Abstract
The conditional fluorescent (flu) mutant of Arabidopsis accumulates the photosensitizer protochlorophyllide in the dark. After a dark-to-light shift, the generation of singlet oxygen, a nonradical reactive oxygen species, starts within the first minute of illumination and was shown to be confined to plastids. Immediately after the shift, plants stopped growing and developed necrotic lesions. These early stress responses of the flu mutant do not seem to result merely from physicochemical damage. Peroxidation of chloroplast membrane lipids in these plants started rapidly and led to the transient and selective accumulation of a stereospecific and regiospecific isomer of hydroxyoctadecatrieonic acid, free (13S)-HOTE, that could be attributed almost exclusively to the enzymatic oxidation of linolenic acid. Within the first 15 min of reillumination, distinct sets of genes were activated that were different from those induced by superoxide/hydrogen peroxide. Collectively, these results demonstrate that singlet oxygen does not act primarily as a toxin but rather as a signal that activates several stress-response pathways. Its biological activity in Arabidopsis exhibits a high degree of specificity that seems to be derived from the chemical identity of this reactive oxygen species and/or the intracellular location at which it is generated.
INTRODUCTION
In plants, reactive oxygen species (ROS) are produced continuously as byproducts of various metabolic pathways that are localized in different cellular compartments. Under steady state conditions, these molecules are scavenged by various antioxidative defense mechanisms (Foyer and Noctor, 2000). The equilibrium between the production and the scavenging of ROS may be perturbed by a number of adverse environmental factors. As a result of such disturbances, intracellular levels of ROS may increase rapidly (Elstner, 1991). Precisely how ROS are involved in mediating the plant's responses to these environmental stress conditions is not known. Generally, ROS have been proposed to affect stress responses in two different ways. They react with a large variety of biomolecules, such as lipids, proteins, and nucleic acids, that are essential to maintain the integrity of cellular structures and thus may cause irreversible damage that can lead to tissue necrosis and ultimately may kill the plant (Rebeiz et al., 1988; Girotti, 2001). On the other hand, ROS have been shown to influence the expression of a number of genes and signal transduction pathways. These observations have been interpreted to suggest that cells have evolved strategies to use ROS as biological stimuli and signals that activate and control various genetic stress-response programs (Dalton et al., 1999).
Depending on the quality of the environmental stress, plants differentially enhance the release of ROS that are either chemically distinct or generated within different cellular compartments (Elstner, 1991; Neill et al., 2002). For instance, during an incompatible plant–pathogen interaction, superoxide anions are produced enzymatically outside the cell and are converted rapidly to hydrogen peroxide, which can cross the plasma membrane. The same ROS also are produced in chloroplasts exposed to high light stress, albeit by a different mechanism. The stress reactions of plants induced by pathogens differ from those induced by high light intensities (Bray et al., 2000; Hammond-Kosack and Jones, 2000). If ROS act as signals that evoke these different stress responses, their biological activities should exhibit a high degree of selectivity and specificity that could be derived from their chemical identities and/or the intracellular locations at which they were generated. The main reason why this proposed selective signaling by a given ROS has remained an attractive idea rather than become proven fact is that it is difficult to attribute a particular stress response to a well-defined ROS whose subcellular origin has been determined in planta (Allan and Fluhr, 1997).
Here, we describe a novel experimental strategy that avoids and/or overcomes some of the difficulties that may obstruct the analysis of the biological activity of ROS. It makes use of the flu mutant of Arabidopsis, which makes it possible to induce the release of singlet oxygen in a controlled and noninvasive manner. The FLU protein is a nucleus-encoded chloroplast protein that plays a key role during the negative feedback control of chlorophyll biosynthesis (Meskauskiene et al., 2001). Inactivation of this protein in the flu mutant leads to the overaccumulation of free protochlorophyllide (Pchlide), which may act as a potent photosensitizer. We present experimental evidence to show that after the excitation of Pchlide by light, singlet oxygen is generated in the plastid and is involved in activating distinct groups of early stress-response genes that are different from those activated by superoxide/hydrogen peroxide. Our results do not support the notion that the early stress reactions of the flu mutant are primarily the result of the cytotoxicity of singlet oxygen; instead, they suggest that this ROS leads to the release of a signal(s) that activates the stress responses of the plant.
RESULTS
FLU-Dependent Control of Chlorophyll Synthesis in Light-Adapted Green Plants
We have exploited the ability of the flu mutant of Arabidopsis to accumulate free Pchlide, a potent photosensitizer, in the dark to identify and analyze ROS-controlled stress responses. In angiosperms kept in the dark, the chlorophyll biosynthesis pathway is blocked after the formation of Pchlide, the immediate precursor of chlorophyllide, because the reduction of Pchlide to chlorophyllide requires light. Once a critical level of Pchlide has been reached in dark-grown plants, further synthesis of Mg2+ porphyrins stops. This regulation has been attributed to the feedback control of δ-aminolevulinic acid synthesis (Figure 1A). The flu mutant of Arabidopsis is defective in this metabolic feedback control. As a result of this mutation, etiolated seedlings no longer are able to restrict the accumulation of Pchlide. When such seedlings are transferred to the light, they bleach and die (Meskauskiene et al., 2001). The flu mutant remains viable, however, when it is kept under continuous light. Under these growth conditions, flu plants mature and produce seeds just like wild-type plants, and no obvious differences between the mutant and the wild type can be observed.
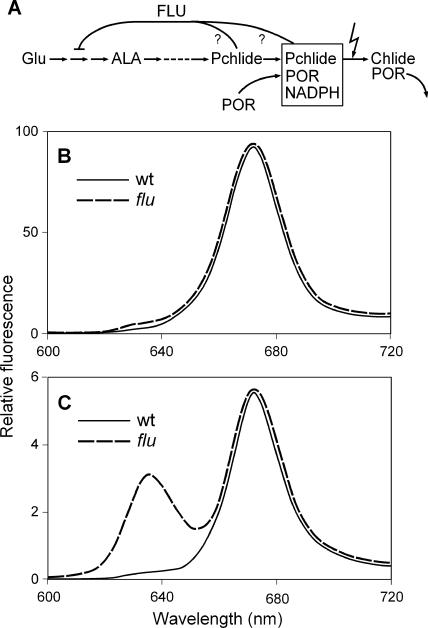
Figure 1.
FLU-Dependent Regulation of Pchlide Synthesis in Arabidopsis.
(A) FLU has been identified as a membrane-bound plastid protein that mediates the feedback control of chlorophyll synthesis (Meskauskiene et al., 2001). As indicated by the question marks, it is not known whether Pchlide acts in its free form or as part of the ternary POR-Pchlide-NADPH complex as an effector of feedback inhibition. ALA, δ-aminolevulinic acid; Chlide, chlorophyllide.
(B) and (C) Pchlide accumulation in dark-adapted, mature wild-type (wt) and flu plants. Plants were grown under continuous light until they were ready to bolt. At this developmental stage, plants were transferred to the dark for the first time. After 8 h, total porphyrins (B) or nonesterified porphyrins (C) were extracted. The fluorescence emission spectra of these samples were recorded using an excitation wavelength of 433 nm.
When wild-type and flu plants were transferred from continuous light to the dark, however, the flu mutant accumulated Pchlide, whereas in wild-type plants, this pigment was not detectable (Figures 1B and 1C). In contrast to etiolated flu seedlings, which contained high levels of Pchlide, mature light-adapted flu plants formed only minor amounts of Pchlide over 8 h in the dark. The accumulation of Pchlide in dark-grown flu mutants was not accompanied by a concomitant increase in the level of the NADPH-Pchlide oxidoreductase protein (POR), which together with Pchlide and NADPH may form a ternary photoactive complex (Sperling et al., 1998). Hence, during dark incubation, Pchlide accumulates in the flu mutant mainly as a free pigment that may act as a potent photosensitizer (Boo et al., 2000).
The Release of Singlet Oxygen in the flu mutant
Upon irradiation, photosensitizing molecules such as Pchlide are capable of generating singlet oxygen by transferring light energy to ground-state (triplet) molecular oxygen, thereby elevating it to the excited singlet state (Gollnick, 1968). The possible release of singlet oxygen in the flu mutant immediately after the onset of illumination was tested in vivo according to Hideg et al. (1998). Leaves from mutant and wild-type plants that had been kept in the dark were infiltrated with dansyl-2,2,5,5-tetramethyl-2,5-dehydro-1H-pyrrole (DanePy). DanePy is a fluorescent compound that after excitation at 345 nm emits fluorescence with a maximum at 532 nm. Singlet oxygen in leaves can be detected via the partial quenching of this fluorescence induced by energy transfer from the fluorophore moiety (dansyl) to the nitroxide (Kalai et al., 1998).
