopsis has 12 PLD genes that are grouped into five classes (Qin and Wang, 2002). The various types of PLDs are regulated differently by Ca2+, polyphosphoinositides, and free fatty acids and display distinguishable substrate selectivity and specificity, suggesting that they are activated differently and may have unique cellular functions (Wang, 2002).
PLDδ is a recently identified PLD with several unique properties (Gardiner et al., 2001; Wang and Wang, 2001). It is activated by oleic acid (Wang and Wang, 2001) and associated with the plasma membrane and the microtubule cytoskeleton (Gardiner et al., 2001; Wang and Wang, 2001). PLDδ mRNA levels are higher in senescent than in young tissues (Wang and Wang, 2001), and the expression of PLDδ increases in response to severe dehydration and high salts (Katagiri et al., 2001). Increases of free unsaturated fatty acids often occur during plant senescence and under adverse conditions. These properties of activation, localization, and expression patterns raise the possibility that PLDδ may have an important function in the cellular response to stress. Here, we show that PLDδ and its derived PA play a critical role in mediating the plant response to H2O2 and plant stress tolerance.
RESULTS
Knockout of the PLDδ Gene Causes the Loss of Oleate-Activated PLD Activity in Arabidopsis
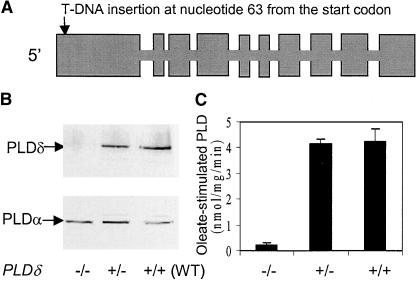
A PLDδ mutant was identified by screening T-DNA insertion lines of Arabidopsis. The T-DNA was inserted near the beginning of the 5′ coding region, 63 nucleotides downstream of the initiation codon (Figure 1A). Homozygous knockout plants had no detectable PLDδ protein (Figure 1B). As a control, the same protein extracts were blotted with a PLDα-specific antibody. Both the PLDδ mutants and the wild type showed similar levels of PLDα (Figure 1B). These data also indicate that the knockout of PLDδ did not alter the expression of PLDα.
Figure 1.
T-DNA Insertion in PLDδ Abolishes the Production of PLDδ and the Oleate-Stimulated PLD Activity in Arabidopsis.
(A) Gene structure of PLDδ and the site of the T-DNA insertion. Boxes represent exons.
(B) Immunoblot analysis of PLDδ (top gel) and PLDα (bottom gel) proteins in PLDδ mutant and wild-type (WT) plants. The symbols −/− and +/− represent plants that are homozygous and heterozygous, respectively, in the knockout locus.
(C) Oleate-stimulated PLDδ activity in PLDδ mutant and wild-type plants.
Both the activity and immunoblot assays used the microsomal fraction of leaf extracts.
Disruption of PLDδ gene function was confirmed further by assaying the oleate-activated PLD activity. Using plant PLDs expressed in Escherichia coli, it was shown previously that PLDδ, but not PLDα1, PLDβ1, or PLDγ1, was activated by oleate (Wang and Wang, 2001). The homozygous PLDδ-knockout plants exhibited no oleate-stimulated PLD activity (Figure 1C), demonstrating that only PLDδ possesses the oleate-activated PLD activity in Arabidopsis. The PLDδ mutant allele cosegregated with kanamycin resistance and susceptibility in a 3:1 ratio, suggesting that the PLDδ mutant contains a single T-DNA insertion in the genome. Introduction of a wild-type PLDδ gene to the knockout mutant plants restored the expression of PLDδ (Figure 2C).
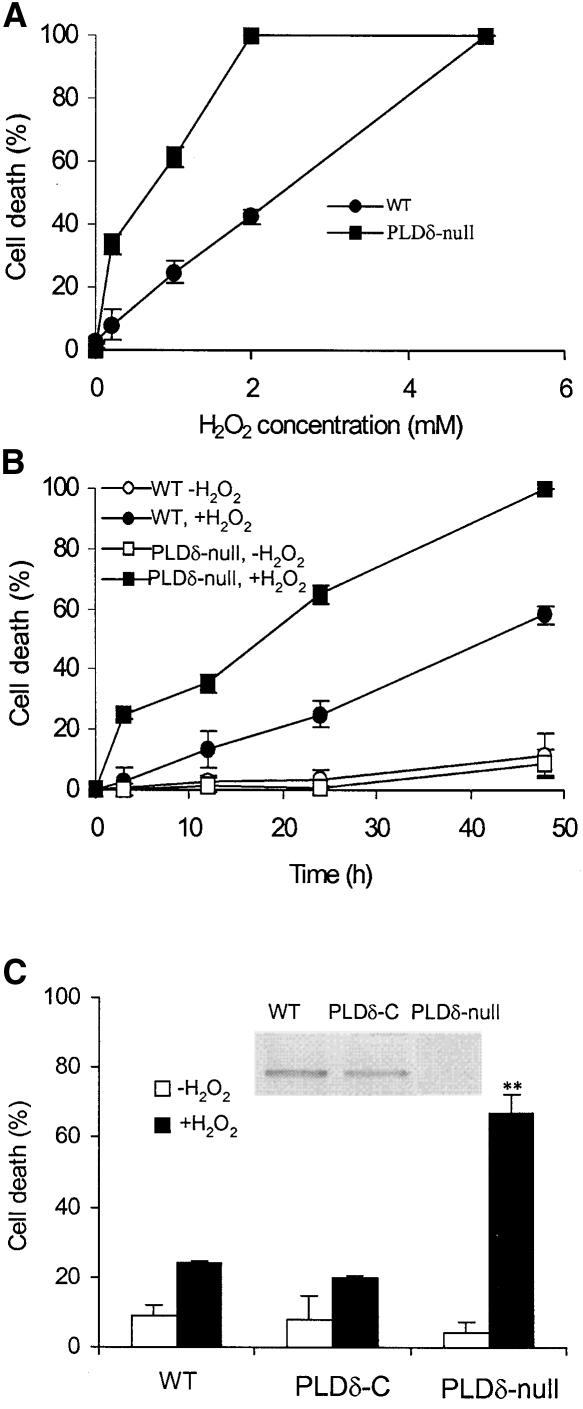
Figure 2.
Increased H2O2-Induced Cell Death in PLDδ-Null Arabidopsis Protoplasts.
(A) Protoplast death as a function of H2O2 concentration. WT, wild type.
(B) H2O2 (1 mM)-induced cell death as a function of incubation time.
(C) Complementation of the PLDδ-null mutant with the wild-type PLDδ gene and restoration of the mutant response to H2O2 to wild type. The inset shows an immunoblot analysis of proteins extracted from wild-type, PLDδ-complemented (PLDδ-C), and PLDδ-null plants with PLDδ-specific antibody. The double asterisks indicate that the mean value is significantly different from that of wild-type protoplasts (P < 0.01).
Dead cells were counted 24 h after H2O2 treatments and are expressed as a percentage of total cells.
