Abstract
The Arabidopsis mutant early in short days4 (esd4) shows extreme early flowering and alterations in shoot development. We have identified ESD4 and demonstrate that it encodes a nuclear protein located predominantly at the periphery of the nucleus. ESD4 contains a segment of >200 amino acids with strong similarity to yeast and animal proteases that are specific for the protein modifier SMALL UBIQUITIN-RELATED MODIFIER (SUMO). ESD4 shows a similar function to these proteases in vitro and processes the precursor of Arabidopsis SUMO (AtSUMO) to generate the mature form. This activity of ESD4 is prevented by mutations that affect the predicted active site of the protease or the cleavage site of the AtSUMO precursor. In yeast, these proteases also recycle SUMO from conjugates, and this appears to be the major role of ESD4 in vivo. This is suggested because esd4 mutants contain less free AtSUMO and more SUMO conjugates than wild-type plants, and a transgene expressing mature SUMO at high levels enhanced aspects of the esd4 phenotype. ESD4 defines an important role for protein modification by AtSUMO in the regulation of flowering.
INTRODUCTION
Protein function is often regulated in vivo by post-translational modifications. In eukaryotes, such modifications frequently occur by the attachment of a small polypeptide to the target protein. Ubiquitin was the first of these polypeptides to be described, and it is attached to substrate proteins by an isopeptide bond formed between a Gly residue at the C terminus of ubiquitin and a Lys residue in the substrate protein. Attachment of a single ubiquitin molecule appears to mediate endocytosis (Katzmann et al., 2001), whereas the formation of polyubiquitin chains directs proteins for degradation by the 26S proteosome (Hershko and Ciechanover, 1998). In addition to ubiquitin, eukaryotes contain proteins that are related in amino acid sequence to ubiquitin and that are involved in protein modification (Hochstrasser, 2000). These proteins include SMALL UBIQUITIN-RELATED MODIFIER (SUMO) and RELATED TO UBIQUITIN1 (RUB1), which also are attached to Lys residues in target proteins via isopeptide bonds. In contrast to SUMO and ubiquitin, which are involved in the modification of a wide range of proteins, RUB1 has only been shown to modify Cullin, a component of SCF (SKP1/Cullin/F-box) E3 ligases, which are enzymes required to attach ubiquitin to specific substrate proteins.
Many proteins modified by SUMO have been described in yeast, mammals, and Drosophila. SUMOylation appears to influence the function of proteins in distinct ways—for example, by altering their cellular location, their activity, or their stability by antagonizing their degradation via ubiquitination and the 26S proteasome (Melchior, 2000; Muller et al., 2001). Examples of proteins modified by SUMO include the mammalian protein PML, whose SUMOylation is required for the formation of subnuclear bodies (Sternsdorf et al., 1997; Muller et al., 1998), the yeast protein PCNA, which is involved in DNA replication and can be modified by SUMO or ubiquitin on the same Lys, suggesting that SUMOylation might protect the protein from degradation (Hoege et al., 2002), and the Drosophila transcription factor BICOID, whose nuclear localization appears to require SUMO (Epps and Tanda, 1998).
Approximately 5% of Arabidopsis genes encode proteins predicted to be involved in the ubiquitin-proteasome system, and regulated protein degradation by ubiquitination is important in many plant processes (Hellmann and Estelle, 2002). By contrast, definitive roles for SUMO in plant protein modification have not been described, although a number of observations suggest that this is likely to be important in many plant processes. The Arabidopsis genome contains genes predicted to encode SUMO isoforms and other components of the SUMO system (Vierstra and Callis, 1999; Kurepa et al., 2003; Lois et al., 2003). In Arabidopsis, the conjugation of AtSUMO1 and AtSUMO2 to target proteins increases in response to stress conditions, particularly heat shock, suggesting a role for SUMO modification in response to these conditions (Kurepa et al., 2003). This was not observed with AtSUMO3, indicating that modification by different AtSUMO isoforms is regulated differentially. Overexpression of AtSUMO1 or AtSUMO2 also reduced responses to the growth regulator abscisic acid, suggesting that SUMO also plays a role in modulating responses to this growth regulator (Lois et al., 2003). Finally, SUMO has been implicated in the response of plants to pathogens. In the yeast two-hybrid system, SUMO was shown to interact with the tomato protein ETHYLENE-INDUCING XYLANASE (EIX), an inducer of plant defense responses, and this interaction appeared to suppress the induction of defense responses by EIX (Hanania et al., 1999). In addition, the Xanthomonas campestris avirulence gene AvrBsT was predicted to encode a SUMO-specific protease that is introduced into the host plant cell during infection (Orth et al., 2000).
Here, we demonstrate that early in short days4 (esd4), an extreme early-flowering mutant of Arabidopsis, is impaired in the regulation of SUMOylation. Previously, we showed that the early flowering of esd4 is attributable to the decreased abundance of the mRNA of the MADS box transcription factor Flowering Locus C (FLC), which is a potent repressor of flowering, as well as to an FLC-independent mechanism (Reeves et al., 2002). In addition, the esd4 mutation has other effects on plant development, including broadening of the silique, alterations in phyllotaxy, and a reduction in stature. We demonstrate that ESD4 encodes a protease that regulates the abundance of SUMO conjugates, suggesting an important role for SUMO metabolism in the control of flowering time.
RESULTS
Isolation of ESD4 Suggests That it Encodes a SUMO-Specific Protease
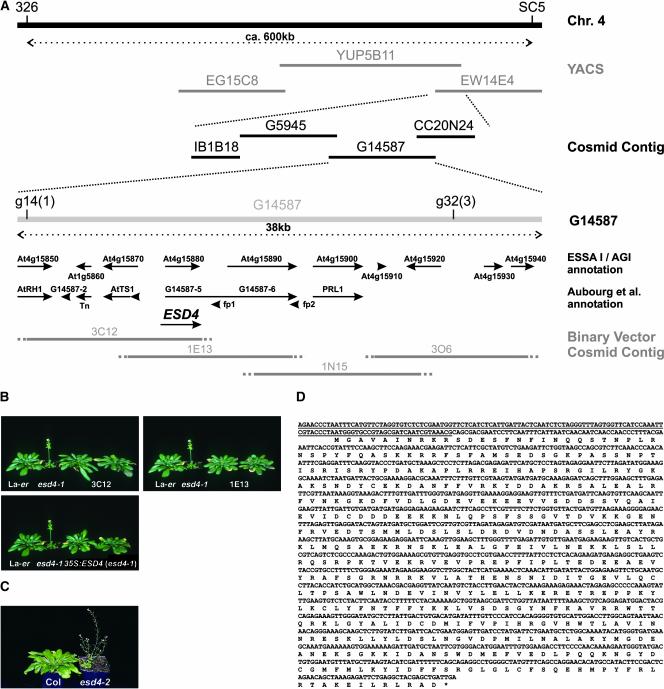
We isolated the ESD4 gene to further understand its role in plant development and the regulation of flowering. Previously, esd4-1 (Landsberg erecta [Ler]) was located to a 2-centimorgan interval on chromosome 4 between markers 326 and SC5 (Reeves et al., 2002). To increase the resolution of the esd4 map position, an additional 691 esd4-like F2 plants were screened with these markers, and ESD4 was located to a 35-kb region (Figure 1A; see Methods) (Arabidopsis Genome Initiative, 2000).
