Abstract
Dynamin-related proteins are high molecular weight GTP binding proteins and have been implicated in various biological processes. Here, we report the functional characterization of two dynamin homologs in Arabidopsis, Arabidopsis dynamin-like 1C (ADL1C) and Arabidopsis dynamin-like 1E (ADL1E). ADL1C and ADL1E show a high degree of amino acid sequence similarity with members of the dynamin family. However, both proteins lack the C-terminal Pro-rich domain and the pleckstrin homology domain. Expression of the dominant-negative mutant ADL1C[K48E] in protoplasts obtained from leaf cells caused abnormal mitochondrial elongation. Also, a T-DNA insertion mutation at the ADL1E gene caused abnormal mitochondrial elongation that was rescued by the transient expression of ADL1C and ADL1E in protoplasts. In immunohistochemistry and in vivo targeting experiments in Arabidopsis protoplasts, ADL1C and ADL1E appeared as numerous speckles and the two proteins colocalized. These speckles were partially colocalized with F1-ATPase-γ:RFP, a mitochondrial marker, and ADL2b localized at the tip of mitochondria. These results suggest that ADL1C and ADL1E may play a critical role in mitochondrial fission in plant cells.
INTRODUCTION
Dynamin is a high molecular weight GTP binding protein that is known to be involved in endocytosis in rat brain (Obar et al., 1990). Since the discovery of dynamin (now called dynamin I) in rat brain cells, numerous isoforms and homologs have been isolated in a variety of eukaryotic organisms from yeast to human (Obar et al., 1990; Rothman et al., 1990; Cook et al., 1994; Dombrowski and Raikhel, 1995; Gammie et al., 1995; Gu and Verma, 1996; Kang et al., 1998; Yoon et al., 1998). A characteristic feature of the dynamin family is the conserved N-terminal GTPase domain. However, the rest of the polypeptide sequence is less well conserved. Although dynamin and its related proteins belong to a family of high molecular weight GTPases, these proteins are known to be involved in very diverse biological processes (Rothman et al., 1990; Jones and Fangman, 1992; Guan et al., 1993; Herskovits et al., 1993; Wilsbach and Payne, 1993; Damke et al., 1994; Gammie et al., 1995; Vallee and Okamoto, 1995; Gu and Verma, 1996; Park et al., 1997).
One group of proteins, which includes dynamin I, ADL6, and Vps1p, has been shown to act at various steps of intracellular trafficking, such as endocytosis (Vallee and Okamoto, 1995) and vacuolar trafficking (Rothman et al., 1990; Jin et al., 2001). Another group of proteins, which includes DLP1/Drp1, Dnm1p, Mgm1p, and ADL2b, has been shown to be involved in the regulation of mitochondrial morphology (Guan et al., 1993; Otsuga et al., 1998; Pitts et al., 1999; Shepard and Yaffe, 1999; Smirnova et al., 2001, Arimura and Tsutsumi, 2002). Mutation of these proteins or expression of dominant-negative mutants results in the elongation or fusion of mitochondria, indicating that these proteins are involved in mitochondrial fission. In addition, ADL2 has been shown to be targeted to the chloroplast and is thought to be involved in biological processes specific to the plastid (Kang et al., 1998; Kim et al., 2001b). Finally, proteins such as phragmoplastin and ADL1A are thought to play important roles in the formation of the cell division plate in plant cells (Gu and Verma, 1996; Kang et al., 2001, 2003).
Of the dynamin-like proteins, most studies have been done with dynamin I (Gout et al., 1993; Herskovits et al., 1993; Damke et al., 1994; Salim et al., 1994; Hinshaw and Schmid, 1995; Shupliakov et al., 1997; Sever et al., 2000), and the detailed mechanism of action for dynamin I is now well understood at the molecular level. Recently, other members of the dynamin family also have been studied at the molecular level (Vater et al., 1992; Fukushima et al., 2001; Kim et al., 2001b; Yoon et al., 2001). These studies indicate that dynamin I and related proteins are composed of multiple functional domains. Numerous studies have addressed the biological function of these domains. The Pro-rich domain has been shown to interact with the SH3 domain of many proteins and is thought to play a critical role in transmitting the regulatory signals mediated by the SH3 domain–containing proteins (Gout et al., 1993; Okamoto et al., 1997; Shupliakov et al., 1997). The pleckstrin homology (PH) domain has been shown to bind to phospholipids and is thought to be responsible for the membrane association of the molecule (Ferguson et al., 1994; Salim et al., 1994; Timm et al., 1994; Shaw, 1996). The assembly domain is located between the PH domain and the Pro-rich domain and is involved in intramolecular and intermolecular interactions between dynamin molecules, which result in self-assembly of these proteins into a high molecular weight homopolymeric form (Muhlberg et al., 1997; Okamoto et al., 1999; Smirnova et al., 1999; Sever et al., 2000). In addition, the assembly domain is proposed to regulate the N-terminal GTPase activity and thereby to function as a GTPase effector domain. Although the PH domain and the Pro-rich domain appear to be absent from ADL1 and ADL2 of plant cells (Dombrowski and Raikhel, 1995; Kang et al., 1998), DLP1/Drp1 of animal cells (Yoon et al., 1998), and yeast Dnm1p (Gammie et al., 1995), these proteins still bind to membranes (Park et al., 1997; Yoon et al., 1998; Kim et al., 2001b). For example, ADL2 has been shown to bind to phosphatidylinositol 4-phosphate (Kim et al., 2001b). Thus, some of these proteins may have a domain for membrane association other than the PH domain.
Here, we describe two new isoforms of Arabidopsis dynamin-like proteins, ADL1C and ADL1E. We present evidence that ADL1C and ADL1E partially localize to mitochondria and are involved in morphogenesis of the mitochondria.
RESULTS
ADL1C and ADL1E Are Present as Punctate Stains
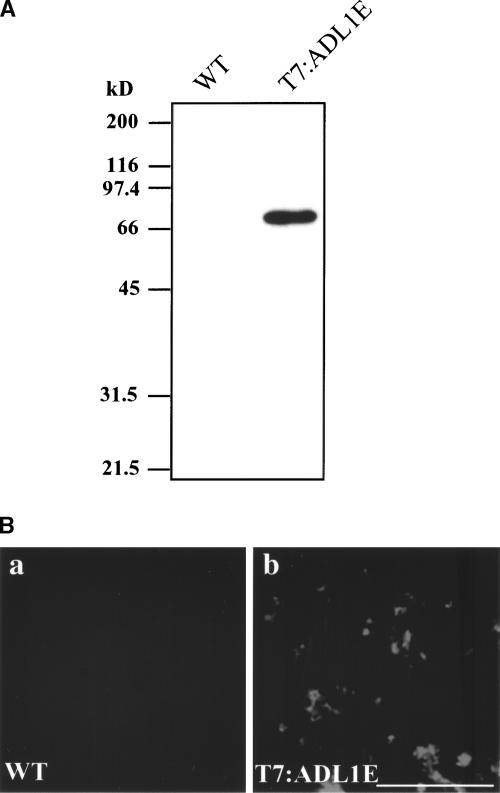
The Arabidopsis genome encodes a large number of dynamin-like proteins (Kang et al., 1998, 2001). Among them, we selected two closely related isoforms, ADL1C and ADL1E, and investigated their localization by immunohistochemistry. Because many isoforms of ADL are present in the Arabidopsis genome, we generated transgenic Arabidopsis plants expressing T7-tagged ADL1E, which can be detected specifically using a monoclonal anti-T7 antibody. The T7:ADL1E construct was introduced into a binary vector and transferred subsequently into plant cells by Agrobacterium tumefaciens–mediated transformation (Clough and Bent, 1998). Transgenic plants were selected on kanamycin plates. To confirm the expression of T7:ADL1E, protein extracts were prepared from the transgenic plants and analyzed by protein gel blot analysis using a monoclonal anti-T7 antibody. As shown Figure 1A, the transgenic plant expressed an immunoreactive protein species that migrated to a position corresponding to a molecular mass of 70 kD. By contrast, in the control extract, this immunoreactive band was not observed, indicating that the antibody specifically detects T7:ADL1E. Immunohistochemistry with root cells of transgenic plants was performed using the monoclonal anti-T7 antibody. As shown in Figure 1B, the anti-T7 antibody signal appeared as speckles throughout the cells. By contrast, no immunoreactivity was observed in control cells, supporting the earlier observation that the monoclonal anti-T7 antibody specifically detects T7:ADL1E.