Ablation of PLDδ Increases the Cell's Sensitivity to H2O2-Induced Cell Death
Under normal growth conditions, PLDδ-knockout plants are indistinguishable from wild-type plants in terms of plant height, number of leaves, days required for flowering and seed maturation, and seed yield. In studying the involvement of PLDδ in cellular processes, we observed that ablation of PLDδ rendered Arabidopsis cells more sensitive to H2O2-induced cell death (Figure 2). When leaf protoplasts were exposed to 0.2 mM H2O2 for 24 h, 35% of PLDδ-null cells died, whereas the death rate was <5% for wild-type cells (Figure 2A). At 2 mM H2O2 for 24 h, all PLDδ-null cells were dead, whereas 60% of wild-type cells were still alive. The apparent H2O2 concentrations for 50% cell death (DC50) for PLDδ-null and wild-type cells were 0.45 and 2.5 mM, respectively (Figure 2A). PLDδ-null cells died much faster than wild-type cells when the time course was determined using 1 mM H2O2. The apparent half-life for PLDδ-null and wild-type cells were 18 and 45 h, respectively (Figure 2B). In the absence of added H2O2, PLDδ-null and wild-type cells displayed no difference in the rate of cell death.
When the PLDδ-knockout mutant was complemented with the PLDδ gene with its own promoter, cells from the complemented plants showed reduced sensitivity to H2O2 and were indistinguishable from wild-type cells (Figure 2C). This result demonstrates that knockout of the PLDδ gene is responsible for the increased H2O2 sensitivity. The H2O2-induced cell death was programmed cell death, as indicated by the positive TUNEL (terminal deoxynucleotidyl transferase–mediated dUTP nick end labeling) staining. After treatment with 1 mM H2O2 for 3 h, ∼85% of the nuclei of PLDδ-null protoplasts were labeled positive with TUNEL that labels free 3′-OH termini of broken DNA strands (Figure 3A). By contrast, under the same conditions, no clear labeling was found in wild-type cells.
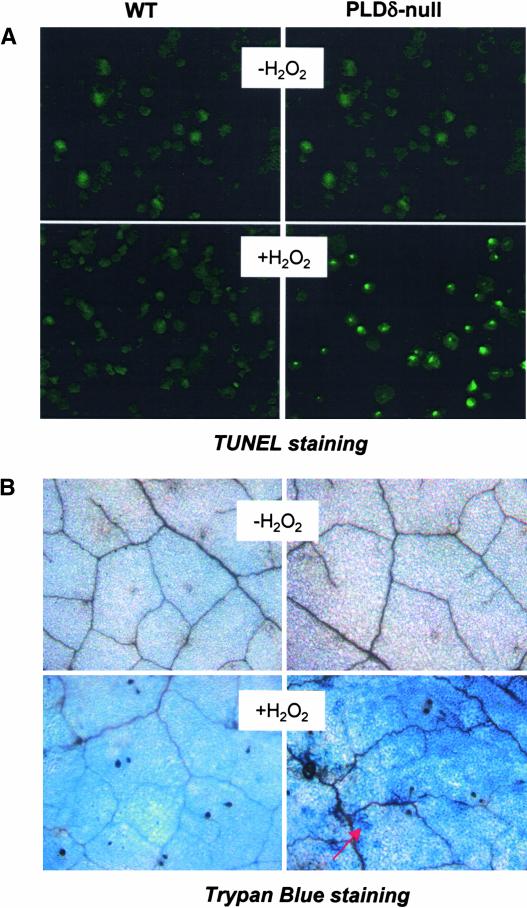
Figure 3.
H2O2-Induced Programmed Cell Death and Increased Cell Death in PLDδ-Null Leaves.
(A) TUNEL staining of H2O2-induced programmed cell death in wild-type (WT) and PLDδ-null protoplasts. 3′-OH groups of DNA were visualized as bright green spots within the nucleus. Protoplasts were incubated with 1 mM H2O2 for 3 h before DNA was labeled using the TUNEL method.
(B) Trypan blue staining of cell death in wild-type and PLDδ-null Arabidopsis leaves. Leaves of 5-week-old plants were infiltrated with 50 mM H2O2 or water. The arrow indicates dead cells.
To determine whether the increased H2O2-induced cell death also occurred in PLDδ-null plants, leaves on plants were infiltrated with various concentrations (10 to 50 mM) of H2O2. No obvious death was noted in wild-type leaves. Plants are able to tolerate high concentrations of H2O2 because of their high H2O2-scavenging activity (Neill et al., 2002a). However, treatment of PLDδ-abrogated leaves with 50 mM H2O2 for 7 days triggered cell death in leaves, which was confirmed by positive trypan blue staining of PLDδ-null, but not wild-type, leaves (Figure 3B).
H2O2 Treatment Activates PLDδ
The involvement of PLDδ in H2O2-promoted cell death was studied further using protoplasts rather than whole plants because of the amenability of protoplasts for lipid labeling and quantitative analysis. The removal of cell walls also reduces the cell's capacity to scavenge H2O2, so protoplasts are more sensitive to H2O2 (Neill et al., 2002a). In addition, the protoplast system provides a synchronized cell system for measuring cellular response. Furthermore, freshly isolated protoplasts have proven to be physiological cell systems for studying the various plant signaling mechanisms (reviewed by Sheen, 2001), including H2O2 response (Kovtun et al., 2000) and PLD in abscisic acid action (Jacob et al., 1999).
To measure the change of PLD activity in response to H2O2 in vivo, protoplasts were prelabeled with fluorescent 1-oleoyl-2-(12-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]dodecanoyl)-glycero-3-phosphocholine (NBD-PC) and then challenged with H2O2 in the presence of 1-butanol. The alcohol was used because PLD can transfer the phosphatidyl moiety to a primary alcohol at the expense of PA, and this activity is specific to PLD, thus providing a unique indicator for PLD activity (Wang, 2002). Immediately after prelabeling and before H2O2 stimulation, no difference was observed in the amounts of NBD-PC– and PLD-derived products—NBD-PA and NBD-phosphatidylbutanol (NBD-PtdBut)—between PLDδ-null and wild-type cells, indicating that the basal metabolism of NBD-PC is not altered by the gene knockout. However, significant increases in PtdBut occurred 30 and 60 min after H2O2 treatment in wild-type but not in PLDδ-null cells (Figure 4A). PA, expressed either as a percentage of total NBD lipids or relative NBD-PA (Figure 4B), also increased in H2O2-treated wild-type cells but not in PLDδ-null cells. The lack of increase in PA and PtdBut in H2O2-treated PLDδ-null cells indicates that, despite the presence of 11 other PLD genes in Arabidopsis, PLDδ is required for the PLD-mediated lipid hydrolysis in response to H2O2.
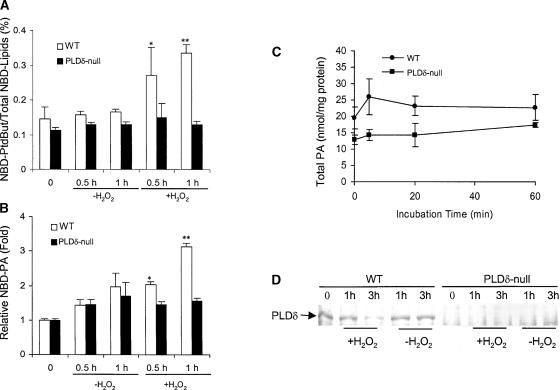
Figure 4.