Figure 1.
Isolation of ESD4.
(A) Map-based cloning of ESD4. ESD4 was positioned between cleaved amplified polymorphic sequence (CAPS) markers 326 and SC5 in a segregating F2 population derived from an esd4 (Ler) × Columbia cross. A yeast artificial chromosome (YAC) and a cosmid contig covering this region had been constructed previously (Bevan et al., 1998). Two new CAPS markers, g14 and g32, were generated that flanked esd4 such that one and three crossovers were identified between esd4 and g14 and g32, respectively. Thus, ESD4 was positioned to a region of ∼35 kb, covered by cosmid G14587. This region has been annotated independently by the Arabidopsis Genome Initiative (2000) and Aubourg et al. (1997). Predicted genes from these annotations are shown as arrows. A binary vector cosmid contig covering the region containing ESD4 was constructed from YAC YUP5B11. The exact end point of each cosmid was not known; therefore, these are shown as dotted lines. Four cosmids (3C12, 1E13, 1N15, and 3O6) were transformed into the esd4 mutant via Agrobacterium-mediated root transformation. Cosmids 3C12 and 1E13 complemented the mutation, indicating that ESD4 is located in the overlap of these cosmids. Subsequent analysis indicated that ESD4 corresponds to At4g15880/G14587-5, although the exact sequence is different from the predicted annotations (see below).
(B) Complementation of esd4-1. Photographs of 7-week-old plants grown under short days. Ler and esd4-1 plants are shown to the left of esd4-1 plants transformed with cosmids 3C12, 1E13, or a 35S::ESD4 cDNA construct. Two independent lines are shown for each construct. esd4-1 plants flowered much earlier than wild-type plants. The transformed plants flower at approximately the same time as Ler and show none of the other effects of esd4-1 (Reeves et al., 2002), indicating complementation of the esd4 mutation.
(C) Early-flowering phenotype caused by the esd4-2 allele. Photographs of 7-week-old plants grown under short days. The esd4-2 mutant flowered with 9.8 ± 0.5 rosette leaves and 2.8 ± 0.7 cauline leaves compared with 51.3 ± 4.6 rosette leaves and 9.3 ± 2.8 cauline leaves for the Columbia wild type.
(D) Sequence and annotation of the ESD4 cDNA. A PCR-based approach was used to identify an ESD4 cDNA. The sequence of the ESD4 cDNA is shown, together with the predicted amino acid sequence of the longest open reading frame. This sequence is slightly longer than the Arabidopsis Genome Initiative At4g15880 prediction. The region of the cDNA deleted in the esd4-1 mutant is underlined. The deletion in esd4-1 is 762 bp long and occurs between and including bases 180,698 and 181,459 of ESD4. The sequence likely represents the full-length ESD4 ORF, because the ORF ends in a translational stop, and 27 bp upstream of the predicted AUG initiation codon there is an in-frame UAG translational stop.
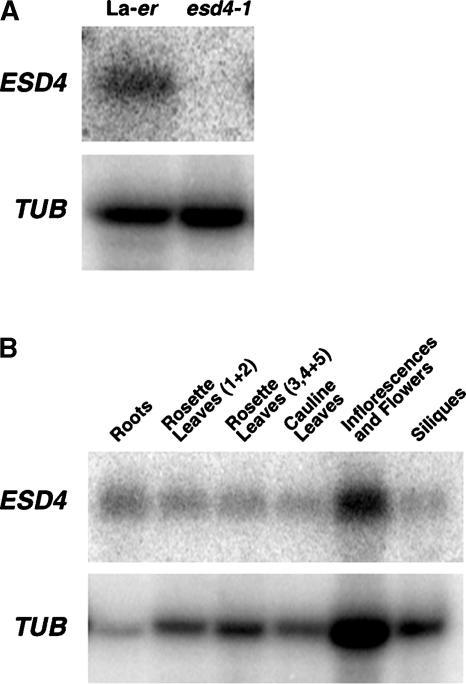
Four overlapping binary vector cosmids (3C12, 1E13, 1N15, and 3O6) spanning this region were introduced into esd4-1 by Agrobacterium tumefaciens–mediated root transformation to identify those that complemented the mutation (Valvekens et al., 1988) (Figure 1). Transgenic esd4-1 mutants carrying 3C12 or 1E13 flowered at a similar time to the wild type and showed none of the pleiotropic phenotypes of esd4-1 (Figure 1) (Reeves et al., 2002). The region of overlap between 3C12 and 1E13 was predicted to encode at least two genes (At4g15870 and At4g15880; Figure 1A). Both genes were amplified from the mutant by PCR and sequenced. At4g15880 was amplified successfully from DNA of wild-type plants but not from esd4-1, suggesting that this region may contain a deletion in esd4-1, consistent with the mutation having been induced with γ-rays. Subsequent analysis revealed a deletion of 762 bp in esd4-1 that removed the first 10 codons from the protein-coding sequence and 732 bp upstream of the predicted ESD4 open reading frame (ORF), a modified form of At4g15880 (see below) (Figures 1A and 1D). No expression of At4g15880 was detected by RNA gel blot analysis in esd4-1 seedlings (Figure 2A), although an mRNA was found in extracts of wild-type plants, suggesting that this ORF might represent ESD4 and that esd4-1 is a null allele.
Figure 2.
Analysis of ESD4 RNA in Mutant and Wild-Type Plants.
(A) RNA gel blot analysis of ESD4 RNA in 10-day-old Ler and esd4-1 seedlings. No ESD4 RNA is detectable in the mutant. β-TUBULIN (TUB) was used as a loading control.
(B) RNA gel blot analysis of ESD4 RNA from different tissues of Ler plants. ESD4 RNA is detectable in all tissues tested. β-TUBULIN was used as a loading control.
Previously, only a single allele of esd4 was isolated. To support our analysis of esd4-1 with a second allele and to strengthen the argument that At4g15880 represents ESD4, the Salk Institute Genomic Analysis Laboratory sequence-indexed insertion line database was screened for an insertion in this ORF. A line carrying a T-DNA insertion within the predicted ESD4 gene was recovered (line SALK_032317). We obtained seeds of this line and identified plants homozygous for the T-DNA insertion. These plants flowered early under both long and short days (Figure 1C) and showed the same pleiotropic effects as the esd4-1 mutation. Thus, we refer to this line as esd4-2.
To confirm the structure of the At4g15880 (ESD4) gene, a cDNA was identified and sequenced (Figure 1D). That this ORF encodes ESD4 was confirmed by demonstrating that expression of this cognate cDNA from the 35S promoter of Cauliflower mosaic virus complemented esd4-1 (Figure 1B). Database searches with the predicted ESD4 protein sequence identified proteins from yeast, mammals, Caenorhabditis elegans, and Drosophila as well as other proteins from Arabidopsis that showed identity with a region of 200 amino acids at the C terminus of ESD4 (Figure 3). These proteins included the human SENTRIN-SPECIFIC PROTEASE1 (SENP1) (Gong et al., 2000), the yeast UBIQUITIN-LIKE PROTEASE1 (ULP1) (Li and Hochstrasser, 1999), and the mouse SMT3-SPECIFIC ISOPEPTIDASE1 (SMT3IP1) (Nishida et al., 2001). This region of ESD4 and the homologous proteins contains residues predicted to form the active site of a Cys protease (Li and Hochstrasser, 1999) (Figure 2). Several of the homologous proteins are proteases involved in regulating the activity of SUMO (Melchior, 2000). A small family of these proteases was described previously in Arabidopsis, although ESD4 was not recognized as a member of this family (Kurepa et al., 2003). These proteases show the endopeptidase activity required to generate mature SUMO from its precursor and the isopeptidase activity that recycles SUMO from conjugates with target proteins.