Figure 1.
Immunohistochemical Localization of ADL1E.
(A) Expression of T7:ADL1E in transgenic plants. Total protein (10 μg) obtained from leaf tissues of nontransgenic wild-type (WT) or transgenic plants was separated on an SDS-polyacrylamide gel and transferred onto nylon polyvinylidene difluoride membranes. The blots were probed with a monoclonal anti-T7 antibody.
(B) Localization of T7:ADL1E. Root protoplasts of nontransgenic wild-type (a) or transgenic (b) Arabidopsis grown in liquid medium for 1 week were prepared for immunohistochemistry. Root tip cells were labeled with the monoclonal anti-T7 antibody, and lissamine rhodamine–labeled anti-mouse IgG was used as the secondary antibody. Bar = 10 μm.
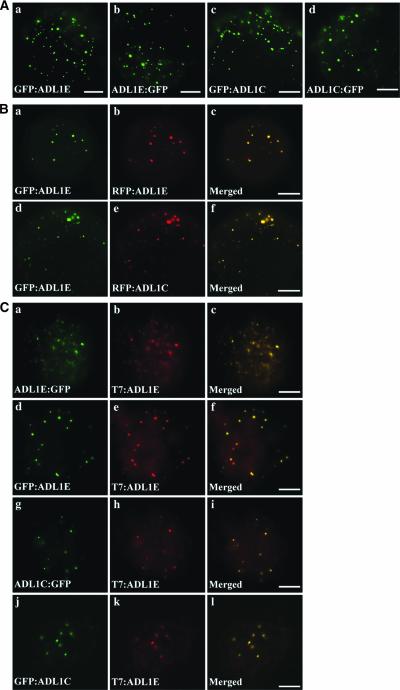
To further define the localization of ADL1E, we performed in vivo targeting experiments in protoplasts. ADL1C and ADL1E were tagged with green fluorescent protein (GFP) or red fluorescent protein (RFP) at either the N or C terminus (Davis and Vierstra, 1998; Heikal et al., 2000). The fusion constructs then were introduced into Arabidopsis protoplasts using the polyethylene glycol–mediated transformation method (Jin et al., 2001), and localization of these fusion proteins was examined by fluorescence microscopy. As shown in Figure 2A, all green fluorescent signals of GFP-fused ADL1C and ADL1E appeared as numerous speckles, as was observed with T7:ADL1E. In addition, ADL1E tagged with GFP at the N terminus also colocalized with RFP-tagged ADL1E at the N terminus (Figures 2Ba to 2Bc). Furthermore, when the GFP:ADL1E and RFP:ADL1C fusion proteins were expressed in Arabidopsis protoplasts, the punctate green fluorescent signals of GFP:ADL1E exactly overlapped the red fluorescent signals of RFP:ADL1C (Figures 2Bd to 2Bf), indicating that ADL1E and ADL1C are colocalized. Next, we examined the colocalization of ADL1C and ADL1E tagged with GFP at the C or N terminus with T7-tagged ADL1E. Protoplasts obtained from transgenic plants harboring T7:ADL1E were transformed with GFP:ADL1C, ADL1C:GFP, ADL1E:GFP, or GFP:ADL1E and fixed at 24 h after transformation. As shown in Figure 2C, green fluorescent signals of all GFP fusion forms of ADL1C or ADL1E closely overlapped the red signals of T7:ADL1E at points of punctate staining. These results strongly suggest that different tags fused to ADL1C or ADL1E at N or C termini do not affect the localization of these proteins and that both ADL1C and ADL1E are targeted to the same organelles, yielding a punctate staining pattern.
Figure 2.
In Vivo Localization of ADL1C and ADL1E.
(A) Patterns of GFP-fused ADL1C and ADL1E. Protoplasts were transformed with the constructs indicated, and the localization of these proteins was examined 24 to 48 h after transformation. Bars = 20 μm.
(B) Colocalization of ADL1C and ADL1E. Protoplasts were transformed with the constructs indicated, and the localization of these fusion proteins was examined. Yellow signal indicates overlap between the green and red fluorescent signals. Bars = 20 μm.
(C) Colocalization of T7-tagged ADL1E with GFP-tagged ADL1C or ADL1E. Protoplasts obtained from transgenic plants harboring T7:ADL1E were transformed with the constructs indicated, and the localization of these proteins was examined by immunohistochemistry. T7:ADL1E was detected from the fixed protoplasts using anti-T7 antibody, whereas GFP-fused proteins were observed directly by green fluorescence. Bars = 20 μm.
Expression of ADL1C[K48E] Causes the Elongation of Mitochondria in Protoplasts
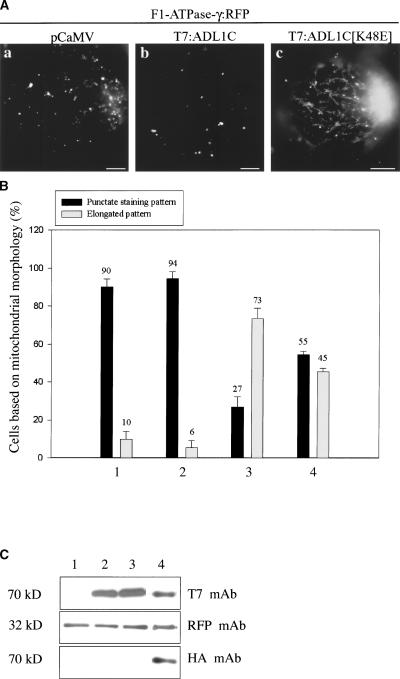
Dynamin-like proteins play important roles in various biological processes. One is in intracellular trafficking, as exemplified by dynamin I in animal cells (Herskovits et al., 1993; Sever et al., 2000) and ADL6 in plant cells (Jin et al., 2001). Another well-characterized role is in the division of mitochondria, as in the case of DLP1/Drp1 in animals (Pitts et al., 1999; Smirnova et al., 2001; Yoon et al., 2001) and Dnm1p in yeast (Otsuga et al., 1998). To investigate the biological role of ADL1C and ADL1E, we used a dominant-negative mutant (GDP-bound form) of ADL1C. Mutation of the first GTP binding motif in many GTP binding proteins, including ADL6 and dynamin I, has been shown to produce a dominant-negative mutant effect through competition with the endogenous proteins (Herskovits et al., 1993; Jin et al., 2001). First, we addressed the possible role in mitochondria. We generated a dominant-negative mutant, ADL1C[K48E], and tagged it with the T7 epitope at the N terminus. In this study, we used F1-ATPase-γ tagged with RFP to visualize mitochondria.
Previously F1-ATPase-γ:GFP was shown to be targeted to mitochondria (Niwa et al., 1999). We replaced the GFP coding region from F1-ATPase-γ:GFP with the RFP coding region to generate F1-ATPase-γ:RFP and found that the RFP form closely overlapped the GFP form in protoplasts, confirming that the RFP form of F1-ATPase-γ:RFP is targeted to mitochondria (data not shown). T7:ADL1C[K48E] was introduced into protoplasts together with the reporter protein F1-ATPase-γ:RFP. As shown in Figure 3Aa, when F1-ATPase-γ:RFP alone was expressed in protoplasts, the red fluorescent signals were present as punctate stains, as expected. Expression of the wild-type T7:ADL1C did not change the pattern of red fluorescent signals (Figure 3Ab). However, when T7:ADL1C[K48E] was introduced into protoplasts together with F1-ATPase-γ:RFP, the majority of protoplasts showed elongated mitochondria, as shown in Figure 3Ac. To quantify the effect of ADL1C[K48E] on mitochondrial morphology, we counted the number of protoplasts based on the mitochondrial morphology. As shown in Figure 3B, in control protoplasts, 90% of protoplasts showed the pattern shown in Figure 3Aa. Also, >94% of protoplasts showed the punctate staining pattern of mitochondria in the presence of T7:ADL1C (Figure 3Ab). However, in the presence of the dominant-negative mutant, T7:ADL1C[K48E], ∼73% of protoplasts showed elongated mitochondria, as shown in Figure 3Ac. Only a minor portion (∼27%) of protoplasts showed the normal punctate staining pattern of mitochondria (Figure 3Ab).