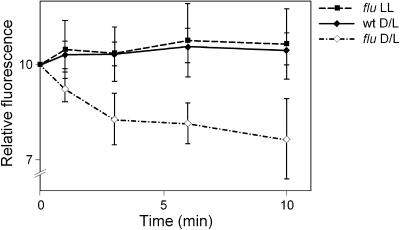
DanePy fluorescence was diminished immediately after illumination in infiltrated leaves from mutant plants that had been kept in the dark for 8 h (Figure 2). Even though during the cutting and subsequent infiltration a wounding stress was imposed on the leaves that has been shown to lead to the generation of hydrogen peroxide (Orozco-Cardenas and Ryan, 1999), the quenching of DanePy fluorescence occurred only in leaves taken from predarkened flu mutants and not from mutants kept in continuous light or from wild-type plants (Figure 2). This result reaffirms the previous finding that DanePy is specific for singlet oxygen (Kalai et al., 1998). The release of hydrogen peroxide was monitored in cut leaves using 3,3′ diaminobenzidine-4HCl (Thordal-Christensen et al., 1997). The formation of hydro- gen peroxide was detected first at 15 min after cutting and continued to increase during the next 15 min, but there was no detectable difference in the staining intensities of cut leaves from flu and wild-type plants (data not shown).
Figure 2.
Release of Singlet Oxygen in the flu Mutant after a Dark/Light Shift.
Wild-type (wt) and flu mutant plants were grown under continuous light until they were ready to bolt. At this stage, plants were transferred to the dark for 8 h (D/L). Cut leaves of these plants were infiltrated with DanePy under green safelight and subsequently illuminated with white light (100 μmol·m−2·s−1). As an additional control, leaves were taken from flu plants that had been kept under continuous illumination (LL). Singlet oxygen trapping was measured as relative quenching of DanePy fluorescence (Hideg et al., 1998).
Stress Responses of the flu Mutant
In animals and human, various stress responses have been attributed to the release of singlet oxygen, which has been proposed to act as a second messenger during exposure to UV-A light and during the treatment of cancers with photodynamic therapy (Klotz, 2002). A signaling role of singlet oxygen in plants also has been discussed (Trebst, 1999), but no stress responses of higher plants have been traced unequivocally back to this ROS, even though singlet oxygen has been shown to be released in photosystem II particles (Macpherson et al., 1993) and in plants under light stress (Hideg et al., 1998; Fryer et al., 2002). Two major stress reactions were observed when dark-grown flu plants were returned to the light: a cell death response and a rapid inhibition of growth. The first signs of cell death in flu plants were detected by trypan blue staining as early as 1 h after the onset of illumination, and visible necrotic lesions began to form 2 to 3 h later. No such stress symptoms were detectable in wild-type control plants subjected to the same dark/light shift (Figure 3A) or in flu mutants kept under continuous light (data not shown). The second stress response was an inhibition of growth of the flu mutant after the dark/light shift, which was revealed by the continuous recording of shoot growth (Figure 3B). The growth rate of flu plants that had started to bolt was reduced almost immediately after the beginning of reillumination and recovered only slowly after ∼12 h of exposure to light. This growth inhibition response was particularly striking when the plants were transferred from continuous illumination to repeated cycles of 8 h of dark and 16 h of light. Whereas flu plants kept under constant light continued to grow like wild-type plants, flu plants exposed to these light/dark cycles stopped growing (Figure 3C). This growth inhibition was reversible: after plants were returned to continuous light, they resumed their normal growth activity (Figure 3D).
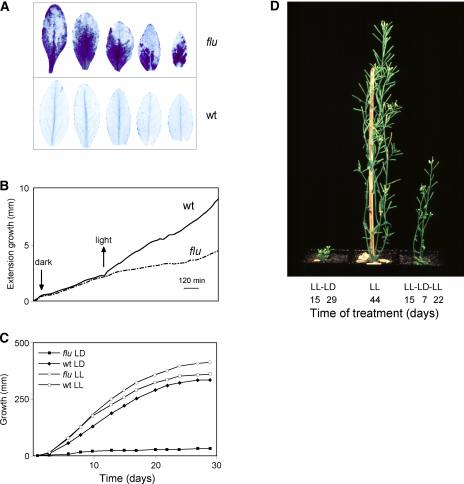
Figure 3.
Stress Reactions of flu Plants after a Dark/Light Shift.
Plants were grown under continuous light until they reached the rosette leaf stage. Then, they were shifted to the dark and reexposed to light after 8 h. Two major stress responses were induced during reillumination: a cell death reaction in rosette leaves of flu (A) and inhibition of plant growth ([B] to [D]).
(A) Twenty-four hours after the beginning of reillumination, rosette leaves of flu and wild-type (wt) plants were cut and stained with trypan blue. The first traces of trypan blue staining of rosette leaves were detectable 1 h after the dark/light shift.
(B) Growth of plants was recorded continuously before and after the dark/light shift. The growth rate of wild-type plants increased during reillumination, whereas in flu plants, growth was inhibited for 12 h, beginning soon after the start of reillumination. After 12 h, the growth rate gradually increased again.
(C) flu plants kept under a dark/light (LD) cycle for several days ceased to grow. Under continuous light (LL), however, the plants grew like wild-type plants.
(D) The light-induced growth inhibition of flu plants was reversible. flu plants resumed growth when they were transferred from the 16-h-light/8-h-dark program to continuous light.
The Intracellular Site of Singlet Oxygen Production
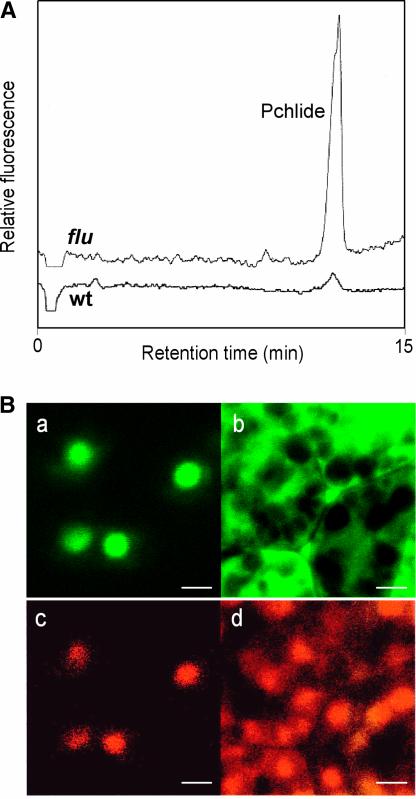
To understand these biological effects, it was important to identify and localize within the cell the tetrapyrrole intermediate(s) that accumulates in the dark and acts as a photosensitizer(s) during the subsequent illumination of the flu mutant. First, tetrapyrroles in the flu mutant and in wild-type controls were extracted and separated by HPLC. Pchlide was the only tetrapyrrole that was accumulated differentially: its concentration in the flu mutant exceeded that in the wild-type control by a factor of 15 (Figure 4A), whereas the concentrations of other intermediates—such as protoporphyrin IX, Mg2+ protoporphyrin IX, and Mg2+ protoporphyrin monomethylester—that also could act as photosensitizers were the same in mutant and wild-type plants (data not shown). The biosynthesis of chlorophyll has been shown to be confined to the plastid compartment (Gomez-Silva et al., 1985). However, intermediates of chlorophyll such as protoporphyrinogen IX (Moeller et al., 2001), and other tetrapyrroles such as phytochromobilin (Terry and Lagarias, 1991), have been shown to be translocated from the plastid to the surrounding cytoplasm.
Figure 4.
Intracellular Localization of the Pchlide That Accumulates in the flu Mutant.
(A) Tetrapyrroles were extracted from etiolated seedlings of wild-type (wt) and flu plants, separated by HPLC, and detected by their fluorescence.
(B) Colocalization of the PORA-GFP fusion protein (a) and free Pchlide (c) in transgenic flu plants. Pchlide was not detected in the cytoplasm outside of the plastids, as indicated by the distribution of the GFP without the PORA signal sequence attached to it ([b] and [d]). Bars = 5 μm.
In mature flu plants grown under continuous light, it was not possible to localize the Pchlide because of the large excess of chlorophyll present, which interfered with the detection of Pchlide. Therefore, we analyzed the distribution of Pchlide in etiolated seedlings of flu, which are devoid of chlorophyll and accumulate much higher levels of Pchlide than wild-type seedlings. Pchlide could be detected with the confocal microscope by its bright red fluorescence. The pigment was confined strictly to the plastid compartment and accumulated in distinct areas (Figure 4Bc). These areas could be identified as prolamellar bodies in transgenic flu plants that expressed the precursor of PORA fused to the green fluorescent protein (GFP) (Figure 4Ba). PORA has been shown to be a major protein constituent of prolamellar bodies (Sperling et al., 1998). When the fluorescence images of Pchlide and PORA-GFP fusion protein distributions were compared, the two components colocalized in the same area of the plastid compartments (Figures 4Ba and 4Bc). GFP without a plastid signal peptide attached to it was excluded from the plastids and accumulated within the cytosol (Figures 4Bb and 4Bd).