H2O2-Induced Activation of PLD in Wild-Type but Not in PLDδ-Null Protoplasts.
(A) H2O2-induced PLD activity measured by the formation of NBD-PtdBut. PtdBut was expressed as percentage of PtdBut fluorescence over the total fluorescence of NBD-(PC+PtdBut+PA). WT, wild type.
(B) H2O2-induced PLD activity measured by the formation of PA. PA was expressed as fold increase over the amount of PA before H2O2 treatments.
In (A) and (B), protoplasts were prelabeled with NBD-PC and then challenged with 1 mM H2O2. Asterisks indicate that the mean value is significantly different from that of the protoplasts without H2O2 treatment at the same time at P < 0.05 (one asterisk) or P < 0.01 (two asterisks).
(C) Total PA in protoplasts as measured by mass spectrometry (n = 5).
(D) Immunoblot analysis of PLDδ after exposure of protoplasts to H2O2 for 1 and 3 h.
Measurements of total cellular PA in protoplasts revealed that the basal level of PA in PLDδ-null cells was ∼70% of that in wild-type cells (Figure 4C). This difference is similar to that in leaves, in which the amounts of PA in PLDδ-null and wild-type plants were 1.5 ± 0.4 and 2.0 ± 0.3 nmol/mg dry weight (n = 5), respectively. PA levels measured on the basis of milligrams of total protoplast protein (Figure 4C) were approximately twofold greater that those measured on the basis of milligrams of dry leaf weight, because proteins constitute ∼20% of dry leaf weight. The PA level alteration caused by the lack of PLDδ in both protoplasts and leaves indicates that PLDδ activity may contribute to the basal cellular level of PA and that the preparation of protoplasts does not increase PLDδ activity specifically. When cells were exposed to H2O2, the total cellular level of PA displayed a small transient increase in wild-type but not in PLDδ-null cells. The extent of the H2O2-induced total PA changes after 1 h of exposure to H2O2 was not as significant as that measured by fluorescence-labeled PA. This finding is not surprising considering the possibility that total PA comprises intracellular PA from both biosynthetic and hydrolytic pools, the latter of which includes PA derived from the hydrolysis of NBD-PC. Moreover, NBD-PC was localized primarily in the plasma membrane, where PLDδ is localized.
The stimulation of PLD activity was not related to changes in PLD proteins. The level of PLDδ protein remained unchanged for the first hour and decreased 3 h after H2O2 treatment (Figure 4D). Immunoblot analysis with a PLDδ-specific antibody detected PLDδ in wild-type but not in PLDδ-null mutant plants. These data suggest that the H2O2-induced formation of PLD reaction products results from the activation of preexisting PLDδ rather than from the synthesis of the enzyme. We then tested whether H2O2 itself might activate PLDδ directly. Coincubation of purified PLDδ (Wang and Wang, 2001) with various concentrations of H2O2 failed to stimulate the PLDδ activity (data not shown).
PLDδ-Derived PA Attenuates H2O2-Induced Cell Death
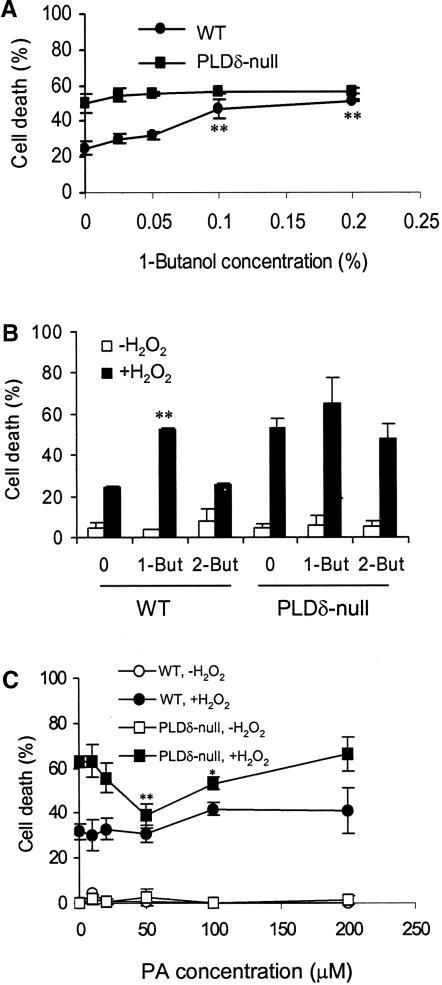
To examine whether the lipid product PA is responsible for decreasing H2O2-promoted cell death, 1-butanol was added together with H2O2 to suppress H2O2-induced PA production. The percentage of cells that died in the wild type increased nearly twofold with increased 1-butanol concentrations, and the optimal stimulation of cell death occurred at 0.1% 1-butanol (Figure 5A). 2-Butanol, which is not a substrate of PLD, exhibited no significant effect on cell death (Figure 5B). By contrast, PLDδ-null cells showed no significant increase in cell death when incubated with 1-butanol (Figure 5A). The lack of a 1-butanol effect was consistent with other results showing that no H2O2-induced activation of PLD occurred in the PLDδ-null cells (Figure 4). These results suggest that the PLDδ-mediated production of PA is involved in reducing H2O2-promoted cell death.
Figure 5.
Effects of 1- and 2-Butanol and PA on H2O2-Promoted Cell Death.
(A) 1-Butanol treatment increased cell death in wild-type (WT) protoplasts.
(B) Specificity of 1-butanol on H2O2-induced cell death. Protoplasts were treated with 0.1% 1- or 2-butanol (But) for 30 min before the addition of 1 mM H2O2.
Double asterisks in (A) and (B) indicate significant differences between 1-butanol–treated and untreated protoplasts at P < 0.01.
(C) PA decreased cell death in PLDδ-null protoplasts. PA was added 30 min before the addition of H2O2. Cell death was counted 24 h after H2O2 treatment. Asterisks indicate significant differences between PA-treated and untreated protoplasts at P < 0.05 (one asterisk) or P < 0.01 (two asterisks).
PA then was supplied to protoplasts to determine whether it might mitigate the H2O2 effect. Protoplasts have the advantage of easy incorporation of PA and other lipids into the cellular membranes, as shown with NBD-PC labeling (Figure 4). Incubation of protoplasts with dioleoyl-PA decreased cell death in PLDδ-null cells (Figure 5C), but incubation with dioleoyl-PC at the same concentration produced no effect on cell death in PLDδ-null or wild-type cells (data not shown). The greatest effect occurred at 50 μM, at which level PLDδ-null and wild-type cells were indistinguishable in their response to H2O2-promoted cell death (Figure 5C). However, PA had no mitigating effect on H2O2-treated wild-type cells. This differential response to the PA treatment between the two genotypes suggests that PA produced in vivo by wild-type cells (Figure 4) is sufficient and likely a better mediator than the added PA. Also, PA lost its mitigating effect on H2O2-induced cell death when the concentration was >100 μM (Figure 5C), suggesting that appropriate cellular concentrations of PA are critical to its function.