Figure 3.
Comparison of ESD4 and AtSUMO with Related Proteins.
(A) Comparison of ESD4 with related proteins from other organisms. The C-terminal region of ESD4 shares identity with SUMO-specific proteases from Homo sapiens (SENP1), Mus musculus (SMT3IP1), and Saccharomyces cerevisiae (ULP1). The sequence alignment of the four proteins is shown, with identical residues in white on a black background and similar residues shaded in gray. Numbers indicate amino acid positions. The arrows mark likely catalytic residues.
(B) Comparison of ESD4 with the predicted protein product of the most closely related Arabidopsis gene. Sequence alignment of ESD4 and At3g06910, the most closely related Arabidopsis protein. Similarity extends over the entire predicted protein sequence. Other Arabidopsis proteins (including At4g00690, At1g10570, and At1g60220) also show significant sequence identity to ESD4, but this is limited to the C-terminal protease domain.
(C) Three Arabidopsis SUMO isoforms compared with SUMO proteins from human and yeast and with ubiquitin from Arabidopsis. Sequence alignment of Arabidopsis SUMO isoforms with human SUMO-1, S. cerevisiae SMT3, and Arabidopsis UBIQUITIN1 (UBQ1).
Comparison of ESD4 with other Arabidopsis sequences revealed that ESD4 is part of a larger gene family. The predicted product of one of these genes (At3g06910) shows a high degree of identity to ESD4 over the entire protein sequence (Figure 3B), whereas another gene (At4g00690) is very similar to ESD4 but the similarity is limited to the predicted protease domain. Three other genes (At1g10570, At1g60220, and At5g60190) show significant identity to the C-terminal region of ESD4, including the residues required to form the active site (Li and Hochstrasser, 1999). In addition, a Basic Local Alignment Search Tool (BLAST) query using ULP1, ULP2, and SENP1 protease domain sequences against predicted Arabidopsis proteins revealed a large number of proteins that contain amino acids that are essential to form the protease active site as well as some proteins that lack one or more of these residues (data not shown).
ESD4 Exhibits SUMO Endopeptidase Activity in Vitro
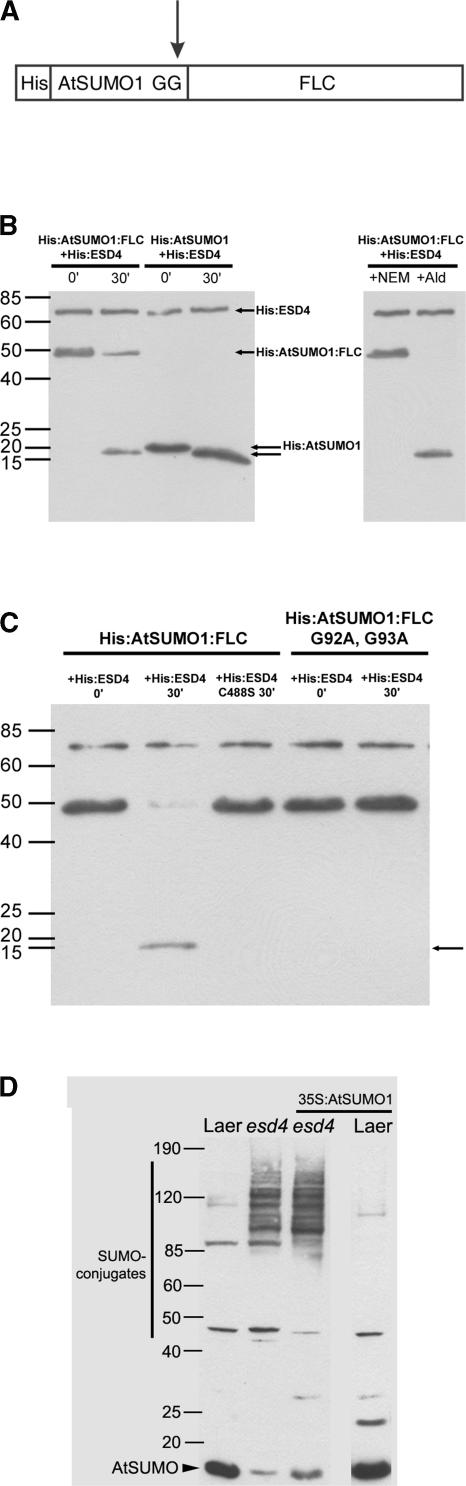
Whether ESD4 shows the SUMO endopeptidase activity characteristic of these proteases was tested in vitro. Three genes predicted to encode SUMO were identified in the Arabidopsis genome, and the cognate cDNAs were isolated (Figure 3C). These were named AtSUMO1 to AtSUMO3, the same nomenclature used independently by Kurepa et al. (2003). These cDNAs were expressed in Escherichia coli with an N-terminal tag of 10 His residues (HIS:AtSUMO). The protease is predicted to cleave HIS:AtSUMO immediately after two highly conserved Gly residues near the C terminus, thereby removing a short peptide and generating mature HIS-AtSUMO (Figures 3C and 4A). To more easily detect this cleavage, the size of the peptide removed by hydrolysis was increased by inserting an unrelated protein at the C terminus of SUMO. FLC was used for this purpose, generating HIS:AtSUMO:FLC. HIS:AtSUMO:FLC was purified from the E. coli extract by nickel-exchange chromatography, and the expected 45-kD protein was detected in the purified extract using an anti-HIS antibody (Figure 4). HIS:ESD4, also purified from E. coli, then was incubated with HIS:AtSUMO1:FLC for 30 min. After incubation, the mixture was separated on a gel and probed with HIS antibody. The HIS:AtSUMO1:FLC protein was either not detected or was present at much lower abundance, and a protein smaller than the expected size for HIS:AtSUMO1 was present. The smaller protein did not appear if HIS:FLC was incubated with HIS:ESD4 (data not shown). These experiments suggested that ESD4 acts in vitro as an AtSUMO1 C-terminal hydrolase.
Figure 4.
Analysis of ESD4 Function in Vitro and in Vivo.
(A) Diagram of the synthetic AtSUMO1 precursor (HIS:AtSUMO1:FLC) used in the in vitro experiments. Ten HIS residues are fused at the N terminus of AtSUMO, and the FLC protein is fused at the C terminus. The arrow indicates that SUMO-specific protease is expected to cleave immediately after two highly conserved Gly residues (Figure 3). FLC is present only to increase the size difference between the precursor and the mature protein.