Enzymatic Peroxidation of Linolenic Acid in the flu Mutant
The development of stress symptoms in the flu mutant after the dark/light shift could be attributable to the cytotoxicity of singlet oxygen or might reflect its more indirect and subtle role as a stress signal (Girotti, 2001; Klotz, 2002). One way to monitor the cytotoxicity of singlet oxygen is to measure the oxidation of polyunsaturated fatty acids. The photosensitizer Pchlide that gives rise to singlet oxygen upon illumination accumulates within the internal membranes of plastids. Singlet oxygen has a short half-life of ∼200 ns in cells (Gorman and Rodgers, 1992), and the distance over which it may interact with other molecules has been calculated to be up to 10 nm (Sies and Menck, 1992). Hence, molecules that are affected by singlet oxygen would be expected to be localized in or close to the plastid membrane. Linolenic acid has been shown to be a preferred target of ROS during lipid peroxidation. It is the most prominent polyunsaturated fatty acid in chloroplast membrane lipids, accounting for up to 80% of total fatty acids (Murakami et al., 2000).
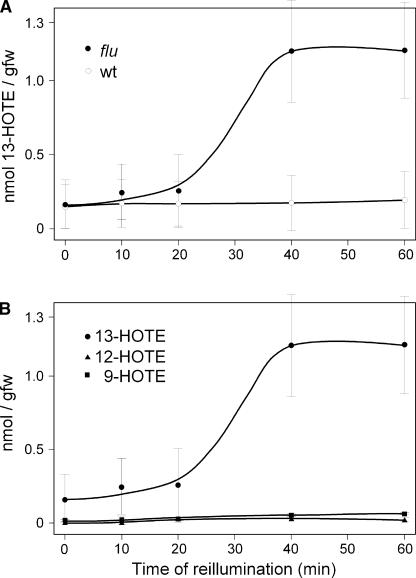
The peroxidation of lipids in flu plants after a dark/light shift was monitored by measuring changes in the amount of the oxygenation products of linolenic acid, hydroperoxy octadecatrieonic acid (HPOTE) and hydroxy octadecatrieonic acid (HOTE), during the first 60 min of illumination. In Arabidopsis, HPOTE and HOTE were present in free and esterified forms. Bound hydroxyperoxides and hydroxides of linolenic acid did not increase dramatically during reillumination, and after 60 min, the levels were only slightly higher than in wild-type controls (data not shown). Free hydroperoxides of linolenic acid represented only a minor fraction of <1% of total oxidized fatty acids in both flu and wild-type plants before reillumination. By 20 min after the beginning of reillumination, the free regiospecific isomer 13-HOTE had begun to accumulate rapidly in flu plants and reached its maximum level after 30 to 60 min of illumination, exceeding that of the wild-type control by ∼10- to 20-fold (Figure 5A). The concentration of the free form of other regioisomers, 9-HOTE and 12-HOTE, remained at a similarly low level throughout reillumination in both wild-type and flu plants (Figure 5B). During reillumination, the concentration of esterified and soluble linolenic acid per gram fresh weight did not change significantly, and levels were comparable in wild-type and flu mutants (data not shown).
Figure 5.
Changes in the Concentrations of the Free Hydroperoxides of Linolenic Acid (HOTE) after a Dark/Light Shift.
(A) Changes in the concentration of the regiospecific isomer 13-HOTE during the first 60 min of reillumination of flu and wild-type (wt) plants. 13-HOTE was separated and identified as described in Methods.
(B) Changes in the concentrations of the three regiospecific isomers 13-HOTE, 12-HOTE, and 9-HOTE during the first 60 min of reillumination of flu.
Each time point represents the mean value ± sd of three independent experiments, and individual time points from each experiment represent average values of 10 plants each. gfw, grams fresh weight.
In flu, HOTE may be generated enzymatically or nonenzymatically by the direct interaction of linolenic acid with singlet oxygen. The enzymatic peroxidation of linolenic acid is catalyzed mainly by lipoxygenases (LOX), leading to different hydroxy derivatives resulting from regiospecific and stereospecific oxygenation. The origin of the HOTE formed in the flu cells could be deduced from its enantiomer composition (Berger et al., 2001). The 13-HOTE that accumulated rapidly during the first 60 min of illumination showed a clear preponderance of the S-isomer over the R-isomer, with the S-enantiomer accounting for >95% of the total (data not shown). The composition of the positional isomers also was consistent with an enzymatic origin for most of the HOTE in the flu mutant. 12-HOTE has been suggested to be a specific marker for nonenzymatic lipid peroxidation (Berger et al., 2001), because no lipoxygenase isoform is known to be able to catalyze the formation of this isomer. Within the fraction of esterified oxylipins, only trace amounts of 12-HOTE were present in both wild-type and flu cells. Furthermore, its level did not increase selectively during the reillumination of flu. Free 12-HOTE was barely detectable: its concentration changed only slightly during reillumination and was similar to the levels in wild-type plants (Figure 5B). Together, these data show that the rapid increase in free HOTE during the reillumination of flu plants kept in the dark for 8 h is attributable almost exclusively to the enzymatic oxygenation of linolenic acid and thus cannot be explained by the cytotoxicity of singlet oxygen.
The Selectivity of Gene Activation in flu Plants after the Dark/Light Shift
After the absorption of photons, photosensitizers such as Pchlide react with molecular oxygen, usually by energy transfer, to generate singlet oxygen. However, a small fraction of superoxide anion radicals may be produced by an electron-transfer reaction (Foote, 1991). The superoxide anion radical can form hydrogen peroxide by spontaneous or enzymatic dismutation. Hence, stress reactions of the flu mutant after the dark/light shift also may be affected by superoxide/hydrogen peroxide. It was not possible to quantitate superoxide/hydrogen peroxide concentrations directly in intact plants. Therefore, an indirect approach was used to estimate the possible contribution of superoxide/hydrogen peroxide to the activation of stress responses in the flu mutant.
The expression of several genes was upregulated rapidly in the flu mutant after the dark/light shift. Some of these genes were identified initially by isolating differentially displayed cDNAs derived from flu plants 20 and 60 min after the beginning of reillumination. They included a drought-induced-like protein (DIL) gene (At4g02200) and a putative c2h2 zinc finger transcription factor (ZP) gene (At5g04340). Changes in the expression of these two genes after the dark/light shift were compared with those that occurred after paraquat treatment of flu plants that had been kept under continuous light. The herbicide paraquat acts as a terminal oxidant of photosystem I, and in the light it reduces oxygen to the superoxide radical, which subsequently dismutates to hydrogen peroxide (Mehler, 1951). The expression of the ascorbate peroxidase I (APXI) gene (At1g07890), which is known to be upregulated by hydrogen peroxide during paraquat treatment and under high-light conditions (Karpinski et al., 1997; Storozhenko et al., 1998), also was measured. The expression of the DIL and ZP genes was upregulated rapidly in flu mutants within the first 30 min of reillumination but not in the paraquat-treated flu plants. Conversely, the expression of the APXI gene was activated in paraquat-treated flu plants under continuous illumination, whereas the expression of this gene was not affected after a dark/light shift (data not shown).
These results suggest that the expression of APXI and DIL and ZP is activated selectively after the release of superoxide/hydrogen peroxide and singlet oxygen, respectively. The expression of APXI, DIL, and ZP was used to define more precisely the light conditions during the reillumination of predarkened flu plants that would exclude the activation of superoxide/hydrogen peroxide–dependent genes and minimize the possible cytotoxicity of singlet oxygen that could be expected to lead to a more general pleiotropic stress response, including secondary effects triggered by other reactive oxygen species. As a result of this optimization, the light intensity during the reillumination of flu plants was kept at 80 to 100 μmol·m−2·s−1.
Under these light conditions, the total number of genes activated differentially after the release of singlet oxygen or superoxide/hydrogen peroxide was assessed by comparing global changes in the gene expression of flu plants subjected to a dark/light shift with those of paraquat-treated flu plants. Total RNAs were prepared at various times after the beginning of reillumination or paraquat treatment. These RNAs were first transcribed into cDNAs and then into biotinylated complementary RNAs that were hybridized to Affymetrix gene chips. These chips contained >22,500 probe sets representing ∼24,000 genes or >95% of the total genome of Arabidopsis. The transcript levels were expressed relative to those in the wild type exposed to a dark/light shift and in flu plants that were sprayed with 0.1% Tween. The gene expression profiles were compared by means of cluster analysis in groups of genes with similar patterns of expression.
