Abstract
Arabidopsis RPS4 belongs to the Toll/interleukin-1 receptor (TIR)–nucleotide binding site (NBS)–Leu-rich repeat (LRR) class of disease resistance (R) genes. Like other family members in different plant species, RPS4 produces alternative transcripts with truncated open reading frames. The dominant alternative RPS4 transcripts are generated by retention of intron 3 or introns 2 and 3, which contain in-frame stop codons and lie downstream of the NBS-encoding exon. We analyzed the biological significance of these alternative transcripts in disease resistance by removing introns 2 and 3, either individually or in combination, from a functional RPS4-Ler (Landsberg erecta) transgene. Removal of one or both introns abolished the function of the RPS4 transgene, whereas expression was not affected. In addition, a truncated RPS4-Ler transgene encoding the putative TIR and NBS domains was not sufficient to confer resistance, suggesting that the combined presence of regular and alternative RPS4 transcripts is necessary for function. Interestingly, we observed partial resistance in transgenic lines expressing both intron-deficient and truncated transgenes. This finding confirms the requirement for regular and alternative RPS4 transcripts and indicates that alternative transcripts function at the protein level rather than as regulatory RNAs. Together with published results on the tobacco N gene, our data suggest that the generation of alternative TIR-NBS-LRR R gene transcripts is of general biological significance across plant species.
INTRODUCTION
Plants are under constant challenge by potential pathogens and have evolved a broad range of mechanisms to limit pathogen growth (Staskawicz et al., 1995; Hammond-Kosack and Jones, 1996). One such mechanism is governed by plant disease resistance (R) genes that specifically recognize pathogen strains expressing cognate avirulence (avr) genes, as described by the gene-for-gene hypothesis (Flor, 1971). Direct or indirect interactions between cognate R and Avr proteins initiate a chain of signaling events to activate plant defense responses that ultimately prevent pathogen replication and movement (Bent, 1996; Baker et al., 1997; Dangl and Jones, 2001; Mackey et al., 2002, 2003; Axtell and Staskawicz, 2003).
The Arabidopsis RPS4 gene specifies disease resistance to Pseudomonas syringae pv tomato strain DC3000 (DC3000) expressing avrRps4 (Gassmann et al., 1999). RPS4 encodes a predicted protein of 1217 amino acids that belongs to the Toll/interleukin-1 receptor (TIR)–nucleotide binding site (NBS)–Leu-rich repeat (LRR) class of R proteins. The TIR domain is homologous with the cytoplasmic domain of the Drosophila Toll and the mammalian interleukin-1 receptors, proteins involved in animal innate immune responses (O'Neill, 2000; Imler and Hoffmann, 2001; Li and Verma, 2002). RPS4 and other Arabidopsis TIR-NBS-LRR (TNL) R genes governing resistance to oomycetes depend on a functional EDS1 (ENHANCED DISEASE SUSCEPTIBILITY) gene (Parker et al., 1996; Falk et al., 1999), whereas coiled-coil (CC)-NBS-LRR R genes do not (Aarts et al., 1998). By analogy to innate immune receptors in animals and because of this specific dependence of TNL R genes on EDS1, the TIR domain of plant R proteins is hypothesized to transduce signals to downstream components in the plant defense signaling pathway (Dinesh-Kumar et al., 1995; Parker et al., 1997; Gassmann et al., 1999).
Another distinguishing feature of TNL R genes from different plant species is their capacity to generate alternative transcripts with truncated open reading frames (ORFs). These encode putative TIR-NBS (TN) proteins that lack the LRR and C-terminal domains (Jordan et al., 2002). By contrast, the Arabidopsis CC-NBS-LRR proteins RPS2, RPM1, and RPS5 are encoded by genes with intronless ORFs. At least three mechanisms for the generation of TN-encoding transcripts have been observed. In the case of the tobacco N gene that confers resistance to Tobacco mosaic virus (TMV), the alternative transcript arises via alternative splicing of a 70-bp exon within intron 3 (Whitham et al., 1994). Arabidopsis RPP5 confers resistance to downy mildew pathogens and appears to produce only a single transcript corresponding to the full-length protein. However, a linked truncated gene family member that is expressed encodes a putative protein with a TN amino acid sequence identical to that of RPP5 but lacking LRRs (Parker et al., 1997; Noël et al., 1999). In the flax L6 gene that confers resistance to flax rust, transcripts retaining intron 3 contain in-frame stop codons (Lawrence et al., 1995). RPS4 appears to be similar to L6, because the predominant alternative RPS4 transcripts retain intron 3 or introns 2 and 3 (Gassmann et al., 1999). These introns lie downstream of the NBS-encoding exon and contain in-frame stop codons, so the alternative RPS4 transcripts also encode putative truncated TN proteins. The conservation of alternative transcript generation suggests that transcript variants play an important role in TNL R gene–mediated disease resistance.
The precise function of alternative R gene transcripts in gene-for-gene resistance remains elusive. To date, the tobacco N gene has been analyzed most carefully. It was shown that the native N gene promoter, the alternative exon within intron 3, and the 3′ untranslated region all are required for the correct regulation of alternative transcript expression and full resistance (Dinesh-Kumar and Baker, 2000). Substitution or deletion of any one of these elements caused a delay in the hypersensitive response on leaves of transgenic tobacco plants inoculated with TMV, permitting TMV to cause a systemic hypersensitive response and mosaic symptoms throughout the plant. Although no direct protein data have been published, it was hypothesized that the truncated N protein encoded by the alternative transcript is required for fast, appropriate, and complete responses to TMV (Dinesh-Kumar and Baker, 2000). By contrast, transgenic flax lines expressing an intronless L6 transgene were indistinguishable from the wild type in flax rust resistance. However, this analysis may be complicated by the presence of an endogenous L allele or highly homologous TNL R genes at other loci that also generate alternative transcripts (Ayliffe et al., 1999). Given the conflicting evidence, it is important to ascertain the significance of alternative TNL R gene transcripts in disease resistance.
Here, we provide evidence that alternative RPS4 transcripts are required for function. We show that the removal of introns 2 and 3 from the functional RPS4-Ler gene abolished avrRps4-specific resistance. In addition, truncated RPS4-Ler gene fragments encoding TN proteins were not sufficient to confer resistance. These data suggest that the combined presence of regular and alternative transcripts is required for RPS4 function. Significantly, we found that expression of both intron-deficient and truncated RPS4 transgenes in double-transgenic plants partially restored avrRps4-specific resistance. Our results indicate that the generation of alternative transcripts may be of general importance for TNL R gene function across plant species.
RESULTS
Alternative RPS4 Transcripts Are Essential for Resistance
To determine whether alternative RPS4 transcripts are required for function, we generated RPS4 gene constructs in which introns 1 and 4 remained but introns 2 and/or 3 were removed. These intron-deficient genes were derived from a genomic clone of the functional RPS4-Ler allele driven by its native promoter. Genomic sequences surrounding intron 2 or 3 were swapped for cDNA sequences that were obtained from Ler RNA by reverse transcriptase–mediated (RT) PCR (see Methods). The resulting intron-deficient RPS4 clones lacking intron 2, intron 3, or both introns 2 and 3 were introduced into the naturally avrRps4-susceptible accession RLD to obtain stable transformants designated RLD-i2r, RLD-i3r, and RLD-i23r, respectively. We used the RPS4 native promoter and stable transgenic lines to closely mimic the natural system. Transformants containing wild-type RPS4-Ler (RLD-R4L) also were generated as a positive control.
