Abstract
A loss-of-function mutation in the Arabidopsis SSI2/FAB2 gene, which encodes a plastidic stearoyl–acyl-carrier protein desaturase, has pleiotropic effects. The ssi2 mutant plant is dwarf, spontaneously develops lesions containing dead cells, accumulates increased salicylic acid (SA) levels, and constitutively expresses SA-mediated, NPR1-dependent and -independent defense responses. In parallel, jasmonic acid–regulated signaling is compromised in the ssi2 mutant. In an effort to discern the involvement of lipids in the ssi2-conferred developmental and defense phenotypes, we identified suppressors of fatty acid (stearoyl) desaturase deficiency (sfd) mutants. The sfd1, sfd2, and sfd4 mutant alleles suppress the ssi2-conferred dwarfing and lesion development, the NPR1-independent expression of the PATHOGENESIS-RELATED1 (PR1) gene, and resistance to Pseudomonas syringae pv maculicola. The sfd1 and sfd4 mutant alleles also depress ssi2-conferred PR1 expression in NPR1-containing sfd1 ssi2 and sfd4 ssi2 plants. By contrast, the sfd2 ssi2 plant retains the ssi2-conferred high-level expression of PR1. In parallel with the loss of ssi2-conferred constitutive SA signaling, the ability of jasmonic acid to activate PDF1.2 expression is reinstated in the sfd1 ssi2 npr1 plant. sfd4 is a mutation in the FAD6 gene that encodes a plastidic ω6-desaturase that is involved in the synthesis of polyunsaturated fatty acid–containing lipids. Because the levels of plastid complex lipid species containing hexadecatrienoic acid are depressed in all of the sfd ssi2 npr1 plants, we propose that these lipids are involved in the manifestation of the ssi2-conferred phenotypes.
INTRODUCTION
Lipids are important structural components of biological membranes. In addition, lipid-derived second messengers participate in signal transduction mechanisms to influence plant growth, development, and responses to environmental cues (Munnik et al., 1998; Somerville et al., 2000; Laxalt and Munnik, 2002; Weber, 2002). Many of these signaling molecules are derived from fatty acids (Weber, 2002). De novo fatty acid synthesis in higher plants occurs solely in the plastids (Ohlrogge et al., 1991), resulting in the synthesis of palmitic acid (16:0)–, stearic acid (18:0)–, and oleic acid (18:1)–acyl-carrier protein (ACP) (McKeon and Stumpf, 1982; Somerville et al., 2000). These are either converted to the plastidic lipids phosphatidylglycerol (PG), monogalactosyldiacylglycerol (MGDG), digalactosyldiacylglycerol (DGDG), and sulfoquinovosyldiacylglycerol by the prokaryotic pathway localized in the plastid inner envelope or exported as CoA thioesters to the cytoplasm, where they enter the eukaryotic lipid biosynthesis pathway, which primarily synthesizes phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), and phosphatidylserine (Somerville et al., 2000; Wallis and Browse, 2002). Some PC species, or fragments thereof, are returned to the plastid, where an acyl-glycerol component is incorporated into complex plastid lipids (Browse et al., 1986; Mongrand et al., 2000). Differences in the enzymes present in the two compartments result in differences in the positions at which 16- and 18-C acyl species are esterified to glycerol, the extent of 16- and 18-C acyl desaturation, and the lipid head group classes formed in the cytoplasm and plastid. The desaturation of 16:0 and 18:1 occurs on 16-C and 18-C fatty acids incorporated into complex lipids. In Arabidopsis, the desaturation of 16:0 to hexadecatrienoic acid (16:3) occurs primarily on plastid-localized complex lipids. Hence, 16:3-containing lipids are found predominantly in the plastid lipid, MGDG (Wallis and Browse, 2002).