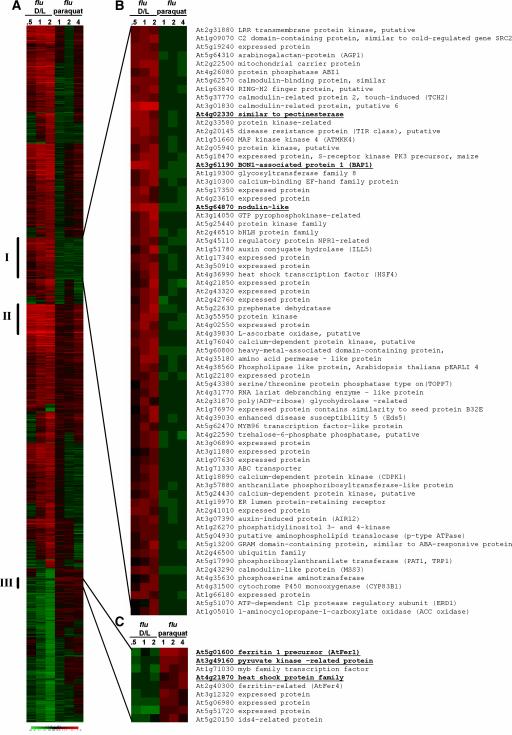
To this end, genes with a 2.5-fold or greater differential expression that were either upregulated or downregulated in at least one experimental condition were selected and analyzed. A total of 1206 genes belonging to this group were identified (Figure 6A; see also supplemental data online). The expression of 70 of these genes was upregulated by 2.5-fold or more in flu during the first 120 min of reillumination but not during the first 4 h of paraquat treatment (Figure 6B, group I; see also supplemental data online). On the other hand, nine genes whose transcript levels were not upregulated in the flu mutant during the first 120 min of reillumination were activated at least 2.5-fold in paraquat-treated flu plants (Figure 6C, group III; see also supplemental data online). A third subcluster (group II) consisted of 31 genes that were upregulated 2.5-fold or more under both experimental conditions (Figure 6A; see also supplemental data online).
Figure 6.
Cluster Analysis of the Expression of Early Stress-Response Genes in the flu Mutant of Arabidopsis after the Release of Singlet Oxygen and Hydrogen Peroxide.
RNA samples were prepared 0.5, 1, and 2 h after the transfer of plants from the dark to the light (flu D/L versus wild-type D/L) and 1, 2, and 4 h after spraying plants with paraquat (flu treated with 20 μM paraquat in 0.1% Tween versus flu treated with 0.1% Tween). Affymetrix gene chips were used that represent ∼24,000 genes. A total of 1206 genes with 2.5-fold or greater differential expression that were either upregulated or downregulated in at least one experimental condition were selected and analyzed (A). One subcluster (group I) represents 70 genes that are upregulated specifically after the release of singlet oxygen, whereas a second subcluster (group III) represents 9 genes whose transcript levels increase selectively after the release of hydrogen peroxide. The members of each of these two groups are shown in (B) and (C), respectively. A third subcluster (group II [A]) consists of 31 genes that are upregulated at least 2.5-fold under both conditions. Each horizontal line displays expression changes for one gene. Degrees of red and green indicate the extent of positive and negative regulation. Gene accession numbers are shown at right of each cluster in (B) and (C). Numbers shown in boldface and underlined indicate genes that were selected for an independent second analysis of mRNA changes by real-time PCR (see Figure 7). Expression data not shown are accessible in the supplemental data online.
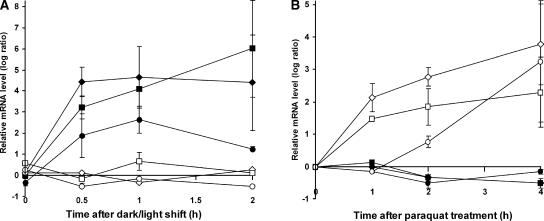
Among the genes that had been implicated by this Affymetrix chip analysis as being induced selectively by either singlet oxygen or superoxide/hydrogen peroxide, three of each group were selected and changes in their transcript levels were quantified independently using real-time PCR to test the reliability of the Affymetrix gene chip analysis (Figure 7). The expression of genes that encode a protein similar to pectinesterase (At4g02330), a nodulin-like protein (At5g64870), and the Bonzai1-associated protein BAP1 (At3g61190) (Hua et al., 2001) was upregulated rapidly in flu mutants within the first 30 to 60 min of reillumination but not in paraquat-treated flu plants (Figure 7). Conversely, the expression of genes that encode ferritin1 (At5g01600) (Petit et al., 2001), a pyruvate kinase–like protein (At3g49160), and a heat-shock protein17–like protein (At4g21870) was activated in paraquat-treated flu plants under continuous illumination but not after a dark/light shift (Figure 7). The apparent lack of activation of superoxide/hydrogen peroxide–specific genes in the flu mutant after a dark/light shift suggests that the concentration of superoxide/hydrogen peroxide in these plants is too low to affect the mutant's early stress responses.
Figure 7.
Activation of Six Selected Early Response Genes in the flu Mutant of Arabidopsis after a Dark/Light Shift or during Paraquat Treatment.
Three of the genes identified by cluster analysis as being upregulated selectively after a dark/light shift (At3g61190, Bonsai-associated protein BAP1 [closed diamonds]; At5g64870, nodulin-like protein [closed squares]; At4g02230, protein similar to pectinesterase [closed circles]) and three of the genes whose transcript levels were shown to increase during paraquat treatment (At5g01600, ferritin1 [open diamonds]; At3g49160, pyruvate kinase–like protein [open squares]; At4g21870, heat-shock protein17–like protein [open circles]) were selected for an independent determination of their transcript levels by real-time quantitative PCR. Results shown are mean values ± sd of four measurements from two independent experiments.
(A) Relative expression levels of these six genes in flu versus wild-type plants after a dark/light shift.
(B) Relative expression levels of these genes in paraquat-treated versus untreated flu plants. These plants were grown in continuous light and were sprayed at the rosette leaf stage either with 20 μM paraquat in 0.1% Tween or with 0.1% Tween alone (untreated plants).
DISCUSSION
Possible Modes of Action of ROS in Plants
Even though most studies agree on the importance of ROS as a mediator of stress defense reactions in plants, few details are known regarding how the increase of ROS concentration is perceived and how this information is translated into signals that direct the plant's response to stress. There are several reasons for this lack of knowledge. In many cases, it is not clear whether the biological activity of ROS is the result of its cytotoxic effect or reflects the more indirect role of a second messenger that signals further cellular responses, or both (Dangl et al., 1996). The term “ROS” embraces chemically distinct oxygen derivatives that may be produced selectively by a particular cellular compartment in response to a specific environmental cue. It is not known to what extent the chemical specificity of these ROS species and the cellular topography of their release may contribute to the multiplicity of stress responses in plants. To date, studies of the biological activities of ROS in plants have been restricted mainly to superoxide and hydrogen peroxide, which are released at the surface of plasma membranes and that are involved in activating defenses against pathogens (Hammond-Kosack and Jones, 2000). Much less is known about the physiological roles of ROS that either are generated in other cellular compartments or are chemically distinct (Foyer and Noctor, 2000). Here, we used the conditional flu mutant of Arabidopsis that releases the nonradical ROS singlet oxygen within the plastid compartment to answer some of these questions.
In animals and human, the biological activity of singlet oxygen has been well documented, and a large number of stress reactions have been described that are closely associated with the release of singlet oxygen, particularly upon exposure to UV-A light and during photodynamic therapy (Klotz, 2002). In plants, singlet oxygen is produced, especially at higher light intensities, in chloroplasts when the absorption of light energy exceeds the capacity for photosynthesis (Nyogi, 1999; Fryer et al., 2002; Hideg et al., 2002). Excited triplet chlorophyll molecules in photosystem II interact with O2 to generate singlet oxygen (Macpherson et al., 1993). At the same time, hyperreduction of the photosynthetic electron carrier chain favors the direct reduction of O2 by photosystem I and the subsequent production of superoxide, hydrogen peroxide, and the hydroxyl radical (Foyer and Noctor, 2000). Many environmental stress conditions limit the ability of a plant to use light energy, so that excessive excitation of the photosystems and enhanced generation of ROS can occur even at moderate light intensities, often causing a reduction in the growth and productivity of plants (Long et al., 1994). Singlet oxygen has been implicated as a mediator of these stress reactions (Trebst, 1999), but its proposed role could not be verified experimentally.
