Abstract
In yeast and animals, the anaphase-promoting complex or cyclosome (APC/C) is an essential ubiquitin protein ligase that regulates mitotic progression and exit by controlling the stability of cell cycle regulatory proteins, such as securin and the mitotic cyclins. In plants, the function, regulation, and substrates of the APC/C are poorly understood. To gain more insight into the roles of the plant APC/C, we characterized at the molecular level one of its subunits, APC2, which is encoded by a single-copy gene in Arabidopsis. We show that the Arabidopsis gene is able to partially complement a budding yeast apc2 ts mutant. By yeast two-hybrid assays, we demonstrate an interaction of APC2 with two other APC/C subunits: APC11 and APC8/CDC23. A reverse-genetic approach identified Arabidopsis plants carrying T-DNA insertions in the APC2 gene. apc2 null mutants are impaired in female megagametogenesis and accumulate a cyclin–β-glucuronidase reporter protein but do not display metaphase arrest, as observed in other systems. The APC2 gene is expressed in various plant organs and does not seem to be cell cycle regulated. Finally, we report intriguing differences in APC2 protein subcellular localization compared with that in other systems. Our observations support a conserved function of the APC/C in plants but a different mode of regulation.
INTRODUCTION
Progression through the eukaryotic cell cycle requires the coordinated destruction of essential cell cycle regulatory proteins by the ubiquitin-dependent pathway (reviewed by King et al., 1996). Protein ubiquitylation is a multistep enzymatic process (reviewed by Ciechanover et al., 2000) that involves at least three enzyme activities. Ubiquitin-activating enzyme (E1) forms a high-energy bond with ubiquitin, which then is transesterified to a ubiquitin-conjugating enzyme (E2). The transfer of ubiquitin to the target protein substrate requires a ubiquitin protein ligase (E3). Polyubiquitylation of the protein substrate is sufficient to target it for degradation by a large ATP-dependent multicatalytic protease, the 26S proteasome (reviewed by Voges et al., 1999).
In yeast and animals, proteolysis during mitosis is essentially assumed by an E3 ligase called the anaphase-promoting complex or cyclosome (APC/C) (reviewed by Harper et al., 2002; Peters, 2002). The first identified target of this multiple-subunit protein complex was cyclin B (reviewed by Irniger, 2002), but many other targets of APC/C-dependent ubiquitylation have been identified subsequently, both in fungi and in animals. Among them are the anaphase inhibitors called securins (Pellman and Christman, 2001), the DNA replication inhibitor geminin (McGarry and Kirschner, 1998), CDC6, which is involved in the initiation of DNA replication (Petersen et al., 2000), chromokinesin Xkid, which is involved in chromosome alignment during metaphase (Funabiki and Murray, 2000), the mitotic spindle-associated protein Ase1p (Juang et al., 1997), and different protein kinases, including polo kinase (Shirayama et al., 1998), Hsl1 (Burton and Solomon, 2000), Nek2A (Hames et al., 2001), and Aurora-A (Littlepage and Ruderman, 2002, and references therein). Furthermore, there is a sequential destruction of these proteins during the cell cycle. For example, cyclin A and Nek2A start to be degraded in prometaphase, cyclin B1 and securin in metaphase, Ase1 in anaphase, and CDC6 during the G1-phase. The proper timing of APC/C's activation and its substrate specificity are regulated, at least in part, by two associated proteins, Cdc20/FIZZY and Cdh1/FIZZY-RELATED (Vodermaier, 2001).
Most of the APC/C targets carry a short peptide motif of nine amino acids called the destruction box (D-box) (Irniger, 2002). Nevertheless, another protein motif that is recognized by the APC/C, the KEN-box, also has been identified (Pfleger and Kirschner, 2000).
The APC/C itself is composed of at least 11 protein subunits, and the three-dimensional structure of the human complex has been solved at a resolution of 24 Å (Gieffers et al., 2001). However, little is known about the function of individual APC/C subunits. Only two of them, APC2 and APC11, are structurally related to components of another E3 class, called the Skp1, CDC53/Cullin, F-box protein (SCF) (Zachariae and Nasmyth, 1999). APC2 is a distant member of the cullin protein family that functions as a scaffold in SCF assembly (Gieffers et al., 2000), whereas APC11 is a RING-H2 finger protein similar to RBX1 that plays a key role in the ubiquitylation reaction (Gmachl et al., 2000).
In plants, APC/C function and regulation have not yet been investigated. Although it has been shown that mitotic cyclins are degraded in a D-box–dependent manner (Genschik et al., 1998, Criqui et al., 2000), a direct demonstration that APC/C is involved in this process is lacking. We have shown previously by computerized analyses that all vertebrate APC/C subunits have counterparts in Arabidopsis, and with the exception of the APC3/CDC27 subunit (see Discussion), all of them are encoded by single-copy genes (Capron et al., 2003). To gain more insight into the function of the APC/C in plants, we have molecularly and genetically characterized the APC2 subunit from Arabidopsis. We found that the apc2 mutants arrest at the gametophytic stage and accumulate a cyclin–β-glucuronidase (GUS) reporter protein. However, the cell cycle phase arrest of the mutant cells and subcellular localization studies suggest differences in regulation between plant and animal APC/C.
RESULTS
Arabidopsis APC2 Is a Member of the Cullin Protein Family
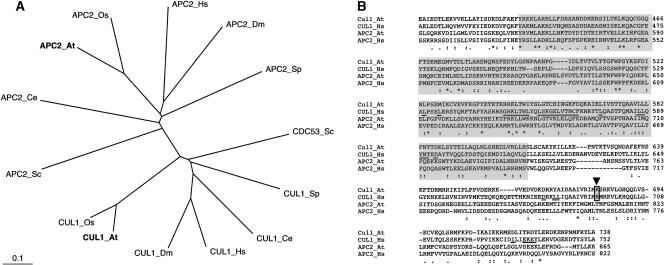
A full-length Arabidopsis APC2 cDNA was reconstituted from an EST clone and a reverse transcriptase–mediated PCR product. Its open reading frame encodes a protein of 865 amino acids. Sequence analysis confirmed the intron/exon structure predicted by the genome project. The plant protein shows sequence conservation throughout the protein sequence with all other APC2 proteins from different organisms (data not shown). It exhibits 61.3, 30.7, and 20.7% sequence identity with a predicted APC2 protein from rice and the APC2 subunits from human and budding yeast, respectively. A phylogenetic tree that includes APC2 subunits from various organisms as well as members of the related CUL1/CDC53 protein family (Figure 1A) shows a clear separation of both protein subfamilies.
Figure 1.
Sequence Analysis of Arabidopsis APC2 and CUL1-Related Proteins.
(A) Phylogenetic tree created with the CLUSTAL W and TreeViewPPC programs, including Arabidopsis APC2 and CUL1 (boldface letters), together with related proteins of Saccharomyces cerevisiae (Sc), Schizosaccharomyces pombe (Sp), Oryza sativa (Os), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), and Homo sapiens (Hs). Branch lengths are proportional to phylogenetic distances.
(B) Alignment of the Arabidopsis and human APC2 and CUL1 C-terminal amino acid sequences covering the conserved cullin domains (boxed in gray). Numbers refer to amino acid positions in the corresponding proteins. Consensus symbols under the alignment are as follows: asterisks indicate identical residues in all sequences; colons and periods indicate conserved and semiconserved substitutions, respectively. The RBX1 binding residues in human CUL1 (Zheng et al., 2002) are underlined. The RUB1/NEDD8 conjugation sites in plant and human CUL1 proteins are boxed and indicated with an arrowhead.
The APC2 and cullin proteins share the highest sequence identities within the so-called cullin homology domain of ∼180 residues (Figure 1B) (for review, see Gieffers et al., 2000). This domain, as well as additional sequences at the C terminus of the CUL1 protein, interacts with the RING-H2 finger protein RBX1/ROC1 (Zheng et al., 2002). SCF and APC/C complexes are known to share a number of structural characteristics (reviewed by Zachariae and Nasmyth, 1999; Harper et al., 2002). Interestingly, certain of the residues in human CUL1, which are involved in the interaction with RBX1/ROC1 (Zheng et al., 2002), also are conserved in the APC2 protein sequences (Figure 1B). Thus, it will be interesting to determine whether APC/C's RING-H2 finger protein counterpart, APC11, also interacts with APC2 in a similar manner.
CUL1 and other members of the cullin family, but not the APC2 proteins, are modified on a conserved Lys residue through the covalent attachment of the ubiquitin-like molecule NEDD8/RUB1 (reviewed by Deshaies, 1999). Like APC2 from animals and yeast, the Arabidopsis APC2 subunit does not carry the neddylation acceptor residue (Figure 1B).