Polyunsaturated fatty acids (PUFAs) released from membrane lipids are precursors for the synthesis of oxylipins, a large family of oxidized lipids. For example, jasmonic acid (JA) and its methyl ester (MeJA) are synthesized from linolenic acid (18:3) via the octadecanoid pathway. In addition, a parallel hexadecanoid pathway in Arabidopsis synthesizes JA-related compounds from 16:3 (Weber, 2002). Oxylipins are important signaling molecules in plant development and defense against wounding, insect feeding, and pathogen infection (Reymond and Farmer, 1998; Weber, 2002). For example, JA is required for pollen development, and several Arabidopsis mutants blocked in JA synthesis or signaling are male sterile as a result of improper pollen development (Feys et al., 1994; McConn and Browse, 1996; Stinzi and Browse, 2000). In addition, JA biosynthesis and signaling mutants exhibit increased susceptibility to fungal pathogens and insects (Penninckx et al., 1996; Staswick et al., 1998; Vijayan et al., 1998). By contrast, JA application activates the expression of the antifungal protein defensin gene PDF1.2 (Penninckx et al., 1998). Moreover, high-level expression of PDF1.2 correlates with increased resistance to the necrotrophic fungus Botrytis cinerea (Berrocal-Lobo et al., 2002).
Salicylic acid (SA) is another important signaling molecule in plants, influencing plant development and defense responses (Shah and Klessig, 1999). The involvement of SA in plant defense has been studied extensively. SA levels are increased in plants resisting pathogen attack, and these increases correlate with the activation of PATHOGENESIS-RELATED (PR) gene expression and resistance (Durner et al., 1997; Shah and Klessig, 1999). In addition, the application of SA activates PR gene expression and disease resistance. Several PR proteins have proven antimicrobial activities, and their expression serves as a molecular marker for the activation of defense responses (Klessig and Malamy, 1994; Hammond-Kosack and Jones, 1996; Hunt and Ryals., 1996; Ryals et al., 1996). Blocking SA biosynthesis in the Arabidopsis eds5 (sid1) and sid2 (eds16) mutants depresses PR expression in pathogen-infected plants and enhances susceptibility to pathogens (Nawrath and Metraux, 1999; Dewdney et al., 2000). Likewise, preventing SA accumulation by expressing the SA-degrading salicylate hydroxylase, which is encoded by the bacterial nahG gene, suppresses the expression of PR genes and confers enhanced susceptibility to pathogens (Gaffney et al., 1993; Delaney et al., 1994). The NPR1 gene is an important regulator of the SA-dependent expression of PR genes and resistance in Arabidopsis. The loss of NPR1 activity affects the SA-inducible expression of PR genes and resistance in npr1/nim1 mutants (Cao et al., 1994; Delaney et al., 1995; Glazebrook et al., 1996; Shah et al., 1997). However, SA signaling also occurs via NPR1-independent mechanism(s) in addition to the NPR1-dependent pathway (Shah et al., 1999, 2001; Clarke et al., 2000).
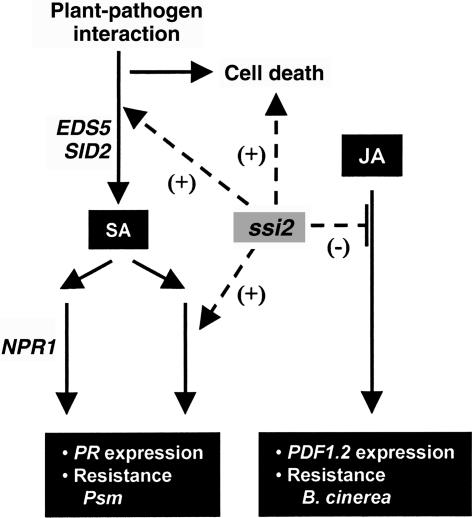
The ssi (suppressor of SA insensitivity) mutants were identified in a screen for mutants that restore SA signaling in the npr1 genetic background (Shah et al., 1999, 2001; Shirano et al., 2002). SA signaling through the NPR1-dependent and -independent pathways is constitutively active in the ssi2 mutant (Shah et al., 2001). PR genes are expressed at increased levels, and resistance to Peronospora parasitica is enhanced in ssi2 and ssi2 npr1 plants (Shah et al., 2001). In addition, enhanced resistance to an avirulent strain of Pseudomonas syringae pv tomato is observed in the ssi2 mutant. The ssi2-conferred PR expression and resistance phenotypes are associated with the accumulation of increased levels of SA. Moreover, increased SA levels are required for the ssi2-conferred PR expression and enhanced disease resistance; PR1 expression and resistance to P. parasitica and P. syringae are depressed in the ssi2 nahG plant (Shah et al., 2001). Crosstalk between SA and JA signaling pathways is important for the fine-tuning of plant defense responses. Both synergistic and antagonistic interactions between SA and JA signaling have been reported (Reymond and Farmer, 1998; Kunkel and Brooks, 2002). Similarly, in the ssi2 mutant, the constitutive activation of SA-dependent defense responses is paralleled by the inability of JA to activate PDF1.2 expression and enhanced susceptibility to B. cinerea (Kachroo et al., 2001). In addition to altering defense responses, ssi2 and the allelic fab2 mutant are dwarf and develop lesions that contain dead cells (Lightner et al., 1994a, 1994b; Shah et al., 2001). Figure 1 summarizes the impact of ssi2 on Arabidopsis defense signaling.