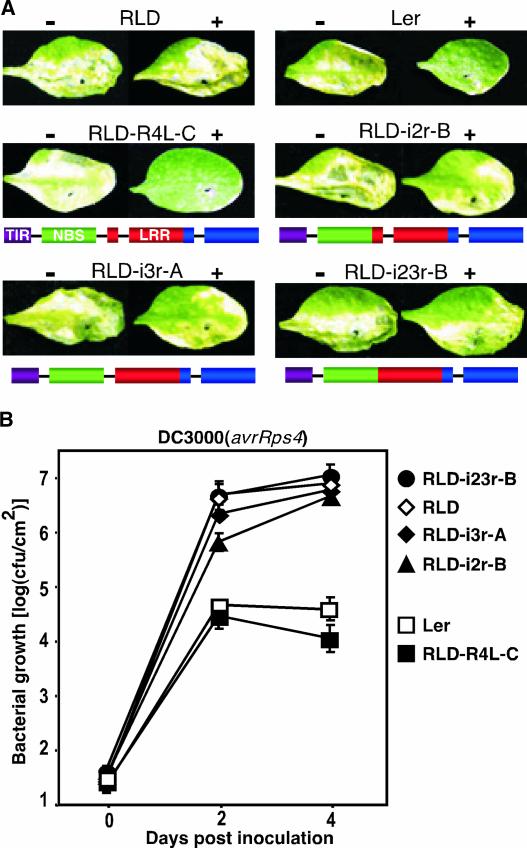
For each transgene, homozygous single-locus transgenic lines were selected from 20 independent transformants for disease and in planta bacterial growth assays. In disease assays, two leaves of similar age on a given seedling were infiltrated with virulent DC3000 or DC3000 expressing avrRps4. In the absence of resistance, disease symptoms appear as chlorosis at 5 days after inoculation. As shown in Figure 1A, both Ler and RLD-R4L were resistant to DC3000(avrRps4), whereas RLD was susceptible. Likewise, RLD-i2r, RLD-i3r, and RLD-i23r plants were fully susceptible to DC3000 (avrRps4), indicating that removal of even one intron severely compromises RPS4 function (Figure 1A). Results similar to those shown in Figure 1A were obtained with all transgenic lines tested (four or five independent homozygous transgenic lines for each transgene; data not shown).
Figure 1.
Alternative RPS4 Transcripts Are Required for Resistance.
(A) Disease assays on transgenic Arabidopsis lines. Leaves of wild-type accessions (top row) and representative transgenic RLD lines (middle and bottom rows) were infiltrated with suspensions of DC3000(empty vector) (−; left leaf in each photograph) or DC3000(avrRps4) (+; right leaf) at a density of 1 × 106 colony-forming units/mL. Disease symptoms were recorded 5 days after inoculation. Plant lines are identified above each photograph. Schemes of wild-type and intron-deficient RPS4-Ler transgenes are shown below the photographs, with black lines representing introns.
(B) In planta bacterial growth of DC3000(avrRps4) in wild-type Ler (open squares) and RLD (open diamonds) and the transgenic RLD lines shown in (A): RLD-R4L-C (closed squares), RLD-i2r-B (closed triangles), RLD-i3r-A (closed diamonds), and RLD-i23r-B (closed circles). Error bars denoting standard deviations are shown where they are larger than the symbols. Plants were vacuum-infiltrated with DC3000(avrRps4) at a density of 5 × 104 colony-forming units (cfu)/mL. Control experiments with DC3000(empty vector) were performed in parallel and showed levels of bacterial growth comparable to that of RLD inoculated with DC3000(avrRps4) for all plant lines. This experiment was repeated twice with similar results.
Because intronic sequences can affect gene expression, we next tested whether the observed differences in resistance phenotypes were caused by differential transgene expression levels. The presence and number of transgenes in transgenic RLD lines were determined by DNA gel blot analysis (Table 1). Also, the overall expression level of transgenes irrespective of splicing at introns 2 and 3 was measured by quantitative RT-PCR using primers flanking intron 1 of RPS4 (Table 1). A cleaved amplified polymorphic sequence marker was used to distinguish transgene RT-PCR products from products arising from the endogenous rps4-RLD gene (see Methods). The cleaved amplified polymorphic sequence–assisted RT-PCR experiments showed that the expression of transgenes largely correlated with the transgene copy number and was similar to endogenous rps4-RLD expression except in two RLD-i23r lines (Table 1). Importantly, transgene expression levels were similar in resistant RLD-R4L and susceptible RLD-i2r, RLD-i3r, and two of four RLD-i23r lines. These data show that the susceptibility of RLD-i2r, RLD-i3r, and RLD-i23r plants reflects the impaired functionality of the intron-deficient RPS4 genes rather than poor transgene expression.
Table 1.
Expression Levels of Intron-Deficient RPS4-Ler Transgenes
| Transgene | Transgenic Line | Insertion Number a |
Expression Level (±sd) b |
Res./ Susc.c |
|---|---|---|---|---|
| RPS4-Ler wild type | RLD-R4L-A | 2 | 2.0 ± 0.2 | Res. |
| RLD-R4L-B | 1 | 1.2 ± 0.2 | ||
| RLD-R4L-Cd | 1 | 1.4 ± 0.3 | ||
| Intron 2 removed | RLD-i2r-A | 1 | 1.6 ± 0.4 | Susc. |
| RLD-i2r-B | 2 | 1.8 ± 0.2 | ||
| Intron 3 removed | RLD-i3r-A | 1 | 1.4 ± 0.1 | Susc. |
| RLD-i3r-B | 1 | 1.3 ± 0.3 | ||
| RLD-i3r-C | 1 | 1.1 ± 0.1 | ||
| Introns 2 and 3 removed |
RLD-i23r-A | 1 | 1.0 ± 0.1 | Susc. |
| RLD-i23r-B | 1 | 1.0 ± 0.1 | ||
| RLD-i23r-C | 2 | 0.1 ± 0.0 | ||
| RLD-i23r-D | 3 | 0.2 ± 0.0 |
Transgene insertion numbers were determined by DNA gel blot analysis.
Transgene expression levels relative to endogenous rps4-RLD expression levels (n = 3).
Res./Susc., resistant/susceptible.
Representative transgenic lines used in in planta bacterial growth assays (Figure 1B) are underlined.
To quantitatively determine the degree of disease resistance, we measured in planta bacterial growth in representative plant lines. The lines RLD-R4L-C, RLD-i2r-B, RLD-i3r-A, and RLD-i23r-B were chosen based on RPS4 transgene expression levels (Table 1). The growth of virulent DC3000 was similar in all plants tested and comparable to the growth of DC3000(avrRps4) in RLD (data not shown). Consistent with qualitative disease assays, Ler and RLD-R4L-C were resistant to DC3000(avrRps4), whereas RLD-i2r-B, RLD-i3r-A, and RLD-i23r-B were fully susceptible compared with the parental RLD line (Figure 1B). Surprisingly, these data show that the removal of one intron alone completely abolishes RPS4 function, indicating that the number of alternative transcripts generated by wild-type RPS4 genes is close to a threshold required for resistance that is not reached by intron-deficient transgenes.