The Arabidopsis APC2 Subunit Interacts with APC11 and CDC23 in a Yeast Two-Hybrid Assay
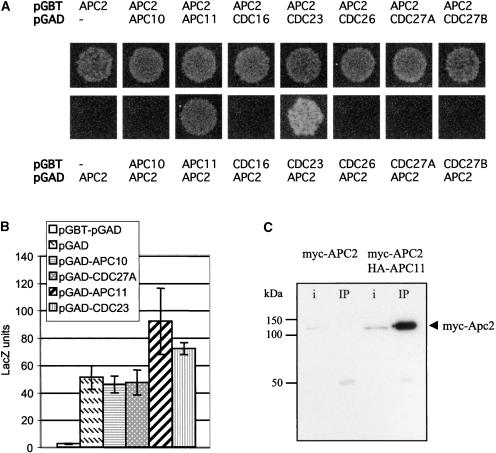
By analogy with CDC53/CUL1 (Zheng et al., 2002), APC2 is believed to form multiple protein–protein interactions inside the APC/C complex. We investigated whether this is the case with certain predicted plant subunits (Capron et al., 2003). Arabidopsis full-length cDNAs of all subunits containing the tetratricopeptide repeat (TPR) repeats (CDC23, CDC16, CDC27A, and CDC27B/ HOBBIT) (Blilou et al., 2002), the Doc domain protein APC10, CDC26, and the RING-H2 finger protein APC11 were isolated and cloned into the pGAD424 and pGBT9 vectors to test for interaction with APC2 in yeast two-hybrid assays (Figure 2). When APC2 was fused to the GAL4 activation domain (pAD-APC2), we observed a clear interaction with APC11 and CDC23 (Figure 2A). Although the pGB-APC2 construct exhibits strong autoactivation of the β-galactosidase reporter gene, it was still possible to confirm these interactions by measuring β-galactosidase activities (Figure 2B).
Figure 2.
The APC2 Protein Interacts with APC11 in Both Yeast and Plant Cells.
(A) Yeast two-hybrid interactions of APC2 with different predicted plant APC/C subunits. The vectors (in boldface) and expressed proteins are indicated. Yeast cells transformed with the different pairs of plasmids were grown for 4 days at 30°C on synthetic complete medium lacking His and adenine.
(B) β-Galactosidase activities of yeast cells transformed with both pGBT and pGAD empty vectors or with pGBT-APC2 cotransformed with the different pGAD vectors as indicated.
(C) APC2 coimmunoprecipitates with APC11. Protein extracts prepared from Arabidopsis protoplasts expressing either myc-APC2 alone or coexpressing both myc-APC2 and HA-APC11 were immunoprecipitated with rat monoclonal anti-HA antibodies, and the complex was purified with protein G magnetic beads. After washing, the eluted proteins were separated by SDS-PAGE and immunoblotted with anti-myc antibody. The efficiency of the immunoprecipitation was verified using a mouse anti-HA monoclonal antibody (data not shown). i, input (one-tenth of the total input); IP, total amount of the immunoprecipitation eluate.
To further investigate the interaction between the APC2 subunit and the RING-H2 finger protein APC11, the proteins were fused to epitope tags and expressed transiently in Arabidopsis protoplasts. Coimmunoprecipitation of myc-tagged APC2 with hemagglutinin (HA)-tagged APC11 was demonstrated (Figure 2C). Thus, it is likely that APC2 and APC11 proteins form a heterodimeric protein complex similar to CUL1/RBX1 (Zheng et al., 2002).
Arabidopsis APC2 and APC11 Are Able to Partially Complement Yeast ts Mutant Strains
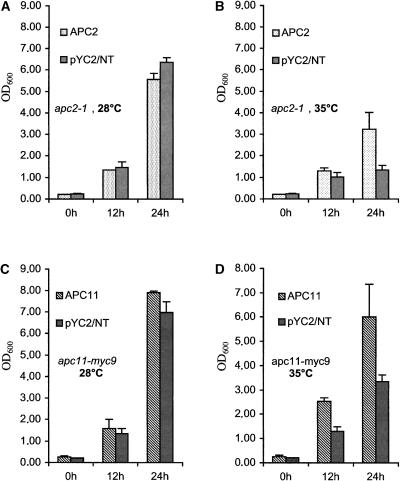
We next determined whether both plant APC/C subunits are able to functionally replace their respective budding yeast orthologs. Full-size cDNAs of both APC2 and APC11 were cloned into a yeast vector under the control of the Gal promoter and transformed into apc2-1 and apc11-myc9 ts yeast mutant strains that grow at 28°C but are restricted at 35°C (Zachariae et al., 1998). Expression of both plant APC/C subunits rescued the yeast mutant strains at the restrictive temperature, whereas the negative control containing the empty vector did not (Figure 3). However, these rescues were only partial, because the final density of the cultures was lower than that at the permissive temperature.
Figure 3.
apc2-1 and apc11-myc9 Yeast Mutant Strains Are Complemented with the Coding Regions of the Corresponding Plant cDNAs.
(A) and (B) Growth at the permissive (A) or restrictive (B) temperature of the apc2-1 yeast strain carrying an empty vector (pYC2/NT) or expressing Arabidopsis APC2 as determined by OD600 measurements.
(C) and (D) Growth at the permissive (C) or restrictive (D) temperature of the apc11-myc9 yeast strain carrying an empty vector (pYC2/NT) or expressing Arabidopsis APC11 as determined by OD600 measurements.
Null Mutation of APC2 Causes Megagametogenesis Arrest
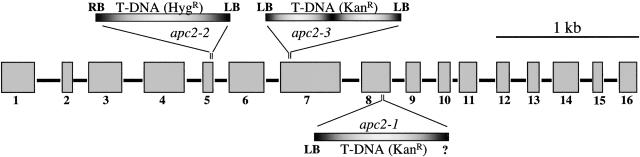
To investigate the function of the Arabidopsis APC2 gene, we used a reverse-genetic approach. The apc2-1 insertion line was identified by PCR screening of 40,000 independent transgenic lines from the Versailles T-DNA collection, whereas the apc2-2 line was obtained by screening 39,700 lines of the recently established Cologne collection (Rios et al., 2002). The apc2-3 line was obtained from the Salk Institute (from the Flanking Sequence Tags project). In all three lines, the T-DNAs were inserted into coding sequences (Figure 4). In apc2-2, one copy of the T-DNA was inserted into the fifth exon, whereas in apc2-3, two copies of the T-DNA were inserted head to head in exon 7. In apc2-1, the T-DNA was inserted into exon 8. In this line, we were able to sequence only one of the T-DNA/plant genomic DNA junctions; nevertheless, the presence of a single T-DNA insertion was confirmed by DNA gel blot analysis (data not shown).
Figure 4.
Scheme of the T-DNA Insertions in the APC2 Gene.
Comparison of the APC2 genomic and cDNA sequences revealed that the coding region of the gene consists of 16 exons (gray boxes) separated by 15 introns. LB indicates the left border of the T-DNAs for which the precise integration sites could be identified. HygR, hygromycin resistance gene; KanR, kanamycin resistance gene (neomycin phosphotransferase); RB, right border.
PCR analysis of the progeny for all three mutant lines after self-fertilization failed to reveal homozygous plants for the T-DNA insertions, indicating that the APC2 gene is essential. Nevertheless, the mutation is recessive, because the heterozygote mutant lines showed a normal sporophytic phenotype throughout their development (data not shown).
Genetic analysis revealed that the percentage of transmission of the mutant alleles in apc2-1/APC2, apc2-2/APC2, and apc2-3/APC2 heterozygous plants after self-fertilization is at most 52% (Table 1), which is consistent with a gametophytic defect. Moreover, open siliques from the self-pollinated apc2-1/APC2 plant showed a large proportion of aborted siblings (∼50% of 296 counted seeds), as illustrated in Figure 5A, indicating that these plants were semisterile. Indeed, analysis of T-DNA transmission through reciprocal backcrosses of the apc2-1 mutant with the wild type showed severe reduction in genetic transmission of the T-DNA only through the female gametes (Table 1). The few kanamycin-resistant plants obtained when the apc2-1 mutant was used as the female parent in the cross with wild-type plants may arise from self-fertilization resulting from imperfect emasculation.
Table 1.