Stress Reactions of the flu Mutant after a Dark/Light Shift
During the reillumination of flu mutants, two major visible stress reactions of the plants could be distinguished: a cell death response and an inhibition of growth. Cell death reactions resulting from oxidative stress have been described frequently and have been associated with the release of ROS (Pourzand and Tyrrell, 1999). The intensity and quality of such cell death reactions can range from necrotic reactions resulting from severe photooxidative damage, which may lead ultimately to the death of the organism, to the tightly controlled hypersensitive reaction, which is restricted locally and is used by a large number of plants as an important element of their defenses against pathogens (Hammond-Kosack and Jones, 2000). In the flu mutant, the contribution of a genetically controlled cell death program to the overall cell death response in leaves may vary depending on the light intensity to which predarkened flu plants have been exposed. Several second-site mutants of flu have been identified that generate similar amounts of singlet oxygen as flu upon reillumination (D. Wagner, D. Przybyla, C. Laloi, and V. Apel, unpublished data). At the light intensity of 100 μmol·m−2·s−1 that was used in most of the experiments, lesion formation in these mutants was suppressed during reillumination. These second-site mutations define a genetic cell death program that is activated in flu after the release of singlet oxygen and that displays some of the hallmarks of programmed cell death described for other organisms, such as DNA fragmentation and its dependence on phytohormones and light (A. Danon and K. Apel, unpublished data).
The second response of flu, inhibition of growth, resembles a stress tolerance strategy used by plants that are exposed to drought, heat, light, or cold stress (Netting, 2000). Under these adverse environmental conditions, plants may pass into a state of minimal metabolic activity that persists until the stress is relieved. Growth inhibition in flu occurs almost instantaneously after a dark/light shift. This response does not seem to result from a gradual reduction in metabolic activity; rather, it is linked more directly to the rapid release of ROS during reillumination. It is conceivable that the release of singlet oxygen in flu is associated closely with the redox control of cell proliferation and the ROS-induced delay of mitotic divisions that has been described for plants and mammalian cells (Wiese et al., 1995; Reichheld et al., 1999).
Selectivity of Singlet Oxygen–Mediated Changes in the Expression of Early Stress-Response Genes of the flu Mutant after a Dark/Light Shift
In chloroplasts of plants under high-light stress, not only singlet oxygen but also superoxide and hydrogen peroxide are generated (Fryer et al., 2002). These latter two ROS, however, do not seem to interfere with the early stress responses of flu after a dark/light shift. During reillumination of the flu mutant, the light intensity was kept at 80 to 100 μmol·m−2·s−1, which is <5% of what has been used in previous studies of high-light stress (Dunaeva and Adamska, 2001). The expression of the DIL and ZP genes was upregulated rapidly, reaching a maximum within the first 60 to 120 min of reillumination, whereas the expression of the APXI gene, which had been shown previously to be activated by hydrogen peroxide during paraquat treatment and under high-light conditions (Karpinski et al., 1997; Storozhenko et al., 1998), did not change. During this early phase of reillumination, no visible lesions were detected in rosette leaves of the mutant. By contrast, in leaves exposed to 600 μmol·m−2·s−1, extensive necrotic lesions appeared within the first 60 to 90 min of illumination, and not only DIL and ZP but also APXI transcripts began to accumulate.
In flu plants kept under continuous light, the expression of APXI, but not of ZP or DIL, was induced after these plants were sprayed with a 20-μM paraquat solution. During the subsequent 12 h, these plants showed no necrotic spots on their leaves. However, in flu plants treated with a 25-fold higher paraquat concentration, extensive lesion formation occurred rapidly. APXI transcripts accumulated, and within the first 4 h of incubation they reached levels threefold higher than in plants treated with the 20-μM paraquat solution. At this higher paraquat concentration, the levels of DIL and ZP transcripts also began to increase. Collectively, these results suggest that at a light intensity of 100 μmol·m−2·s−1, the expression of DIL and ZP is activated selectively after the release of singlet oxygen. However, under more severe stress conditions, such as higher light intensity during the dark/light shift-experiment or a higher paraquat concentration, this selectivity of gene activation is partially lost and the formation of necrotic lesions within the rosette leaves is enhanced drastically.
The selectivity of singlet oxygen–mediated changes in the expression of early stress-response genes was confirmed by comparing global changes in the gene expression of flu plants subjected to a dark/light shift under the lower light intensity with those of flu plants kept under continuous light and treated with the 20-μM paraquat solution. Among 1206 genes with a 2.5-fold or greater differential expression that were either upregulated or downregulated in at least one experimental condition relative to the controls, 70 were found to be upregulated at least 2.5-fold in flu during the first 30 to 120 min of reillumination but not during the first 4 h of paraquat treatment. Genes were identified as being activated specifically by singlet oxygen only if their expression levels in paraquat-treated plants were equal to or lower than that in mock-sprayed control plants. Genes whose expression levels in paraquat-treated plants exceeded that of the control plants were excluded from the group of singlet oxygen–specific genes. For example, DIL and ZP, both of which had been shown by quantitative PCR to be upregulated rapidly and selectively after the release of singlet oxygen, were not found within the subcluster of singlet oxygen–specific genes. The ratios of expression levels of these two genes in paraquat-treated plants versus control plants were slightly greater than 1 (1.4 and 1.5, respectively, after 4 h of incubation). Because of the stringency of the criteria used for the classification of genes, the number of genes that are activated selectively after the release of singlet oxygen probably is much greater than is indicated by the number of genes assigned to group I.
It is reassuring that genes in group III that had been found previously to be activated by superoxide/hydrogen peroxide were shown here to be unaffected by singlet oxygen. For instance, the ferritin1 gene was upregulated rapidly during paraquat treatment but did not respond to the release of singlet oxygen. Ferritin1 is found in chloroplasts (Briat and Lobréaux, 1997; Petit et al., 2001). After an increase in the intracellular concentration of hydrogen peroxide, it plays an important role in keeping Fe2+ levels at a minimum, prohibiting the reduction of hydrogen peroxide that leads to hydroxyl radicals, one of the most noxious radicals, which causes severe oxidative damage in plants. The apparent lack of activation of this and other known superoxide/hydrogen peroxide–specific genes in flu strongly supports the notion that superoxide/ hydrogen peroxide levels were too low to influence the mutant's stress responses after a dark/light shift. This conclusion also was supported by our finding that in flu only the 13LOX but not the 9LOX pathway was activated rapidly after the dark/light shift. The 9LOX pathway had been reported to be upregulated during the hypersensitive response (Rusterucci et al., 1999), a plant defense reaction triggered by hydrogen peroxide (Alvarez et al., 1998) that did not affect the 13LOX pathway.
It is remarkable that during the paraquat treatment of flu, only nine genes were found to be activated selectively, whereas the number of genes activated after the dark/light shift was much greater. Also, changes in gene expression after the release of singlet oxygen occurred much more rapidly and drastically than in paraquat-treated plants. These differences could be attributable to the fact that singlet oxygen was generated almost instantaneously within chloroplasts after flu plants were transferred from the dark to the light, whereas paraquat was applied externally to the leaves at a low concentration. This might have caused a delay not only in the release of superoxide/hydrogen peroxide but also in the stress responses induced by these ROS. However, some of the group-III genes activated during paraquat treatment responded after 60 min, indicating that the delay in superoxide/hydrogen peroxide–mediated changes of gene expression also may reveal inherent differences in the extent and the kinetics of stress responses triggered by singlet oxygen or superoxide/hydrogen peroxide.
Possible Functional Implication of Singlet Oxygen–Mediated Oxidation of Linolenic Acid
Singlet oxygen is known to nonenzymatically oxidize many organic molecules, including membrane lipids (Klotz, 2002). In this way, it may disrupt normal cell functions and indirectly trigger a broad range of pleiotropic stress responses. In animal and human cells, the release of singlet oxygen during UV-A light treatment has been linked to the severe photooxidative damage of membrane lipids (Girotti, 2001), and nonenzymatic lipid peroxidation also has been shown to play a major role in plants during the late stages of leaf senescence (Berger et al., 2001). On the other hand, the importance of fatty acid metabolites formed enzymatically in various aspects of signaling is well documented. Prostaglandins derived from arachidonic acid and jasmonic acid derived from linolenic acid were among the first specific signaling species to be identified in animals and plants, respectively (Blée, 2002; Tang et al., 2002). As shown here, oxygenation derivatives of linolenic acid, by far the most prominent polyunsaturated fatty acid of chloroplast membrane lipids, start to accumulate rapidly in the flu mutant after the dark/light shift. The oxidation of linolenic acid is not caused by direct interaction with singlet oxygen but instead occurs enzymatically. Thus, the development of stress symptoms in the flu mutant seems not to be attributable to cell damage caused by singlet oxygen but rather appears to result from the more indirect role of this ROS as a stress signal.