Intron-Deficient Transgene Transcripts Are Not Spliced Aberrantly
One possible explanation for our findings is that the removal of intron 2 or 3 activates cryptic splice sites, leading exclusively to aberrant RPS4 mRNAs that do not encode the full-length RPS4 protein. We used RT-PCR with primers flanking introns 2 and 3 to analyze the sizes and sequences of all transcript variants from wild-type accessions and transgenic lines. A nucleotide polymorphism in exon 3 between rps4-RLD and RPS4-Ler allowed us to distinguish between endogenous and transgene transcripts by sequencing individual transcript clones (see Methods).
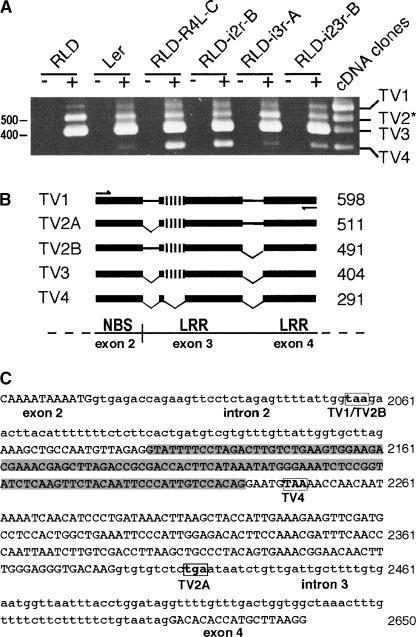
As shown in Figure 2A, multiple transcript variants (TV) were detected in all plant lines. Control experiments without reverse transcriptase showed that all products represented RPS4 transcripts, not genomic DNA contamination (Figure 2A). Wild-type RLD showed alternative transcripts arising from the endogenous rps4-RLD gene similar to wild-type Ler (Figure 2A, lanes 2 and 4), explaining why individual transcripts were not absent in transgenic lines expressing intron-deficient transgenes. Sequencing revealed that TV1 retained introns 2 and 3, whereas TV2 was composed of two transcript species of similar size lacking either intron 2 (TV2A) or intron 3 (TV2B) (Figure 2B). TV3, corresponding to the regular transcript with both introns 2 and 3 removed, was the most abundant form in all lines/plants tested (Figure 2A). TV4, the smallest detectable transcript, was present at similar levels in resistant RLD-R4L-C and susceptible RLD-i2r-B plants and was detected weakly in all lines, including wild-type RLD and Ler plants. An additional cryptic intron within exon 3 was found to be spliced out in TV4 (Figures 2B and 2C). Removal of this cryptic intron caused a frameshift in the open reading frame, potentially producing a third truncated protein containing mostly TN domains (Figure 2C). TV2B and TV4 had not been described previously.
Figure 2.
Analysis of RPS4 Transcript Variants.
(A) RT-PCR analysis of transgene transcripts with primers flanking introns 2 and 3. Reverse transcription was performed with RNA isolated from the indicated plant lines in the presence (+) or absence (−) of reverse transcriptase. In the far-right lane, clones of the five transcript variants (TVs) shown in (B) were used as a template in a single PCR assay for direct size comparison. Numbers at left denote DNA size standards in base pairs. Individual TVs are indicated at right. The asterisk indicates that the band labeled TV2 was composed of two bands of similar size.
(B) Schemes of TVs identified by RT-PCR. The structure of each TV within the amplified region was determined by sequencing. Exons are shown as black boxes. Retained introns are shown as black lines, and spliced introns are indicated by diagonal lines. The cryptic intron within exon 3 is represented by vertical lines. PCR primers used in (A) are indicated by arrows. Numbers next to the TV names indicate the expected amplified size of each TV in base pairs.
(C) Nucleotide sequence of the variably spliced region in RPS4. Exon sequences are represented by uppercase letters, and intron sequences are represented by lowercase letters. The cryptic intron within exon 3 is shaded. In-frame stop codons are boxed for each specific TV. Numbers at right denote nucleotide positions of the full-length genomic RPS4-Ler sequence.
Importantly, sequencing of individual TV3 clones from all transgenic plants showed that intron-deficient transgenes in all transgenic lines gave rise to RPS4-Ler mRNAs that were not spliced aberrantly and thus encoded full-length RPS4-Ler protein. These expression and transcript analyses showed that RLD-i2r, RLD-i3r, and RLD-i23r lines were susceptible despite the presence of full-length RPS4-Ler protein-encoding transcripts. Therefore, we concluded that alternative RPS4 transcripts are essential for resistance. In addition, sequencing of individual TV2A and TV2B clones in RLD-i2r and RLD-i3r lines showed that intron-deficient transgenes with only one intron removed still generated transcripts retaining the other intron. Although we cannot exclude altered frequencies of retention, this finding indicated that the retention of intron 2 or 3 did not require the presence of both introns in the pre-mRNA.
Characterization of rps4-RLD
As proposed in the study of L6, redundancy can hinder the functional characterization of alternative TNL R transcripts (Ayliffe et al., 1999). Specifically, the L6 allele L9 and L6-homologous TNL genes at the M locus were proposed to substitute for alternative L6 transcript function in transgenic Ward and Trac2 flax lines, respectively (Ayliffe et al., 1999). Our data from intron-deficient RPS4 transgenes showed that there is no complementation of alternative transcript function by the endogenous rps4-RLD allele or RPS4 homologs (Figures 1A and 1B). The rps4-RLD allele does not contain major alterations in the nucleotide sequence (Gassmann et al., 1999). It is expressed and gives rise to alternative transcripts (Figure 2A, lane 2). Comparison of the putative rps4-RLD amino acid sequence with four functional RPS4 alleles revealed two unique amino acid changes, N195D in the NBS region and Y950H in the C-terminal non-LRR region (Gassmann et al., 1999). Neither polymorphism affects conserved R protein motifs. To better understand why endogenous rps4-RLD alternative transcripts do not complement intron-deficient transgenes, we set out to conclusively establish the molecular lesion(s) in rps4-RLD.