Genetic Analysis of Different APC2 Insertion Alleles and Molecular Complementation of the apc2-1 Mutant
| Hypothesis
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Progeny
|
χ2
|
|||||||
| Genotype | No. of KanR:KanS or HygR:HygS Plants |
df | R/S | T-DNA Transmission Percent |
R/S = 1 | R/S = 2 | R/S = 3 | Threshold |
| apc2-1 +/− selfing | 1796:1814 | 23 | 0.99 | 49 | 29.7 | 441 | 1176 | 35 |
| apc2-2 +/− selfing | 1867:2769 | 15 | 0.67 | 40 | 185 | 1464 | 2994 | 25 |
| apc2-3 +/− selfing | 2049:1878 | 25 | 1.09 | 52 | 37 | 404 | 1131 | 37.6 |
| apc2-1 +/− × WT | 24:785 | 11 | 0.031 | 2.9 | 716 | 1477 | 2239 | 19.6 |
| WT × apc2-1 +/− | 629:700 | 9 | 0.90 | 47 | 6 | 226 | 546 | 16.9 |
| apc2-1 +/− (pCAMB-APC2)-5 | 1634:914 | 7 | 1.79 | 64 | 204 | 7.9 | 161 | 14 |
| apc2-1 +/− (pCAMB-APC2)-7 | 1013:475 | 3 | 2.13 | 68 | 194 | 1.4 | 38 | 7.8 |
HygR/HygS, hygromycin resistant/hygromycin sensitive; KanR/KanS, kanamycin resistant/kanamycin sensitive; R, resistant; S, sensitive; WT, wild type.
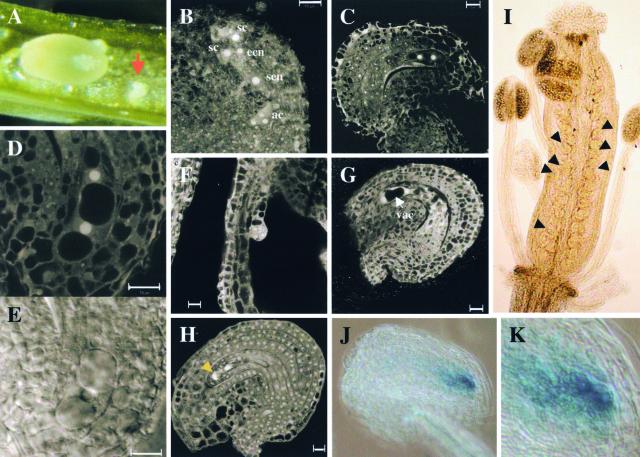
Figure 5.
Phenotypes of the apc2 Mutants and Cyclin-GUS Reporter Protein Accumulation in the apc2-2 Arrested Mutant.
(A) Mature silique of a selfed heterozygous apc2/APC2 plant. Seeds derived from female gametophytes carrying the mutant apc2 allele abort, as indicated by the arrow. The aborted ovules are shrunken and white, whereas the big wild-type ovules are green.
(B) to (E) CLSM and differential interference contrast images from the apc2-1 mutant at stage FG6. Images were taken from the same silique.
(B) CLSM image of a seven-cell ovule.
(C) CLSM image showing an arrest after the megaspore underwent its first mitotic division.
(D) and (E) CLSM (D) and differential interference contrast (E) images of an arrested female gametophyte at higher magnification.
(F) and (G) CLSM images of the apc2-1 mutant during early embryogenesis. Images were taken from the same silique.
(F) CLSM image of an embryo at the dermatogen stage.
(G) CLSM image showing a degenerated female gametophyte with two visible nuclei separated by an important vacuole.
(H) CLSM image from the apc2-2 mutant arrested at the one-nucleus stage, whereas other embryo sacs, from the same silique, are at stage FG6. The arrowhead points to the degenerating megaspores.
(I) to (K) Cyclin-GUS reporter protein accumulation in pAL10 apc2-2 plants.
(I) Histochemical analysis of an inflorescence from a pAL10 apc2-2 line. The arrowheads point to several ovules showing GUS staining.
(J) GUS staining of cyclin A3–GUS reporter protein activity in an arrested female gametophyte at the two-nucleus stage.
(K) Higher magnification of (J).
ac, antipodal cells; ecn, egg cell nucleus; sc, synergid cell, sen, secondary nucleus; vac, vacuole.
To determine the nature of the defect in apc2 female gametophyte mutants, we analyzed embryo sac formation at different developmental stages in apc2-1/APC2 and apc2-2/APC2 plants using confocal laser scanning microscopy (CLSM). It is well known that embryo sacs develop synchronously inside the same silique. From these observations, we found for the apc2-1 mutant that 100 of 278 (36%) female gametophytes were arrested at the two-nucleus stage (Figures 5B to 5E), indicating that APC2 loss of function impairs primarily megagametogenesis after the first mitotic division. Furthermore, the female gametophyte mutants arrested in interphase and not in mitotic prophase, because the nucleoli clearly were visible (Figures 5D and 5E). It is noteworthy that we never observed an arrest during metaphase. When analyzed at later stages, after fertilization of the nonmutated ovules (Figure 5F), the mutant gametophytes degenerated entirely (Figure 5G). Based on the genetic analysis and on the observation that ∼50% of the seeds were aborted in siliques of self-pollinated heterozygous mutant plants (see above), it is likely that some of the mutant gametes also arrest at later stages and may even be able to develop into embryo sacs.
Similarly, we observed by CLSM a 26% female gametophyte arrest (75 of 286) in the apc2-2 mutant. Nevertheless, in this mutant, the female gametophytes arrested either at the one-nucleus stage (Figure 5H) or at the two-nucleus stage (data not shown), indicating an earlier arrest of the mutant. This mutant also showed a more severe reduction in genetic transmission of the T-DNA (Table 1), suggesting reduced transmission of the mutation through the pollen. However, this issue was not explored.
Our observations strongly suggest that the disruption of the APC2 gene primarily affects megagametogenesis. This possibility was investigated further by genetic complementation. A genomic fragment spanning from 2048 bp upstream of the ATG to 432 bp downstream of the stop codon was cloned into a vector carrying the HPT gene, which confers hygromycin resistance (pCAMB-APC2). Hygromycin-resistant plants were obtained from transformation of heterozygous apc2-1 mutant plants (here referred to as the T0 generation), and their self-progeny were scored for kanamycin resistance (T2 generation). If the female gametophyte lethality were complemented successfully by the introduced construct, we would expect to recover kanamycin-resistant progeny at greater frequency in these lines. In the case of a single integration of the transgene that segregates independently of the APC2 locus, we would expect a 2:1 segregation ratio of kanamycin resistant:kanamycin sensitive in the T2 generation. Indeed, 11 of 29 independent transformants displayed such a kanamycin resistant:kanamycin sensitive segregation ratio (two of them are shown in Table 1), indicating that functional complementation occurred in these lines. Moreover, in the progeny of these lines, we obtained lines that were 100% kanamycin resistant and thus homozygous for the mutation. This finding was confirmed further by PCR analysis (data not shown).
The Arrested apc2 Mutants Accumulate a Cyclin-GUS Fusion Reporter Protein
Mitotic cyclins were the first demonstrated APC/C substrates (reviewed by Harper et al., 2002; Peters, 2002). These proteins carry a specific sequence element in their N-terminal regions termed the D-box that is required for their degradation (Glotzer et al., 1991). We have shown previously that reporter proteins containing the N-terminal domains of either A3-type or B1-type mitotic cyclins are actively degraded in a D-box–dependent manner in plant cells (Genschik et al., 1998).
To investigate whether the apc2 mutants accumulate D-box–containing proteins, the 5′ part of the cyclin A3 cDNA encoding the N-terminal domain with the D-box (Genschik et al., 1998) was fused to the GUS gene and expressed under the control of the gametophyte-specific BnSKP1 g1 promoter (Drouaud et al., 2000). This construct in binary vector, called pAL101, was introduced in wild-type Arabidopsis (Wassilewskija ecotype), and T1 transformants were selected. These plants were self-fertilized, and T2 plants homozygous for the insertion were selected. None of these plants displayed GUS staining during different stages of female gametophyte development (data not shown). However, when the T2 plants were fertilized with pollen from the apc2-2 mutant and apc2-2/APC2 pAL101/pAL101 plants were selected, GUS staining was observed in the embryo sacs (Figure 5I). The GUS staining was present only in the arrested megagametophytes (Figures 5J and 5K), indicating that the apc2-2 mutant is unable to degrade D-box–containing proteins.