Oxidation of polyunsaturated fatty acids and the release of oxylipins are catalyzed by lipoxygenases and lipases, respectively (Feussner and Wasternack, 2002). In the case of the redox-controlled activation of a specific lipoxygenase that uses free linolenic acid as a substrate, one would expect to see a transient decline in either esterified or free linolenic acid at the onset of reillumination of flu plants, but this was not observed. Alternatively, a specific lipase may be the target of redox control. Lipases have been implicated previously as key regulators of stress reactions that control the release of signaling molecules from membrane lipids (Ishiguro et al., 2001). Esterified linolenic acids that have been oxidized may serve as a storage form for oxylipins that can be released rapidly and act as signals after such a lipase has been activated. The free 13-HOTE that accumulates in the flu mutant after the dark/light shift shows a clear preponderance of the S-enantiomer, whereas the fraction of esterified 13-HOTE consists of variable amounts of both S- and R-enantiomers. Thus, any lipase that might be involved in the release of free 13-HOTE would have to exhibit a high degree of selectivity and recognize only one of the two stereoisomers of 13-HOTE as a substrate.
Activation of either a lipoxygenase or a lipase would explain the upregulation of 13-HOTE accumulation. However, these enzymes cannot be the only target of redox control. After the release of singlet oxygen, the immediate and transient activation of a large number of nuclear genes precedes the accumulation of oxylipins. Such changes in nuclear activities imply a rapid exchange of signals between the chloroplast, as the likely site at which singlet oxygen is being generated, and the nucleus. Because of the short half-life of singlet oxygen, it seems unlikely that this ROS leaves the plastid compartment and directly controls nuclear gene activities. Instead, singlet oxygen could be expected to interact with components that are localized more closely to its site of origin, such as lipids or fatty acids of chloroplast membranes. In this way, singlet oxygen might generate nonenzymatically peroxy derivatives with enhanced stability that could disseminate to other subcellular areas and trigger multiple stress responses. Such singlet oxygen–specific derivatives of fatty acids have not been observed in plants. However, a precedent for such a lipid-derived second messenger has been described in mammalian cells. There, singlet oxygen has been implicated in triggering the nonenzymatic generation of ceramide from sphingomyelin, which then causes the activation of the transcription factor AP-2 (Grether-Beck et al., 2000). Signaling cascades have been identified in both animals and plants that are activated rapidly after the release of hydrogen peroxide (Kovtun et al., 2000; Klotz, 2002). It remains to be determined whether any of these signal transduction pathways is involved in transmitting signals derived selectively from singlet oxygen.
METHODS
Plant Materials
For the cultivation of mature plants, seeds of Arabidopsis thaliana ecotype Landsberg erecta (Ler) were sown on soil under continuous light (80 to 100 μmol·m−2·s−1). Immediately before the experiment, the plants were transferred to the dark for 8 h and subsequently reexposed to light. Seedlings were cultivated by germinating surface-sterilized seeds on plates of Murashige and Skoog (1962) agar, in some cases supplemented with 0.5% sucrose. Plated seeds were either kept in the dark for 4 days or moved to continuous light (80 to 100 μmol·m−2·s−1) for 5 days, transferred to the dark for 15 h, and then reilluminated for various times.
Growth Measurements
Growth of the main shoot axis was determined either by measuring the length of the shoot daily for 2 to 3 weeks or by using an extensometer device coupled to a laser deflection system. Briefly, a light, stiff metal rod was balanced on the tip of a growing apex and connected via a pivot to a laser-movement sensor (Micro-Epsilon, St. Gallen, Switzerland). Thus, upward movement of the rod in response to extension growth of the shoot apex was coupled to deflection of a laser focused on the opposite end of the rod. The weight of the rod on the tip of the apex was <3 mg. Angular laser deflection was quantified as extension growth using transformation software supplied by the manufacturer, allowing noninvasive continuous measurement of shoot growth over a period of up to 48 h.
Differential Display
Wild-type and flu plants were grown under continuous light until they reached the rosette leaf stage. After 8 h in the dark, the plants were reilluminated for 20 and 60 min, respectively, and rosette leaves were harvested. Total RNA was extracted as described by Melzer et al. (1990). For reverse transcription and random PCR, the RNAmap System (Genhunter Corp., Nashville, TN) was used according to the manufacturer's instructions. The PCR products amplified by a dT12MN anchor primer and a 10mer arbitrary primer were separated electrophoretically on a 6% polyacrylamide/50% urea gel. Differentially displayed cDNA fragments were reamplified using the corresponding primers and screened using a reverse RNA gel blot technique (Zegzouti et al., 1997). Total cDNAs from reilluminated wild-type and flu plants were used as probes. Differentially hybridizing DNA fragments were cloned into the pBluescript SK+ vector (Stratagene, Amsterdam, The Netherlands) and sequenced. The differential expression of the transcripts was confirmed by RNA gel blot analysis.
Oligonucleotide Microarray Expression and Cluster Analysis
All plants were grown under continuous light until they reached the rosette leaf stage. For the analysis of changes in the expression of early stress-response genes of the flu mutant after a dark/light shift, wild-type and flu plants were kept in the dark for 8 h and then reilluminated for 30, 60, and 120 min before the rosette leaves were harvested. For the analysis of changes in the expression of genes after paraquat treatment, flu plants were sprayed either with a solution of 20 μM paraquat (methyl viologen; Sigma) in 0.1% Tween or with Tween alone, and rosette leaves were harvested at 1, 2, and 4 h after spraying. For each sample, the rosette leaves of five to six plants were collected for RNA extraction. Total RNAs from two separate biological experiments were pooled for the preparation of cDNA and the subsequent synthesis of biotin-labeled complementary RNA as recommended by Affymetrix (Santa Clara, CA).
Hybridization to the Affymetrix GeneChip Arabidopsis ATHI Genome Array, which contains >22,500 probe sets representing ∼24,000 genes, detection of labeled complementary RNA using streptavidin-phycoerythrin, and reading of the arrays using a confocal scanner (Affymetrix) were performed according to the manufacturer's instructions. The Affymetrix Microarray Suite 5.0 program was used to normalize microarray data to negative controls and to identify genes that show a reliable level of RNA, giving a detection call of P (present), M (marginal), and A (absent). The overall intensity of all probe sets of each array was scaled to 100, so the hybridization intensity of all 12 arrays was equivalent. To identify genes upregulated or downregulated in flu either after a dark/light shift or in response to paraquat treatments, comparisons were performed with “baseline” samples (i.e., either the wild-type Ler after a dark/light shift or flu plants treated with 0.1% Tween, respectively).
For the comparative analysis of gene activation in flu plants after the dark/light shift versus paraquat treatment, only genes that met the following criteria were considered: (1) the genes should be expressed in all of six different experimental time points (P in the Affymetrix nomenclature); and (2) the signals should be changed by at least 2.5-fold or greater (“difference” call) relative to the baseline sample. Based on these criteria, 1206 genes were selected for cluster analysis. Hierarchical clustering was performed using EPCLUST software (http://ep.ebi.ac.uk/EP/EPCLUST/), and the setting was modified until functionally significant clusters were reached.
The reliability of the expression data was supported by the following experiments: (1) 87% of a set of >500 genes taken from the set of 1206 genes that had been identified as upregulated or downregulated showed the same regulation of expression as in a previous independent experiment performed with RNA samples analyzed on Affymetrix GeneChips representing 8200 genes; (2) for both experiments (dark/light shift and paraquat treatment), an expression kinetic rather than a single time point was analyzed; and (3) the expression profiles of six genes from subcluster groups I and III were confirmed by real-time quantitative PCR performed with cDNAs from separate biological experiments.
Real-Time PCR
RNAs were treated with RQ1 RNase-Free DNase (Promega) and reverse-transcribed using random hexamers and SuperScript II RNase H− Reverse Transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations. Quantitative real-time PCR was performed with the ABI PRISM 7700 Sequence Detection System (Applied Biosystems, Foster City, CA).
Construction and Detection of the GFP-PORA Fusion Protein in Vivo
The plasmid pPORA.1.0 carrying the PORA cDNA of Arabidopsis (Armstrong et al., 1995) was modified by inserting internal EcoRI and NcoI recognition sites upstream of the start codon and downstream of the stop codon, respectively. The PCR product was cut with EcoRI and NcoI and ligated into the multicloning site of the pSH9 vector between the 35S promoter and the terminal polyadenylation site. The enhanced green fluorescent protein (EGFP) cDNA was excised from the pCL60 plasmid (Clontech, Palo Alto, CA) with NcoI and inserted into the modified pSH9 plasmid containing the PORA cDNA. Competent Escherichia coli cells (DH5α) were transformed with this plasmid. After propagation, the amplified cDNA was isolated and partially digested with HindIII. The fragment containing the 35S promoter, the Ω element, PORA, EGFP, and the terminal polyadenylation site was ligated into the binary vector pCAMBIA 3300 containing a phosphinothricin gene as a selectable marker. Competent cells of Agrobacterium tumefaciens C58 were transformed with the binary vector and then used for stable transformation of flu Ler plants.