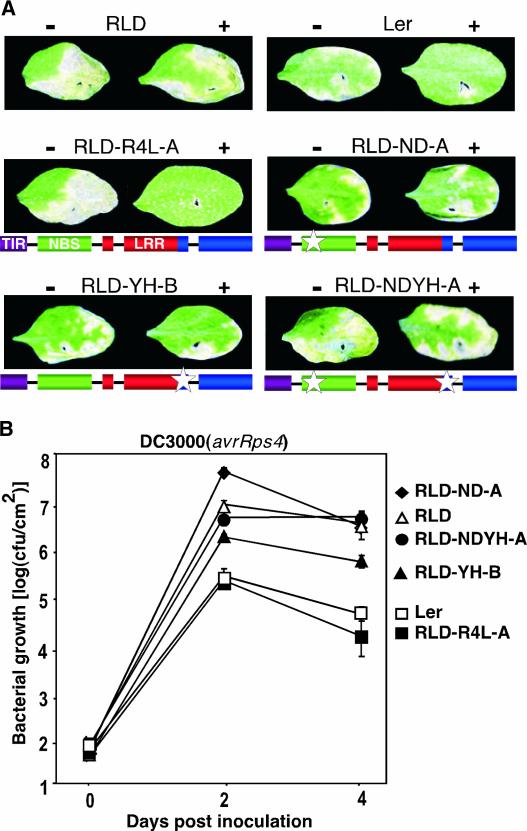
Genomic RLD DNA surrounding the two unique amino acid changes in rps4-RLD was amplified and swapped into RPS4-Ler (see Methods). The resulting chimeric RPS4 clones with the amino acid changes N195D, Y950H, and N195D plus Y950H were introduced into RLD plants to obtain stable transformants designated RLD-ND, RLD-YH, and RLD-NDYH, respectively. As shown in Figure 3A, RLD-ND, RLD-YH, and RLD-NDYH plants were susceptible to DC3000(avrRps4), indicating that either one of the amino acid changes compromised RPS4 function. Similar results were obtained with all transgenic lines tested (four or five independent transgenic lines for each transgene). Disease symptoms appeared to be less severe on RLD-YH than on RLD, RLD-ND, and RLD-NDYH, suggesting that RPS4(Y950D) retained residual function.
Figure 3.
A Single Amino Acid Change Specific to rps4-RLD Is Sufficient to Abolish RPS4 Function.
(A) Disease assays on transgenic Arabidopsis lines expressing chimeric RPS4 genes. Leaves of wild-type accessions (top row) and representative transgenic RLD lines (middle and bottom rows) were infiltrated with DC3000(empty vector) (−; left leaf in each photograph) or DC3000(avrRps4) (+; right leaf) as described in the legend to Figure 1A. Plant lines are identified above each photograph. Schemes of wild-type and chimeric RPS4-Ler transgenes are shown below the photographs. The positions of amino acid polymorphisms specific to rps4-RLD are indicated by white stars.
(B) In planta bacterial growth of DC3000(avrRps4) in wild-type Ler (open squares) and RLD (open triangles) and the transgenic RLD lines shown in (A): RLD-R4L-A (closed squares), RLD-ND-A (closed diamonds), RLD-YH-B (closed triangles), and RLD-NDYH-A (closed circles). Error bars denoting standard deviations are shown where they are larger than the symbols. Control experiments with DC3000(empty vector) were performed in parallel and showed levels of bacterial growth comparable to that of RLD inoculated with DC3000(avrRps4) for all plant lines. This experiment was repeated twice with similar results. cfu, colony-forming units.
We again measured the expression level of transgenes by quantitative RT-PCR using specific primers spanning intron 1 of RPS4 (Table 2). As with intron-deficient transgenes, the expression of chimeric transgenes largely correlated with the transgene copy number and was similar to endogenous rps4-RLD expression. Similarly, transgene expression levels were comparable in resistant RLD-R4L and susceptible RLD-ND, RLD-YH, and RLD-NDYH lines, again suggesting impaired transgene function in susceptible lines. In planta bacterial growth was measured in the representative lines RLD-R4L-A, RLD-ND-A, RLD-YH-B, and RLD-NDYH-A (Figure 3B). The growth of virulent DC3000 was similar in all plants tested (data not shown). Ler and RLD-R4L-A were resistant to DC3000(avrRps4), whereas RLD-ND-A, RLD-YH-B, and RLD-NDYH-A were susceptible at various levels (Figure 3B). The growth of DC3000(avrRps4) in RLD-ND-A and RLD-NDYH-A was similar to that in parental RLD plants and to the in planta growth of virulent DC3000. By contrast, the growth of DC3000(avrRps4) was ∼10-fold lower in RLD-YH-B than in RLD-ND-A, RLD-NDYH-A, and RLD plants and was equally lower than the growth of virulent DC3000 in RLD-YH-B. These findings were consistent with the qualitative disease assay data (Figure 2A).
Table 2.
Expression Levels of Chimeric RPS4-Ler Transgenes
| Transgene | Transgenic Line | Insertion Number a |
Expression Level (±sd) b |
Res./ Susc.c |
|---|---|---|---|---|
| Wild type | RLD-R4L-Ad,e | 2 | 2.0 ± 0.2 | Res. |
| Amino acid change N195D |
RLD-ND-A | 2 | 2.1 ± 0.2 | Susc. |
| RLD-ND-B | 1 | 1.5 ± 0.3 | ||
| RLD-ND-C | 1 | 1.1 ± 0.2 | ||
| Amino acid change Y950H |
RLD-YH-A | 1 | 1.3 ± 0.1 | Susc. |
| RLD-YH-B | 2 | 2.2 ± 0.4 | ||
| RLD-YH-C | 1 | 1.2 ± 0.1 | ||
| RLD-YH-D | 1 | 1.4 ± 0.2 | ||
| N195D and Y950H | RLD-NDYH-A | 1 | 2.0 ± 0.2 | Susc. |
Transgene insertion numbers were determined by DNA gel blot analysis.
Transgene expression levels relative to endogenous rps4-RLD expression levels (n = 3).
Res./Susc., resistant/susceptible.
Data for RLD-R4L-A are taken from Table 1.
Representative transgenic lines used in in planta bacterial growth assays (Figure 3B) are underlined.
Together, these data indicate that the amino acid polymorphisms N195D and Y950H specific to rps4-RLD are sufficient to account for the nonfunctionality of rps4-RLD. Importantly, in the context of this study, the N195D polymorphism would be present in truncated proteins encoded by alternative rps4-RLD transcripts. Because RPS4-Ler and rps4-RLD transcript sequences are >99% identical and expressed at similar levels, this finding indicates that alternative RPS4 transcripts function at the protein level rather than as regulatory RNAs.
The Combination of Intron-Deficient and Truncated Transgenes Partially Reconstitutes RPS4 Function
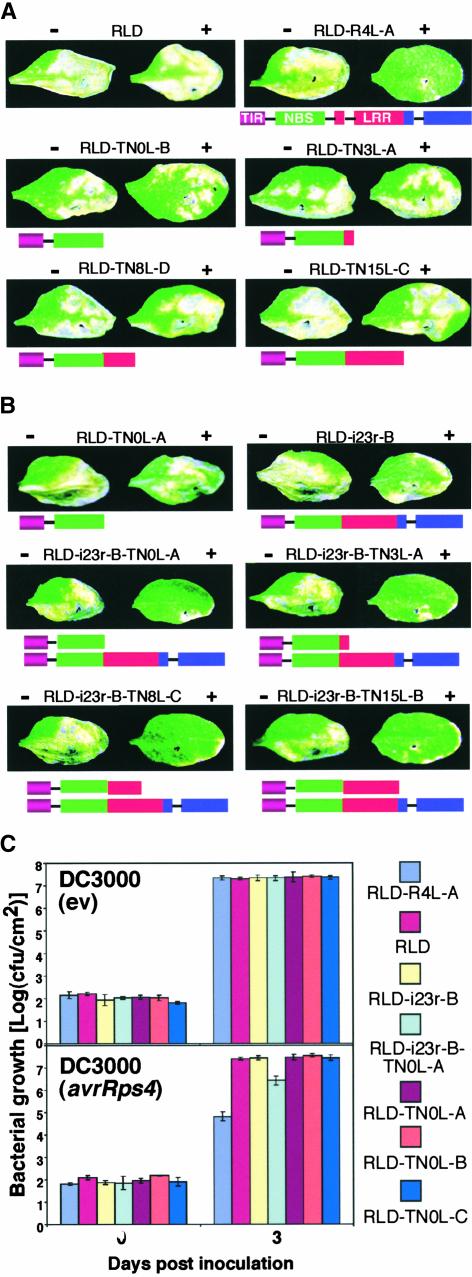
Our results with only a single intron removed from RPS4-Ler (Figures 1A and 1B) indicated that the number of alternative transcripts may need to reach a certain threshold for RPS4 to function. To manipulate the number of alternative transcripts directly, we constructed four truncated TN-encoding RPS4-Ler fragments. Two of them had ORFs that were truncated in a similar region as the naturally occurring alternative RPS4 transcripts and encoded putative proteins lacking LRRs (abbreviated TN0L) or having 3 of 15 LRRs (TN3L). The other two extended the ORF toward the end of the LRR-encoding region with 8 (TN8L) and all 15 (TN15L) LRRs (Figure 4A). As expected, disease assays on homozygous RLD transformants with these constructs showed disease symptoms comparable to those on RLD (Figure 4A).