Figure 3.
Elongation of Mitochondria in the Presence of Overexpressed ADL1C[K48E].
(A) Protoplasts were transformed with F1-ATPase-γ:RFP plus pCaMV (a), F1-ATPase-γ:RFP plus T7:ADL1C (b), or F1-ATPase-γ:RFP plus T7:ADL1C[K48E] (c), and the fluorescence emitted by RFP was detected 24 to 48 h after transformation. Bars = 20 μm.
(B) Quantification of protoplasts based on mitochondrial morphology. Protoplasts were transformed with F1-ATPase-γ:RFP plus pCaMV (bars 1), F1-ATPase-γ:RFP plus T7:ADL1C (bars 2), F1-ATPase-γ:RFP plus T7:ADL1C[K48E] (bars 3), or F1-ATPase-γ:RFP plus HA:ADL1C plus T7:ADL1[K48E] (bars 4). The number of protoplasts was counted based on the mitochondrial morphology at 48 h after transformation. The punctate staining pattern (shown in [Aa] and [Ab]) was considered normal, whereas the elongated pattern (shown in [Ac]) was considered abnormal. More than 30 randomly selected protoplasts were counted at each time for each condition. The transformation experiment was performed three times with identical conditions.
(C) Protein gel blot analysis. Protein extracts were obtained from protoplasts transformed with F1-ATPase-γ:RFP plus pCaMV (lane 1), F1-ATPase-γ:RFP plus T7:ADL1C (lane 2), F1-ATPase-γ:RFP plus T7:ADL1C[K48E] (lane 3), or F1-ATPase-γ:RFP plus HA:ADL1C plus T7:ADL1[K48E] (lane 4) and examined by protein gel blot analysis using the monoclonal anti-T7 (T7 mAb), anti-HA (HA mAb), and anti-RFP (RFP mAb) antibodies for T7-tagged ADL1C, HA-tagged ADL1C, and F1-ATPase-γ:RFP, respectively. pCaMV was included in the transformation as a control DNA.
Next, we examined whether an increase in the amount of wild-type ADL1C could suppress the effect of the dominant-negative mutant, ADL1C[K48E], on mitochondrial morphology. Protoplasts were cotransformed with three constructs, HA: ADL1C, T7:ADL1C[K48E], and F1-ATPase-γ:RFP, and mitochondrial morphology was examined at 48 h after transformation. At that time, wild-type ADL1C expression was detected by protein gel blot analysis, because it was tagged with the different hemagglutinin (HA) epitope. As shown in Figure 3B, we still observed abnormal elongation of mitochondria in protoplasts transformed with both HA:ADL1C and T7:ADL1C[K48E]. However, the proportion of cells with elongated mitochondria was reduced to 45% in the presence of both HA:ADL1C and T7:ADL1C[K48E] compared with 73% in the presence of ADL1C[K48E] alone, suggesting that coexpression of wild-type ADL1C partially suppresses the effect of ADL1C[K48E] on mitochondrial morphology. These results strongly suggest that ADL1C and ADL1E may be involved in the fission of mitochondria, as are other dynamin-like proteins localized at the mitochondria (Otsuga et al., 1998; Pitts et al., 1999; Smirnova et al., 2001; Yoon et al., 2001). To confirm that the wild-type and mutant forms of ADL1C as well as F1-ATPase-γ:RFP were expressed in the protoplasts, protein extracts were prepared and used for protein gel blot analysis using anti-T7, anti-HA, or anti-RFP antibodies. As shown in Figure 3C, the wild-type and mutant forms of ADL1C were expressed at nearly the same level. Also, F1-ATPase-γ:RFP was at a nearly equal level.
Mitochondria Are Elongated in Mutant Plants with a T-DNA Insertion at the ADL1E Locus
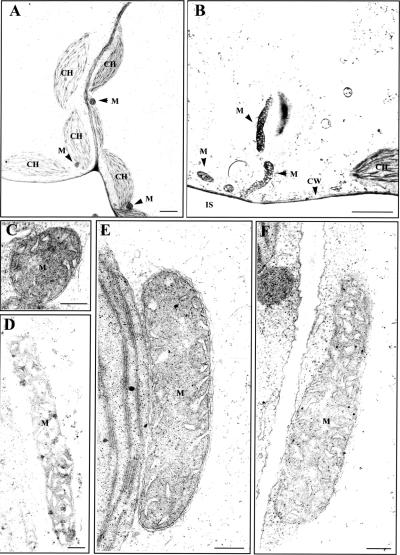
To obtain independent evidence for the role of ADL1C and ADL1E in mitochondrial morphology, we searched the collection of SALK T-DNA insertion lines for a T-DNA insertion mutation at ADL1C or ADL1E and found a mutant line (SALK-060080) that has a T-DNA insertion at the ADL1E gene. In the mutant plant, a T-DNA is reported to be inserted at the 14th exon of ADL1E. Total genomic DNA was obtained from homozygous plants and used to verify the presence of the T-DNA at ADL1E. To confirm the presence of T-DNA, PCR was performed using genomic DNA prepared from homozygotes as a template, with a set of ADL1E-specific and left-border primers or a set of ADL1E-specific primers. As expected, a T-DNA–specific band but not an ADL1E band was obtained from the PCR (data not shown), confirming that a T-DNA was inserted into ADL1E. adl1e mutant plants did not have any noticeable morphological alterations or growth defects (data not shown). To examine the morphology of mitochondria at the electron microscopic level, ultrathin sections were prepared from leaf tissues of adl1e and wild-type plants. Transmission electron microscopy revealed that a large proportion of mitochondria (36%) were abnormally elongated in adl1e mutant plants compared with wild-type plants (Figure 4, Table 1), confirming the morphological change observed in protoplasts expressing the mutant form of ADL1C. However, other organelles, such as chloroplasts, were normal in adl1e mutants.
Figure 4.
Mitochondria Are Elongated in adl1e Mutants.
Ultrathin sections prepared from leaf tissues of wild-type ([A] and [C]) and adl1e mutant ([B] and [D] to [F]) plants were examined by transmission electron microscopy. CH, chloroplast; CW, cell wall; IS, intercellular space; M, mitochondria. Bars = 2 μm for (A) and (B) and 200 nm for (C) to (F).
Table 1.
Morphology of Mitochondria in Wild-Type and adl1e Mutant Plants
| Plant Material
|
||
|---|---|---|
| Mitochondrial Group | Wild Type | adl1e Mutant |
| Elongated (%) | 0 | 36 |
| Normal (%) | 100 | 64 |
Mitochondria were divided into two groups, normal and elongated forms, based on the length of the mitochondria. Normal morphology indicates mitochondria with an oval shape, as shown in Figure 4c. In the elongated form, the length of the mitochondria is longer than threefold that of normal oval-shaped mitochondria. Data were obtained from 87 samples chosen randomly from serial sections at magnifications of ×30,000 to ×50,000.