Figure 1.
Impact of the ssi2 Mutant on Defense Signaling in Arabidopsis.
SA and JA are important signaling molecules in plant defense. JA signaling is required for the pathogen-activated expression of PDF1.2 and resistance to the necrotrophic pathogen B. cinerea. SA signaling is required for the pathogen-activated expression of the PR genes and resistance to Psm. The EDS5 and SID2 genes are required for SA synthesis; the loss-of-function eds5 and sid2 mutations block SA synthesis. SA signaling is activated via both NPR1-dependent and -independent mechanisms. In addition to SA, an unknown pathogen-activated factor is required for signaling through the NPR1-independent pathway. The loss of the SSI2-encoded stearoyl-ACP desaturase activity in the ssi2 mutant has pleiotropic effects on plant defense responses. The ssi2 mutant allele promotes (+) the spontaneous development of lesions containing dead cells, the accumulation of increased SA levels, and the constitutive expression of NPR1-dependent and -independent defense mechanisms, which confer high-level expression of PR genes and enhanced resistance to Psm. By contrast, the ssi2 mutant interferes with (−) the ability of JA/MeJA to activate PDF1.2 expression and exhibits enhanced susceptibility to B. cinerea. 18:1 application restores the JA-inducible expression of PDF1.2 in the ssi2 mutant plant (Kachroo et al., 2001), suggesting a role for an 18:1-derived factor, which is limiting in the ssi2 mutant plant, in promoting JA signaling.
The ssi2 phenotypes are caused by a deficiency in a plastidic stearoyl-ACP desaturase activity (Kachroo et al., 2001), suggesting the involvement of lipids and lipid signaling in plant development and in modulating SA- and JA-dependent defense mechanisms. The FAB2 (SSI2)-encoded stearoyl-ACP desaturase preferentially desaturates 18:0-ACP to yield 18:1-ACP (Shanklin and Somerville, 1991; Shanklin and Cahoon, 1998). Compared with those in wild-type plants, 18:0 levels were increased and 18:1 levels were depressed in the ssi2 mutant. Moreover, coapplication of 18:1 restored JA-activated PDF1.2 expression in the ssi2 mutant, suggesting that 18:1 deficiency, rather than the increased 18:0 level, was responsible for the inability of the ssi2 mutant to express PDF1.2 (Kachroo et al., 2001). However, the role of altered lipid composition in other ssi2-conferred phenotypes was unclear.
To further elucidate the role of lipids in the ssi2-conferred growth and developmental phenotypes and defense responses, we conducted a suppressor screen in the ssi2 npr1 genetic background for mutants that lack ssi2-conferred dwarfing and PR1 expression. The sfd1, sfd2, and sfd4 mutant alleles suppress the ssi2-conferred dwarfing and lesion development, the NPR1-independent expression of PR1, and resistance to Pseudomonas syringae pv maculicola (Psm). In addition, sfd1 overcomes the ssi2-conferred block on MeJA activation of PDF1.2 expression. sfd4 contains a mutation in the 16:1/18:1 ω6 desaturase gene FAD6. Moreover, levels of the PUFA 16:3 were depressed in all sfd ssi2 npr1 plants, suggesting a role for PUFA-derived signals in promoting the ssi2-conferred plant developmental and defense response phenotypes.