Figure 4.
The Combined Presence of Regular and Alternative RPS4 Transcripts Is Required for Function.
(A) Disease assays as described in the legend to Figure 1A on RLD lines expressing single truncated RPS4-Ler transgenes encoding putative TN proteins with no LRR (TN0L) or 3 (TN3L), 8 (TN8L), or 15 (TN15L) of 15 LRRs. Plant lines are identified above each photograph. Schemes of truncated RPS4-Ler transgenes are shown below the photographs.
(B) Partial reconstitution of RPS4 function in RLD lines expressing both intron-deficient and truncated RPS4-Ler transgenes. Plant lines are identified above each photograph. Schemes of intron-deficient and truncated RPS4-Ler transgenes are shown below the photographs. RLD-i23r-B has one intron-deficient RPS4-Ler transgene insertion and was the recipient line.
(C) In planta bacterial growth of DC3000(empty vector [ev]) (top graph) and DC3000(avrRps4) (bottom graph) in wild-type RLD and representative single- and double-transgenic lines. Plant lines are indicated at right. Error bars denote standard deviations. This experiment was repeated twice with similar results. cfu, colony-forming units.
To determine whether the combined presence of intron-deficient and truncated transgenes confers resistance, we transformed the TN-encoding fragments of RPS4-Ler into the RLD-i23r-B line and selected homozygous double-transgenic RLD lines with varying TN transgene insertion numbers (Table 3). Interestingly, double-transgenic lines showed partial resistance with each of the truncated constructs (9 of 11 homozygous lines tested; Table 3, Figure 4B). We measured the degree of resistance in representative lines by in vivo bacterial growth experiments (Figure 4C). All lines were equally susceptible to virulent DC3000, indicating that the expression of truncated RPS4 did not constitutively activate plant defenses. Consistent with the disease assays, the growth of DC3000(avrRps4) was ∼10-fold lower in RLD-i23r-B-TN0L-A double-transgenic plants than in the corresponding single-transgenic plants and equally lower than the growth of virulent DC3000 in RLD-i23r-B-TN0L-A. This resistance clearly was partial, because the growth of DC3000(avrRps4) in RLD-R4L-A was ∼30-fold lower than in RLD-i23r-B-TN0L-A. Nevertheless, similar inhibition of DC3000 (avrRps4) growth in RLD-i23r-B-TN0L-A was measured in two additional experiments and was statistically highly significant in each experiment (analysis of variance; P < 0.05).
Table 3.
Transgene Combinations and Copy Numbers in Double-Transgenic RLD Lines
| Recipient Line | Truncated Transgenea |
Double- Transgenic Line |
Insertion Numberb |
Part. Res./ Susc.c |
|---|---|---|---|---|
| RLD-i23r-B | TN0L | TN0L-Ad | 2 | Part. Res. |
| TN0L-B | 2 | |||
| RLD-i23r-B | TN3L | TN3L-A | 2 | Part. Res. |
| TN3L-B | 4 | Susc. | ||
| RLD-i23r-B | TN8L | TN8L-A | 2 | Part. Res. |
| TN8L-B | 1 | Susc. | ||
| TN8L-C | 5 | Part. Res. | ||
| TN8L-D | N.D.e | Part. Res. | ||
| RLD-i23r-B | TN15L | TN15L-A | 2 | Part. Res. |
| TN15L-B | 2 | |||
| TN15L-C | 1 |
Truncated transgenes encode putative TN proteins with no LRR (TN0L) or 3 (TN3L), 8 (TN8L), or 15 (TN15L) of 15 LRRs.
Truncated transgene insertion numbers were determined by DNA gel blot analysis.
Part. Res./Susc., partially resistant/susceptible.
A representative double-transgenic line used in in planta bacterial growth assays (Figure 4C) is underlined.
N.D., not determined.
Importantly, these results show directly that the combined presence of RPS4 full-length and truncated TN-encoding transcripts is required and sufficient for (partial) RPS4 function. The truncated RPS4-Ler transcript in RLD-i23r-B-TN0L-A does not resemble naturally occurring alternative transcripts in that the 3′ half of RPS4, including introns 2 and 3, is missing. However, like the natural alternative transcripts, it encodes a truncated TN protein. This finding again suggests that alternative transcripts function at the protein level rather than as regulatory RNAs.
DISCUSSION
In this study, we have shown that alternative RPS4 transcripts are essential for the function of this R gene. Surprisingly, removal of only one of the two alternatively spliced introns abolished RPS4 function. The characterization of RPS4 transcript variants occurring in wild-type and transgenic lines revealed a new cryptic intron in exon 3. Although rare, removal of this intron also leads to ORF truncation. All of the altered transgenes, including the one lacking both introns 2 and 3, retained the capacity to generate alternative transcripts at some lower frequency, yet all of them failed to confer measurable resistance. We further showed that RPS4 function was partially reconstituted when intron-deficient and truncated RPS4 transgenes were present in the same plant. This finding is a significant advance toward directly showing that the combined presence of transcript variants is required for resistance. Together, these data suggest that resistance requires a minimal number of alternative RPS4 transcripts that is not reached by intron-deficient transgenes. Furthermore, full resistance may depend on finely balanced or tightly regulated numbers of regular and alternative RPS4 transcripts.
Together with the previous study on alternative N gene transcripts (Dinesh-Kumar and Baker, 2000), our data indicate that alternative TNL R gene transcripts that encode putative TN proteins are of general functional significance across plant species. However, unlike with RPS4, partial resistance in the form of a systemic hypersensitive response was observed with an N cDNA that should only generate a full-length N protein–encoding transcript (Dinesh-Kumar and Baker, 2000). This suggests that N has less stringent requirements for a wild-type transcript ratio than does RPS4. Alternatively, viral replication could lead to a higher elicitor concentration within plant cells than that generated by the bacterial delivery of avirulence proteins into host cells, thus activating a partially functional resistance pathway. Finally, tobacco plants may harbor an N gene homolog that can partially substitute for alternative N transcript function.