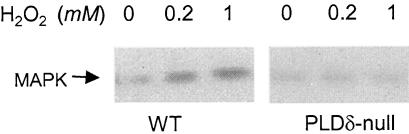
Knockout of PLDδ Diminishes the H2O2-Induced Activation of Mitogen-Activated Protein Kinase Activity
The mitogen-activated protein kinases (MAPKs) have been implicated in various cellular processes, including plant programmed cell death and response to H2O2 (Zhang and Klessig, 1997; Kovtun et al., 2000; Neill et al., 2002a). H2O2 has been shown to activate MAPK cascades in various tissues, although how such activation is achieved is unknown (Kovtun et al., 2000; Neill et al., 2002b). To determine whether the PLDδ function is linked to MAPK activation, an in-gel kinase assay was conducted to compare MAPK activity between PLDδ-null and wild-type protoplasts in response to H2O2 (Figure 6). One major MAPK band at ∼49 kD was observed (Figure 6), and the size was similar to those reported for plant MAPKs (Zhang and Klessig, 1997; Kovtun et al., 2000). PLDδ-null and wild-type cells had similar levels of basal MAP activity but differed in H2O2-induced kinase activity. In wild-type cells, the kinase activity increased at 0.2 and 1 mM H2O2. However, H2O2 failed to increase kinase activity in PLDδ-null cells (Figure 6).
Figure 6.
Ablation of H2O2-Promoted MAPK Activity in PLDδ-Null Arabidopsis.
Autoradiogram from an in-gel kinase assay using protein extracts from wild-type (WT) and PLDδ-null Arabidopsis protoplasts. Myelin basic proteins were embedded on a 10% polyacrylamide gel as the substrate. After electrophoresis, proteins were renatured on the gel and assayed for phosphorylation activity using γ-32P-ATP.
Knockout of PLDδ Increases Plant Sensitivity to Stress Damage
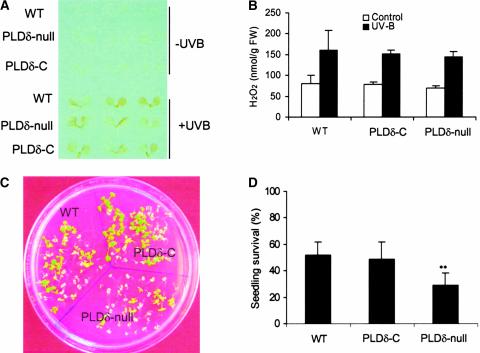
H2O2 is known to accumulate under various stress conditions (Prasad et al., 1994; Sharma et al., 1996; Bowler and Fluhr, 2000; Breusegem et al., 2001; Mittler, 2002; Neill et al., 2002b). We tested how the increased sensitivity to H2O2 would affect the plant's ability to produce H2O2 and cope with stresses (Figure 7). PLDδ-null and wild-type plants were challenged with UV-B irradiation. This treatment induced H2O2 production, as monitored by both 3,3′-diaminobenzidine staining and H2O2 measurement (Figures 7A and 7B). The levels of accumulation for H2O2 were indistinguishable between PLDδ-null and wild-type plants, indicating that the loss of PLDδ did not compromise the plant's ability to produce H2O2 under stress. However, PLDδ-null plants exhibited less tolerance than wild-type plants to UV-B irradiation, and the number of PLDδ-null plants surviving after these treatments was less than half the number of wild-type and PLDδ-complemented plants (Figures 7C and 7D).
Figure 7.
UV-B Light–Induced H2O2 Production and Seedling Death in Wild-Type, PLDδ-Null, and PLDδ-Complemented Arabidopsis.
(A) UV-B light–induced H2O2 production as measured by 3,3′-diaminobenzidine staining of 10-day-old seedlings. PLDδ-C, PLDδ-complemented; WT, wild type.
(B) UV-B light–induced H2O2 production as measured by the Amplex red hydrogen peroxide/peroxidase assay kit. FW, fresh weight.
(C) Increased death of Arabidopsis seedlings after exposure to UV-B light.
(D) Quantitation of UV-B light–induced seedling death. Double asterisks indicate that the mean value is significantly different from that of wild-type plants at P < 0.01.
DISCUSSION
Phospholipid hydrolysis increases under various stress conditions (Chapman, 1998; Laxalt and Munnik, 2002; Wang, 2002). H2O2 is a reactive oxygen species that increases under different stresses and has multifaceted functions in plant stress responses (Prasad et al., 1994; Sharma et al., 1996; Bowler and Fluhr, 2000; Breusegem et al., 2001; Mittler, 2002; Neill et al., 2002b; Dat et al., 2003; Kwak et al., 2003; Yoshioka et al., 2003). However, little is known about the cellular function of lipid hydrolysis and the cellular components that mediate the plant response to H2O2. Recently, PLDζ1 was found to be involved in both the initiation and maintenance of root hair morphogenesis (Ohashi et al., 2003); on the other hand, NADPH oxidase and reactive oxygen species also were required for root hair development (Foreman et al., 2003). However, the connection between phospholipid signaling and H2O2 function has not been elucidated.
The results of this study demonstrate that the PLDδ-mediated hydrolysis of phospholipids plays a positive role in the plant response to H2O2, thus providing a link between membrane phospholipid hydrolysis and the signaling of the plant response to oxidative stress. The activation of PLDδ and its derived PA function to decrease H2O2-induced programmed cell death and increase plant stress tolerance. The anti-cell death effect of PLDδ has been documented in freshly isolated protoplasts (Figures 2 and 3A) and in leaves on plants (Figure 3B) and also is reflected in stress tolerance in Arabidopsis seedlings (Figure 7). It should be noted that although most of these studies used 1 mM H2O2, the pro-cell death effect of H2O2 begins at a lower concentration, because the DC50 for applied H2O2 in the PLDδ-null cell is 0.45 mM (Figure 2A). In addition, the effective DC50 for H2O2 in the cell should be lower than that because cells have a high capacity to metabolize H2O2 by several scavenging enzymes (Neill et al., 2002a). These considerations aside, the DC50 for H2O2 is still within the physiological range reported for Arabidopsis cells, in which the steady state H2O2 levels varied from 60 μM to 7 mM (Karpinski et al., 1999; Veljovic-Jovanovic et al., 2001).