RESULTS
sfd Mutant Alleles Suppress the ssi2-Conferred Dwarfing and Lesion Development and the NPR1-Independent Expression of the PR1 Gene
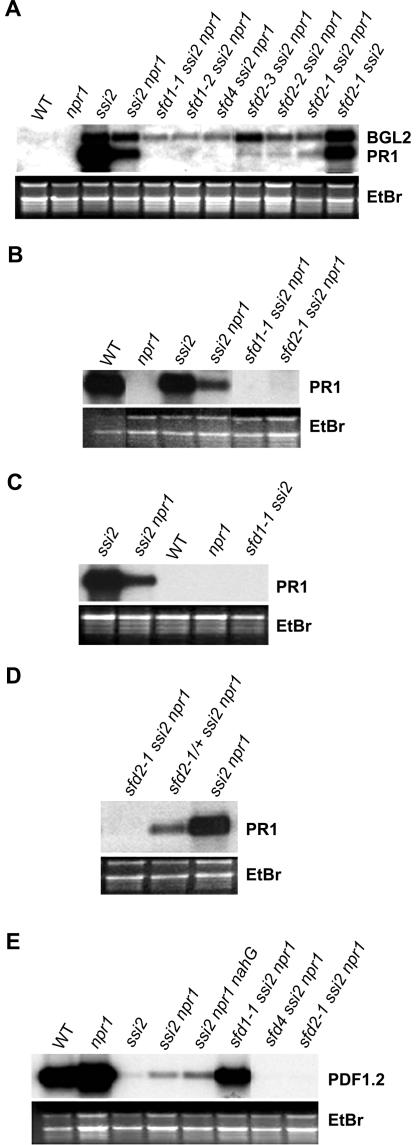
To isolate suppressors of the stearoyl-ACP desaturase–deficient ssi2 mutation, seeds from ssi2 npr1 plants were mutagenized with ethyl methanesulfonate as described previously (Shah et al., 1997). Four-week-old M2 progeny of these ethyl methanesulfonate–mutagenized M1 seeds were screened for large plants and for the loss of ssi2-conferred constitutive PR1 expression. Seven sfd mutants were identified among the 4000 M2 plants screened. These sfd mutants define four complementation groups: sfd1 (two alleles: sfd1-1 and sfd1-2), sfd2 (three alleles: sfd2-1, sfd2-2, and sfd2-3), sfd3, and sfd4. Mutants in the sfd1, sfd2, and sfd4 complementation groups were characterized further. As shown in Figures 2A and 3A, mutants in the sfd1, sfd2, and sfd4 complementation groups ameliorate the ssi2-conferred dwarf phenotype and depress the ssi2-conferred NPR1-independent expression of the PR1 gene. The suppressive effect of these sfd alleles on the ssi2-conferred BGL2 expression was less pronounced.
Figure 2.
Comparison of Morphological and Cell Death Phenotypes of the sfd ssi2 npr1 Mutants.
(A) Comparison of the morphology of 4-week-old npr1, ssi2 npr1, sfd1-1 ssi2 npr1, sfd1-2 ssi2 npr1, sfd2-1 ssi2 npr1, sfd2-1/+ ssi2 npr1, sfd2-2 ssi2 npr1, and sfd4 ssi2 npr1 plants. The sfd2-1/+ ssi2 npr1 plant is heterozygous for the sfd2-1 mutant allele. All plants were photographed from the same distance.
(B) Light microscopy of trypan blue–stained leaves from npr1, ssi2 npr1, sfd1-1 ssi2 npr1, sfd2-1 ssi2 npr1, and sfd4 ssi2 npr1 plants. The arrows mark areas containing intensely stained dead cells in ssi2 npr1. All photographs were taken at the same magnification.
Figure 3.
Defense Gene Expression in the sfd Mutants.
(A) Comparison of PR1 and BGL2 expression in leaves of 4-week-old soil-grown wild-type (WT), npr1, ssi2, ssi2 npr1, sfd1-1 ssi2 npr1, sfd1-2 ssi2 npr1, sfd4 ssi2 npr1, sfd2-3 ssi2 npr1, sfd2-2 ssi2 npr1, sfd2-1 ssi2 npr1, and sfd2-1 ssi2 plants.