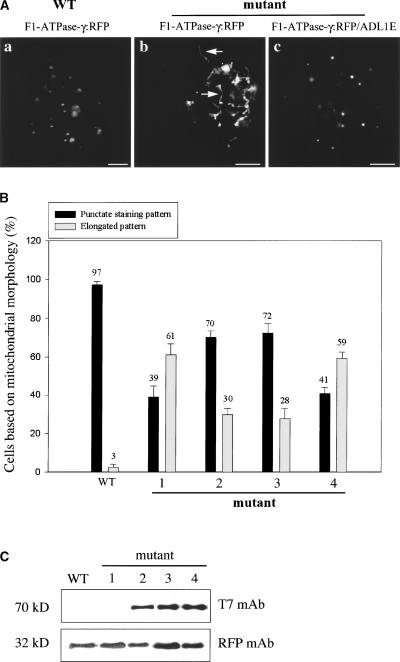
Next, we examined whether the abnormal mitochondrial morphology of the adl1e mutant could be rescued by transiently expressing ADL1E in protoplasts. To address this question, we first examined the morphology of mitochondria in protoplasts prepared from the leaf tissues of mutant plants using fluorescence microscopy. Protoplasts were transformed with F1-ATPase-γ:RFP, and mitochondria were visualized. When the morphology of mitochondria was examined by fluorescence microscopy, the majority of protoplasts obtained from adl1e mutants had elongated mitochondria (Figure 5Ab). In wild-type plants, the punctate staining pattern of mitochondria was observed (Figure 5Aa). When this effect was quantified, 39% of ad11e mutant cells had a punctate staining pattern and 61% had elongated mitochondria, whereas in wild-type plants, >97% of cells had the punctate staining pattern. This result further confirmed the transmission electron microscopy observations. Next, we examined whether the elongated mitochondria could be rescued by the transient expression of T7:ADL1E. Protoplasts prepared from mutant plants were transformed with T7:ADL1E together with F1-ATPase-γ:RFP, and mitochondrial morphology was observed after transformation. As shown in Figure 5Ac, at 48 h after transformation, the majority of transformed cells exhibited the punctate staining pattern (Figure 5Ac). When quantified, cells with the punctate staining pattern were increased to 70%; concomitantly, cells with elongated mitochondria were decreased to 30% (Figure 5B).
Figure 5.
Abnormal Morphology of Mitochondria in adl1e Mutants Can Be Rescued by Transient Expression of ADL1C and ADL1E in Protoplasts.
(A) Complementation of adl1e by transiently expressed ADL1E. Protoplasts were prepared from adl1e mutant plants and transformed with F-ATPase-γ:RFP alone or together with T7:ADL1E. As a control, protoplasts obtained from wild-type (WT) plants were transformed with F1-ATPase-γ:RFP. The fluorescence emitted by RFP was examined at various time points after transformation. Bars = 20 μm.
(B) Quantification of morphological change. Protoplasts obtained from wild-type and adl1e mutant plants were transformed with F1-ATPase-γ:RFP (bars WT and 1), F1-ATPase-γ:RFP plus T7:ADL1E (bars 2), F1-ATPase-γ:RFP plus T7:ADL1C (bars 3), and F1-ATPase-γ:RFP plus T7:ADL1C[K48E] (bars 4). The number of protoplasts was counted based on the mitochondrial morphology indicated by RFP fluorescence. The mitochondria shown in (Aa) and (Ac) were considered to be in a punctate staining pattern, whereas the mitochondria shown in (Ab) were considered to be in an elongated pattern. More than 50 transformed cells were counted at each time point in triplicate experiments. The numbers and error bars indicate means and standard deviations, respectively.
(C) Expression of T7:ADL1C and T7:ADL1E in protoplasts. Protein extracts were prepared from protoplasts transformed with F-ATPase-γ:RFP alone (lanes WT and 1) or together with T7:ADL1E (lane 2), ADL1C (lane 3), or ADL1C[K48E] (lane 4) and analyzed by protein gel blot analysis with anti-RFP (RFP mAb) and anti-T7 (T7 mAb) antibodies.
To investigate whether ADL1C could rescue the abnormal morphology of mitochondria in protoplasts obtained from adl1e mutants, protoplasts obtained from adl1e mutants were transformed with T7:ADL1C. As shown in Figure 5B, 72% of cells showed the punctate staining pattern in the presence of T7:ADL1C, indicating that overexpressed ADL1C can complement the loss of ADL1E in the mutant as efficiently as ADL1E itself. This result further confirms that ADL1C is functionally equivalent to ADL1E with respect to their ability to maintain mitochondrial morphology. As a control, we examined the effect of the dominant-negative mutant of ADL1C, ADL1C[K48E], on mitochondrial morphology by introducing it into protoplasts obtained from adl1e mutants. As shown in Figure 5B, 59% of cells showed the elongated mitochondrial pattern, indicating that the dominant-negative mutant could neither rescue the abnormal mitochondrial morphology nor provoke any additional detrimental effects on mitochondrial morphology when examined by fluorescence microscopy. This result further supports the notion that ADL1C and ADL1E are involved in the maintenance of normal mitochondrial morphology. The expression of T7:ADL1C, T7:ADL1E, T7:ADL1C[K48E], and F1-ATPase-γ:RFP in protoplasts was confirmed by protein gel blot analysis with anti-T7 and anti-GFP antibodies (Figure 5C).
ADL1C Partially Colocalizes with F1-ATPase-γ:RFP and ADL2b
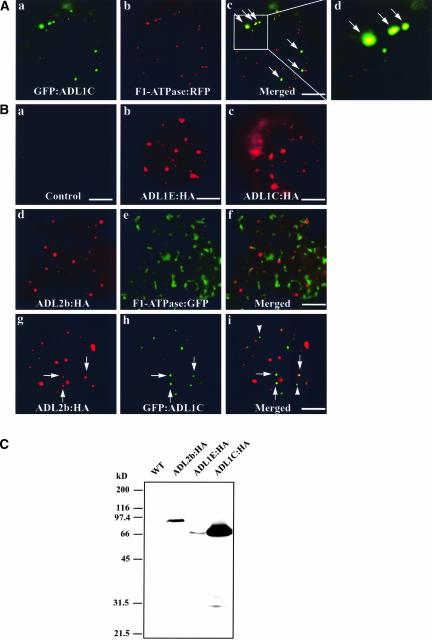
The results described above strongly suggest that ADL1C and ADL1E may localize to mitochondria, as in the case of DLP1/Drp1 in animals (Pitts et al., 1999; Smirnova et al., 2001; Yoon et al., 2001), Dnm1p in yeast (Otsuga et al., 1998), and ADL2b in plants (Arimura and Tsutsumi, 2002). To examine this possibility, protoplasts were transformed with F1-ATPase-γ:RFP and GFP:ADL1C and the localization of these proteins was examined. As shown in Figures 6Aa to 6Ad), both GFP:ADL1C and F1-ATPase-γ:RFP were present as punctate staining patterns. Interestingly, ∼20% (±7%, n = 900) of the green fluorescent GFP:ADL1C signal colocalized with the red punctate stains of F1-ATPase-γ:RFP, indicating that a fraction of ADL1C-positive speckles may represent mitochondria.
Figure 6.
GFP:ADL1C Partially Colocalizes with F1-ATPase-γ:RFP and ADL2b.
(A) Localization of ADL1C at mitochondria. Protoplasts were transformed with the indicated constructs, and the localization of these proteins was examined 24 to 48 h after transformation. Arrows indicate overlap of green and red fluorescent signals. Bars = 20 μm.
(B) Partial colocalization of ADL1C with ADL2b in the mitochondria. Protoplasts were transformed with the indicated constructs, and the localization of the encoded proteins was examined by immunohistochemistry. HA-tagged proteins were detected with a monoclonal anti-HA antibody. GFP-tagged proteins were observed directly by green fluorescence. Arrows indicate the overlap between ADL2b:HA and GFP:ADL1C. Arrowheads indicate GFP:ADL1C-positive speckles that did not colocalize with ADL2b:HA. Bars = 20 μm.
(C) Protein gel blot analysis of transiently expressed ADL isoforms. Protein extracts were prepared from protoplasts transformed with the indicated constructs and used for protein gel blot analysis using anti-HA antibody. WT, untransformed.