(B) Cleavage of HIS:AtSUMO1:FLC by HIS:ESD4 in vitro as analyzed on a protein gel blot probed with antibody against the HIS epitope tag. The gel at left shows the results of incubating HIS:ESD4 with either HIS:AtSUMO1:FLC or HIS:AtSUMO1 for 0 or 30 min at 37°C. The bottom, unlabeled arrow indicates the novel product of the reactions incubated for 30 min and is the size expected for mature HIS:AtSUMO1. The gel at right shows the effects of incubating HIS:ESD4 with HIS:AtSUMO1:FLC for 30 min at 37°C in the presence of the ubiquitin inhibitor aldehyde (Ald) or 5 mM N-ethylmaleimide (NEM). N-Ethylmaleimide prevents the appearance of mature SUMO and therefore inhibits the reaction, whereas aldehyde does not.
(C) Effect of amino acid substitutions in HIS:ESD4 or HIS:AtSUMO1:FLC. Lanes 1 and 2 show both wild-type proteins incubated for 0 or 30 min. Lane 3, HIS:ESD4(C488S) incubated with HIS:AtSUMO1:FLC for 30 min. Lanes 4 and 5, HIS:AtSUMO1:FLC(G92A G93A) incubated with HIS:ESD4 for 0 or 30 min. Both mutations prevent the production of mature HIS:AtSUMO.
(D) SUMO conjugate patterns in wild-type, esd4, and transgenic Arabidopsis plants. From left to right: Ler, esd4-1, transgenic esd4-1 overexpressing mature AtSUMO1, and transgenic Ler overexpressing mature AtSUMO1. Total proteins were transferred to a filter and probed with AtSUMO1 antibody. Similar results were obtained from plants overexpressing mature AtSUMO2 or mature AtSUMO3 and from plants overexpressing the precursors of AtSUMO1, AtSUMO2, or AtSUMO3.
Previously, SUMO C-terminal hydrolase was distinguished from deubiquitinating enzymes based on their differential sensitivity to protease inhibitors. The sensitivity of HIS:ESD4 to these inhibitors was tested by assessing their effect on the in vitro cleavage of HIS:AtSUMO1:FLC. The activity of HIS:ESD4 was blocked by a thiol reagent (Cys protease inhibitor) and by N-ethylmaleimide (Figure 4B), which inhibited the activity of yeast ULP1 (Li and Hochstrasser, 1999), but not by ubiquitin aldehyde, which is a specific inhibitor of deubiquitinating protease (Figure 4B). Similarly, pepstatin A, an Asp protease inhibitor, and benzamidine HCl, a trypsin protease inhibitor, did not affect HIS:ESD4 activity (data not shown). Thus, ESD4 is a Cys protease with endopeptidase activity similar to that of yeast ULP1 (Li and Hochstrasser, 1999). Cleavage of HIS:AtSUMO2:FLC and HIS:AtSUMO3:FLC by HIS:ESD4 also occurred in vitro, so under these conditions, ESD4 was not specific to a particular AtSUMO isoform.
To test the specificity of this reaction further, mutant forms of HIS:ESD4 and HIS:AtSUMO:FLC were generated and tested in reactions similar to those described above. ESD4 contains all four residues identified as being conserved between adenovirus protease and ULP1-related enzymes and predicted to be essential for protease activity (Figure 3A) (Li and Hochstrasser, 1999). To determine whether the conserved Cys residue (Cys-488) is required for the observed HIS:ESD4 activity in vitro, it was converted to a Ser. The HIS:ESD4(C488S) protein was purified from E. coli extracts and tested for its ability to cleave the HIS:AtSUMO1:FLC substrate. The mutant protein did not show SUMO C-terminal hydrolase activity, although the protein was detected using anti-HIS antibody, indicating that this Cys is essential for ESD4 activity and that the cleavage of HIS:AtSUMO1:FLC is not caused by another protease present in the reaction (Figure 4C). In another experiment designed to test the specificity of this reaction, a mutation was generated within the HIS:AtSUMO1:FLC. The C-terminal hydrolase activity is predicted to cleave SUMO immediately after the conserved Gly residues (residues 92 and 93 in AtSUMO1) close to the C terminus, which, as illustrated in the crystal structure of the ULP1:SUMO complex, are recognized by conserved amino acids within the active site of the protease (Mossessova and Lima, 2000). To determine whether these Gly residues are required for the cleavage of HIS:AtSUMO1:FLC observed in vitro, they were converted to Ala. No cleavage of HIS:AtSUMO1 (G92A G93A):FLC was detected in vitro (Figure 4C). Therefore, cleavage of HIS:AtSUMO1:FLC in vitro requires the Gly residues shown previously to be essential for SUMO maturation, suggesting that it occurs precisely as predicted for the C-terminal hydrolysis of SUMO. This conclusion also is supported by the size of the cleaved product, which is slightly smaller than the HIS:AtSUMO1 precursor synthesized in E. coli (Figure 4B).
Free SUMO Levels Are Reduced and the Abundance of SUMO Conjugates Is Increased in esd4 Mutants
Whether ESD4 affects SUMOylation in vivo was tested using rabbit polyclonal antibodies raised against HIS:AtSUMO1 and HIS:AtSUMO3. As a result of the high degree of identity between these proteins (Figure 2), both antibodies recognized all three SUMO isoforms made in E. coli. In protein extracts of wild-type plants, a protein of the size expected for AtSUMO was detected with the anti-AtSUMO1 antibody. In addition, several larger proteins that may correspond to conjugates between AtSUMO and target proteins were detected. In esd4-1 extracts, putative SUMO conjugates were increased dramatically in abundance compared with wild-type plants (Figure 4D). Furthermore, the abundance of free SUMO was reduced in esd4-1 mutant plants compared with that in the wild type (Figure 4D). These results support a role for ESD4 in SUMO metabolism in vivo and suggest that its major function is to recycle AtSUMO from conjugates, causing an accumulation of conjugated AtSUMO and a reduced abundance of the free form. The involvement of a SUMO protease predominantly in recycling SUMO from conjugates was proposed for ULP2 of yeast (Schwienhorst et al., 2000).
Effect of the Overexpression of SUMO or SUMO Precursor from a Transgene in the esd4-1 Mutant Background
Comparisons of SUMO and SUMO conjugate levels in wild-type and esd4-1 mutant plants suggested that the major role of ESD4 in vivo is to recycle SUMO from conjugates. To examine this possibility further, we tested whether overexpression of the mature or precursor forms of SUMO affected the phenotype of esd4-1 mutants. Of particular interest was whether the expression of mature forms of SUMO reduced the severity of the mutant phenotype, as might be expected if the protease acts as an endopeptidase required to generate mature SUMO from the precursor. 35S promoter fusions to AtSUMO1, AtSUMO2, and AtSUMO3 as well as to truncated open reading frames encoding each of the mature forms were constructed (see Methods). All six constructs were introduced into wild-type and esd4-1 mutant plants.