(B) Comparison of PR1 expression in leaves of 4-week-old wild-type, npr1, ssi2, ssi2 npr1, sfd1-1 ssi2 npr1, and sfd2-1 ssi2 npr1 plants 48 h after treatment with 500 μM SA.
(C) Comparison of PR1 expression in leaves of 4-week-old soil-grown ssi2, ssi2 npr1, wild-type, npr1, and sfd1-1 ssi2 plants.
(D) Comparison of PR1 expression in sfd2-1 ssi2 npr1, sfd2-1/+ ssi2 npr1, and ssi2 npr1 plants.
(E) Comparison of PDF1.2 expression in leaves of 4-week-old wild-type, npr1, ssi2, ssi2 npr1, ssi2 npr1 nahG, sfd1-1 ssi2 npr1, sfd4 ssi2 npr1, and sfd2-1 ssi2 npr1 plants 48 h after treatment with 5 μM MeJA dissolved in 0.1% ethanol.
All RNAs were resolved on denaturing gels, transferred to Nytran Plus membranes (Schleicher & Schuell), and probed for the indicated genes. Gel loading was monitored by photographing the ethidium bromide–stained gel (EtBr) before transferring the RNA to a Nytran Plus membrane.
Compared with the wild type and npr1, leaves of all sfd ssi2 npr1 plants were lighter green. Leaves of the ssi2 and ssi2 npr1 mutant plants develop macroscopic lesions containing dead cells (Shah et al., 2001). However, leaves of all sfd1 ssi2 npr1 and sfd2 ssi2 npr1 plants and the sfd4 ssi2 npr1 plant do not exhibit macroscopic lesions. We examined trypan blue–stained leaves from the sfd1-1 ssi2 npr1, sfd2-1 ssi2 npr1, and sfd4 ssi2 npr1 plants to determine whether they had microscopic lesions containing dead cells. Trypan blue stains dead cells dark blue. As shown in Figure 2B, unlike the ssi2 npr1 plant, but like the npr1 control, leaves of the sfd1-1 ssi2 npr1, sfd2-1 ssi2 npr1, and sfd4 ssi2 npr1 plants lacked intensely stained dead cells.
sfd1 and sfd4 Suppress the ssi2-Conferred SA Accumulation
Previously, we had shown that the ssi2-conferred accumulation of increased SA is not required for cell death and dwarfing (Shah et al., 2001). Dwarfing, cell death, and SA accumulation may be activated independently by ssi2. Alternatively, dwarfing and/or the activation of cell death in the ssi2 mutant may stimulate SA accumulation. Therefore, we measured the total SA plus SA-glucoside levels in leaves of sfd1-1 ssi2 npr1, sfd2-1 ssi2 npr1, and sfd4 ssi2 npr1 plants. As shown in Table 1, the total SA content of sfd1-1 ssi2 npr1 and sfd4 ssi2 npr1 plants was comparable to the total SA level in wild-type and npr1 plants and 25- to 50-fold lower than the total SA level in the ssi2 npr1 plant. By contrast, the total SA content in the sfd2-1 ssi2 npr1 plant was ∼13-fold higher than the total SA level in wild-type and npr1 plants and one-quarter of that in the ssi2 npr1 plant.
Table 1.
Total SA Content in sfd Plants
| Genotype | Total SA ± sd (μg/g FW) |
|---|---|
| Wild type | 0.5 ± 0.2 |
| npr1 | 0.5 ± 0.1 |
| ssi2 npr1 | 24.0 ± 1.8 |
| sfd1-1 ssi2 npr1 | 0.5 ± 0.1 |
| sfd2-1 ssi2 npr1 | 6.6 ± 1.3 |
| sfd4 ssi2 npr1 | 0.9 ± 0.3 |
Total SA values for each line are averages of five samples ± sd. All values are corrected based on a 73% recovery. FW, fresh weight.