Expression of APC2 in Arabidopsis Plants and Suspension Cells
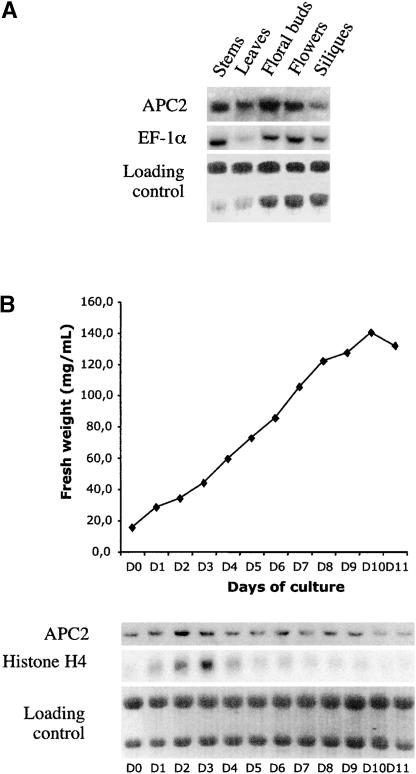
RNA gel blot analysis showed that the APC2 gene encodes a single transcript that is present in different organs of Arabidopsis plants (Figure 6A). Consistent with the mutant phenotype, we observed the expression of the APC2 gene in immature flowers. Indeed, APC2 RNA was detected in ovules by in situ hybridization (data not shown). Because the APC/C is involved in the control of the cell cycle, we investigated the expression of the gene during different growth phases of suspension-cultured cells. Whereas the histone H4 transcripts showed higher expression levels during days 2 and 3 of the culture (corresponding to an enriched cell population at the G1/S transition), the APC2 transcript was found at a relatively constant level (Figure 6B). Thus, the APC2 gene, like CDC27A (Blilou et al., 2002), does not seem to be cell cycle regulated.
Figure 6.
RNA Gel Blot Analysis of APC2 Expression.
(A) Total RNA was isolated from different organs of Arabidopsis plants, and RNA gel blot analysis was performed using successive hybridizations with different probes, as indicated.
(B) Samples were taken at different days of subculture from an Arabidopsis cell suspension culture and used for fresh weight measurement and RNA analysis. RNA gel blot analysis was performed using successive hybridizations with different probes, as indicated.
Subcellular Localization of APC2 and CDC27A Proteins in Interphase and during Mitosis
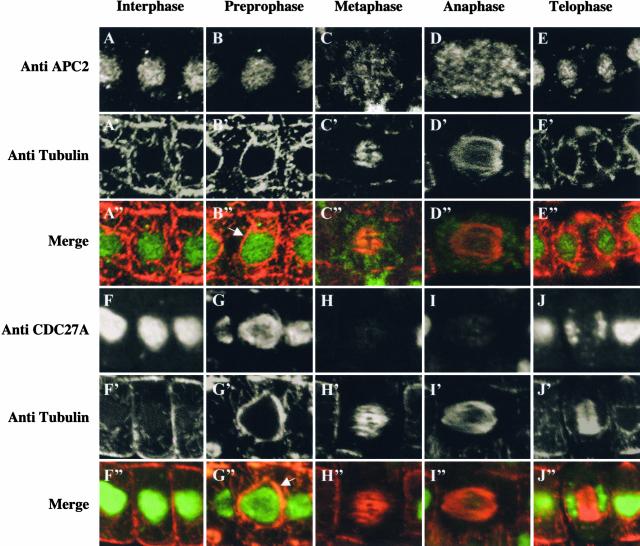
To study the subcellular localization of the APC/C, we raised polyclonal antibodies against the Arabidopsis APC2 and CDC27A subunits. Both antibodies gave only a single signal on protein gel blots that was specific, as demonstrated by competition experiments (data not shown). Control immunolocalization experiments using preimmune anti-APC2 or anti-CDC27A serum did not show any detectable staining above background levels (data not shown). During interphase, both proteins were found mainly in the nuclei of Arabidopsis root meristems (Figures 7A and 7F). In prophase, the nuclear envelope is surrounded by a cage of microtubules—the so-called “prophase spindle” (Figures 7B and 7G). At this stage, both proteins are still nuclear. However, in metaphase and anaphase, the APC2 signal is diffuse and not enriched on the mitotic spindle or the condensed chromosomes (Figures 7C and 7D). By contrast, the CDC27A protein is not or is barely detectable in metaphase and during anaphase, suggesting that it is degraded or inaccessible to the antibodies (Figures 7H and 7I). As soon as cells are in telophase and reform their nuclear envelopes, both APC/C subunits are localized again to the nucleus (Figures 7E and 7J).
Figure 7.
Subcellular Localization of APC2 and CDC27A in Arabidopsis Root Meristematic Cells in Interphase and during Mitosis.
The coimmunolocalization studies were performed using, as primary antibodies, either the rabbit anti-APC2 antibody or the rabbit anti-CDC27A antibody and the mouse anti-α-tubulin antibody, together with their corresponding secondary antibodies (goat anti-rabbit fluorescein isothiocyanate and goat anti-mouse tetramethyl rhodamine isothiocyanate, respectively). The fluorescence produced by the secondary antibodies was visualized in interphase ([A] to [A′′] and [F] to [F′′]), preprophase ([B] to [B′′] and [G] to [G′′]), metaphase ([C] to [C′′] and [H] to [H′′]), anaphase ([D] to [D′′] and [I] to [I′′]), and telophase ([E] to [E′′] and [J] to [J′′]), as indicated. In the merge images ([A′′] to [J′′]), the APC/C subunits are represented in green and α-tubulin is represented in red. The arrows indicate the microtubules assembled at the nuclear surface in preprophase and prophase.
DISCUSSION
In plants, the functions of putative APC/C subunits are poorly understood. Recently, a computerized analysis allowed us to identify homologs in Arabidopsis for all 11 vertebrate APC/C subunits (Capron et al., 2003). Here, we present a molecular and genetic characterization of the APC2 subunit from Arabidopsis. Both sequence homology and functional complementation of a budding yeast thermosensitive mutant indicate that the plant gene is an APC2 ortholog. Moreover, we demonstrated that the plant APC2 protein interacts physically with the APC11 subunit, both in a yeast two-hybrid assay and in vivo. Using protein binding assays, it was found previously that a C-terminal fragment of human APC2 was able to interact with APC11, although the full-length APC2 failed in these assays (Tang et al., 2001). Thus, it is likely that APC2 and APC11 proteins form a heterodimeric protein complex similar to CUL1/RBX1 (Zheng et al., 2002) that may act as the catalytic center of the APC/C enzyme. The APC2 protein is a distant member of the cullin family (reviewed by Gieffers et al., 2000), whose founding member is Caenorhabditis elegans CUL1 (Kipreos et al., 1996). The exact function of this protein in the APC/C complex is unknown, but based on CUL1, it is believed to play an essential structural role as a scaffold protein that brings together different components of the complex. However, how the different APC/C subunits interact with each other remains unknown.
In addition to the interaction with the APC11 subunit, we also observed an interaction, at least in yeast, between the APC2 subunit and the TPR repeat–containing protein CDC23. In budding yeast, it has been found that the CDC23 subunit interacts with CDC27 (Lamb et al., 1994), whereas human APC10/DOC1 binds directly to APC11 (Tang et al., 2001). Nevertheless, our understanding of APC/C's quaternary organization is still far from complete.
In Saccharomyces cerevisiae and Schizosaccharomyces pombe, mutations in various APC/C subunits cause a failure of the release of sister chromatid cohesion and block the cells during the metaphase/anaphase transition (Hirano et al., 1988; Samejima and Yanagida, 1994; Irniger et al., 1995; Zachariae et al., 1996, 1998; Kramer et al., 1998). In C. elegans, mutations of different APC/C subunits cause arrest in the metaphase I/anaphase I transition and in the completion of meiosis (Furuta et al., 2000; Golden et al., 2000; Davis et al., 2002; Shakes et al., 2003). In Xenopus oocytes, the injection of specific antibodies against the CDC27 subunit also produces an arrest in meiosis, but at metaphase II (Peter et al., 2001). It was shown recently that the Drosophila APC2 subunit is encoded by the morula gene (Kashevsky et al., 2002). When this gene is mutated, both the endocycles and the mitotic cycles are impaired. Thus, the strong mutant alleles cause lethality in late larval development, and in the brain, an increased number of mitotic cells was seen that were arrested predominantly in metaphase (Reed and Orr-Weaver, 1997).
In plants, mutants in a putative APC/C subunit, CDC27B, have been reported (Blilou et al., 2002). These hobbit mutants were isolated initially because they were impaired in root meristem formation (Willemsen et al., 1998), but later they were shown to be affected more generally in the maintenance of cell division (Blilou et al., 2002). However, in Arabidopsis, the CDC27 subunit is encoded by two different genes: one that is now termed CDC27A, because its amino acid sequence is slightly closer to that of the APC3/CDC27 proteins, and CDC27B/HOBBIT. The existence of two related CDC27 genes may explain why the hobbit mutants are impaired only in postembryonic cell division.