Previously, ADL2b was shown to localize to mitochondria (Arimura and Tsutsumi, 2002). We compared the localization of ADL1C with that of ADL2b. First, we confirmed the localization of ADL2b at the tip of mitochondria. ADL2b was tagged with HA at the C terminus and introduced into protoplasts. The localization of these proteins then was examined by immunohistochemistry using a monoclonal anti-HA antibody. ADL2b produced a punctate staining pattern (Figure 6B), as shown previously (Arimura and Tsutsumi, 2002). We examined the colocalization of ADL2b with F1-ATPase-γ:RFP, the mitochondrial marker, in protoplasts. The majority of ADL2b closely overlapped the red fluorescent signals of F1-ATPase-γ:RFP or was present side by side with F1-ATPase-γ:RFP (Figures 6Bd to 6Bf). Thus, these results confirm the localization of ADL2b to the mitochondria.
Next, we compared the localization of GFP:ADL1C and ADL2b:HA. Both proteins were expressed transiently in protoplasts, and their localization was determined by immunohistochemistry. As shown in Figures 6Bg to 6Bi, both ADL2b:HA and GFP:ADL1C gave punctate staining patterns. The punctate stains of ADL2b:HA and GFP:ADL1C appeared to consist of two different groups: one group showing overlap between ADL2b:HA and GFP:ADL1C (Figures 6Bg to 6Bi, arrows) and the other group showing no overlap (Figures 6Bg to 6Bi, arrowheads). When quantified, ∼20% (±10%, n = 1200) of green punctate GFP:ADL1C closely overlapped the red punctate stains of ADL2b. The partial colocalization of ADL1C with ADL2b was expected, because ∼20% of GFP:ADL1C-positive speckles overlapped the mitochondrial marker, F1-ATPase-γ:RFP. To confirm the expression of these proteins, protein extracts were prepared from transformed protoplasts and used to detect their expression. As shown in Figure 6C, protein gel blot analysis with the anti-HA antibody clearly demonstrated that all of these proteins were expressed and that the antibody is specific to these proteins.
DISCUSSION
Many isoforms of dynamin-like proteins are present in the Arabidopsis genome. Already, several of them have been studied and shown to be involved in seemingly different biological processes (Kang et al., 1998, 2003; Jin et al., 2001). In this study, we investigated two Arabidopsis dynamin-like proteins, ADL1C and ADL1E.
Mitochondria are present in different forms depending on the organism. Usually, plant cell mitochondria have a small, oval shape. Overexpression of ADL1C[K48E] in Arabidopsis leaf protoplasts caused a dramatic elongation of the mitochondria, similar to that seen in the presence of dominant-negative mutants of DLP1/Drp1 and ADL2b (Pitts et al., 1999; Smirnova et al., 2001; Arimura and Tsutsumi, 2002). In addition, adl1e plants also had abnormally elongated mitochondria that were rescued by the transient expression of ADL1C and ADL1E in protoplasts. It is known that mitochondrial morphology is maintained by two opposing processes, fission and fusion of individual mitochondria (Sesaki and Jensen, 1999). Thus, the elongation of mitochondria in the presence of ADL1C[K48E] in protoplasts, and the absence of ADL1E in the mutant plants, suggest that ADL1C and ADL1E may play a role in the fission process, as is the case with other mitochondrial dynamin-like proteins. Mutations in DLP1/Drp1 in animal cells, ADL2b in plant cells (Arimura and Tsutsumi, 2002), and Dnm1p in yeast cells (Otsuga et al., 1998; Pitts et al., 1999; Smirnova et al., 2001; Arimura and Tsutsumi, 2002) cause the elongation of mitochondria as a result of defects in fission.
Immunohistochemistry experiments demonstrated that ADL1C and ADL1E gave punctate staining patterns and colocalized at the speckles. Interestingly ∼20% of the speckles localized to mitochondria. Thus, the localization of ADL1C and ADL1E at mitochondria further supports the notion that these proteins may play a role in the maintenance of mitochondrial morphology. However, interestingly, only a portion of the ADL1C-positive speckles localized to mitochondria, indicating that ADL1C and ADL1E may localize to at least two different organelles. The localization of a dynamin-like protein to two different organelles is not unique. In fact, ADL1A and phragmoplastin also have been shown to localize to two different sites depending on the different cell types or different stages of a cell (Gu and Verma, 1997; Kang et al., 2003). In animal cells, DLP1, a dynamin-like protein, also has been shown to exhibit punctate staining, the majority of which is associated with the endoplasmic reticulum; the rest is localized to mitochondria (Yoon et al., 1998; Pitts et al., 1999). Also, in the primitive red algae Cyanidioschyzon merolae, CnDnm2, which is a dynamin-like protein involved in chloroplast division, appears in cytoplasmic patches just before chloroplast division and is recruited to the cytosolic side of the chloroplast division site to form a ring in the late stage of division (Miyagishima et al., 2003). Thus, it is possible that cytosolic ADL1C and ADL1E may be stored at different organelles and then recruited to the mitochondria when they are necessary for processes such as mitochondrial division. However, we cannot exclude the possibility that nonmitochondrial ADL1C or ADL1E also may play a role in different biological processes. In the case of the dynamin-like protein DLP1, it plays an important role in the fission of mitochondria in animal cells (Yoon et al., 1998). In addition, it is essential for the normal distribution and morphology of the endoplasmic reticulum (Pitts et al., 1999).
The dynamin-like proteins of mitochondria that have been identified in diverse species from yeast to human (Otsuga et al., 1998; Yoon et al., 1998; Pelloquin et al., 1999; Smirnova et al., 2001; Arimura and Tsutsumi, 2002) can be divided into two groups based on their localization. One group of proteins, which includes Dnm1p and DLP1/Drp1, is associated with the outer membrane. These proteins are thought to be involved in mediating the fission of the outer envelope membrane during division. By contrast, Msp1p in fission yeast and OPA in animal cells are localized within the mitochondrion proper (Pelloquin et al., 1999; Olichon et al., 2002). Together, these results strongly suggest that all eukaryotic organisms may have two different groups of dynamin-like proteins involved in mitochondrial division. Furthermore, they raise the possibility that dynamin-like proteins localized to the outer envelope membrane may be involved in the fission of that envelope membrane, whereas those localized at the inner envelope membrane may be play a role in its fission.
In support of this notion, both OPA and Msp1p localized within mitochondria also have been shown to play an important role in the maintenance of mitochondria (Delettre et al., 2000). Also, in budding yeast, another dynamin-like protein, Mgm1p, that has been shown to function in mitochondrial morphogenesis also localizes to an intermembrane space (Guan et al., 1993; Wong et al., 2000). Thus, it is intriguing that two different isoforms of dynamin-like proteins localize to mitochondria in plants, although their localization patterns clearly are different; the majority of ADL2b localizes to the tip of mitochondria, whereas ∼20% of ADL1C localizes to mitochondria. At present, the exact localization of these plant mitochondrial dynamin-like proteins in mitochondria is not understood. One possibility is that one of them may localize to the inner membrane and the other localizes to the outer envelope membrane. However, this hypothesis needs to be confirmed.
Recently, it was shown that ADL1A and ADL1E are essential for polar cell expansion and cell plate biogenesis (Kang et al., 2003). However, here, we clearly demonstrated that ADL1E is involved in the maintenance of mitochondrial morphology. One possible explanation for these results is that ADL1E may be involved in more than one process in the cell. In fact, this notion is in agreement with the expression pattern of ADL1E, which is expressed in various tissues, including mature leaf cells. In the leaf cell, ADL1E must be involved in a process that is different from that in cell plate biogenesis. In fact, there are other examples of a single dynamin-like protein being involved in multiple processes. DLP1 in animal cells and Vps1p in yeast cells, which are involved in mitochondrial division and vacuolar trafficking at the trans-Golgi network, respectively, also are involved in peroxisome biogenesis (Hoepfner et al., 2001; Koch et al., 2003). Also, DLP1 is essential for the normal distribution and morphology of the endoplasmic reticulum (Pitts et al., 1999). In animal cells, dynamin 2, which is involved in the generation of a vesicle at the trans-Golgi network or at the plasma membrane (Takei et al., 1996; McNiven, et al., 2000), has been shown to play an important role in actin dynamics (Schafer et al., 2002) and actin comet formation (Orth et al., 2002).