To determine if the loss of ssi2-conferred NPR1-independent PR1 expression in sfd1-1 ssi2 npr1 is attributable to the lack of SA accumulation, we examined PR1 expression in leaves of the SA-treated sfd1-1 ssi2 npr1 plant. SA-treated wild-type, ssi2, and ssi2 npr1 plants served as positive controls for monitoring the efficacy of SA application in this experiment, and the npr1 mutant served as a negative control. As shown in Figure 3B, SA was ineffective at restoring ssi2-conferred PR1 expression in the sfd1-1 ssi2 npr1 plant. Likewise, SA application was ineffective at restoring PR1 expression in the sfd2-1 ssi2 npr1 (Figure 3B) and sfd4 ssi2 npr1 plants (data not shown). By contrast, the SA-treated wild-type and ssi2 control plants expressed high levels of PR1. Similar results were obtained with the application of benzothiadiazole, a functional analog of SA (data not shown). Hence, another ssi2-modulated factor(s), in addition to SA, is required for the NPR1-independent PR1 expression observed in the ssi2 mutant.
sfd1, sfd2, and sfd4 Suppress the ssi2-Conferred NPR1-Independent Resistance to Psm
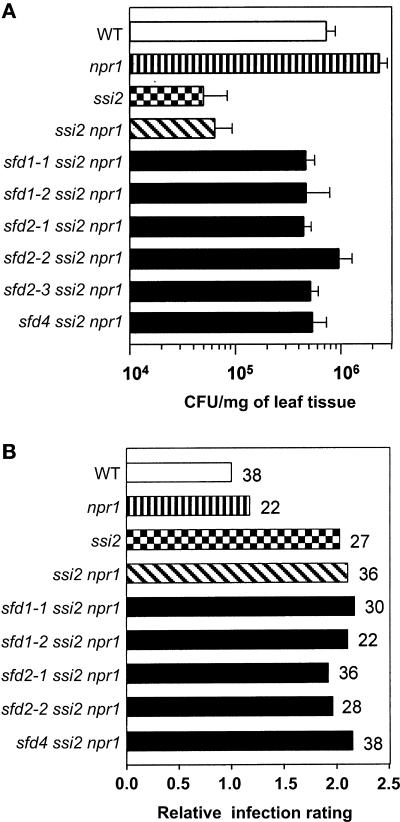
We had shown previously that the activation of SA signaling in the ssi2 npr1 plant confers enhanced resistance to P. syringae pv tomato DC3000 containing the avirulence gene avrRpt2 and the oomycete Peronospora parasitica Emco5 (Shah et al., 2001). Similarly, as shown in Figure 4A, compared with wild-type and npr1 plants, the ssi2 npr1 double mutant plant exhibited enhanced resistance to Psm. Bacterial numbers at 3 days after inoculation in the ssi2 npr1 plant were 20-fold lower than those in the npr1 plant. Therefore, we tested if the sfd1, sfd2, and sfd4 mutant alleles could suppress the ssi2-conferred NPR1-independent resistance to Psm. Bacterial numbers were monitored in sfd1-1 ssi2 npr1, sfd1-2 ssi2 npr1, sfd2-1 ssi2 npr1, sfd2-2 ssi2 npr1, sfd2-3 ssi2 npr1, and sfd4 ssi2 npr1 plants. As shown in Figure 4A, bacterial numbers in sfd1-1 ssi2 npr1, sfd1-2 ssi2 npr1, sfd2-1 ssi2 npr1, sfd2-2 ssi2 npr1, sfd2-3 ssi2 npr1, and sfd4 ssi2 npr1 plants at 3 days after inoculation were 8- to 10-fold higher than those in the ssi2 npr1 plant.
Figure 4.
Growth of Pathogen in the sfd Mutants.
(A) Bacterial numbers in the sfd mutants. Psm (OD600 = 0.0002) was infiltrated into the abaxial surfaces of leaves of wild-type (WT), npr1, ssi2, ssi2 npr1, sfd1-1 ssi2 npr1, sfd1-2 ssi2 npr1, sfd2-1 ssi2 npr1, sfd2-2 ssi2 npr1, sfd2-3 ssi2 npr1, and sfd4 ssi2 npr1 plants with a needleless syringe. Leaf discs were harvested from the inoculated leaves at 3 days after inoculation, weighed, and ground in 10 mM MgCl2, and the bacterial numbers were titered. Each sample contained five leaf discs. The bacterial numbers, presented as colony-forming units (CFU) per mg of leaf tissue, represent the average of five samples ± sd.