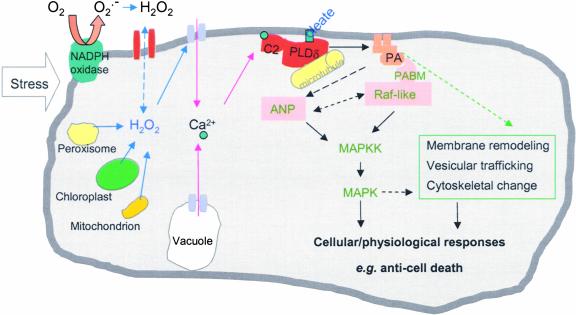
Comparative measurements of the PLD-specific product PtdBut in wild-type and PLDδ-null cells demonstrate that PLDδ is activated specifically in response to H2O2, despite the presence of 11 other PLD genes in Arabidopsis. Although the exact mechanism for the H2O2-specific activation of PLDδ remains to be elucidated, our results and those of other researchers have provided important insights into the unique activation and function of PLDδ. Immunoblot analysis suggests that the H2O2 activation of PLDδ likely results from a modulation of preexisting PLDδ (Figure 4D). Our testing of purified PLDδ with H2O2 suggests that H2O2 may not interact with and activate PLDδ directly. One possible effector for the H2O2-induced activation of PLDδ might be the increase in cytosolic Ca2+. H2O2 and various stresses have been shown to activate the Ca2+ channel, stimulating a transient increase in the cytosolic Ca2+ level (Figure 8) (Pei et al., 2000; Murata et al., 2001; Zhang et al., 2001). Ca2+ has been shown to stimulate PLDδ activity by increasing its affinity for its lipid substrate (Qin et al., 2002), and it binds to the Ca2+/phospholipid binding C2 domain that resides near the N terminus of PLDδ (Zheng et al., 2000; Wang, 2002) (Figure 8). On the other hand, several other PLDs also contain the C2 domain (Qin and Wang, 2002); thus, the specificity of PLDδ activation also should require other properties that are unique to the enzyme.
Figure 8.
Proposed Model Depicting the Activation and Function of PLDδ and PA in the Cell's Response to H2O2.
Stress stimulates the cellular production of H2O2 that activates PLDδ associated with the plasma membrane and the microtubule cytoskeleton. Potential mediators in the activation include an increase in cytosolic Ca2+ and oleic acid in the membrane. Such interactions increase the affinity of PLDδ to its substrates, stimulating lipid hydrolysis and PA production. PA may bind to target proteins, such as Raf-like MAPK kinase kinase (MAPKK), that contain a PA binding motif (PABM). Such binding may lead to the activation of MAPK cascades, and PA also may be involved directly in the regulation of ANP. In addition, PA may function by modulating membrane trafficking and remodeling. These interactions modulate the cell's ability to respond to oxidative stress and decrease cell death. Dashed lines denote undocumented, hypothetical interactions.
One such unique property is the stimulation of PLDδ by oleic acid (Wang and Wang, 2001). This study has established unequivocally that PLDδ is solely responsible for the oleate-stimulated PLD activity (Figure 1). Such oleate-stimulated PLD activity also has been identified in mammalian cells, but the gene for this activity remains elusive (Liscovitch et al., 2000). Oleate has been shown to reduce apoptosis in animal cells (Hardy et al., 2000). In Arabidopsis, deficiency in oleate synthesis results in an increase in cell death (Kachroo et al., 2001). This raises the intriguing possibility that PLDδ may serve as a direct target of oleate (Figure 8), which might underlie in part oleate's anti-cell death activity. In addition, PLDδ is distinctively different from other Arabidopsis PLDs in its intracellular localization and patterns of expression. Unlike the other PLDs examined, PLDδ is associated with the plasma membrane, binds to tubulin, and is expressed highly in stressed and senescent tissues (Figure 8) (Gardiner et al., 2001; Katagiri et al., 2001; Wang and Wang, 2001). These distinct properties provide the cellular and biochemical bases for the specific activation and unique function of PLDδ in the plant response to H2O2. For example, its membrane association indicates that PLDδ-derived PA will be localized on the plasma membrane. This site of PA production may explain why the process can be mimicked by exogenous PA, because it can be incorporated easily into the plasma membrane.
Several lines of evidence indicate that it is PLDδ-derived PA that mediates the anti-cell death effect promoted by H2O2. First, wild-type cells, which produced a higher level of PA than PLDδ-null mutant cells in response to H2O2, were more tolerant to H2O2-induced cell death than were mutant cells (Figure 4). In addition, the estimated rate of oleate-stimulated PLDδ activity in vitro (4 nmol PA·mg−1 microsomal protein·min−1; Figure 1C) is sufficient to account for the actual amount of PA in vivo (∼1 nmol PA·mg−1 total protoplast protein·min−1; Figure 4C). Second, 1-butanol, which decreases PA production by competing with water to be the hydroxyl donor, resulted in more cell death in the presence of H2O2 (Figures 5A and 5B). Third, the addition of PA improved cell survival in PLDδ-null cells that failed to produce PA in response to H2O2 (Figure 5C). This information is important for determining how the activation of a PLD mediates cellular function, because in addition to PA, the activation of PLD also generates water-soluble head groups and results in a change in membrane lipid composition attributable to the hydrolysis of common membrane lipids (Welti et al., 2002).
PA has been shown to regulate various cellular functions in multiple ways (Ghosh et al., 1996; Liscovitch et al., 2000; Rizzo et al., 2000; Fang et al., 2001; Jones and Huanun, 2002; Wang, 2002). It may serve as a direct activator or inhibitor of enzymes involved in signal transduction and/or function as a membrane anchor for assembling signaling complexes. The direct targets of PA identified in animal cells include protein kinases (Ghosh et al., 1996; Fang et al., 2001) and protein phosphatases (Jones and Huanun, 2002). The molecular target of PA has not been identified in plants (Munnik, 2001; Wang, 2002). Pharmacological data suggest that PA is an upstream regulator of MAPK in plant-wounding signal transduction (Lee et al., 2001). The present study showed that ablation of PLDδ suppressed the H2O2 induction of MAPK activity (Figure 6). This finding raises the possibility that the activation of PLDδ and its derived PA are required for the H2O2 stimulation of the MAPK activity that is regulated by upstream kinases. In animal cells, PA has been shown to recruit Raf1, a MAPK kinase kinase, to receptors and to help assemble signaling complexes on the plasma membrane (Rizzo et al., 2000). We have observed that PA binds to an Arabidopsis Raf-like MAPK kinase kinase and that the binding occurs through a PA binding motif that is ∼50% identical to the amino acid sequence of the animal Raf1 (Y. Sang and X. Wang, unpublished data). In plants, the non-Raf MAPK kinase kinase, ANP, has been shown to be involved in the plant response to H2O2, and this kinase is important for increasing plant stress tolerance (Kovtun et al., 2000). ANP does not contain the PA binding motif found in Raf-like MAPK kinase kinase. However, it is possible that PLDδ/PA may regulate ANP directly through some other mechanism of interaction and/or through the modulation of Raf-like kinases (Figure 8).
In addition, PLDδ/PA also may protect cells through their functions in membrane biogenesis, vesicular trafficking, and cytoskeletal rearrangement (Figure 8). PLDδ has been shown to be associated with the microtubule cytoskeleton (Gardiner et al., 2001). A recent study showed that Arabidopsis PLDβ interacts with actin (Kusner et al., 2003). PA also is a known stimulator of phosphatidylinositol-4-phosphate kinase that synthesizes phosphoinositol-4,5-bisphosphate, an important regulator of cytoskeletal movement and membrane trafficking (Liscovitch et al., 2000). Furthermore, PA is fusogenic and also a substrate for glycerolipid synthesis. Whether and how PLDδ and PA affect these cellular processes require further investigations. The present finding that PLDδ is required for an active cellular response to H2O2, decreasing cell death and increasing stress tolerance, indicates that such investigation will be important to revealing the networks of signaling cascades in the plant response to reactive oxygen species and stresses.