All of these transgenes had similar effects on conjugate profiles and on the phenotypes of the plants (Figures 4D and 5). In the esd4-1 mutant, the expression of mature AtSUMO1 from the 35S promoter increased the abundance of conjugates to an even higher level (Figure 4D), and similar effects were observed with the other five transgenes. By contrast, the abundance of conjugates was not increased in transgenic Ler plants expressing mature AtSUMO1 (Figure 4D) or carrying the other transgenes. The transgenes also had more severe phenotypic effects on esd4-1 mutants than on wild-type plants. The esd4-1 mutants expressing mature AtSUMO1 were dramatically shorter and dwarfed compared with the progenitor mutant (Figure 5), and similar phenotypes were created by the overexpression of the other AtSUMO transgenes. By contrast, overexpression of AtSUMO or the mature form in wild-type plants did not dramatically alter their phenotype (Figure 5), consistent with previous observations (Lois et al., 2003). Analysis of the transgenic plants supported the suggestion that the esd4 phenotype is caused by impaired recycling of SUMO from conjugates and therefore is enhanced by increasing the abundance of these conjugates in transgenic plants overexpressing AtSUMO. Furthermore, the reduced efficiency of the processing of mature SUMO from the precursor is unlikely to contribute greatly to the esd4 phenotype, because expression of the mature form did not reduce the effect of the mutation.
Figure 5.
Overexpression of AtSUMO1 Enhances the Mutant Phenotype of esd4-1 Plants.
(A) Comparison of the phenotypes of Ler (plant A1), esd4-1 (plant A2), transgenic esd4-1 expressing mature AtSUMO1 from the 35S promoter (plant A3), and transgenic Ler expressing mature AtSUMO1 from the 35S promoter (plant A4).
(B) Close-up of transgenic plant A3 shown in (A). The stake = 1 cm. Similar results were obtained with plants overexpressing mature AtSUMO2, mature AtSUMO3, precursor AtSUMO1, precursor AtSUMO2, and precursor AtSUMO3.
ESD4 Is Localized to the Nucleus and Appears Predominantly at the Periphery of the Nucleus
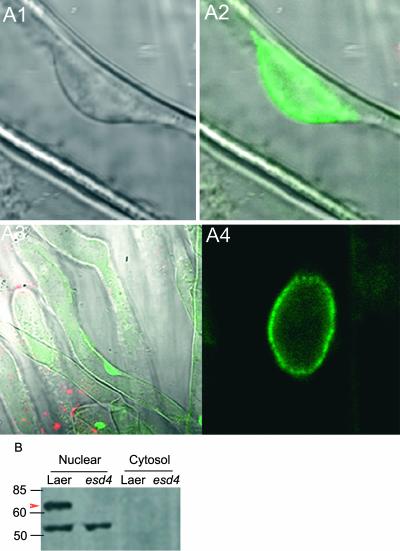
In yeast and animal cells, different isoforms of SUMO proteases differ in their subcellular locations. To test this for ESD4, an ESD4:green fluorescent protein (GFP) fusion protein was expressed in an esd4-1 mutant from the 35S promoter. The 35S::ESD4:GFP transgene fully complemented all aspects of the esd4-1 mutant phenotype. The ESD4:GFP fusion protein was visualized in root cells by confocal microscopy. The protein was detected specifically in the nucleus (Figure 6A). The subnuclear location of the protein was examined at higher magnification in root hair cells and in the root epidermis (Figure 6A). In both cases, the protein was detected at highest abundance at the periphery of the nucleus, apparently in association with the nuclear envelope.
Figure 6.
Subcellular Localization of the ESD4:GFP Fusion Protein.
(A) Confocal microscopy images of the location of ESD4:GFP in esd4-1 mutants carrying the 35S::ESD4:GFP transgene. ESD4:GFP, which complements the esd4-1 mutation, is localized predominantly to the periphery of the nucleus.
(A1) Transmissible light image of the nucleus of a root hair cell.
(A2) The nucleus shown in (A1), with the image collected in the 500- to 530-nm range. The ESD4:GFP signal is located at the periphery of the nucleus.
(A3) The root hair cell shown in (A1) and (A2) at lower magnification, with the image collected in the 500- to 530-nm range. ESD4:GFP is located in the nucleus.
(A4) Nucleus of a root epidermal cell, with the image collected in the 500- to 530-nm range. The ESD4:GFP signal is located at the periphery of the nucleus.
(B) Detection of ESD4 protein in nuclear extracts. Nuclear and cytosolic proteins from 10-day-old seedlings of Ler and esd4-1 were probed with an antibody against ESD4. The arrowhead indicates the candidate ESD4 protein of the expected size (65 kD) that is absent in esd4. The smaller protein is presumed to represent cross-reaction of the antibody.
In addition, an antibody against HIS:ESD4 was raised in rabbits. Protein gel blots were prepared using nuclear protein extracts made from wild-type and esd4-1 seedlings (see Methods). The antibody detected a protein of the expected size for ESD4 (∼65 kD) in nuclear extracts of wild-type plants but not in esd4-1 mutants (Figure 6). This finding further indicated that ESD4 is located in the nucleus.
Expression Pattern of ESD4 and Effects of Its Overexpression on Transgenic Plants
The esd4-1 mutant has a pleiotropic phenotype, suggesting that ESD4 may be required in a variety of plant tissues. ESD4 mRNA expression was examined by RNA gel blot analysis and was present in seedlings, leaves, shoots, flowers, and roots of wild-type plants (Figure 2B). Many flowering-time genes show a diurnal pattern of expression (Mouradov et al., 2002; Simpson and Dean, 2002), so this was tested for ESD4. However, the abundance of the ESD4 mRNA was constant through a 24-h cycle (data not shown), indicating that it did not exhibit a diurnal rhythm in its expression.
Many flowering-time genes identified by loss-of-function mutations also have an effect on flowering when overexpressed (Mouradov et al., 2002); therefore, we examined the effect of overexpressing ESD4 from the 35S promoter of Cauliflower mosaic virus in wild-type Ler plants. Plants carrying the 35S::ESD4 transgene contained increased levels of the ESD4 mRNA (data not shown), but they flowered at the same time as Ler controls under long and short days, suggesting that ESD4 activity is saturated at wild-type levels of expression (data not shown).
DISCUSSION
Mutations in the Arabidopsis ESD4 gene were shown previously to cause extreme early flowering and to affect other aspects of shoot development (Reeves et al., 2002). We show here that ESD4 encodes a protein with similarity to yeast and animal proteases that process the ubiquitin-like protein SUMO. These proteases exhibit both endopeptidase activity that generates the mature form of SUMO from its precursor and isopeptidase activity that recycles SUMO from conjugates with substrate proteins. We have shown that ESD4 is related in function to these proteases and propose that in vivo it acts predominantly as an isopeptidase. This notion is supported by the observations that in esd4 mutants the abundance of free AtSUMO is reduced, the abundance of conjugates is increased, and expression of mature SUMO does not reduce the severity of the esd4 phenotype but rather enhances it. Although genes that encode SUMO and other components of the SUMO system have been identified in the Arabidopsis genome (Vierstra and Callis, 1999; Kurepa et al., 2003; Lois et al., 2003) and plants overexpressing SUMO showed altered responses to abscisic acid (Lois et al., 2003), no mutations that impair the function of these genes and implicate SUMO in particular processes have been described. Our analysis of ESD4 demonstrates the importance of the SUMO system in plant development and indicates particularly that SUMO conjugates regulated by ESD4 play an important role in the control of flowering time.