Lack of Functional Substitution by rps4-RLD or RPS4 Homologs
As proposed in the study of L6, redundancy may hinder the functional characterization of alternative TNL R transcripts (Ayliffe et al., 1999). However, our data from intron-deficient RPS4 transgenes show that there was no complementation of alternative transcript function by the endogenous rps4-RLD allele (discussed below) or RPS4 homologs. High-stringency DNA gel blot analysis had indicated that RPS4 is a single-copy gene (Gassmann et al., 1999). Using the Basic Local Alignment Search Tool (BLAST) algorithm (Altschul et al., 1997), we took advantage of the completed Arabidopsis genome sequence to determine the degree of sequence identity between RPS4 and its closest paralogs. With the TN-encoding nucleotide sequence of RPS4 as a query, only one putative TNL R gene, At5g45060, showed sequence similarity across the whole query sequence (75% nucleotide identity). The corresponding putative TN protein showed 63% amino acid sequence identity (73% similarity) to the RPS4 TN region. Four additional putative proteins were similar in the TN region in the range of 44 to 50% identity (61 to 66% similarity). It is not known whether any of these putative genes actually are subject to alternative splicing.
As a comparison, we aligned the TN-encoding sequence of flax L6 with L9 and M, the two genes that were proposed to substitute for alternative L6 transcript function in transgenic Ward and Trac2 flax lines, respectively (Ayliffe et al., 1999). L6 showed 97% nucleotide sequence identity to L9 and 82% to M. Amino acid sequence identity was 95 and 74% and similarity was 97 and 83%, respectively. This finding shows that sequence similarity between RPS4 and At5g45060 in Arabidopsis is in the same range as the sequence similarity between L6 and M in flax, yet the effect of intron removal on resistance gene function is very different for L6 and RPS4. Possibly, an uncharacterized TNL gene at the M locus showing greater sequence identity with L6 functions in transgenic Trac2 lines. Further study of the At5g45060 gene transcript profile may help us determine the sequence requirements for alternative RPS4 transcript functions. Apart from overall sequence similarity, our analysis of the endogenous rps4-RLD transcript demonstrates that very subtle changes can abolish R gene function. Consequently, it is difficult to predict the functional redundancy of alternative TNL R gene transcripts based on sequence alone.
Alternative Transcripts: Regulatory RNAs or mRNAs Encoding Truncated Proteins?
Although we established a stringent requirement of alternative RPS4 transcripts for resistance, the mechanism by which these alternative transcripts function is unknown. These transcript variants could act as regulatory RNAs to regulate the turnover or accumulation of fully spliced RPS4 transcripts. Alternatively, they could encode truncated RPS4 proteins that may regulate the activity of full-length RPS4. Although not conclusive, we provide preliminary evidence that favors the second model. First, endogenous rps4-RLD transcripts in our transgenic plants failed to substitute for alternative RPS4-Ler transcript function, despite the fact that rps4-RLD and RPS4-Ler sequences are >99% identical to each other. However, the N195D amino acid polymorphism in the putative NBS domain that distinguishes rps4-RLD from functional RPS4 protein abolished function when it was introduced into RPS4-Ler. This amino acid polymorphism would be present in the truncated rps4-RLD protein encoded by alternative rps4-RLD transcripts.
Conversely, truncated RPS4-Ler transgenes that do not resemble naturally occurring alternative RPS4 transcripts were found to partially complement the function of an intron-deficient RPS4-Ler transgene. Note that we did not directly test truncated transgenes with the N195D polymorphism in double-transgenic plants because of the complete lack of complementation by the endogenous rps4-RLD gene, which gives rise to the same spectrum of alternative transcripts as functional RPS4 alleles, in all of our transgenic lines. Future experiments will critically address the existence and stability of functional and nonfunctional truncated RPS4 proteins. Interpretation of our data is complicated by the apparent requirement for a finely balanced ratio of RPS4 transcripts for full resistance, and a regulatory RNA role cannot be excluded at this time. Clearly, investigation of transcript ratios during the infection process and detection of truncated and full-length RPS4 protein are essential next steps. Our success in reconstituting the RPS4 system with two transgenes will allow us to differentially tag the two proteins and vary the expression level of each protein for future biochemical assays.
Based on the observation that alternative TNL R gene transcripts largely encode TN proteins and on the assumption that these truncated proteins are required for resistance, it has been suggested that the LRR domain negatively regulates full-length TNL R protein function (Jordan et al., 2002). We tested four different truncated RPS4-Ler genes that varied in the number of LRRs they encode (from 0 to all 15 LRRs) and noticed no differences in function. If the alternative transcripts function at the protein level, our data would suggest that the LRRs are inconsequential for truncated RPS4 function. Instead, the C-terminal non-LRR region, which is conserved among a subgroup of TNL proteins across plant species (Dodds et al., 2001) and is missing in all of our truncated RPS4-Ler constructs, may negatively regulate full-length RPS4 function. Truncated RPS4 proteins could alleviate this self-inhibition by forming dimers with full-length RPS4 and therefore facilitate activation in a subpopulation of RPS4 resistance complexes. This model would be consistent with the observation that the truncated RPS4-Ler constructs by themselves did not constitutively activate resistance. It may be significant that L6, for which no requirement for alternative transcripts could be discerned, does not possess a C-terminal non-LRR domain, whereas N and RPS4 do (Dodds et al., 2001).
Interestingly, the putative truncated TN proteins resemble adapter proteins such as MyD88 that function in animal innate immunity. MyD88 contains a TIR domain and a second protein–protein interaction domain, the death domain, which allows it to function as an adapter between the interleukin-1 or Toll-like receptor TIR domains and downstream kinase complexes (O'Neill, 2000; Tauszig-Delamasure et al., 2002). In contrast to the limited number of such adapter proteins in animals that interact with diverse receptors, plants may generate specific TN proteins via alternative splicing that interact with only identical or very closely related TIR domains.
METHODS
Plants, Bacterial Strains, and Disease Inoculations
Pseudomonas syringae pv tomato strain DC3000 (DC3000) containing the empty vector pVSP61 or expressing avrRps4 on plasmid pV316-1A were grown as described previously (Hinsch and Staskawicz, 1996). Arabidopsis thaliana accessions RLD and Landsberg erecta (Ler) were grown in a TC-30 environmental growth chamber (Conviron, Winnipeg, Manitoba, Canada) at 24°C and 70% humidity with an 8-h-light/16-h-dark cycle. Disease assays were performed on 5-week-old plants by syringe infiltration with DC3000(empty vector) or DC3000(avrRps4) resuspended to an OD600 of 0.001 (≈1 × 106 colony-forming units/mL) in 10 mM MgCl2. Disease symptoms appeared as chlorosis in the inoculated leaves at 3 to 5 days after inoculation. In planta bacterial growth assays were performed as described (Whalen et al., 1991). Briefly, 5-week-old plants were vacuum-infiltrated with a bacterial suspension of 5 × 104 colony-forming units/mL. Four tissue samples (in triplicate for each time point) were combined for a total of 0.5 cm2 of leaf tissue, ground in 10 mM MgCl2, and plated out in serial dilutions on selective medium at the time points indicated in the figures, as described (Hinsch and Staskawicz, 1996). Statistical analysis of bacterial growth in double-transgenic lines was performed using the analysis of variance procedure in the SAS software package (SAS Institute, Cary, NC).