Interestingly, ADL1E and ADL1C colocalized when they were expressed transiently in Arabidopsis leaf protoplasts. In addition, ADL1C complemented the loss of ADL1E with respect to the maintenance of mitochondrial morphology in leaf cells. Together, these results strongly suggest that these two proteins may be involved in the same or very similar biological processes. However, this result raises an intriguing question: why do adl1e mutant plants show elongated mitochondria in leaf cells despite the presence of wild-type ADL1C? Our results showing abnormal mitochondrial morphology in leaf cells of adl1e mutants are in contrast to the results of Kang et al. (2003), who reported that mitochondria in the heart- and torpedo-shaped embryos of the double mutant ADL1A;E are normal. One explanation for this discrepancy could be that these genes are expressed in a tissue-specific manner. For example, in heart- and torpedo-shaped embryos, the expression level of ADL1C may be high enough to compensate for the loss of ADL1E. By contrast, in leaf cells, ADL1C expression may be low and ADL1E expression may be high. Thus, the loss of ADL1E may have a more profound effect on mitochondrial morphology in leaf cells. In fact, it has been shown that ADL1A and ADL1E are expressed differentially. Similarly, in animal cells, three closely related dynamin isoforms are expressed in a tissue-specific manner (Schmid et al., 1998). However, at present, the expression pattern of ADL1C remains unknown. Further studies are necessary to prove this hypothesis.
METHODS
Growth of Plants
Arabidopsis thaliana (ecotype Columbia) was grown either on Murashige and Skoog (1962) plates at 20°C in a culture room or in a greenhouse under conditions of 70% RH and a 16-h-light/8-h-dark cycle.
Screening of cDNAs Encoding Dynamin-Like Proteins
Probes for screening of cDNAs encoding dynamin-like proteins in Arabidopsis were amplified by PCR using primers designed from the nucleotide sequence information deposited in the EST database. The primers were 5′-GGAGTCTGTTATCAGGA-3′ (ADL1E-5), 5′-AGCTGGACCTCTTAAAG-3′ (ADL1E-3), 5′-CCCATTGCAGTAGATTCA-3′ (ADL1C-5), and 5′-GGCACGAGTGCAGAACT-3′ (ADL1C-3). To obtain full-length cDNA clones, a λ cDNA library was screened using the PCR products as hybridization probes after labeling with 32P. The inserts of positive clones were excised as pBluescript+ clones, and nucleotide sequences were determined by PCR sequencing using the dideoxy dye terminator method (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol.
Construction of Plasmids
The chimeric fusion constructs ADL1E:GFP, GFP:ADL1E, ADL1C:GFP, and GFP:ADL1C were generated for targeting experiments using standard recombinant technology. The full-length ADL1E and ADL1C cDNAs without the termination codons were prepared by PCR. The primers for the PCR amplification were the T3 primer and gene-specific primers (ADL1E-end, 5′-CTCGAGTCTTACCCAAGCAACAG-3′; ADL1C-end, 5′-CCTCGAGCTTCCAAGCCACTGCAT-3′) and were ligated to the N terminus of the GFP coding region. To generate the chimeric genes GFP:ADL1E and GFP:ADL1C, the ADL cDNAs were amplified by PCR with the T7 primer and gene-specific primers (ADL1E-ATG, 5′-CCCGGGTGAGAGTTTGATTGG-3′; ADL1C-ATG, 5′-CCCGGGCATGGCGACGATGAAAAG-3′) and ligated in frame to the N terminus of GFP. ADL1C[K48E] was generated by PCR using the primers 5′-CTCAAGAACCGAAGACTCTCCAGA-3′ and 5′-GGTCAGAGTTCTGGAGAGTCTTCG-3′. To add the T7 epitope at the N terminus of ADL1E, ADL1C, and ADL1C[K48E], the coding regions were inserted in frame at the BamHI site of pET21a (Novagen, Madison, WI). Hemagglutinin (HA):ADL1C was generated by PCR using primers 5′-ATGGCCTACCCATACGATGTTCCAGATTACGCTTCCATGGCGACGATGAAAAGT-3′ and ADL1C-end. ADL2b:HA was generated by PCR using primers 5′-ATGTCCGTCGACGATCTCCCT-3′ and 5′-TCAGGAAGCGTAATCTGGAACATCGTATGGGTAGGCCATATGAAGCCGTCCGTTCCC-3′. The T7- or HA-tagged coding regions were transferred subsequently downstream of the 35S promoter of Cauliflower mosaic virus.
Generation of Transgenic Plants
Transgenic Arabidopsis plants expressing T7-tagged ADL1E were generated with the binary vector pBIT7:ADL1E using the floral dip method (Clough and Bent, 1998). Kanamycin-resistant plants were selected from the T1 generation and used for immunohistochemistry or protein gel blot analysis.
Screening of Homozygote Plants with T-DNA Insertion at ADL1E
Seeds (mutant line SALK-060080) obtained from ABRC (Ohio State University, Columbus) were planted on kanamycin plates, and kanamycin-resistant plants were transferred to soil to set seeds. Seeds were harvested separately from individual plants. Subsequently, the seeds were planted again on kanamycin plates, and genomic DNA was prepared from two true leaves of individual plants. PCR was performed with genomic DNA using two ADL1E-specific primers or one ADL1E-specific primer and one left-border–specific primer.
Immunohistochemistry
For immunohistochemistry with root tissues, monoclonal antibodies were incubated with the fixed Arabidopsis cells at 4°C overnight as described previously (Paris et al., 1996; Jiang and Rogers, 1998). Anti-T7 monoclonal antibody was purchased from Novagen and used at a concentration of 10 μg/mL. The primary antibodies were detected with Cy5- or rhodamine-conjugated anti-mouse secondary antibody from Jackson ImmunoResearch Laboratories (West Grove, PA). For immunohistochemistry with protoplasts, transformed protoplasts were placed onto poly-l-Lys–coated glass slides and fixed with 3% paraformaldehyde in a fixing buffer (10 mM Hepes, pH 7.2, 154 mM NaCl, 125 mM CaCl2, 2.5 mM maltose, and 5 mM KCl) for 1 h at room temperature. The fixed cells were incubated with rat monoclonal anti-HA or rabbit anti-VSR antibodies at 4°C overnight and washed three times with TSW buffer (10 mM Tris-HCl, pH 7.4, 0.9% NaCl, 0.25% gelatin, 0.02% SDS, and 0.1% Triton X-100).
Subsequently, the cells were incubated with Cy3-conjugated goat anti-rabbit IgG or tetramethyl rhodamine isothiocyanate-conjugated anti-rat IgG (Zymed, San Francisco, CA) secondary antibodies. Data then were processed using Adobe Photoshop (Mountain View, CA). Images were collected by fluorescence microscopy, as described above, or by confocal microscopy. All confocal fluorescence images were collected using a Bio-Rad MRC 1024 system (Hercules, CA) with the following parameters: ×60 objective, ×1 zoom, 1250 gain, 2.2 iris, 0 background, and 0.348 pixel size. A Cy5/rhodamine software program was used to collect images under conditions in which no crossover between rhodamine and Cy5 emissions occurs, and the two images were collected sequentially from the same optical section. The Cy5 images were pseudocolored green, and the rhodamine images were pseudocolored red.
Ultrastructural Analysis and Immunogold Labeling
To visualize the detailed structure of mitochondrial morphology, leaf tissues were fixed for 4 h with 2% paraformaldehyde and 2% glutaraldehyde in cacodylate buffer, pH 7.2, rinsed with the same buffer, and postfixed with 1% osmium tetroxide in cacodylate buffer for 1 h. After dehydration, specimens were embedded in London Resin White (London Resin Co., London, UK). Ultrathin sections (40 to 60 nm thick) collected on uncoated nickel grids (300 mesh) were stained with 4% uranyl acetate and examined using a transmission electron microscope (JEOL 1200) at 60 to 80 kV.