In contrast to hobbit mutants, we show here that a null mutation in a subunit of the APC/C arrests the first mitotic division after meiosis during megagametogenesis. Consistently, a null mutation in the Arabidopsis CDC16 gene also impairs female gametophyte development (V. Sundaresan, personal communication). It is likely that the APC/C also is essential for meiosis in plants, but because of some residual activity sufficient to complete meiosis, only mitotic arrest after meiosis was observed in our mutants.
Unlike APC/C mutants from other organisms (see above), which result in meiotic or mitotic metaphase arrest, we never observed such an arrest in the Arabidopsis apc2 mutants. This is surprising, because the APC/C is known to promote the transition from metaphase to anaphase by ubiquitylation of the securin proteins in budding yeast (Cohen-Fix et al., 1996), fission yeast (Funabiki et al., 1996), and Drosophila (Leismann et al., 2000). Degradation of the securin protein indirectly activates the separase protease that targets a subunit of the cohesin complex, causing the release of sister chromatid cohesion (reviewed by Pellman and Christman, 2001). One possible explanation is that plants do not have securin-like proteins. To date, orthologs of securins have not been reported in plants, but it should be noted that these proteins are poorly conserved. Nevertheless, this possibility seems unlikely because APC/C-dependent ubiquitylation in plants is controlled by the spindle checkpoint (reviewed by Criqui and Genschik, 2002), which involves the securin proteins in other organisms. Moreover, it was shown that when tobacco BY2 cells are treated with the proteasome inhibitor MG132, they arrest in metaphase (Genschik et al., 1998). Another possible explanation is that the stability of plant securins is controlled by the proteasome, but in an APC/C-independent manner. Finally, it is possible that maternally provided APC/C is sufficient to allow the first mitotic division after meiosis in the apc2 mutants. Moreover, it seems that the maternal contribution of the APC/C allows successful microgametogenesis in most cases. This could be explained by the larger cytosolic volume of the embryo sac compared with the microspore and the pollen.
An important point is to understand where, at the subcellular level, APC/C-dependent ubiquitylation occurs. However, this issue remains controversial. In budding yeast, it was shown by indirect immunofluorescence that different APC/C subunits were localized in the nucleus and that none of them accumulated at the spindle pole bodies or on the mitotic spindles (Zachariae et al., 1996). By contrast, the Aspergillus nidulans CDC27 subunit concentrated on the spindle pole bodies (Mirabito and Morris, 1993). In mammalian cells, the largest subunit of the complex, APC1/Tsg24, has been found to be associated with the centromeres (Jörgensen et al., 1998), whereas CDC16 and CDC27 proteins were found concentrated on both the centrosomes and the spindle during mitosis (Tugendreich et al., 1995). However, in Drosophila, CDC16 and CDC27 were associated only weakly with the mitotic spindle (Huang and Raff, 1999). Furthermore, a green fluorescent protein (GFP)–tagged CDC27 subunit in Drosophila embryos was able to bind to mitotic chromatin, whereas the GFP-CDC16 protein was not (Huang and Raff, 2002). Thus, it was concluded that the subcellular localization of the APC/C subunits varies within and between organisms.
To our knowledge, none of the plant APC/C subunits have been localized previously. It is particularly challenging to address this issue, because the subcellular localization of the mitotic cyclin B, a well-known target of the APC/C, is different in plant and animal cells just before it is destroyed (Criqui et al., 2001). Whereas animal cyclin B is bound mainly to the mitotic spindle, the plant cyclin remains associated on the chromosomes during metaphase. Using specific antibodies, we immunolocalized the APC2 and CDC27A subunits in Arabidopsis. In contrast to those in Drosophila (Huang and Raff, 2002), both plant subunits were found in the nucleus of cells in interphase. During mitosis, the APC2 subunit was found diffuse and not enriched on the mitotic spindle or the chromosomes. This finding suggests that neither the spindle nor the condensed chromatin forms APC/C-dependent ubiquitylation centers in plants, as hypothesized for animal cells.
Curiously, we observed that the signal of the CDC27A protein disappeared, or at least decreased strongly, during metaphase and anaphase. There are two possible explanations for this finding. Either the protein is not accessible to the antibodies, suggesting some rearrangement of the APC/C during mitosis, or the protein is degraded. Interestingly, it was found that the CDC27-related protein CDC27B/HOBBIT is cell cycle regulated and that its mRNAs peak at approximately the G2/M-phase (Blilou et al., 2002). Thus, the plant APC/C may exchange its CDC27 subunit during mitosis.
METHODS
Cloning of the Different Arabidopsis Anaphase-Promoting Complex or Cyclosome cDNAs
The Arabidopsis thaliana APC2 full-length cDNA clone was obtained using an incomplete EST missing the 5′ end of the sequence. The 5′ end was obtained by reverse transcriptase–mediated PCR using the Smart RACE cDNA amplification kit (Clontech, Palo Alto, CA) and primer 5′-ATC- AGCCTCGACAGGGTCCGGTTC-3′ for the reverse transcription step. The RNAs were extracted from floral buds, and mRNAs were purified using the PolyATract mRNA Isolation System III (Promega). The 5′ and 3′ sequences were subcloned by PCR using primers 5′-AAATTTCTCGAGATGGAAGCTTTAGGTTCCT-3′ and 5′-AAATTTCAGCTGGATCTA- TTGCCCGA-3′. The cDNA of EST clone AV563067 was used for PCR amplification with primers 5′-AAATTTCAGCTGGTGTATTTCTGGAAG- CA-3′ and 5′-AAATTTGAGCTCCTACTTCTTTAGCAAATACA-3′. The two PCR fragments then were assembled through the PvuII site and cloned into the pGEM-T vector (Promega). Sequence analysis of the cloned fragment revealed that PCR amplification caused an amino acid substitution (Val to Ala) at position 373. The CDC26 cDNA clone was obtained by PCR amplification on genomic Arabidopsis DNA using primers 5′-ATCACTTTGTATCGAAGAAATGTTGAGAAG-3′ and 5′-GGAGAGTGG- TGAGACGGAGATGAGCAGCGG-3′ and then introduced into pGEM-T vector (Promega).
The other APC/C subunits were amplified directly by PCR from an Arabidopsis ecotype Columbia cell suspension cDNA library in the pACT2 vector (kindly provided by Csaba Koncz, Max-Planck Institut für Züchtungsforschung, Köln, Germany). The oligonucleotides used in these reactions were 5′-AGAGATATGGCGACAGAGT-3′ and 5′-TCA- TCTCAGTGTTGAATAAGT-3′ for APC10 (At2g18290), 5′-ATGGCTTTT- GATGGTTGTTG-3′ and 5′-TTACTCTTTGAACTGCCATTC-3′ for APC11 (At3g05870), 5′-ATGAGGGAAGAAATTGAG-3′ and 5′-ACTGACCAATTCCTAGCAGAG-3′ for CDC16 (At1g78770), 5′-ATGGCTTCTAAAGAGTGTTGC-3′ and 5′-GGAACAGTACAGCTAAATAGG-3′ for CDC23 (At3g48150), 5′-ATGATGGAGAATCTACTGGC-3′ and 5′-TTAGGGTTAGTCCACAAG-3′ for CDC27A (At3g16320), and 5′-ATGGAAGCTATG- CTTGTGGA-3′ and 5′-TCACGGGCTCTCATCGATCTC-3′ for CDC27B/HOBBIT (At2g20000). All PCR fragments were introduced into EcoRV-digested pBluescript KSII +.
Cloning of the Cyclin A3–GUS Reporter Gene Downstream of a Promoter Expressed in Gametophytes and Cloning of the Gene to Complement the apc2 Mutant
The first 137 amino acids of tobacco an A-type cyclin (Nicta;CycA3;1) (Genschik et al., 1998) containing the destruction box was fused upstream of the GUS reporter gene in such a way that the GUS coding sequence is in translational fusion with the cyclin. A NotI-NotI DNA fragment isolated from plasmid pAL51 containing the BnSKP1 g1 promoter and the 5′ untranslated region (Drouaud et al., 2000), the cyclin A3–GUS reporter gene, and the 19S terminator of Cauliflower mosaic virus were cloned into the NotI sites of the pEC2 binary plasmid (Cartea et al., 1998), generating the pAL101 plasmid.
To complement the apc2-1 mutant, the APC2 genomic region spanning the region from 2048 bp upstream of the ATG to 432 bp downstream of the stop codon was amplified by PCR using oligonucleotides 5′-CCCGGGGAATTCGTAGCGGATTGGTAGTTGCTAGCTG-3′ and 5′-CCCGGGGGATCCCATAAGTAATGTGTAGCGGTTCTGC-3′. The resulting fragment was digested with EcoRI and BamHI and subsequently cloned into the pCAMBIA1380 vector (CAMBIA, Canberra, Australia), resulting in plasmid pCAMB-APC2, and sequenced.