RPS4 Constructs and Plant Transformations
DNA subcloning, plasmid extraction, and electrophoresis were performed according to standard procedures (Ausubel et al., 1987). A genomic clone of RPS4-Ler was obtained by digesting a cosmid subclone of YAC ABI14A7 (Gassmann et al., 1999) with XhoI and SpeI and subcloning the 6-kb RPS4-Ler fragment into pBluescript KS+ (Stratagene, La Jolla, CA). The resulting clone was digested completely with SpeI and partially with AvaI, blunted, and religated. This removed 570 bp at the 3′ end containing an NdeI and a PpuMI site, thus rendering NdeI and PpuMI within the RPS4-Ler open reading frame unique. All additional sequence swaps were performed with this pBluescript RPS4-Ler clone, which includes 510 bp of native 5′ untranslated region and 865 bp of 3′ untranslated region sequences.
To generate intron-deficient constructs of RPS4-Ler, total RNA isolated from Ler plants was used as a template for reverse transcriptase–mediated (RT) PCR using random hexamer primers. First-strand cDNA was amplified with Pfu DNA polymerase (Stratagene) using primers flanking intron 2 (forward, 5′-AACGCTGGAACTTCCTCAGG-3′; reverse, 5′-CTGTAGGGCAGCTTAAGGTC-3′) and intron 3 (forward, 5′-CAAGGGAAGTTGATCTGAAG-3′; reverse, 5′-TCCTTAAAAGTTGAGCAGCC-3′). Amplified regions included the unique restriction sites Bsu36I and SalI flanking intron 2 and SalI and NdeI flanking intron 3. Individual cloned PCR products were sequenced to verify correct splicing and the absence of amplification errors. Clones were digested with the indicated restriction enzymes and swapped into the RPS4-Ler construct. Constructs were verified by PCR for the absence of introns and by sequencing of cloning junctions.
Amino acid exchange constructs were obtained in a similar manner. With RLD genomic DNA as a template, rps4-RLD sequences containing the amino acid polymorphisms were amplified using Pfu DNA polymerase. For sequences containing the N195D polymorphism, the primers 5′-CAAACCTAGACACAGTAGTG-3′ (forward) and 5′-CTTGGACAACTCGCCTAAGA-3′ (reverse) were used, and for Y950H, the primers 5′-AGTCAGCTTTCTCAACTTAA-3′ (forward) and 5′-TCCAATGAAGACATGGTCTA-3′ (reverse) were used. The amplified regions included the unique restriction sites BglII and Bsu36I flanking N195D and NdeI and PpuMI flanking Y950H. Note that N195D is the only amino acid difference between RLD and Ler in the amplified region. The amplified region surrounding Y950H contains a second amino acid polymorphism at position 1004 (Ile in Ler, Val in RLD, Columbia-0, Wassilewskija-0, and Poppelsdorf-1). Because this polymorphism is shared between rps4-RLD and the functional alleles in the other accessions, we did not expect this polymorphism to influence the function of RPS4. Individual PCR clones were verified by sequencing. Clones were digested with the indicated restriction enzymes and swapped into the RPS4-Ler construct. Constructs were verified using cleaved amplified polymorphic sequence markers that distinguish RPS4-Ler from rps4-RLD in the swapped regions and by sequencing of cloning junctions. To generate truncated constructs, RPS4-Ler lacking both introns 2 and 3 was digested with SacII (in polylinker) and within the gene with BsaI (partial), SalI, AlwnI (partial), or NdeI, blunted, and religated.
Using XhoI and SacI, RPS4-Ler constructs were subcloned into the binary plant transformation vectors pCLD04541 (Bancroft et al., 1997) (full-length constructs) and pCAMBIA1200 (truncated constructs). The truncated open reading frames run into stop codons in pCAMBIA1200 within 10 codons. Transgenic Arabidopsis lines were generated by the floral dipping method (Clough and Bent, 1998) using Agrobacterium tumefaciens strain GV3101. Transformants were screened on half-strength Murashige and Skoog (1962) medium (Invitrogen, Carlsbad, CA) containing 50 μg/mL kanamycin (pCLD04541) or 12.5 μg/mL hygromycin (pCAMBIA1200). Single-locus transgenic lines were selected by scoring for the segregation of selective markers in the T2 and T3 generations. Transgene insertion numbers were determined by DNA gel blot analysis with the left border region of pCLD04541 or with sequence within the pCAMBIA1200 hptII hygromycin resistance gene as a probe using Magna nylon transfer membranes (Osmonics, Inc., Westborough, MA) and a nonradioactive CDP-Star detection kit (Amersham Pharmacia Biotech, Piscataway, NJ).
RT-PCR Analysis of RPS4 Transcripts
Total RNA was isolated using Tri Reagent (Sigma Chemical Co., St. Louis, MO). After RNA treatment with DNaseI, all RT-PCR experiments were performed using SuperScriptII reverse transcriptase (Invitrogen) for first-strand cDNA synthesis with random hexamer primers according to the manufacturer's instructions. For the overall determination of transgene expression levels, we developed a variation of the competitive PCR approach that allows quantitative measurement of target mRNAs (Gilliland et al., 1990). Briefly, we took advantage of the endogenous rps4-RLD allele in transgenic plants as an internal competitor. This ensured equal treatment of target and competitor templates in all steps of the RT-PCR process and robust quantification of transgene expression levels. A primer pair flanking intron 1 was used for PCR (forward, 5′-CAGCATCAGGTGTTCATCAA-3′; reverse, 5′-TTCCCTCCTTACCCTCCTTA-3′). The exponential PCR amplification phase was determined empirically. RT-PCR products were digested with XhoI (which cuts RLD DNA within the amplified region but not Ler DNA), separated on a 1.5% agarose gel, and stained with ethidium bromide. Band intensities of XhoI-treated RT-PCR products were quantified using an AlphaImager 2200 digital imaging system (Alpha Innotech, San Leandro, CA). Relative transgene expression levels were determined with the amount of RT-PCR product arising from the endogenous RLD gene as an internal standard. Three replicate assays were performed with independently isolated RNA.
To display RPS4 transcript variants, nesting forward primers upstream of intron 2 (distal, 5′-AACTTGCACAGAGCCCCAGC-3′; proximal, 5′-CTGTGGCTCCATCAACACAT-3′) and nesting reverse primers downstream of intron 3 (distal, 5′-TCCTTAAAAGTTGATCAGCC-3′; proximal, 5′-GATCGACCCACCTTAAGCAT-3′) were used for PCR. RT-PCR products were separated on 1.5% agarose gels for size comparison. TV3 and TV4 bands were cut from a gel and cloned into the pGEM-T Easy cloning vector (Promega, Madison, WI). To enrich for TV1 and TV2A transcripts, a proximal reverse primer with a 3′ end within intron 3 (5′-GCATGGTGTGTCCTATTACAG-3′) was used; for TV2B, a proximal forward primer with a 3′ end within intron 2 (5′-ATGGTGAGACCAGAAGTTCC-3′) was used. PCR products were cloned directly into pGEM-T Easy. Three to six clones for each transcript were sequenced to determine transcript origin based on a single nucleotide polymorphism between RLD and Ler within the amplified region. TV3 clones were analyzed in all transgenic lines shown in Figure 2A, whereas TV1 clones were analyzed only in RLD-R4L-C, TV2A clones were analyzed in RLD-R4L-C and RLD-i2r-B, TV2B clones were analyzed in RLD-i3r-A, and TV4 clones were analyzed in Ler, RLD-R4L-C, and RLD-i23r-B. Although not quantitative, this approach enabled a qualitative verification of all possible transgene-derived transcripts in transgenic lines.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Walter Gassmann, gassmannw@missouri.edu.