(B) B. cinerea disease ratings in the sfd mutants. Leaves of 4-week-old soil-grown wild-type, npr1, ssi2, ssi2 npr1, sfd1-1 ssi2 npr1, sfd1-2 ssi2 npr1, sfd2-1 ssi2 npr1, sfd2-2 ssi2 npr1, and sfd4 ssi2 npr1 plants were inoculated with spores of B. cinerea. Four days later, plants were scored for the extent of spreading necrosis. Leaves from each line were grouped based on the extent of necrosis. A four-step grading system was used. Leaves with <25%, 25 to 50%, 50 to 75%, and 75 to 100% of leaf area exhibiting necrosis were given scores of 0.25, 0.5, 0.75, and 1, respectively. The leaves in each category were counted, multiplied by the score, and divided by the total number of leaves inoculated (given next to each bar) to give an infection rating for each line. The “relative infection rating” for each line was calculated as the ratio of the infection rating for the line to the infection rating of the wild type.
sfd2 Does Not Suppress the ssi2-Conferred NPR1-Dependent Expression of PR1
The ssi2 mutant activates SA signaling through the NPR1-dependent pathway in addition to via the NPR1-independent mechanism (Shah et al., 2001). Therefore, we tested if the sfd1-1 and sfd2-1 mutant alleles could suppress ssi2-conferred PR1 expression in the wild-type NPR1-containing sfd1-1 ssi2 and sfd2-1 ssi2 plants. As shown in Figure 3C, the sfd1-1 allele effectively suppressed ssi2-conferred constitutive PR1 expression in the sfd1-1 ssi2 plant. By contrast, the sfd2-1 allele was not very effective at blocking ssi2-conferred PR1 expression in the sfd2-1 ssi2 double mutant (Figure 3A). Compared with the PR1 transcript levels in the ssi2 mutant plant, only a slight reduction in the accumulation of PR1 transcript, probably as a result of the suppression of the NPR1-independent pathway, was observed in the sfd2-1 ssi2 plant, suggesting that sfd2 specifically affects NPR1-independent signaling.
sfd1 Bypasses the ssi2-Conferred Block on the JA-Activated Expression of PDF1.2
The loss of SSI2 activity in the ssi2 mutant prevents the JA-activated expression of the PDF1.2 gene (Kachroo et al., 2001). Kloek et al. (2001) have shown that JA signaling antagonizes SA signaling in plant defense, including the expression of PR1 and resistance to P. syringae. The lack of ssi2-conferred PR1 expression in the sfd1 ssi2 npr1, sfd2 ssi2 npr1, and sfd4 ssi2 npr1 plants could result from the restoration of JA signaling. Therefore, we examined the expression of the PDF1.2 gene in MeJA-treated leaves of the sfd1-1 ssi2 npr1, sfd2-1 ssi2 npr1, and sfd4 ssi2 npr1 plants. MeJA-treated leaves of wild-type and npr1 plants served as positive controls for this experiment, and ssi2, ssi2 npr1, and ssi2 npr1 nahG plants served as negative controls. As shown previously (Kachroo et al., 2001), MeJA application conferred high-level expression of the PDF1.2 gene in leaves of wild-type and npr1 plants but not in ssi2, ssi2 npr1, and ssi2 npr1 nahG plants (Figure 3E). The presence of the sfd1-1 allele restored PDF1.2 expression in MeJA-treated leaves of the sfd1-1 ssi2 npr1 plant (Figure 3E). However, MeJA was unable to activate PDF1.2 expression in the leaves of sfd2-1 ssi2 npr1 and sfd4 ssi2 npr1. As expected, control plants treated with ethanol, which was used to dilute the MeJA, did not activate the expression of PDF1.2 (data not shown).