Transient Expression in Protoplasts and Immunoprecipitation
For transient expression in Arabidopsis suspension cultured cells, the APC11 and APC2 cDNAs were cloned into the pRT104-derived vectors (Topfer et al., 1989) with 3×HA and 3×myc epitopes, respectively, as N-terminal fusions. Protoplasts from Arabidopsis ecotype Columbia were transfected using the polyethylene glycol–mediated method (Kiegerl et al., 2000). For the immunoprecipitation, 0.5 mg of protein extract was incubated with 500 ng of rat monoclonal anti-HA antibody 3F10 (Roche, Mannheim, Germany) and 50 μL of Protein G MicroBeads (Miltenyi Biotec, Köln, Germany) for 16 h at 4°C. The washing and elution of immune complexes were performed according to the manufacturer's recommendations. The eluates were loaded on SDS-PAGE gels, and proteins were transferred to a polyvinylidene difluoride membrane. The efficiency of the immunoprecipitation was verified using the mouse monoclonal 12CA5 anti-HA antibody. myc-tagged APC2 was detected with the 9E10 mouse monoclonal antibody (Roche).
Yeast Two-Hybrid Techniques and Yeast Complementation Experiments
The cDNAs that encode the different subunits were introduced into the pGAD424 and pGBT9 yeast two-hybrid vectors (Clontech). The yeast two-hybrid analysis was performed using the GAL4-based Matchmaker Two-Hybrid System (Clontech) according to the manufacturer's protocol. For the transformations, the PJ69-4a strain (James et al., 1996) was used. Positive interactions were selected on synthetic complete medium lacking His and adenine. The β-galactosidase activities were assayed as described by Gindullis et al. (1999).
For the budding yeast mutant complementation experiments, the coding sequence of AtAPC11 was amplified by PCR with the primers 5′-ATATATTCTAGAATGAAAGTCAAGATCTTG-3′ and 5′-TATATAGGATCC- TTACTCTTTGAACTGCCA-3′ to generate a product with XbaI and BamHI restriction sites. This product was subcloned in p416GALS using these sites and then excised using HindIII and BamHI. The digested AtAPC11 then was inserted into pYC/2NT-A (Invitrogen, Carlsbad, CA) opened with the same enzymes, yielding the pYC2/NT APC11. For AtAPC2, its coding sequence was amplified by PCR with the primers 5′-ATATATGGATCCATGGAAGCTTTAGGTTCC-3′ and 5′-TATATAGAATTCCTACTTCTTTAGCAAATA-3′, and the resulting product was digested with BamHI and EcoRI and cloned in pYC2/NT-A (Invitrogen) using the same restriction sites, giving the pYC2/NT APC2. The yeast strain W373 (apc11-myc9) (Zachariae et al., 1996) was transformed with either pYC2/NT APC11 or pYC2/NT-A and cultured on SD-tryptophan/-uracil (SD-T/U) with glucose as a carbon source. The strain K6966 (apc2-1) (Zachariae et al., 1996) was transformed with either pYC2/NT APC2 or pYC/2NT-A and kept on SD-leucine/-histidine/-uracil (SD-L/H/U) with glucose as a carbon source. The yeast then were transferred in liquid SD-T/U or SD-L/H/U with 1% raffinose and 2% galactose for W373 and K6966, respectively, and grown for 48 h at 28°C with vigorous shaking. The cultures then were transferred to fresh medium, and their OD600 values were adjusted to 0.2. All cultures were split in two, with half of the volume placed at 28°C and the other half at 35°C. Growth was monitored by measurements at OD600. This experiment was performed using three different transformants for each construction.
RNA Gel Blot and Reverse Transcriptase–Mediated PCR Analyses
RNA was extracted from young leaves using the Trizol reagent (Gibco BRL). RNA gel blot analysis was performed with 20 μg of total RNA per lane. The RNA gel blot procedure has been described by Genschik et al. (1998). The histone H4 probe corresponds to the 196-bp restriction fragment AccI-DdeI of the coding region of the gene H4A748 (Chaboute et al., 1987). The integrity and the amount of RNA applied to each lane were verified by ethidium bromide staining and hybridization with a probe encoding translation elongation factor 1α (EF-1α) (cDNA clone 232A19T7, gene At1g07920).
Plant Transformation and Selection
Three-week-old Arabidopsis (Wassilewskija ecotype) plants were transformed by in planta transformation (Bechtold et al., 1993) with Agrobacterium tumefaciens strain C58 harboring pAL101 plasmid and strain GV3101 harboring pCAMB-APC2 plasmid.
For transformation with the pAL101 construct, T0 plants were sown on sand and watered with Basta-containing nutrient solution. Forty Basta-resistant plants (T1) then were selected, grown in the greenhouse, and observed for GUS activity in inflorescences. Plants were self-fertilized to obtain T2 seeds. Basta-resistant T2 plants from T1 plants were crossed as the female with the hygromycin-resistant apc2-2 mutant. Hybrid seeds were sterilized and sown in vitro on medium as described by Estelle and Somerville (1987) and supplemented with hygromycin (20 mg/L), and among them, Basta-resistant plants were selected by PCR analysis using Basta primers (5′-CCGTACCGAGCCGCAGGAAC-3′ and 5′-ATCTCGGTGACGGGCAGGAC-3′). Ten to 18 plants per cross were submitted to GUS assay.
For segregation analysis of the mutants, seeds were surface-sterilized and plated onto medium (half-strength Murashige and Skoog [1962] salts, 1% sucrose, and 0.9% agar, pH 5.7) supplemented with 50 mg/L kanamycin (mutants apc2-1 and apc2-3) or 15 mg/L hygromycin (mutant apc2-2). After 2 days at 4°C, the seeds were grown under 12-h-light/12-h-dark cycles at 22°C. The kanamycin or hygromycin phenotypes (resistant or sensitive) were scored after 2 weeks.
Histochemical Analysis
Histochemical assays for GUS activity were performed on inflorescences (closed bud to mature fruit). Inflorescences were removed and placed in ice. Samples were infiltrated for 2.5 min under vacuum (−93.3 kPa) in fixation buffer (chloroform:ethanol:water, 3:6:1 [v/v/v]). Three washes then were performed in wash buffer (100 mM sodium phosphate buffer, pH 7.0, 0.5 mM ferrocyanide, 0.5 mM ferricyanide, and 0.1% Triton X-100); during the second wash, the samples were infiltrated under vacuum (−93.3 kPa) for 5 min. Finally, X-Glu buffer (100 mM sodium phosphate buffer, pH 7.0, 0.5 mM ferrocyanide, 0.5 mM ferricyanide, 0.1% Triton X-100, and 1.6 mM 5-bromo-4-chloro-3-indolyl-β-glucuronic acid diluted in DMSO) was added, and samples were infiltrated for 30 min under vacuum (−93.3 kPa) and placed at 37°C overnight. The next day, the GUS buffer was replaced with 95% ethanol to discolor the tissues.
Intact Silique Analysis and Whole-Mount Preparation of Ovules
Siliques were prepared as described by Christensen et al. (1997) with the following modifications: individual pistils or whole floral stems were fixed in 4% glutaraldehyde diluted in 15 mM Sorenen's buffer (4.66 mM Na2HPO4·2H2O and 2.06 mM KH2PO4, pH 7.2) for 2 h at room temperature, with the first half-hour under vacuum. The samples were dehydrated in graded ethanol baths (50, 70, 90, and 100%) for 10 min each. The samples then were cleared in toluene, mounted in Immersol 518F (Zeiss, Jena, Germany), and dissected, and the slides were sealed using nail polish. Confocal laser scanning microscopy was performed using a Zeiss LSM510 laser scanning confocal microscope with helium-neon laser excitation at 543 nm, and emitted light was collected through a 560-nm Long Pass filter using a Plan APOCHROMAT (63 × 1.4 oil differential interference contrast). The images are presented either as single sections or three-dimensional reconstituted images. Transmitted light images were taken using differential interference contrast (Nomarski) optics and helium-neon laser illumination at 543 nm.
Antibodies and Immunofluorescence
A peptide of 14 amino acids located in the C-terminal domain of the protein ([C]FGSMALERIHNTLK, where [C] indicates an extra Cys) was synthesized, linked to KLH carrier protein, and used to immunize rabbits. The antiserum was immunoaffinity purified against the same peptide bound to Sepharose matrix.