Accession Numbers
The GenBank accession number for the TNL R gene (locus At5g45060) is NM_123874, that for the corresponding putative protein is NP_199319, and that for the vector pCAMBIA1200 is AF234292.
Acknowledgments
We thank Dipanwita Saha and Nkemdi Anyanwu for technical assistance, Mark Ellersieck for assistance with statistical analyses, and Jim Schoelz and Sharon Pike for critical reading of the manuscript. This work was supported by University of Missouri Research Board Grant RB 01-133 and U.S. Department of Agriculture/National Research Initiative Competitive Grant 2002-35319-12639 to W.G.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.013474.
References
- Aarts, N., Metz, M., Holub, E., Staskawicz, B.J., Daniels, M.J., and Parker, J.E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 10306–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.G., and Struhl, K. (1987). Current Protocols in Molecular Biology. (New York: Greene Publishing Associates and Wiley-Interscience).
- Axtell, M.J., and Staskawicz, B.J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112, 369–377. [DOI] [PubMed] [Google Scholar]
- Ayliffe, M.A., Frost, D.V., Finnegan, E.J., Lawrence, G.J., Anderson, P.A., and Ellis, J.G. (1999). Analysis of alternative transcripts of the flax L6 rust resistance gene. Plant J. 17, 287–292. [DOI] [PubMed] [Google Scholar]
- Baker, B., Zambryski, P., Staskawicz, B., and Dinesh-Kumar, S.P. (1997). Signaling in plant-microbe interactions. Science 276, 726–733. [DOI] [PubMed] [Google Scholar]
- Bancroft, I., Love, K., Bent, E., Sherson, S., Lister, C., Cobett, C., Goodman, H.M., and Dean, C. (1997). A strategy involving the use of high redundancy YAC subclone libraries facilitates the contiguous representation in cosmid and BAC clones of 1.7 Mb of the genome of the plant Arabidopsis thaliana. Weeds World 4, 1–9. [Google Scholar]
- Bent, A.F. (1996). Plant disease resistance genes: Function meets structure. Plant Cell 8, 1757–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Dinesh-Kumar, S.P., and Baker, B.J. (2000). Alternatively spliced N resistance gene transcripts: Their possible role in tobacco mosaic virus resistance. Proc. Natl. Acad. Sci. USA 97, 1908–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh-Kumar, S.P., Whitham, S., Choi, D., Hehl, R., Corr, C., and Baker, B. (1995). Transposon tagging of tobacco mosaic virus resistance gene N: Its possible role in the TMV-N-mediated signal transduction pathway. Proc. Natl. Acad. Sci. USA 92, 4175–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N., Lawrence, G.J., and Ellis, J.G. (2001). Six amino acid changes confined to the leucine-rich repeat β-strand/β-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell 13, 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk, A., Feys, B.J., Frost, L.N., Jones, J.D.G., Daniels, M.J., and Parker, J.E. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 96, 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor, H.H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–296. [Google Scholar]
- Gassmann, W., Hinsch, M.E., and Staskawicz, B.J. (1999). The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 20, 265–277. [DOI] [PubMed] [Google Scholar]
- Gilliland, G., Perrin, S., and Bunn, H.F. (1990). Competitive PCR for quantitation of mRNA. In PCR Protocols: A Guide to Methods and Applications, M.A. Innis, D.H. Gelfand, J.J. Sninsky, and T.J. White, eds (San Diego, CA: Academic Press), pp. 60–69.
- Hammond-Kosack, K.E., and Jones, J.D.G. (1996). Resistance gene-dependent plant defense responses. Plant Cell 8, 1773–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsch, M., and Staskawicz, B.J. (1996). Identification of a new Arabidopsis disease resistance locus, RPS4, and cloning of the corresponding avirulence gene, avrRps4, from Pseudomonas syringae pv. pisi. Mol. Plant-Microbe Interact. 9, 55–61. [DOI] [PubMed] [Google Scholar]
- Imler, J.L., and Hoffmann, J.A. (2001). Toll receptors in innate immunity. Trends Cell Biol. 11, 304–311. [DOI] [PubMed] [Google Scholar]
- Jordan, T., Schornack, S., and Lahaye, T. (2002). Alternative splicing of transcripts encoding Toll-like plant resistance proteins: What's the functional relevance to innate immunity? Trends Plant Sci. 7, 392–398. [DOI] [PubMed] [Google Scholar]
- Lawrence, G.J., Finnegan, E.J., Ayliffe, M.A., and Ellis, J.G. (1995). The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N. Plant Cell 7, 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q.T., and Verma, I.M. (2002). NF-kappa B regulation in the immune system. Nat. Rev. Immunol. 2, 725–734. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Belkhadir, Y., Alonso, J.M., Ecker, J.R., and Dangl, J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112, 379–389. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Holt, B.F., Wiig, A., and Dangl, J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108, 743–754. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Noël, L., Moores, T.L., van der Biezen, E.A., Parniske, M., Daniels, M.J., Parker, J.E., and Jones, J.D.G. (1999). Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11, 2099–2111. [PMC free article] [PubMed] [Google Scholar]
- O'Neill, L. (2000). The Toll/interleukin-1 receptor domain: A molecular switch for inflammation and host defence. Biochem. Soc. Trans. 28, 557–563. [DOI] [PubMed] [Google Scholar]
- Parker, J.E., Coleman, M.J., Szabò, V., Frost, L.N., Schmidt, R., van der Biezen, E., Moores, T., Dean, C., Daniels, M., and Jones, J.D.G. (1997). The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the Toll and interleukin-1 receptors with N and L6. Plant Cell 9, 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J.E., Holub, E.B., Frost, L.N., Falk, A., Gunn, N.D., and Daniels, M.J. (1996). Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8, 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz, B.J., Ausubel, F.M., Baker, B.J., Ellis, J.G., and Jones, J.D.G. (1995). Molecular genetics of plant disease resistance. Science 268, 661–667. [DOI] [PubMed] [Google Scholar]
- Tauszig-Delamasure, S., Bilak, H., Capovilla, M., Hoffmann, J.A., and Imler, J.L. (2002). Drosophila MyD88 is required for the response to fungal and Gram-positive bacterial infections. Nat. Immunol. 3, 91–97. [DOI] [PubMed] [Google Scholar]
- Whalen, M.C., Innes, R.W., Bent, A.F., and Staskawicz, B.J. (1991). Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3, 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham, S., Dinesh-Kumar, S.P., Choi, D., Hehl, R., Corr, C., and Baker, B. (1994). The product of the tobacco mosaic virus resistance gene N: Similarity to Toll and the interleukin-1 receptor. Cell 78, 1101–1115. [DOI] [PubMed] [Google Scholar]