Comparison of ESD4 with SUMO Proteases of Yeast and Animals
SUMO-specific proteases appear to have distinct functions in yeast (Li and Hochstrasser, 1999, 2000; Schwienhorst et al., 2000). Two yeast genes, ULP1 and ULP2, encode these enzymes, and ULP1 is essential for viability. In vitro, this enzyme shows SUMO endopeptidase activity, which generates mature SUMO from its precursor, and ulp1 mutations can be complemented partially by the expression of mature forms of SUMO, indicating that reduction of endopeptidase activity contributes to the ulp1 phenotype. On the other hand, ulp2 mutants show a slow growth phenotype and accumulate much higher levels of SUMO conjugates, which appear to be responsible for the ulp2 phenotype (Li and Hochstrasser, 2000; Schwienhorst et al., 2000). Mutations in these genes are not simply additive, but the ulp1 mutation partially suppressed the growth defect of ulp2 mutants (Li and Hochstrasser, 2000). This finding suggests that these two proteases differ in the balance of their activities, with ULP1 predominantly showing C-terminal endopeptidase activity and ULP2 acting mainly as an isopeptidase, so that ulp1 suppresses ulp2 by reducing the abundance of mature SUMO available to become incorporated into conjugates. Our data suggest that ESD4 may play a role similar to that of ULP2 in regulating that abundance of SUMO conjugates.
The yeast proteins also differ in their location within the cell. ULP2 is localized throughout the nucleus, whereas ULP1 appears predominantly at the periphery of the nucleus (Li and Hochstrasser, 2000). Localization to the periphery of the nucleus also is shown by the animal SUMO protease SENP2 (Hang and Dasso, 2002; Zhang et al., 2002). Our data suggest that ESD4 may play a role similar to that of ULP2 in regulating the abundance of SUMO conjugates, but its location in the nucleus is more similar to that of SENP2 and ULP1.
There are several genes present in the Arabidopsis genome that encode proteins related to ESD4 and that also may encode SUMO proteases (Figure 3B) (Kurepa et al., 2003). None of these proteins has been tested in vitro for activity against SUMO, and mutations in these genes have not been described, but they may have functions distinct from those of ESD4. For example, they may differ in their balance of C-terminal endopeptidase and isopeptidase functions, as described for ULP1 and ULP2 in yeast, they might not be located with ESD4 in the nucleus but in other intracellular locations, as shown for different isoforms of these enzymes present in yeast cells (Li and Hochstrasser, 2000), or they may be expressed at different times or locations. Genetic and biochemical data similar to those described here for ESD4 will be required to characterize the other Arabidopsis proteins.
The Effect of Modification by SUMO on Protein Function
The identification of ESD4 as a SUMO-specific protease identifies a role for SUMOylation in the control of flowering time and the other aspects of plant development affected by the mutation. No proteins modified by SUMO have been identified definitively in plants. However, SUMO was suggested previously to play a role in phytochrome A signal transduction by modifying the LAF1 (LONG AFTER FAR-RED LIGHT1) protein (Ballesteros et al., 2001). Mutations in LAF1 impair responses to far-red light that are activated by phytochrome A, and the LAF1 protein contains a consensus sequence for the attachment of SUMO, which when altered prevents the formation of characteristic nuclear speckles formed by wild-type LAF1.
An increasing number of substrates for SUMO are known in yeast and human cells (reviewed by Hochstrasser, 2000, 2001). The attachment of SUMO may alter the function of the modified protein in different ways—for example, by altering its affinity for other proteins (reviewed by Hochstrasser, 2000, 2001) or by antagonizing the attachment of ubiquitin and thereby stabilizing the target protein (Hoege et al., 2002).
ESD4 is located at the periphery of the nucleus, as has been shown for components of the SUMO system in yeast and plant cells and for several targets of SUMOylation.
Modification by SUMO has been implicated in nucleocytoplasmic partitioning in animal cells. For example, the GTP binding protein Ran is required for the movement of proteins through nuclear pores (Gorlich and Kutay, 1999), and its activity is stimulated by RAN GTPase-activating protein (RanGAP). Modification of the C terminus of RanGAP by SUMO enables the protein to become attached to the nuclear pore via interaction with a nucleoporin, RanBP2/Nup358 (Matunis et al., 1998), and this protein was shown recently to act as a SUMO E3 ligase that attaches SUMO to RanGAP (Pichler et al., 2002). These observations implicate SUMOylation in nuclear transport in animal cells, although this specific mechanism is unlikely to be involved in plants, because plant RanGAP lacks the C-terminal domain of the animal protein that contains the attachment site for SUMO (Rose and Meier, 2001). Nevertheless, RanGAP is only one link between SUMOylation and nucleocytoplasmic partitioning in animal cells. Another recently described example is the transcriptional repressor TEL, which is present in the nucleus in its unmodified form but exported actively when SUMOylated (Wood et al., 2003). Further understanding of the function of ESD4 and its possible role in these processes will require the identification of the SUMO substrates whose abundances differ between the mutant and the wild type.
The Role of ESD4 in the Control of Flowering Time and Other Aspects of Plant Development
The esd4-1 mutation was recovered initially in a genetic screen for plants that flower much earlier than wild-type plants under short-day conditions (Reeves et al., 2002). Arabidopsis is a facultative long-day plant that flowers much earlier under short days than long days. The esd4-1 mutant exhibits an extreme early-flowering phenotype in short days and flowers only slightly earlier under long days. The original allele was identified in the Ler accession, but identification of esd4-2 demonstrates that ESD4 is similarly important in the control of flowering in the Columbia accession. The early-flowering phenotype is caused in part by reduction in the expression level of the MADS box transcription factor FLC (Reeves et al., 2002), an inhibitor of flowering (Michaels and Amasino, 1999; Sheldon et al., 1999), but it also was proposed to act independently of FLC because its effect on flowering time was more severe than would be expected if it were caused by the partial reduction in FLC expression. Recently, another protein associated with the periphery of the nucleus also was implicated in flowering-time control. Mutations in HASTY alter flowering time and impair or inactivate the Arabidopsis ortholog of mammalian EXPORTIN5, which is involved in nucleocytoplasmic transport (Bollman et al., 2003). Although there may be a relationship between the function of this protein and ESD4, there are distinct differences between the effects of the corresponding mutations on flowering time. For example, although esd4 mutants flower earlier than wild-type plants under both long and short days (Reeves et al., 2002), the hasty mutant flowers later than the wild type under short days.
In addition to its effects on flowering, the esd4 mutation affects plant height, the shape of the silique, and the positioning of cauline leaves on the shoot, suggesting that SUMOylation plays roles in other aspects of plant development and that these also are regulated by ESD4. A wider role for protein modification by SUMO also is suggested by other recent studies in which plants overexpressing AtSUMO were shown to be impaired in their response to the plant growth regulator abscisic acid (Lois et al., 2003) and in which the abundance of proteins conjugated to AtSUMO1 or AtSUMO2 was demonstrated to increase on exposure of the plant to stress (Kurepa et al., 2003).
Analysis of the esd4 mutant suggests that the ESD4 protease regulates the abundance of many SUMO conjugates. Identification of the substrates will allow analysis of whether different aspects of the esd4 phenotype are caused by misregulation of the abundance of a single SUMO conjugate or whether each phenotypic character is caused by misregulation of a different substrate. Furthermore, at present, it is unclear whether particular phenotypes, such as the extreme early-flowering phenotype of esd4, are attributable to the accumulation of SUMOylated conjugates or to the reduced SUMOylation of substrate proteins. Our analysis suggests that the major role of ESD4 in vivo is as an isopeptidase and therefore that target conjugates would accumulate, but this defect also would reduce the pool of free SUMO and consequently may indirectly reduce the abundance of other SUMO conjugates. The identification of SUMOylated proteins whose abundance differs between the mutant and the wild type and analysis of the function of the corresponding genes by reverse genetics will allow these issues to be addressed.