For Arabidopsis CDC27A, a specific amino acid region (amino acids 256 to 372) excluding TPR domains was amplified by PCR and cloned into PQE31 vector containing an N-terminal 6× His tag (Qiagen, Valencia, CA). Recombinant protein was purified according to the manufacturer's recommendations. Polyclonal serum was obtained after three subsequent immunizations in two different rabbits. Sera were tested to determine affinity and specificity using ELISA and protein gel blot analysis (data not shown) and purified against the recombinant protein using Affigel-10 (Bio-Rad). The best serum was used in further immunolocalization experiments (1:200).
The immunolocalization procedure was described by Lauber et al. (1997). Primary antibodies were used at the following concentrations: affinity-purified anti-APC2, 1:100; affinity-purified anti-CDC27A, 1:200; anti-α-tubulin, 1:200 (NeoMarkers, Fremont, CA). Fluorochrome-conjugated secondary antibodies (Jackson Immunoresearch, West Grove, PA) were used at the following concentrations: goat anti-rabbit fluorescein isothiocyanate, 1:200; goat anti-mouse tetramethyl rhodamine isothiocyanate, 1:200. Confocal laser scanning was performed with an inverse fluorescence microscope (Leica, Wetzlar, Germany).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact P. Genschik, pascal.genschik@ibmp-ulp.u-strasbg.fr.
Accession Numbers
The accession number for the incomplete expressed sequence tag for APC2 is AV563067. DDBJ/EMBL/GenBank accession numbers for the sequences shown in Figure 1 are as follows: NP_013228 (for APC2_Sc), CAD62574 (for APC2_Sp), AAN10196 (for APC2_At), CAD41391 (for APC2_Os), NP_498762 (for APC2_Ce), AAM97765 (for APC2_Dm), NP_037498 (for APC2_Hs), Q12018 (for CDC53_Sc), NP_594259 (for Cul1_Sp), AAK76704 (for CUL1_At), AKK53839 (for CUL1_Os), Q17389 (for CUL1_Ce), AAD33676 (for CUL1_Dm), and NP_003583 (for CUL1_Hs).
Acknowledgments
We thank Wolfgang Zachariae for providing the yeast mutant strains, the ABRC for providing the EF-1α clone, Philippe Hammann for DNA sequencing, and Université Louis Pasteur de Strasbourg, Centre National de la Recherche Scientifique, Association pour la Recherche contre le Cancer, La Ligue Nationale Contre le Cancer, and Région Alsace for funding the confocal microscope. O.S. was funded by European Union Framework 5 Contract HPRN-CT-2002-00333. K.F. was supported by a PhD fellowship of the French Government and by the European Community ECCO QLG2-99-00454 research network program. Part of this work also was funded by the French plant genomic program Génoplante.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.013847.
References
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316, 1194–1199. [Google Scholar]
- Blilou, I., Frugier, F., Folmer, S., Serralbo, O., Willemsen, V., Wolkenfelt, H., Eloy, N.B., Ferreira, P.C., Weisbeek, P., and Scheres, B. (2002). The Arabidopsis HOBBIT gene encodes a CDC27 homolog that links the plant cell cycle to progression of cell differentiation. Genes Dev. 16, 2566–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton, J.L., and Solomon, M.J. (2000). Hsl1p, a Swe1p inhibitor, is degraded via the anaphase-promoting complex. Mol. Cell. Biol. 20, 4614–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capron, A., Okresz, L., and Genschik, P. (2003). First glance at the plant APC/C, a highly conserved ubiquitin-protein ligase. Trends Plant Sci. 8, 83–89. [DOI] [PubMed] [Google Scholar]
- Cartea, M.E., Migdal, M., Galle, A.M., Pelletier, G., and Guerche, P. (1998). Comparison of sense and antisense methodologies for modifying the fatty acid composition of Arabidopsis thaliana oilseed. Plant Sci. 136, 181–194. [Google Scholar]
- Chaboute, M.E., Chaubet, N., Philipps, G., Ehling, M., and Gigot, C. (1987). Genomic organization and nucleotide sequences of two histone H3 and two histone H4 genes of Arabidopsis thaliana. Plant Mol. Biol. 8, 179–191. [DOI] [PubMed] [Google Scholar]
- Christensen, C.A., King, E.J., Jordan, J.R., and Drews, G.N. (1997). Megagametogenesis in Arabidopsis wild type and Gf mutant. Sex. Plant Reprod. 10, 49–64. [Google Scholar]
- Ciechanover, A., Orian, A., and Schwartz, A.L. (2000). Ubiquitin-mediated proteolysis: Biological regulation via destruction. Bioessays 22, 442–451. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix, O., Peters, J.M., Kirschner, M.W., and Koshland, D. (1996). Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 10, 3081–3093. [DOI] [PubMed] [Google Scholar]
- Criqui, M.C., and Genschik, P. (2002). Mitosis in plants: How far we have come at the molecular level? Curr. Opin. Plant Biol. 5, 487–493. [DOI] [PubMed] [Google Scholar]
- Criqui, M.C., Parmentier, Y., Derevier, A., Shen, W.H., Dong, A., and Genschik, P. (2000). Cell cycle-dependent proteolysis and ectopic overexpression of cyclin B1 in tobacco BY2 cells. Plant J. 24, 763–773. [DOI] [PubMed] [Google Scholar]
- Criqui, M.C., Weingartner, M., Capron, A., Parmentier, Y., Shen, W.H., Heberle-Bors, E., Bogre, L., and Genschik, P. (2001). Sub-cellular localisation of GFP-tagged tobacco mitotic cyclins during the cell cycle and after spindle checkpoint activation. Plant J. 28, 569–581. [DOI] [PubMed] [Google Scholar]
- Davis, E.S., Wille, L., Chestnut, B.A., Sadler, P.L., Shakes, D.C., and Golden, A. (2002). Multiple subunits of the Caenorhabditis elegans anaphase-promoting complex are required for chromosome segregation during meiosis I. Genetics 160, 805–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies, R.J. (1999). SCF and cullin/ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Drouaud, J., Marrocco, K., Ridel, C., Pelletier, G., and Guerche, P. (2000). A Brassica napus skp1-like gene promoter drives GUS expression in Arabidopsis thaliana male and female gametophytes. Sex. Plant Reprod. 13, 29–35. [Google Scholar]
- Estelle, M.A., and Somerville, C.R. (1987). Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol. Gen. Genet. 206, 200–206. [Google Scholar]
- Funabiki, H., and Murray, A.W. (2000). The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell 102, 411–424. [DOI] [PubMed] [Google Scholar]
- Funabiki, H., Yamano, H., Kumada, K., Nagao, K., Hunt, T., and Yanagida, M. (1996). Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature 381, 438–441. [DOI] [PubMed] [Google Scholar]
- Furuta, T., Tuck, S., Kirchner, J., Koch, B., Auty, R., Kitagawa, R., Rose, A.M., and Greenstein, D. (2000). EMB-30: An APC4 homologue required for metaphase-to-anaphase transitions during meiosis and mitosis in Caenorhabditis elegans. Mol. Biol. Cell 11, 1401–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik, P., Criqui, M.C., Parmentier, Y., Derevier, A., and Fleck, J. (1998). Cell cycle–dependent proteolysis in plants: Identification of the destruction box pathway and metaphase arrest produced by the proteasome inhibitor MG132. Plant Cell 10, 2063–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieffers, C., Dube, P., Harris, J.R., Stark, H., and Peters, J.M. (2001). Three-dimensional structure of the anaphase-promoting complex. Mol. Cell 7, 907–913. [DOI] [PubMed] [Google Scholar]
- Gieffers, C., Schleiffer, A., and Peters, J.M. (2000). Cullins and cell cycle control. Protoplasma 211, 20–28. [Google Scholar]
- Gindullis, F., Peffer, N.J., and Meier, I. (1999). MAF1, a novel plant protein interacting with matrix attachment region binding protein MFP1, is located at the nuclear envelope. Plant Cell 11, 1755–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer, M., Murray, A.W., and Kirschner, M.W. (1991). Cyclin is degraded by the ubiquitin pathway. Nature 349, 132–138. [DOI] [PubMed] [Google Scholar]
- Gmachl, M., Gieffers, C., Podtelejnikov, A.V., Mann, M., and Peters, J.M. (2000). The RING-H2 finger protein APC11 and the E2 enzyme UBC4 are sufficient to ubiquitinate substrates of the anaphase-promoting complex. Proc. Natl. Acad. Sci. USA 97, 8973–8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden, A., Sadler, P.L., Wallenfang, M.R., Schumacher, J.M., Hamill, D.R., Bates, G., Bowerman, B., Seydoux, G., and Shakes, D.C. (2000). Metaphase to anaphase (mat) transition-defective mutants in Caenorhabditis elegans. J. Cell Biol. 151, 1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hames, R.S., Wattam, S.L., Yamano, H., Bacchieri, R., and Fry, A.M. (2001). APC/C-mediated destruction of the centrosomal kinase Nek2A occurs in early mitosis and depends upon a cyclin A-type D-box. EMBO J. 20, 7117–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, J.W., Burton, J.L., and Solomon, M.J. (2002). The anaphase-promoting complex: It's not just for mitosis any more. Genes Dev. 16, 2179–2206. [DOI] [PubMed] [Google Scholar]