Protein Preparation and Protein Gel Blot Analysis
To prepare cell extracts, transformed protoplasts were subjected to repeated freeze-thaw cycles in ice-cold homogenization buffer (25 mM Hepes, pH 7.7, 1 mM MgCl2, 250 mM sucrose, and 1 mM DTT) supplemented with protease inhibitors (1 μg/mL pepstatin A, 1 μg/mL aprotinin, 0.5 μg/mL leupeptin, and 100 μM phenylmethylsulfonyl fluoride), followed by brief sonication for 3 s at 50% output (repeated three times). Whole-cell extracts were subjected to low-speed centrifugation (8000g) at 4°C for 5 min to remove cellular debris. Whole-cell extracts were assayed by protein gel blot analysis using monoclonal anti-GFP antibody (Clontech, Palo Alto, CA), monoclonal anti-T7 antibody (Novagen), or monoclonal anti-HA antibody.
In Vivo Expression of Green and Red Fluorescent Protein Fusion Constructs
Plasmids were purified using Qiagen (Valencia, CA) columns according to the manufacturer's protocol. The fusion constructs were introduced into Arabidopsis protoplasts prepared from whole seedlings by polyethylene glycol–mediated transformation as described previously (Jin et al., 2001; Kim et al., 2001a). Protein expression was monitored at various times after transformation, and images were captured with a cooled charge-coupled device camera using a Zeiss Axioplan fluorescence microscope (Jena, Germany). The filter sets used were XF116 (exciter, 474AF20; dichroic, 500DRLP; emitter, 510AF23), XF33/E (exciter, 535DF35; dichroic, 570DRLP; emitter, 605DF50), and XF137 (exciter, 540AF30; dichroic, 570DRLP; emitter, 585ALP) (Omega, Inc., Brattleboro, VT) for GFP, RFP, and autofluorescence of chlorophyll, respectively. Data then were processed using Adobe Photoshop, and the images were rendered in pseudocolor.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Inhwan Hwang, ihhwang@postech.ac.kr.
Acknowledgments
The T-DNA insertion mutant at the ADL1E gene was obtained from the collection of SALK T-DNA insertion lines (Ohio Stock Center). This work was supported by Grant M10116000005-02F0000-00310 from the Creative Research Initiatives Program of the Ministry of Science and Technology (Korea). L.J. is supported by grants from the Research Grants Council of Hong Kong (CUHK4156/01M, CUHK4260/02M, and CUHK4307/03M) and by Area of Excellence.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.015222.
References
- Arimura, S.S., and Tsutsumi, N. (2002). A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division. Proc. Natl. Acad. Sci. USA 99, 5727–5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cook, T.A., Urrutia, R., and McNiven, M.A. (1994). Identification of dynamin 2, an isoform ubiquitously expressed in rat tissues. Proc. Natl. Acad. Sci. USA 91, 644–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damke, H., Baba, T., Warnock, D.E., and Schmid, S.L. (1994). Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J. Cell Biol. 127, 915–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, S.J., and Vierstra, R.D. (1998). Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol. Biol. 36, 521–528. [DOI] [PubMed] [Google Scholar]
- Delettre, C., et al. (2000). Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 26, 207–210. [DOI] [PubMed] [Google Scholar]
- Dombrowski, J.E., and Raikhel, N.V. (1995). Isolation of a cDNA encoding a novel GTP-binding protein of Arabidopsis thaliana. Plant Mol. Biol. 28, 1121–1126. [DOI] [PubMed] [Google Scholar]
- Ferguson, K.M., Lemmon, M.A., Schlessinger, J., and Sigler, P.B. (1994). Crystal structure at 2.2 Å resolution of the pleckstrin homology domain from human dynamin. Cell 79, 199–209. [DOI] [PubMed] [Google Scholar]
- Fukushima, N.H., Brisch, E., Keegan, B.R., Bleazard, W., and Shaw, J.M. (2001). The GTPase effector domain sequence of the Dnm1p GTPase regulates self-assembly and controls a rate-limiting step in mitochondrial fission. Mol. Biol. Cell 12, 2756–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie, A.E., Kurihara, L.J., Vallee, R.B., and Rose, M.D. (1995). DNM1, a dynamin-related gene, participates in endosomal trafficking in yeast. J. Cell Biol. 130, 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout, I., Dhand, R., Hiles, I.D., Fry, M.J., Panayoutou, G., Das, P., Truong, O., Totty, N.F., Husan, J., Booker, G.W., Campbell, I.D., and Waterfield, M.D. (1993). The GTPase dynamin binds to and is activated by a subset of SH3 domains. Cell 75, 25–36. [PubMed] [Google Scholar]
- Gu, X., and Verma, D.P. (1997). Dynamics of phragmoplastin in living cells during cell plate formation and uncoupling of cell elongation from the plane of cell division. Plant Cell 9, 157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, X., and Verma, D.P.S. (1996). Phragmoplastin, a dynamin-like protein associated with cell plate formation in plants. EMBO J. 15, 695–704. [PMC free article] [PubMed] [Google Scholar]
- Guan, K., Farh, L., Marshall, T.K., and Deschenes, R.J. (1993). Normal mitochondrial structure and genome maintenance in yeast requires the dynamin-like product of the MGM1 gene. Curr. Genet. 24, 141–148. [DOI] [PubMed] [Google Scholar]
- Heikal, A.A., Hess, S.T., Baird, G.S., Tsien, R.Y., and Webb, W.W. (2000). Molecular spectroscopy and dynamics of intrinsically fluorescent proteins: Coral red (dsRed) and yellow (Citrine). Proc. Natl. Acad. Sci. USA 97, 11996–12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskovits, J.S., Burgess, C.C., Obar, R.A., and Vallee, R.B. (1993). Effects of mutant rat dynamin on endocytosis. J. Cell Biol. 122, 565–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw, J.E., and Schmid, S.L. (1995). Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature 374, 190–192. [DOI] [PubMed] [Google Scholar]
- Hoepfner, D., van den Berg, M., Philippsen, P., Tabak, H.F., and Hettema, E.H. (2001). A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 155, 979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, L., and Rogers, J.C. (1998). Integral membrane protein sorting to vacuoles in plant cells: Evidence for two pathways. J. Cell Biol. 143, 1183–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, J.B., Kim, Y.A., Kim, S.J., Lee, S.H., Kim, D.H., Cheong, G.W., and Hwang, I. (2001). A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13, 1511–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, B.A., and Fangman, W.L. (1992). Mitochondrial DNA maintenance in yeast requires a protein containing a region related to the GTP-binding domain of dynamin. Genes Dev. 6, 380–389. [DOI] [PubMed] [Google Scholar]
- Kang, B.-H., Busse, J.S., and Bednarek, S.Y. (2003). Members of the Arabidopsis dynamin-like gene family, ADL1, are essential for plant cytokinesis and polarized cell growth. Plant Cell 15, 899–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, B.-H., Busse, J.S., Dickey, C., Rancour, D.M., and Bednarek, S.Y. (2001). The Arabidopsis cell plate-associated dynamin-like protein, ADL1Ap, is required for multiple stages of plant growth and development. Plant Physiol. 126, 47–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, S.G., Jin, J.B., Piao, H.L., Jang, H.J., Lim, J.H., and Hwang, I. (1998). Molecular cloning of an Arabidopsis cDNA encoding a dynamin-like protein that is localized to plastids. Plant Mol. Biol. 38, 437–447. [DOI] [PubMed] [Google Scholar]
- Kim, D.H., Eu, Y.J., Yoo, C.M., Kim, Y.W., Pih, K.T., Jin, J.B., Kim, S.J., Stenmark, H., and Hwang, I. (2001. a). Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13, 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.W., Park, D.S., Park, S.C., Kim, S.H., Cheong, G.W., and Hwang, I. (2001. b). Arabidopsis dynamin-like 2 that binds specifically to phosphatidylinositol 4-phosphate assembles into a high-molecular weight complex in vivo and in vitro. Plant Physiol. 127, 1243–1255. [PMC free article] [PubMed] [Google Scholar]