METHODS
Isolation of a PLDδ-Knockout Mutant and Genetic Complementation
A PLDδ-knockout mutant was isolated from the Wassilewskija ecotype of Arabidopsis thaliana. The mutant was identified by screening T-DNA insertion lines according to the protocol of the Arabidopsis Gene Knockout Research Facility at the University of Wisconsin (Sussman et al., 2000). The loss of PLDδ function was confirmed by the absence of PLDδ's protein, enzymatic activity (Figure 1), and transcript (data not shown).
To make up for the loss of PLDδ, the PLDδ gene with its own 1.5-kb 5′ untranslated region was cloned from wild-type plants by PCR. The gene with its own promoter was ligated to the agrobacterial binary vector pBin19 and introduced into homozygous PLDδ-knockout plants via Agrobacterium tumefaciens–mediated transformation using floral dipping. The T-DNAs used in PLDδ-knockout and pBin19 plants carried the selection marker for kanamycin and hygromycin resistance, respectively. Thus, the PLDδ-complemented plants were selected by their resistance to both antibiotics. The complementation plants were confirmed by PCR for the presence of the original T-DNA insertion allele and the introduced wild-type PLDδ gene as well as by detection for the production of the PLDδ protein (Figure 2C).
Plant Growth and Protoplast Isolation
Seeds of PLDδ-null mutant and wild-type plants were sown in soil and treated at 4°C for 2 days. Plants were grown in a growth chamber under 14-h-day/10-h-night and 23/18°C cycles. The expanded leaves from 4- to 5-week-old plants were used to isolate protoplasts based on a protocol described previously (Kovtun et al., 2000).
Cell Death Assays
Freshly isolated protoplasts (5 × 105) were incubated in the dark with various concentrations of H2O2 or other reagents as indicated in Figures 4 and 5. Cells were stained with 0.5 mg/mL fluorescein diacetate for 15 min, and total and live cells were counted with a fluorescence microscope. When the effect of added phosphatidic acid (PA) on cell death was examined, dioleoyl-PA and other phospholipids were emulsified in the incubation solution by sonication before use. Suspended lipids were added to protoplasts, and the lipid-protoplast mixture was incubated with gentle agitation at 22°C for 30 min before H2O2 treatment. Incorporation of phospholipids into protoplast membranes was verified using 1-oleoyl-2-NBD (12[(7nitro-2-1,3-benzoxadiazol-4-yl)amino]dodecanoyl)-PA and NBD-phosphatidylcholine (NBD-PC). The presence of the fluorescent PA or PC in membranes was detected by fluorescence microscopy and analyzed as described below.
Programmed cell death was determined by the TUNEL (terminal deoxynucleotidyl transferase–mediated dUTP nick end labeling) assay using an in situ cell death kit (Roche Molecular Biochemicals, Mannheim, Germany). Protoplasts were collected, and the free 3′-OH groups in single- and double-stranded DNA were labeled according to the manufacturer's instructions. Labeled DNA showing bright green fluorescence was visualized by fluorescence microscopy. To test H2O2's effects on cell death in Arabidopsis plants, the first true leaves from 5-week-old plants were infiltrated using a needleless syringe with either 10 to 50 mM H2O2 or water. Leaves were excised from plants at 1, 2, and 7 days after infiltration and stained with a lactophenol–trypan blue solution (Rate et al., 1999).
Protein Extraction and Immunoblot Analysis of PLDs
Total protein from wild-type, PLDδ-null, and PLDδ-complemented leaves was isolated as described previously (Wang and Wang, 2001). Protein contents were determined using a dye binding assay. Equal amounts of protein (20 μg/lane) were separated by SDS-PAGE analysis and then transferred onto polyvinylidene difluoride filters. The filters were blotted with PLDδ- or PLDα-specific antibodies followed by incubation with a second antibody conjugated to alkaline phosphatase. The PLD proteins were made visible by staining the blot showing phosphatase activity (Wang and Wang, 2001).
Phospholipid Labeling and Assays of Oleate-Stimulated PLD Activities
NBD-PC in chloroform was dried under a stream of nitrogen and suspended in the incubation solution by sonication before use. Protoplasts (5 × 106/mL) were incubated with 0.5 mg/mL NBD-PC for 80 min on ice and then transferred to room temperature (22°C) for 10 min (Jacob et al., 1999). In vivo PLD activity was determined according to the production of phosphatidylbutanol (PtdBut). 1-Butanol (0.1%, v/v) was added with 1 mM H2O2 (or water) to NBD-PC–labeled protoplasts (∼3 × 105) and incubated in a glass tube at 22°C for the indicated time (Figure 4). Hot isopropanol (75°C) was added and then incubated for 10 min at 75°C to inactivate PLD. Lipids were extracted and separated on thin layer chromatography plates (silica G) with chloroform:methanol:NH4OH (65:35:5) as described previously (Ryu and Wang, 1996). NBD-PC, NBD-PA, and NBD-PtdBut were well separated and visualized under UV illumination. The spots were scraped, extracted with chloroform:methanol:water (5:5:1), and quantitated with a fluorescence spectrophotometer at 460 nm (excitation) and 534 nm (emission). In vitro PLDδ activity was measured in the presence of oleate as described previously (Wang and Wang, 2001) using proteins extracted from Arabidopsis leaves.
Total PA Measurement
Total lipids were extracted from Arabidopsis leaves and leaf protoplasts and then analyzed using electron spray ionization tandem mass spectrometry (ESI-MS/MS). Hot isopropanol (75°C) was added to protoplasts after the treatments to inactivate PLD activity. The processes of lipid extraction, ESI-MS/MS analysis, and quantification followed the procedure detailed previously (Welti et al., 2002). Total proteins from equivalent amounts of protoplasts were measured using a dye binding assay.
In-Gel Kinase Activity Assay
Protein was extracted from protoplasts with a buffer (100 mM HEPES, pH 7.5, 5 mM EDTA, 5 mM EGTA, 10 mM DTT, 10 mM Na3VO4, 10 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 5 μg/mL antipain, 10% glycerol, 7.5% polyvinylpolypyrrolidone) using the method of Zhang and Klessig (1997) with minor modifications. Protoplasts were sonicated twice for 15 s each in a 1.5-mL microcentrifuge tube followed by centrifugation at 18,000g for 20 min. Proteins (16 μg/lane) in the supernatant were subjected to electrophoresis on 10% SDS-polyacrylamide gels embedded with 0.25 mg/mL myelin basic protein as the substrate. Proteins were renatured on the gel and assayed for kinase activity. The reaction buffer for kinase activity contained 25 mM Tris, pH 7.5, 2 mM EGTA, 12 mM MgCl2, 1 mM DTT, 0.1 mM Na3VO4, 200 nM ATP, and 50 μCi of γ-32P-ATP (300 Ci/mmol). The gel was dried and exposed to x-ray film.