Transgenic plants were selected on Murashige and Skoog (1962) agar plates containing phosphinothricin (25 mg/L). The green fluorescence of GFP and the red fluorescence of protochlorophyllide (Pchlide) were monitored using a confocal laser scanning microscope (TCS-NT; Leica, Wetzlar, Germany) with krypton/argon laser excitation. GFP and Pchlide were excited in intact cotyledons of etiolated flu seedlings at an excitation wavelength of 488 nm, and GFP and Pchlide were detected at emission wavelengths set at 510 to 530 nm and 630 to 700 nm, respectively.
Extraction and Measurement of Tetrapyrroles
Tetrapyrroles were extracted from plant samples with 80% acetone supplemented with ammonia to a final concentration of 0.0083% (v/v) overnight at 4°C in the dark. Pchlide and chlorophyllide contents of green plants were determined according to Yoshida et al. (1995). The fluorescence emission spectra (600 to 720 nm) of total acetone extracts and hexane-washed extracts were recorded at room temperature with the LS50 luminescence spectrophotometer (Perkin-Elmer, Rotkreuz, Switzerland) set at an excitation wavelength of 433 nm. For HPLC analysis, tetrapyrroles were extracted with 90% acetone and 10% 0.1 M NH4OH in water. Porphyrins were separated on a C18 reverse-phase silica-gel column (Nucleosil ODS 5 μm, 250 × 4.6 mm; Machery Nagel, Duren, Germany) and were detected by their fluorescence using the following excitation/emission wavelengths: 430/630 nm for Pchlide, 416/594 nm for Mg2+ protoporphyrin IX and Mg2+ protoporphyrin monomethylester, and 404/630 nm for protoporphyrin IX, as described by La Rocca et al. (2001).
Extraction and Detection of Lipid Peroxidation Products by HPLC
Oxidized fatty acids were extracted using a modification of the method of Weichert et al. (2002). For the analysis of esterified fatty acids, the solvent was removed and 333 μL of a mixture of toluene and methanol (1:1, v/v) and 167 μL of 0.5 mM sodium methoxide were added. As internal standards, triheptadecanoate and triricinoleate were added. After incubation of the samples for 20 min, 0.5 mL of 1 M NaCl and 50 μL of HCl (37%, v/v) were added, and the fatty acid methyl esters (FAMEs) were extracted twice each with 0.75 mL of hexane. The combined organic phases were evaporated to dryness under nitrogen, and the FAMEs were dissolved in 10 μL of acetonitrile. For the analysis of unesterified fatty acid derivatives, the solvent was removed and the sample was dissolved in 400 μL of methanol. As internal standards, heptadecanoic acid and 13γ-hydroxylinolenic acid were added. To 40 μL of this solution, 10 μL of an EDAC solution [1 mg of 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide and 10 μL of methanol] was added and incubated for 2 h.
After adding 200 μL of Tris buffer (0.1 M Tris-HCl, pH 7.5), the FAMEs were extracted twice each with 1 mL of hexane. The combined organic phases were evaporated to dryness under nitrogen, and the corresponding FAMEs were redissolved in 10 μL of acetonitrile and analyzed using a gas chromatograph/flame ion detector.
Singlet Oxygen Determination and Trypan Blue Staining
Singlet oxygen was detected in leaves as described by Hideg et al. (1998). Dead cells were identified by staining with lacto-phenol trypan blue as described by Keogh et al. (1980).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact K. Apel, klaus.apel@ipw.biol.ethz.ch.
Supplementary Material
Acknowledgments
We are indebted to Dieter Rubli for artwork, to Martha Geier-Bächtold for editorial assistance, to K. Hideg and T. Kalai for the gift of DanePy, to the Functional Genomics Center in Zurich for support of the Affymetrix microarray analysis, and to Paul Hardy for correcting the language. This work was supported by grants from the Swiss Federal Institute of Technology (Zurich) and the Swiss National Science Foundation.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.014662.
Footnotes
Online version contains Web-only data.
References
- Allan, A.C., and Fluhr, R. (1997). Two distinct sources of elicited reactive oxygen species in tobacco epidermal cells. Plant Cell 9, 1559–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez, M.E., Pennell, R.I., Meijer, P.J., Ishikawa, A., Dixon, R.A., and Lamb, C. (1998). Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92, 773–784. [DOI] [PubMed] [Google Scholar]
- Armstrong, G.A., Runge, S., Frick, G., Sperling, U., and Apel, K. (1995). Identification of NADPH:protochlorophyllide oxidoreductase A and B: A branched pathway for light-dependent chlorophyll biosynthesis in Arabidopsis thaliana. Plant Physiol. 108, 1505–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, S., Weichert, H., Porzel, A., Wasternack, C., Kühn, H., and Feussner, I. (2001). Enzymatic and non-enzymatic lipid peroxidation in leaf development. Biochim. Biophys. Acta 1533, 266–276. [DOI] [PubMed] [Google Scholar]
- Blée, E. (2002). Impact of phyto-oxylipins in plant defense. Trends Plant Sci. 7, 315–321. [DOI] [PubMed] [Google Scholar]
- Boo, J.C., Lee, K.P., and Jung, J. (2000). Rice plants with a high protochlorophyllide accumulation show oxidative stress in low light that mimics water stress. J. Plant Physiol. 157, 405–411. [Google Scholar]
- Bray, E.A., Bailey-Serres, J., and Weretilnyk, E. (2000). Responses to abiotic stresses. In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: American Society of Plant Physiologists), pp. 1158–1203.
- Briat, J.-F., and Lobréaux, S. (1997). Iron transport and storage in plants. Trends Plant Sci. 2, 187–193. [Google Scholar]
- Dalton, T.P., Shertzer, H.G., and Puga, A. (1999). Regulation of gene expression by reactive oxygen. Annu. Rev. Pharmacol. Toxicol. 39, 67–101. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., Dietrich, R.A., and Richberg, M.H. (1996). Death don't have no mercy: Cell death programs in plant–microbe interactions. Plant Cell 8, 1793–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaeva, M., and Adamska, I. (2001). Identification of genes expressed in response to light-stress in leaves of Arabidopsis thaliana using RNA differential display. Eur. J. Biochem. 268, 5521–5529. [DOI] [PubMed] [Google Scholar]
- Elstner, E.F. (1991). Mechanisms of oxygen activation in different compartments of plant cells. In Active Oxygen/Oxidative Stress in Plant Metabolism, E.J. Pelland and K.L. Steffen, eds (Rockville, MD: American Society of Plant Physiologists), pp. 13–25.
- Feussner, I., and Wasternack, C. (2002). The lipoxygenase pathway. Annu. Rev. Plant Biol. 53, 275–297. [DOI] [PubMed] [Google Scholar]
- Foote, C.S. (1991). Definition of type I and type II photosensitized oxidation. Photochem. Photobiol. 54, 659. [DOI] [PubMed] [Google Scholar]
- Foyer, C.H., and Noctor, G. (2000). Oxygen processing in photosynthesis: Regulation and signaling. New Phytol. 146, 359–388. [Google Scholar]
- Fryer, M.J., Oxborough, K., Mullineaux, P.M., and Baker, N.R. (2002). Imaging of photo-oxidative stress responses in leaves. J. Exp. Bot. 53, 1249–1254. [PubMed] [Google Scholar]
- Girotti, A.W. (2001). Photosensitized oxidation of membrane lipids: Reaction pathways, cytotoxic effects and cytoprotective mechanisms. J. Photochem. Photobiol. B 63, 103–113. [DOI] [PubMed] [Google Scholar]
- Gollnick, K. (1968). Type II photooxygenation reactions in solution. Adv. Photochem. 6, 1–122. [Google Scholar]
- Gomez-Silva, B., Timko, M.P., and Schiff, J.A. (1985). Chlorophyll biosynthesis from glutamate or 5-aminolevulininate in intact Euglena chloroplasts. Planta 165, 12–22. [DOI] [PubMed] [Google Scholar]
- Gorman, A.A., and Rodgers, M.A. (1992). Current perspectives of singlet oxygen detection in biological environments. J. Photochem. Photobiol. B 14, 159–176. [DOI] [PubMed] [Google Scholar]
- Grether-Beck, S., Bonizzi, G., Schmitt-Brenden, H., Felsner, I., Timmer, A., Sies, H., Johnson, J.P., Piette, J., and Krutmann, J. (2000). Non-enzymatic triggering of the ceramide signalling cascade by solar UVA radiation. EMBO J. 19, 5793–5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack, K., and Jones, J.D.G. (2000). Responses to plant pathogens. In Biochemistry and Molecular Biology of Plants, B.B., Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: American Society of Plant Physiologists), pp. 1102–1156.