- Hirano, T., Hiraoka, Y., and Yanagida, M. (1988). A temperature-sensitive mutation of the Schizosaccharomyces pombe gene nuc2+ that encodes a nuclear scaffold-like protein blocks spindle elongation in mitotic anaphase. J. Cell Biol. 106, 1171–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J., and Raff, J.W. (1999). The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells. EMBO J. 18, 2184–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J.Y., and Raff, J.W. (2002). The dynamic localisation of the Drosophila APC/C: Evidence for the existence of multiple complexes that perform distinct functions and are differentially localised. J. Cell Sci. 115, 2847–2856. [DOI] [PubMed] [Google Scholar]
- Irniger, S. (2002). Cyclin destruction in mitosis: A crucial task of Cdc20. FEBS Lett. 532, 7–11. [DOI] [PubMed] [Google Scholar]
- Irniger, S., Piatti, S., Michaelis, C., and Nasmyth, K. (1995). Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell 81, 269–278. [DOI] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. (1996). Genomic libraries and a host designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jörgensen, P.M., Brundell, E., Starborg, M., and Hoog, C. (1998). A subunit of the anaphase-promoting complex is a centromere-associated protein in mammalian cells. Mol. Cell. Biol. 18, 468–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juang, Y.L., Huang, J., Peters, J.M., McLaughlin, M.E., Tai, C.Y., and Pellman, D. (1997). APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science 275, 1311–1314. [DOI] [PubMed] [Google Scholar]
- Kashevsky, H., Wallace, J.A., Reed, B.H., Lai, C., Hayashi-Hagihara, A., and Orr-Weaver, T.L. (2002). The anaphase promoting complex/cyclosome is required during development for modified cell cycles. Proc. Natl. Acad. Sci. USA 99, 11217–11222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiegerl, S., Cardinale, F., Siligan, C., Gross, A., Baudouin, E., Liwosz, A., Eklöf, S., Till, S., Bögre, L., Hirt, H., and Meskiene, I. (2000). SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress–induced MAPK, SIMK. Plant Cell 12, 2247–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, R.W., Deshaies, R.J., Peters, J.M., and Kirschner, M.W. (1996). How proteolysis drives the cell cycle. Science 274, 1652–1659. [DOI] [PubMed] [Google Scholar]
- Kipreos, E.T., Lander, L.E., Wing, J.P., He, W.W., and Hedgecock, E.M. (1996). cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell 85, 829–839. [DOI] [PubMed] [Google Scholar]
- Kramer, K.M., Fesquet, D., Johnson, A.L., and Johnston, L.H. (1998). Budding yeast RSI1/APC2, a novel gene necessary for initiation of anaphase, encodes an APC subunit. EMBO J. 17, 498–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, J.R., Michaud, W.A., Sikorski, R.S., and Hieter, P.A. (1994). Cdc16p, Cdc23p and Cdc27p form a complex essential for mitosis. EMBO J. 13, 4321–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber, M.H., Waizenegger, I., Steinmann, T., Schwarz, H., Mayer, U., Hwang, I., Lukowitz, W., and Jürgens, G. (1997). The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 139, 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leismann, O., Herzig, A., Heidmann, S., and Lehner, C.F. (2000). Degradation of Drosophila PIM regulates sister chromatid separation during mitosis. Genes Dev. 14, 2192–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlepage, L.E., and Ruderman, J.V. (2002). Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev. 16, 2274–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry, T.J., and Kirschner, M.W. (1998). Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93, 1043–1053. [DOI] [PubMed] [Google Scholar]
- Mirabito, P.M., and Morris, N.R. (1993). BIMA, a TPR-containing protein required for mitosis, localizes to the spindle pole body in Aspergillus nidulans. J. Cell Biol. 120, 959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Pellman, D., and Christman, M.F. (2001). Separase anxiety: Dissolving the sister bond and more. Nat. Cell Biol. 3, E207–E209. [DOI] [PubMed] [Google Scholar]
- Peter, M., Castro, A., Lorca, T., Le Peuch, C., Magnaghi-Jaulin, L., Doree, M., and Labbe, J.C. (2001). The APC is dispensable for first meiotic anaphase in Xenopus oocytes. Nat. Cell Biol. 3, 83–87. [DOI] [PubMed] [Google Scholar]
- Peters, J.M. (2002). The anaphase-promoting complex: Proteolysis in mitosis and beyond. Mol. Cell 9, 931–943. [DOI] [PubMed] [Google Scholar]
- Petersen, B.O., Wagener, C., Marinoni, F., Kramer, E.R., Melixetian, M., Denchi, E.L., Gieffers, C., Matteucci, C., Peters, J.M., and Helin, K. (2000). Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev. 14, 2330–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger, C.M., and Kirschner, M.W. (2000). The KEN box: An APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 14, 655–665. [PMC free article] [PubMed] [Google Scholar]
- Reed, B.H., and Orr-Weaver, T.L. (1997). The Drosophila gene morula inhibits mitotic functions in the endo cell cycle and the mitotic cell cycle. Development 124, 3543–3553. [DOI] [PubMed] [Google Scholar]
- Rios, G., et al. (2002). Rapid identification of Arabidopsis insertion mutants by non-radioactive detection of T-DNA tagged genes. Plant J. 32, 243–253. [DOI] [PubMed] [Google Scholar]
- Samejima, I., and Yanagida, M. (1994). Bypassing anaphase by fission yeast cut9 mutation: Requirement of cut9+ to initiate anaphase. J. Cell Biol. 127, 1655–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakes, D.C., Sadler, P.L., Schumacher, J.M., Abdolrasulnia, M., and Golden, A. (2003). Developmental defects observed in hypomorphic anaphase-promoting complex mutants are linked to cell cycle abnormalities. Development 130, 1605–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama, M., Zachariae, W., Ciosk, R., and Nasmyth, K. (1998). The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 17, 1336–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Z., Li, B., Bharadwaj, R., Zhu, H., Ozkan, E., Hakala, K., Deisenhofer, J., and Yu, H. (2001). APC2 Cullin protein and APC11 RING protein comprise the minimal ubiquitin ligase module of the anaphase-promoting complex. Mol. Biol. Cell 12, 3839–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topfer, R., Matzeit, V., Gronenborn, B., Schell, J., and Steinbiss, H.H. (1989). A set of plant expression vectors for transcriptional and translational fusions. Nucleic Acids Res. 15, 5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugendreich, S., Tomkiel, J., Earnshaw, W., and Hieter, P. (1995). CDC27Hs colocalizes with CDC16Hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell 81, 261–268. [DOI] [PubMed] [Google Scholar]
- Vodermaier, H.C. (2001). Cell cycle: Waiters serving the Destruction machinery. Curr. Biol. 11, R834–R837. [DOI] [PubMed] [Google Scholar]
- Voges, D., Zwickl, P., and Baumeister, W. (1999). The 26S proteasome: A molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68, 1015–1068. [DOI] [PubMed] [Google Scholar]
- Willemsen, V., Wolkenfelt, H., de Vrieze, G., Weisbeek, P., and Scheres, B. (1998). The HOBBIT gene is required for formation of the root meristem in the Arabidopsis embryo. Development 125, 521–531. [DOI] [PubMed] [Google Scholar]
- Zachariae, W., and Nasmyth, K. (1999). Whose end is destruction: Cell division and the anaphase-promoting complex. Genes Dev. 13, 2039–2058. [DOI] [PubMed] [Google Scholar]
- Zachariae, W., Schwab, M., Nasmyth, K., and Seufert, W. (1998). Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science 282, 1721–1724. [DOI] [PubMed] [Google Scholar]
- Zachariae, W., Shin, T.H., Galova, M., Obermaier, B., and Nasmyth, K. (1996). Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science 274, 1201–1204. [DOI] [PubMed] [Google Scholar]
- Zheng, N., et al. (2002). Structure of the Cul1-Rbx1-Skp1-F box-Skp2 SCF ubiquitin ligase complex. Nature 416, 703–709. [DOI] [PubMed] [Google Scholar]