- Koch, A., Thiemann, M., Grabenbauer, M., Yoon, Y., McNiven, M.A., and Schrader, M. (2003). Dynamin-like protein 1 is involved in peroxisomal fission. J. Biol. Chem. 278, 8597–8605. [DOI] [PubMed] [Google Scholar]
- McNiven, M.A., Kim, L., Krueger, E.W., Orth, J.D., Cao, H., and Wong, T.W. (2000). Regulated interactions between dynamin and the actin-binding protein cortactin modulate cell shape. J. Cell Biol. 151, 187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima, S.Y., Nishida, K., Mori, T., Matsuzaki, M., Higashiyama, T., Kuroiwa, H., and Kuroiwa, T. (2003). A plant-specific dynamin-related protein forms a ring at the chloroplast division site. Plant Cell 15, 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlberg, A.B., Warnock, D.E., and Schmid, S.L. (1997). Domain structure and intramolecular regulation of dynamin GTPase. EMBO J. 16, 6676–6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Niwa, Y., Hirano, T., Yoshimoto, K., Shimizu, M., and Kobayashi, H. (1999). Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J. 18, 455–463. [DOI] [PubMed] [Google Scholar]
- Obar, R.A., Collins, C.A., Hammarback, J.A., Shpetner, H.S., and Vallee, R.B. (1990). Molecular cloning of the microtubule-associated mechanochemical enzyme dynamin reveals homology with a new family of GTP-binding proteins. Nature 347, 256–261. [DOI] [PubMed] [Google Scholar]
- Okamoto, P.M., Herskovits, J.S., and Vallee, R.B. (1997). Role of the basic, proline-rich region of dynamin in Src homology 3 domain binding and endocytosis. J. Biol. Chem. 272, 11629–11635. [DOI] [PubMed] [Google Scholar]
- Okamoto, P.M., Tripet, B., Litowski, J., Hodges, R.S., and Vallee, R.B. (1999). Multiple distinct coiled-coils are involved in dynamin self-assembly. J. Biol. Chem. 274, 10277–10286. [DOI] [PubMed] [Google Scholar]
- Olichon, A., et al. (2002). The human dynamin-related protein OPA1 is anchored to the mitochondrial inner membrane facing the inter-membrane space. FEBS Lett. 523, 171–176. [DOI] [PubMed] [Google Scholar]
- Orth, J.D., Krueger, E.W., Cao, H., and McNiven, M.A. (2002). The large GTPase dynamin regulates actin comet formation and movement in living cells. Proc. Natl. Acad. Sci. USA 99, 167–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga, D., Keegan, B.R., Brisch, E., Thatcher, J.W., Hermann, G.J., Bleazard, W., and Shaw, J.M. (1998). The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 143, 333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris, N., Stanley, C.M., Jones, R.L., and Rogers, J.C. (1996). Plant cells contain two functionally distinct vacuolar compartments. Cell 85, 563–572. [DOI] [PubMed] [Google Scholar]
- Park, J.M., Kang, S.G., Pih, K.T., Jang, H.J., Piao, H.L., Yoon, H.W., Cho, M.J., and Hwang, I. (1997). A dynamin-like protein, ADL1, is present in membranes as a high-molecular-mass complex in Arabidopsis thaliana. Plant Physiol. 115, 763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelloquin, L., Belenguer, P., Menon, Y., Gas, N., and Ducommun, B. (1999). Fission yeast Msp1 is a mitochondrial dynamin-related protein. J. Cell Sci. 112, 4151–4161. [DOI] [PubMed] [Google Scholar]
- Pitts, K.R., Yoon, Y., Krueger, E.W., and McNiven, M.A. (1999). The dynamin-like protein DLP1 is essential for normal distribution and morphology of the endoplasmic reticulum and mitochondria in mammalian cells. Mol. Biol. Cell 10, 4403–4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman, J.H., Raymond, C.K., Gilbert, T., O'Hara, P.J., and Stevens, T.H. (1990). A putative GTP binding protein homologous to interferon-inducible Mx proteins performs an essential function in yeast protein sorting. Cell 61, 1063–1074. [DOI] [PubMed] [Google Scholar]
- Salim, K., et al. (1994). Dynamin binds to SH3 domains of phospholipase C gamma and GRB-2. J. Biol. Chem. 269, 16009–16014. [PubMed] [Google Scholar]
- Schafer, D.A., Weed, S.A., Binns, D., Karginov, A.V., Parsons, J.T., and Cooper, J.A. (2002). Dynamin2 and cortactin regulate actin assembly and filament organization. Curr. Biol. 12, 1852–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, S.L., McNiven, M.A., and De Camilli, P. (1998). Dynamin and its partners: A progress report. Curr. Opin. Cell Biol. 10, 504–512. [DOI] [PubMed] [Google Scholar]
- Sesaki, H., and Jensen, R.E. (1999). Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147, 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever, S., Damke, H., and Schmid, S.L. (2000). Dynamin:GTP controls the formation of constricted coated pits, the rate limiting step in clathrin-mediated endocytosis. J. Cell Biol. 150, 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, G. (1996). The pleckstrin homology domain: An intriguing multifunctional protein module. Bioessays 18, 35–46. [DOI] [PubMed] [Google Scholar]
- Shepard, K.A., and Yaffe, M.P. (1999). The yeast dynamin-like protein, Mgm1p, functions on the mitochondrial outer membrane to mediate mitochondrial inheritance. J. Cell Biol. 144, 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shupliakov, O., Löw, P., Grabs, D., Gad, H., Chen, H., David, C., Takei, K., De Camilli, P., and Brodin, L. (1997). Synaptic vesicle endocytosis impaired by disruption of dynamin-SH3 domain interactions. Science 276, 259–263. [DOI] [PubMed] [Google Scholar]
- Smirnova, E., Griparic, L., Shurland, D.L., and van der Bliek, A.M. (2001). Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell 12, 2245–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova, E., Shurland, D.L., Newman-Smith, E.D., Pishvaee, B., and van der Bliek, A.M. (1999). A model for dynamin self-assembly based on binding between three different protein domains. J. Biol. Chem. 274, 14942–14947. [DOI] [PubMed] [Google Scholar]
- Takei, K., Mundigl, O., Daniell, L., and De Camilli, P. (1996). The synaptic vesicle cycle: A single vesicle budding step involving clathrin and dynamin. J. Cell Biol. 133, 1237–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm, D., Salim, K., Gout, I., Guruprasad, L., Waterfield, M., and Blundell, T. (1994). Crystal structure of the pleckstrin homology domain from dynamin. Nat. Struct. Biol. 1, 782–788. [DOI] [PubMed] [Google Scholar]
- Vallee, R.B., and Okamoto, P.M. (1995). The regulation of endocytosis: Identifying dynamin's binding partners. Trends Cell Biol. 5, 43–47. [DOI] [PubMed] [Google Scholar]
- Vater, C.A., Raymond, C.K., Ekena, K., Howald-Stevenson, I., and Stevens, T.H. (1992). The VPS1 protein, a homolog of dynamin required for vacuolar protein sorting in Saccharomyces cerevisiae, is a GTPase with two functionally separable domains. J. Cell Biol. 119, 773–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsbach, K., and Payne, G.S. (1993). Vps1p, a member of the dynamin GTPase family, is necessary for Golgi membrane protein retention in Saccharomyces cerevisiae. EMBO J. 12, 3049–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, E.D., Wagner, J.A., Gorsich, S.W., McCaffery, J.M., Shaw, J.M., and Nunnari, J. (2000). The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J. Cell Biol. 151, 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, Y., Pitts, K.R., Dahan, S., and McNiven, M.A. (1998). A novel dynamin-like protein associates with cytoplasmic vesicles and tubules of the endoplasmic reticulum in mammalian cells. J. Cell Biol. 140, 779–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, Y., Pitts, K.R., and McNiven, M.A. (2001). Mammalian dynamin-like protein DLP1 tubulates membranes. Mol. Biol. Cell 12, 2894–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]