- Hideg, É., Barta, C., Kalai, T., Vass, I., Hideg, K., and Asada, K. (2002). Detection of singlet oxygen and superoxide with fluorescent sensors in leaves under stress by photo inhibition or UV radiation. Plant Cell Physiol. 43, 1154–1164. [DOI] [PubMed] [Google Scholar]
- Hideg, É., Kalai, T., Hideg, K., and Vass, I. (1998). Photoinhibition of photosynthesis in vivo results in singlet oxygen production: Detection via nitroxide-induced fluorescence quenching in broad bean leaves. Biochemistry 37, 11405–11411. [DOI] [PubMed] [Google Scholar]
- Hua, J., Grisafi, P., Cheng, S.-H., and Fink, G.R. (2001). Plant growth homeostasis is controlled by the Arabidopsis BON1 and BAP1 genes. Genes Dev. 15, 2263–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro, S., Kawai-Oda, A., Ueda, J., Nishida, I., and Okada, K. (2001). The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13, 2191–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalai, T., Hideg, É., Vass, I., and Hideg, K. (1998). Double (fluorescent and spin) sensors for detection of reactive oxygen species in the thylakoid membrane. Free Radical Biol. Med. 24, 649–652. [DOI] [PubMed] [Google Scholar]
- Karpinski, S., Escobar, C., Karpinska, B., Creissen, G., and Mullineaux, P.M. (1997). Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 9, 627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh, R.C., Deverall, B.J., and McLeod, S. (1980). Comparison of histological and physiological responses to Phakopsora pachyrhizi in resistant and susceptible soybean. Trans. Br. Mycol. Soc. 74, 329–333. [Google Scholar]
- Klotz, L.-O. (2002). Oxidant-induced signaling: Effects of peroxynitrite and singlet oxygen. Biol. Chem. 383, 443–456. [DOI] [PubMed] [Google Scholar]
- Kovtun, Y., Chiu, W.-L., Tena, G., and Sheen, J. (2000). Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 97, 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rocca, N., Rascio, N., Oster, U., and Rüdiger, W. (2001). Amitrole treatment of etiolated seedlings leads to deregulation of tetrapyrrole synthesis and to reduced expression of Lhc and RbcS genes. Planta 213, 101–108. [DOI] [PubMed] [Google Scholar]
- Long, S.P., Humphries, S., and Falkowski, P.G. (1994). Photoinhibition of photosynthesis in nature. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45, 633–662. [Google Scholar]
- Macpherson, A.N., Telfer, A., Barber, J., and Truscott, T.G. (1993). Direct detection of singlet oxygen from isolated photosystem II reaction centres. Biochim. Biophys. Acta 1143, 301–309. [Google Scholar]
- Mehler, A.H. (1951). Studies on reactions of illuminated chloroplasts. II. Stimulation and inhibition of the reaction with molecular oxygen. Arch. Biochem. Biophys. 33, 339–351. [DOI] [PubMed] [Google Scholar]
- Melzer, S., Majewski, D.M., and Apel, K. (1990). Early changes in gene expression during the transition from vegetative to generative growth in the long-day plant Sinapis alba. Plant Cell 2, 953–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskauskiene, R., Nater, M., Goslings, D., Kessler, F., op den Camp, R., and Apel, K. (2001). FLU: A negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98, 12826–12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller, S.G., Kunkel, T., and Chua, N.H. (2001). A plastidic ABC protein involved in intercompartmental communication of light signaling. Genes Dev. 15, 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami, J., Tsujama, M., Kobayashi, Y., Kodama, H., and Iba, K. (2000). Trienoic fatty acids and plant tolerance of high temperature. Science 287, 476–479. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Neill, N., Desikan, R., and Hancock, J. (2002). Hydrogen peroxide signaling. Curr. Opin. Plant Biol. 5, 388–395. [DOI] [PubMed] [Google Scholar]
- Netting, A.G. (2000). pH, abscisic acid and the integration of metabolism in plants under stressed and non-stressed conditions: Cellular responses to stress and their implication for plant water relations. J. Exp. Bot. 51, 147–158. [DOI] [PubMed] [Google Scholar]
- Nyogi, K.K. (1999). Photoprotection revisited: Genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 333–359. [DOI] [PubMed] [Google Scholar]
- Orozco-Cardenas, M., and Ryan, C.A. (1999). Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc. Natl. Acad. Sci. USA 96, 6553–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit, J.-M., Briat, J.-F., and Lobréaux, S. (2001). Structure and differential expression of the four members of the Arabidopsis thaliana ferritin gene family. Biochem. J. 359, 575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourzand, C., and Tyrrell, R.M. (1999). Apoptosis, the role of oxidative stress and the example of solar UV radiation. Photochem. Photobiol. 70, 380–390. [PubMed] [Google Scholar]
- Rebeiz, C.A., Montazer-Zouhoor, A., Mayasich, J.M., Tripathy, B.C., Wu, S.-M., and Rebeiz, C. (1988). Photodynamic herbicides: Recent developments and molecular basis of selectivity. CRC Crit. Rev. Plant Sci. 6, 385–436. [Google Scholar]
- Reichheld, J.-P., Vernoux, T., Lardon, F., van Montagu, M., and Inzé, D. (1999). Specific checkpoints regulate plant cell cycle progression in response to oxidative stress. Plant J. 17, 647–656. [Google Scholar]
- Rusterucci, C., Montillet, J.-L., Aguel, J.-P., Battesti, C., Alonso, B., Knoll, A., Bessoules, J.-J., Etienne, P., Suty, L., Blein, J.-P., and Triantaphylidiès, C. (1999). Involvement of lipoxygenase-dependent production of fatty acid hydroperoxides in the development of the hypersensitive cell death induced by cryptogein on tobacco leaves. J. Biol. Chem. 51, 36446–36455. [DOI] [PubMed] [Google Scholar]
- Sies, H., and Menck, C.F. (1992). Singlet oxygen induced DNA damage. Mutat. Res. 275, 367–375. [DOI] [PubMed] [Google Scholar]
- Sperling, U., Franck, F., van Cleve, B., Frick, G., Apel, K., and Armstrong, G. (1998). Etioplast differentiation in Arabidopsis: Both PORA and PORB restore the prolamellar body and photoactive protochlorophyllide F655 to the cop1 photomorphogenic mutant. Plant Cell 10, 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storozhenko, S., de Pauw, P., van Montagu, M., Inzé, D., and Kushnir, S. (1998). The heat-shock element is a functional component of the Arabidopsis APX1 gene promoter. Plant Physiol. 118, 1005–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, D.G., La, E., Kern, J., and Kehrer, J.P. (2002). Fatty acid oxidation and signaling in apoptosis. Biol. Chem. 383, 425–442. [DOI] [PubMed] [Google Scholar]
- Terry, M.J., and Lagarias, J.C. (1991). Holophytochrome assembly: Coupled assay for phytochromobilin synthase in organello. J. Biol. Chem. 266, 22215–22221. [PubMed] [Google Scholar]
- Thordal-Christensen, H., Zhang, Z., Wei, Y., and Collinge, D.B.C. (1997). Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley powdery mildew interaction. Plant J. 11, 1187–1194. [Google Scholar]
- Trebst, A. (1999). Singlet oxygen in photosynthesis. In Different Pathways through Life: Biochemical Aspects of Plant Biology and Medicine, A. Denke and K. Dornisch, eds (Munich, Germany: Lincom Europe), pp. 125–142.
- Weichert, H., Kolbe, A., Kraus, A., Wasternack, C., and Feussner, I. (2002). Metabolic profiling of oxylipins in germinating cucumber seedlings: Lipoxygenase-dependent degradation of triacylglycerols and biosynthesis of volatile aldehydes. Planta 215, 612–619. [DOI] [PubMed] [Google Scholar]
- Wiese, A.G., Pacifici, R.E., and Davies, K.J. (1995). Transient adaptation to oxidative stress in mammalian cells. Arch. Biochem. Biophys. 318, 231–240. [DOI] [PubMed] [Google Scholar]
- Yoshida, K., Chen, R.-M., Tanaka, A., Teramoto, H., Tanaka, R., Timko, M.P., and Tsuji, H. (1995). Correlated changes in the activity, amount of protein, and abundance of transcript of NADPH:protochlorophyllide oxidoreductase and chlorophyll accumulation during greening of cucumber cotyledons. Plant Physiol. 109, 231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegzouti, H., Marty, C., Jones, B., Bouquin, T., Latché, A., Pech, J.C., and Bouzayen, M. (1997). Improved screening of cDNAs generated by mRNA differential display enables the selection of true positives and the isolation of weakly expressed messages. Plant Mol. Biol. Rep. 15, 236–245. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.