METHODS
Plant Material and Growth Conditions
The Arabidopsis thaliana esd4-1 mutant (Reeves et al., 2002) and the growth conditions used (Reeves and Coupland, 2001) were described previously. The homozygous esd4-2 mutant was in the Columbia ecotype and identified as segregating in T3 seeds provided by the Salk Institute Genomic Analysis Laboratory via the Nottingham Arabidopsis Stock Centre. For in vivo analysis of ESD4 function, plants were grown in cabinets under true long days of 16 h of light with no daylength extension.
Agrobacterium tumefaciens–Mediated Arabidopsis Root Transformation
Cosmids containing DNA from the vicinity of ESD4 were mobilized into Agrobacterium tumefaciens C58C1. The T-DNA was introduced into esd4-1 plants using standard procedures (Valvekens et al., 1988).
RNA Gel Blot Analysis of mRNA Abundance
RNA extraction, RNA gel blot analysis, and hybridization were as described previously (Suarez-Lopez et al., 2001). Hybridization signals were assessed using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Construction of HIS:AtSUMO:FLC and HIS:ESD4 Plasmids
The plasmid pET19b (Novagen, Madison, WI) was used for the expression of His10-recombinant proteins in Escherichia coli BL21. Binary vectors containing fusions of the 35S promoter to the mature forms of AtSUMO1/2/3 were constructed as follows. A GATEWAY Entry clone (Invitrogen, Carlsbad, CA) containing the AtSUMO open reading frame was made by first amplifying a fragment extending from the ATG codon to the end of the codon, including the second Gly residue that defines the cleavage site of SUMO. The primers also were designed to incorporate a stop codon after the Gly and included attB1 and attB2 sequences (Invitrogen), which facilitated recombination into the donor vector via a BP reaction (Invitrogen, Gateway Technology, catalog nos. 11821-014 and 12535-019, version C, www.invitrogen.com). The resulting entry clone was used in an LR reaction with the destination binary vector pAlligator-2 (F. Parcy, Institut des Sciences du Végétal, CNRS, Gif-sur-Yvette, France). This generated a translational fusion between the 3xHA tag and the mature form of AtSUMO at the N terminus of AtSUMO and placed the translational fusion downstream of the 35S promoter. Binary vectors overexpressing the precursor forms of SUMO were made in a similar way, except that the entire open reading frames of SUMO, including the region after the Gly residues, were amplified by PCR.
Purification of Recombinant Proteins
Expression of recombinant proteins was induced by isopropylthio-β-galactoside for 4 h at 28°C. Harvested cells were resuspended in buffer I (250 mM Tris-HCl, pH 8.0, 25% sucrose, and 2 mg/mL lysozyme), diluted in buffer II (300 mM Tris-HCl, pH 8.0, 75 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 2 mM DTT, and PIC [a protein inhibitor cocktail for E. coli; Sigma]), and incubated for 10 min on ice. After brief sonication, cell debris were removed by spinning down at 12,000 rpm for 30 min. The supernatant was filtered, and buffer III (50 mM NaH2PO4, 10 mM Tris, pH 8.0, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 5 mM 2-mercaptoethonol, 5% sucrose, PIC for plants, and 0.05% Tween 20) was changed for buffer II with a desalting column (Econo-Pac 10DG column; Bio-Rad). Purification of HIS:AtSUMO1:FLC, HIS:AtSUMO1 (G92A G93A):FLC, HIS:ESD4, and HIS:ESD4(C448S) was achieved using nickel–nitrilotriacetic acid agarose beads (Qiagen, Valencia, CA) according to the manufacturer's instructions.
In Vitro Analysis of ESD4 Protease Activity
HIS:AtSUMO substrates (500 ng per lane for SDS-PAGE) were incubated with HIS:ESD4 (1 μg per lane for SDS-PAGE) for 30 min at 37°C. Reactions were stopped by adding SDS-PAGE sampling buffer. For the inhibitor study, substrates with inhibitors were placed on ice for 10 min before adding ESD4. The inhibitors used were 1 μM ubiquitin aldehyde (Sigma) and 5 mM N-ethylmaleimide (ICN, Costa Mesa, CA). Standard protocols were used for SDS-PAGE, and proteins were detected by protein gel blot analysis with an antibody against His6 (Pierce).
In Vivo Analysis of ESD4 Protease Activity
Ten-day-old seedling material (0.5 g) was ground in liquid nitrogen. Five hundred microliters of lysis buffer (50 mM Tris-HCl, pH 8.0, 120 mM NaCl, 0.2 mM sodium orthovanadate, 100 mM NaF, 10% glycerol, 0.2% Triton X-100, 5 mM DTT, 1× PIC, and 5 mM N-ethylmaleimide) was added, and samples were shaken end-over-end for 1 h at 4°C. Samples were spun at 20,000g for 20 min at 4°C, and the concentration of proteins in the supernatant was determined. One hundred micrograms of protein was loaded onto an SDS-PAGE gel, and the protein gel blot was probed with the AtSUMO1 antibody using the Supersignal West Pico Chemiluminescent Substrate for HRP system (Pierce).
Localization of ESD4:GFP
35S::ESD4:GFP was constructed using the GATEWAY recombination system (Invitrogen) as follows. An entry clone containing the ESD4 open reading frame was made by first amplifying a fragment extending from the ATG codon to the end of the ESD4 open reading frame but omitting the stop codon. The primers used included attB1 and attB2 sequences (Invitrogen), which facilitated recombination into the donor vector via a BP reaction. The resulting entry clone was used in an LR reaction with the destination binary vector p35S::GW::GFP (F. Turck, Max-Planck-Institut für Züchtungsforschung) to generate the C-terminal fusion of GFP to ESD4. This was used for transformation into esd4-1 mutants to demonstrate that ESD4:GFP complemented the mutant phenotype, and the cells were imaged using a Zeiss LSM 510 Meta confocal laser scanning microscope (Jena, Germany). The stomata guard cells, hypocotyl cells, and root cells of the 10-day-old transgenic 35S::GFP:ESD4 seedlings grown on Murashige and Skoog (1962) agar plates were analyzed. Confocal images were collected using a ×63 or a ×40 oil-immersion lens. GFP fluorescence was excited with a 488-nm argon laser, and images were collected in the 500- to 530-nm range. In all cases, transmissible light images were collected simultaneously.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact George Coupland, coupland@mpiz-koeln.mpg.de.
Accession Numbers
The EMBL Nucleotide Sequence Database accession number for ESD4 is AJ582719. Accession numbers for the sequences shown in Figure 3 are AF149770 (human SENP1), AF194031 (mouse SMT3IP1), NC_001148 (Saccharomyces cerevisiae ULP1), U67122 (human SUMO-1), U33057 (S. cerevisiae SMT3), and AY035148 (Arabidopsis UBIQUITIN1).