Stress Treatments
Seeds of wild-type, PLDδ-null, and PLDδ-complemented plants were surface-sterilized with 70% ethanol and 1.2% sodium hypochlorite and planted on a plate containing half-strength Murashige and Skoog (1962) mineral salts, pH 5.7, with 0.8% agar. Each plate contained 50 seeds of each of the three genotypes. Plates were kept at 4°C for 2 days, and then seedlings were grown under 36 μmol·m−2·s−1 light/dark (12/12 h) at 23°C for 2 weeks. These 2-week-old seedlings were illuminated with 3 μmol·m−2·s−1 UV-B light at 23°C for 3 h. Some of the UV-B light–treated seedlings were used to measure H2O2 production immediately after irradiation, and others were grown for 7 days before they were scored for survival ratio.
H2O2 Measurements
Production of H2O2 in plants was measured by staining plants with 3,3′-diaminobenzidine and by assaying H2O2 concentrations in the leaves. Briefly, 3 h after UV illumination, whole plants were submerged into a 2.8-mM 3,3′-diaminobenzidine solution, pH 5.5, for 2 h and then boiled in 96% ethanol for 10 min. For quantification, H2O2 was extracted from leaves according to the method described previously (Rao et al., 2000). Extract was diluted 10-fold and then measured for H2O2 concentration with an Amplex red hydrogen peroxide/peroxidase assay kit (Molecular Probes, Eugene, OR).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Xuemin Wang, wangs@ksu.edu.
Acknowledgments
We thank Jen Sheen for the protocol and advice regarding protoplast preparation. We also thank Weiqi Li and Todd Williams for mass spectrometry–based lipid analysis. This work was supported by grants from the National Science Foundation (IBN-9808279 and MCB-0110979) and the U.S. Department of Agriculture (2001-35304-10087). This is contribution 03-276-J from the Kansas Agricultural Experiment Station.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.013961.
References
- Alvarez, M.E., Pennel, R.I., Meijer, P.J., Ishikawa, A., Dixon, R.A., and Lamb, C. (1998). Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92, 773–784. [DOI] [PubMed] [Google Scholar]
- Austin-Brown, S.L., and Chapman, K.D. (2002). Inhibition of phospholipase Dα by N-acylethanolamines. Plant Physiol. 129, 1892–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke, P.C., and Jones, R.L. (2001). Cell death of barley aleurone protoplasts is mediated by reactive oxygen species. Plant J. 25, 19–29. [DOI] [PubMed] [Google Scholar]
- Bowler, C., and Fluhr, R. (2000). The role of calcium and activated oxygens as signals for controlling cross-tolerance. Trends Plant Sci. 5, 241–245. [DOI] [PubMed] [Google Scholar]
- Breusegem, F.V., Vranova, E., Dat, J.F., and Inze, D. (2001). The role of active oxygen species in plant signal transduction. Plant Sci. 161, 405–414. [Google Scholar]
- Chapman, K.D. (1998). Phospholipase activity during plant growth and development and in response to environmental stress. Trends Plant Sci. 3, 419–426. [Google Scholar]
- Dat, J.F., Pellinen, R., Van De Cotte, B., Langebartels, C., Kangasjarvi, J., Inze, D., and Van Breusegem, F. (2003). Changes in hydrogen peroxide homeostasis trigger an active cell death process in tobacco. Plant J. 33, 621–632. [DOI] [PubMed] [Google Scholar]
- Delledonne, M., Zeier, J., Marocco, A., and Lamb, C. (2001). Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc. Natl. Acad. Sci. USA 98, 13454–13459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Y., Vilella-Bach, M., Bachmann, R., Flanigan, A., and Chen, J. (2001). Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science 294, 1942–1945. [DOI] [PubMed] [Google Scholar]
- Foreman, J., Demidchik, V., Bothwell, J.H., Mylona, P., Miedema, H., Torres, M.A., Linstead, P., Costa, S., Brownlee, C., Jones, J.D., Davies, J.M., and Dolan, L. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422, 442–446. [DOI] [PubMed] [Google Scholar]
- Gardiner, J.C., Harper, J.D.I., Weerakoon, H.D., Collings, D.A., Ritchie, S., Gilroy, S., Cyr, R.J., and Marc, J.A. (2001). A 90-kD phospholipase D from tobacco binds to microtubules and the plasma membrane. Plant Cell 13, 2143–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, S., Strum, J.C., Sciorra, V.A., Daniel, L., and Bell, R.M. (1996). Raf-1 kinase possesses distinct binding domains for phosphatidylserine and phosphatidic acid: Phosphatidic acid regulates the translocation of Raf-1 in 12-O-tetradecanoylphorbol-13-acetate-stimulated Madin-Darby canine kidney cells. J. Biol. Chem. 271, 8472–8480. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T. (1996). Programmed cell death: A way of life for plants. Proc. Natl. Acad. Sci. USA 93, 12094–12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy, S., Langelier, Y., and Prentki, M. (2000). Oleate activates phosphatidylinositol 3-kinase and promotes proliferation and reduces apoptosis of MDA-MB-231 breast cancer cells, whereas palmitate has opposite effects. Cancer Res. 60, 6353–6358. [PubMed] [Google Scholar]
- Jacob, T., Ritchie, S., Assmann, S.M., and Gilroy, S. (1999). Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc. Natl. Acad. Sci. USA 96, 12192–12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.A., and Huanun, Y.A. (2002). Tight binding inhibition of protein phosphatase-1 by phosphatidic acid: Specificity of inhibition by the phospholipid. J. Biol. Chem. 277, 15530–15538. [DOI] [PubMed] [Google Scholar]
- Kachroo, P., Shanklin, J., Shah, J., Whittle, E.J., and Klessig, D.F. (2001). A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc. Natl. Acad. Sci. USA 98, 9448–9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski, S., Reynolds, H., Karpinska, B., Wingsle, G., Creissen, G., and Mullineaux, P. (1999). Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 284, 654–657. [DOI] [PubMed] [Google Scholar]
- Katagiri, T., Takahashi, S., and Shinozaki, K. (2001). Involvement of a novel Arabidopsis phospholipase D, AtPLDδ, in dehydration-inducible accumulation of phosphatidic acid in stress signaling. Plant J. 26, 595–605. [DOI] [PubMed] [Google Scholar]
- Kovtun, Y., Chiu, W.-H., Tena, G., and Sheen, J. (2000). Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 97, 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusner, D.J., Barton, J.A., Qin, C., Wang, X., and Iyer, S.S. (2003). Evolutionary conservation of physical and functional interactions between phospholipase D and actin. Arch. Biochem. Biophys. 412, 231–241. [DOI] [PubMed] [Google Scholar]
- Kwak, J.M., Mori, I.C., Pei, Z.M., Leonhardt, N., Torres, M.A., Dangl, J.L., Bloom, R.E., Bodde, S., Jones, J.D., and Schroeder, J.I. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22, 2623–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxalt, A.M., and Munnik, T. (2002). Phospholipid signalling in plant defense. Curr. Opin. Plant Biol. 5, 332–338. [DOI] [PubMed] [Google Scholar]
- Lee, S., Hirt, H., and Lee, Y. (2001). Phosphatidic acid activates a wound-activated MAPK in Glycine max. Plant J. 26, 479–486. [DOI] [PubMed] [Google Scholar]