Acknowledgments
We thank the Nottingham Arabidopsis Stock Centre (http://nasc.nott.ac.uk) for providing us with seeds developed at the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu). Francois Parcy and Franzsiska Turck kindly provided GATEWAY vectors. We are grateful to Aidyn Mouradov, who performed the confocal microscopy and provided the images, and to Jürgen Dohmen (Universität zu Köln) for many helpful suggestions. Y.-F.F. was supported by China Scholarship Fellowship 98326080 and by a Max Planck Society Fellowship.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.015487.
References
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Aubourg, S., Takvorian, A., Cheron, A., Kreis, M., and Lecharny, A. (1997). Structure, organization and putative function of the genes identified within a 23.9-kb fragment from Arabidopsis thaliana chromosome IV. Gene 199, 241–253. [DOI] [PubMed] [Google Scholar]
- Ballesteros, M.L., Bolle, C., Lois, L.M., Moore, J.M., Vielle-Calzada, J.P., Grossniklaus, U., and Chua, N.H. (2001). LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev. 15, 2613–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan, M., et al. (1998). Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 391, 485–488. [DOI] [PubMed] [Google Scholar]
- Bollman, K.M., Aukerman, M.J., Park, M.-Y., Hunter, C., Berardini, T.Z., and Poethig, R.S. (2003). HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development 130, 1493–1504. [DOI] [PubMed] [Google Scholar]
- Epps, J.L., and Tanda, S. (1998). The Drosophila semushi mutation blocks nuclear import of bicoid during embryogenesis. Curr. Biol. 8, 1277–1280. [DOI] [PubMed] [Google Scholar]
- Gong, L., Millas, S., Maul, G.G., and Yeh, E.T. (2000). Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J. Biol. Chem. 275, 3355–3359. [DOI] [PubMed] [Google Scholar]
- Gorlich, D., and Kutay, U. (1999). Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Hanania, U., Furman-Matarasso, N., Ron, M., and Avni, A. (1999). Isolation of a novel SUMO protein from tomato that suppresses EIX-induced cell death. Plant J. 19, 533–541. [DOI] [PubMed] [Google Scholar]
- Hang, J., and Dasso, M. (2002). Association of the human SUMO-1 protease SENP2 with the nuclear pore. J. Biol. Chem. 277, 19961–19966. [DOI] [PubMed] [Google Scholar]
- Hellmann, H., and Estelle, M. (2002). Plant development: Regulation by protein degradation. Science 297, 793–797. [DOI] [PubMed] [Google Scholar]
- Hershko, A., and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M. (2000). Evolution and function of ubiquitin-like protein-conjugation systems. Nat. Cell Biol. 2, E153–E157. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M. (2001). SP-RING for SUMO: New functions bloom for a ubiquitin-like protein. Cell 107, 5–8. [DOI] [PubMed] [Google Scholar]
- Hoege, C., Pfander, B., Moldovan, G.L., Pyrowolakis, G., and Jentsch, S. (2002). RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141. [DOI] [PubMed] [Google Scholar]
- Katzmann, D.J., Babst, M., and Emr, S.D. (2001). Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106, 145–155. [DOI] [PubMed] [Google Scholar]
- Kurepa, J., Walker, J.M., Smalle, J., Gosink, M.M., Davis, S.J., Durham, T.L., Sung, D.-Y., and Vierstra, R.D. (2003). The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. J. Biol. Chem. 278, 6862–6872. [DOI] [PubMed] [Google Scholar]
- Li, S.J., and Hochstrasser, M. (1999). A new protease required for cell-cycle progression in yeast. Nature 398, 246–251. [DOI] [PubMed] [Google Scholar]
- Li, S.J., and Hochstrasser, M. (2000). The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 20, 2367–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois, L.M., Lima, C.D., and Chua, N.-H. (2003). Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis. Plant Cell 15, 1347–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis, M.J., Wu, J., and Blobel, G. (1998). SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol. 140, 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior, F. (2000). SUMO: Nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16, 591–626. [DOI] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova, E., and Lima, C.D. (2000). Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5, 865–876. [DOI] [PubMed] [Google Scholar]
- Mouradov, A., Cremer, F., and Coupland, G. (2002). Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell 14 (suppl.), S111.–S130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, S., Hoege, C., Pyrowolakis, G., and Jentsch, S. (2001). SUMO, ubiquitin's mysterious cousin. Nat. Rev. Mol. Cell Biol. 2, 202–210. [DOI] [PubMed] [Google Scholar]
- Muller, S., Matunis, M.J., and Dejean, A. (1998). Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 17, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Nishida, T., Kaneko, F., Kitagawa, M., and Yasuda, H. (2001). Characterization of a novel mammalian SUMO-1/Smt3-specific isopeptidase, a homologue of rat axam, which is an axin-binding protein promoting β-catenin degradation. J. Biol. Chem. 276, 39060–39066. [DOI] [PubMed] [Google Scholar]
- Orth, K., Xu, Z., Mudgett, M.B., Bao, Z.Q., Palmer, L.E., Bliska, J.B., Mangel, W.F., Staskawicz, B., and Dixon, J.E. (2000). Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science 290, 1594–1597. [DOI] [PubMed] [Google Scholar]
- Pichler, A., Gast, A., Seeler, J.S., Dejean, A., and Melchior, F. (2002). The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108, 109–120. [DOI] [PubMed] [Google Scholar]
- Reeves, P.H., and Coupland, G. (2001). Analysis of flowering time control in Arabidopsis by comparison of double and triple mutants. Plant Physiol. 126, 1085–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves, P.H., Murtas, G., Dash, S., and Coupland, G. (2002). early in short days 4, a mutation in Arabidopsis that causes early flowering and reduces the mRNA abundance of the floral repressor FLC. Development 129, 5349–5361. [DOI] [PubMed] [Google Scholar]
- Rose, A., and Meier, I. (2001). A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc. Natl. Acad. Sci. USA 98, 15377–15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwienhorst, I., Johnson, E.S., and Dohmen, R.J. (2000). SUMO conjugation and deconjugation. Mol. Gen. Genet. 263, 771–786. [DOI] [PubMed] [Google Scholar]
- Sheldon, C.C., Burn, J.E., Perez, P.P., Metzger, J., Edwards, J.A., Peacock, W.J., and Dennis, E.S. (1999). The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, G.G., and Dean, C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296, 285–289. [DOI] [PubMed] [Google Scholar]
- Sternsdorf, T., Jensen, K., and Will, H. (1997). Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J. Cell Biol. 139, 1621–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Lopez, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120. [DOI] [PubMed] [Google Scholar]
- Valvekens, D., Van Montagu, M., and Van Lijsebettens, M. (1988). Agrobacterium tumefaciens mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 85, 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra, R.D., and Callis, J. (1999). Polypeptide tags, ubiquitous modifiers for plant protein regulation. Plant Mol. Biol. 41, 435–442. [DOI] [PubMed] [Google Scholar]
- Wood, L.D., Irvin, B.J., Nucifora, G., Luce, K.S., and Hiebert, S.W. (2003). Small ubiquitin-like modifier conjugation regulates nuclear export of TEL, a putative tumor suppressor. Proc. Natl. Acad. Sci. USA 100, 3257–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Saitoh, H., and Matunis, M.J. (2002). Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell. Biol. 22, 6498–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]