- Lee, S., Suh, S., Kim, S., Crain, R.C., Kwak, J.M., Nam, H.-G., and Lee, Y. (1997). Systemic elevation of phosphatidic acid and lysophospholipid levels in wounded plants. Plant J. 12, 547–556. [Google Scholar]
- Levine, A., Tenhaken, R., Dixon, R., and Lamb, C. (1994). H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79, 583–593. [DOI] [PubMed] [Google Scholar]
- Liscovitch, M., Czarny, M., Fiucci, G., and Tang, X. (2000). Phospholipase D: Molecular and cell biology of a novel gene family. Biochem. J. 345, 401–415. [PMC free article] [PubMed] [Google Scholar]
- Meijer, H.J., ter Riet, B., van Himbergen, J.A., Musgrave, A., and Munnik, T. (2002). KCl activates phospholipases D at two different concentration ranges: Distinguishing between hyperosmotic stress and membrane depolarization. Plant J. 31, 51–59. [DOI] [PubMed] [Google Scholar]
- Mittler, R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, 405–410. [DOI] [PubMed] [Google Scholar]
- Munnik, T. (2001). Phosphatidic acid: An emerging plant lipid second messenger. Trends Plant Sci. 6, 227–233. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Murata, Y., Pei, Z.M., Mori, I.C., and Schroeder, J.L. (2001). Abscisic acid activation of plasma membrane Ca2+ channels in guard cells requires cytosolic NAD(P)H and is differentially disrupted upstream and downstream of reactive oxygen species production in abi1-1 and abi2-1 protein phosphatase 2C mutants. Plant Cell 13, 2513–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill, S.J., Desikan, R., Clarke, A., Hurst, R.D., and Hancock, J.T. (2002. a). Hydrogen peroxide and nitric oxide as signaling molecules in plants. J. Exp. Bot. 53, 1237–1247. [PubMed] [Google Scholar]
- Neill, S.J., Desikan, R., and Hancock, J. (2002. b). Hydrogen peroxide signaling. Curr. Opin. Plant Biol. 5, 388–395. [DOI] [PubMed] [Google Scholar]
- Ohashi, Y., Oka, A., Rodrigues-Pousada, R., Possenti, M., Ruberti, I., Morelli, G., and Aoyama, T. (2003). Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science 300, 1427–1430. [DOI] [PubMed] [Google Scholar]
- Park, K.Y., Jung, J.Y., Park, J., Hwang, J.U., Kim, Y.W., Hwang, I., and Lee, Y. (2003). A role for phosphatidylinositol 3-phosphate in abscisic acid-induced reactive oxygen species generation in guard cells. Plant Physiol. 132, 92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, Z.M., Murata, Y., Benning, G., Thomine, S., Klusener, B., Allen, G.J., Grill, E., and Schroeder, J.L. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signaling in guard cells. Nature 46, 731–734. [DOI] [PubMed] [Google Scholar]
- Prasad, T.K., Anderson, M.D., Martin, B.A., and Stewart, C.R. (1994). Evidence for chilling-induced oxidative stress in maize seedlings and a regulatory role for hydrogen peroxide. Plant Cell 6, 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, C., and Wang, X. (2002). The Arabidopsis phospholipidase D family: Characterization of a calcium-independent and phosphatidylcholine-selective PLDζ1 with distinct regulatory domains. Plant Physiol. 128, 1057–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, C., Wang, C., and Wang, X. (2002). Kinetic analysis of Arabidopsis phospholipases Dδ: Substrate preference and mechanism of activation by Ca2+ and phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 277, 49685–49690. [DOI] [PubMed] [Google Scholar]
- Rao, M.V., Lee, H., Creelman, R.A., Mullet, J.E., and Davis, K.R. (2000). Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 12, 1633–1646.11006337 [Google Scholar]
- Rate, D.N., Cuenca, J.V., Bowman, G.R., Guttman, D.S., and Greenberg, J.T. (1999). The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11, 1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, D., Yang, H., and Zhang, S. (2002). Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J. Biol. Chem. 277, 559–565. [DOI] [PubMed] [Google Scholar]
- Rizzo, M.A., Shome, K., Watkins, S.C., and Romero, G. (2000). The recruitment of Raf-1 to membranes is mediated by direct interaction with phosphatidic acid and is independent of association with Ras. J. Biol. Chem. 275, 23911–23918. [DOI] [PubMed] [Google Scholar]
- Ryu, S.B., and Wang, X. (1996). Activation of phospholipase D and the possible mechanism of activation in wound-induced lipid hydrolysis in castor bean leaves. Biochim. Biophys. Acta 1303, 243–250. [DOI] [PubMed] [Google Scholar]
- Sharma, Y.K., León, J., Raskin, I., and Davis, K.R. (1996). Ozone-induced responses in Arabidopsis thaliana: The role of salicylic acid in the accumulation of defense-related transcripts and induced resistance. Proc. Natl. Acad. Sci. USA 93, 5099–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen, J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127, 1466–1475. [PMC free article] [PubMed] [Google Scholar]
- Sussman, M.R., Amasino, R.M., Young, J.C., Krysan, P.J., and Austin-Phillips, S. (2000). The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol. 124, 1465–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veljovic-Jovanovic, S.D., Pignocchi, C., Noctor, G., and Foyer, C.H. (2001). Low ascorbic acid in the vtc-1 mutant of Arabidopsis is associated with decreased growth and intracellular redistribution of the antioxidant system. Plant Physiol. 127, 426–435. [PMC free article] [PubMed] [Google Scholar]
- Wang, C., and Wang, X. (2001). A novel phospholipase D of Arabidopsis that is activated by oleic acid and associated with the plasma membrane. Plant Physiol. 127, 1102–1112. [PMC free article] [PubMed] [Google Scholar]
- Wang, C., Zien, C.A., Afitlhile, M., Weilt, R., Hildebrand, D.F., and Wang, X. (2000). Involvement of phospholipase D in wound-induced accumulation of jasmonic acid in Arabidopsis. Plant Cell 12, 2237–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. (2002). Phospholipase D in hormonal and stress signaling. Curr. Opin. Plant Biol. 5, 408–414. [DOI] [PubMed] [Google Scholar]
- Welti, R., Li, W., Li, M., Sang, Y., Biesiada, H., Zhou, H.E., Rajashekar, C.B., Williams, T.D., and Wang, X. (2002). Profiling membrane lipids in plant stress responses: Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 277, 31994–32002. [DOI] [PubMed] [Google Scholar]
- Yoshioka, H., Numata, N., Nakajima, K., Katou, S., Kawakita, K., Rowland, O., Jones, J.D., and Doke, N. (2003). Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15, 706–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., and Klessig, D.F. (1997). Salicylic acid activates a 48-kD MAPK kinase in tobacco. Plant Cell 9, 809–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Zhang, L., Dong, F., Gao, J., Galbraith, D.W., and Song, C.-P. (2001). Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiol. 126, 1438–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, L., Krishnamoorthi, R., Zolkiewski, M., and Wang, X. (2000). Distinct Ca2+ binding properties of novel C2 domains of plant phospholipase D alpha and beta. J. Biol. Chem. 275, 19700–19706. [DOI] [PubMed] [Google Scholar]