ssi2-Conferred Susceptibility to B. cinerea Is Unaffected by sfd1, sfd2, and sfd4
An increased susceptibility to B. cinerea parallels the inability of the ssi2 mutant to express the PDF1.2 gene in response to JA application (Kachroo et al., 2001). Therefore, we tested whether the restoration of JA-inducible PDF1.2 expression in the sfd1-1 ssi2 npr1 plant is associated with the reestablishment of resistance to B. cinerea. As shown in Figure 4B, compared with the wild-type plant, the sfd1-1 ssi2 npr1 plant retained the ssi2-conferred enhanced susceptibility to infection with B. cinerea. The relative infection rating of sfd1-1 ssi2 npr1 was comparable to those of ssi2 npr1 and ssi2 plants. Similarly, the sfd1-2 ssi2 npr1 plant retained the ssi2-conferred enhanced susceptibility to B. cinerea. The relative infection ratings for sfd2-1 ssi2 npr1, sfd2-2 ssi2 npr1, and sfd4 ssi2 npr1 plants were comparable to that of the ssi2 npr1 plant as well.
sfd1, sfd2, and sfd4 Modify the Lipid Profile of the ssi2 Mutant
It was hypothesized previously that decreased levels of 18:1 contribute directly or indirectly to the ssi2 phenotypes (Kachroo et al., 2001). Alternatively, high levels of 18:0 in membrane lipids of the ssi2 mutant might activate one or more ssi2-conferred phenotypes. The sfd-mediated suppression of the ssi2-conferred phenotypes could result from restoration or other alterations in the lipid composition of the mutant plants. Therefore, we compared the fatty acid composition of polar lipids from sfd1-1 ssi2 npr1, sfd2-1 ssi2 npr1, and sfd4 ssi2 npr1 plants with those of wild-type, npr1, and ssi2 npr1 plants. As shown previously (Kachroo et al., 2001), the mol % of 18:0 in ssi2 npr1 was sixfold higher and that of 18:1 was one-third of the level in the npr1 plant (Table 2). The mol % of 18:0 in sfd1-1 ssi2 npr1 was comparable to that in ssi2 npr1, whereas the mol % of 18:0 in the sfd2-1 ssi2 npr1 and sfd4 ssi2 npr1 plants were slightly lower than that in ssi2 npr1. However, these levels still were fivefold higher than the mol % of 18:0 in the npr1 plant. Hence, it seems unlikely that increased 18:0 mol % is responsible for the ssi2 phenotypes.
Table 2.
Fatty Acyl Profile of Polar Lipids in sfd Plants
| Genotype | 16:0 | 16:1 | 16:2 | 16:3 | 18:0 | 18:1 | 18:2 | 18:3 |
|---|---|---|---|---|---|---|---|---|
| Wild type | 15.7 ± 0.5 | 4.3 ± 0.1 | 0.8 ± 0.1 | 15.5 ± 0.7 | 1.4 ± 0.1 | 2.5 ± 0.4 | 14.1 ± 0.5 | 45.3 ± 0.6 |
| npr1 | 18.4 ± 1.4 | 5.6 ± 0.7 | 1.0 ± 0.2 | 14.0 ± 1.2 | 1.9 ± 0.5 | 2.7 ± 0.5 | 14.5 ± 0.6 | 41.1 ± 1.8 |
| ssi2 npr1 | 13.9 ± 0.7 | 3.4 ± 0.4 | 0.6 ± 0.1 | 12.4 ± 1.0 | 12.6 ± 0.4 | 1.0 ± 0.1 | 15.5 ± 0.4 | 40.4 ± 0.7 |
| sfd1-1 ssi2 npr1 | 13.6 + 2.6 | 5.8 ± 0.7 | 0.3 ± 0.1 | 7.3 ± 0.8 | 13.2 ± 1.9 | 2.4 ± 0.6 | 16.3 ± 0.6 | 40.3 ± 3.5 |
| sfd2-1 ssi2 npr1 | 20.0 ± 2.1 | 4.9 ± 0.5 | 0.2 ± 0.1 | 2.2 ± 0.2 | 10.0 ± 0.6 | 2.1 ± 0.3 | 15.2 ± 0.5 | 45.2 ± 2.7 |
| sfd4 ssi2 npr1 | 16.0 ± 1.4 | 15.9 ± 0.6 | 0.3 ± 0.2 | 1.3 ± 1.6 | 9.6 ± 0.8 | 17.7 ± 2.8 | 14.6 ± 0.4 | 24.3 ± 1.2 |
| sfd2-1 ssi2 | 19.3 ± 1.6 | 4.0 ± 0.4 | 0.3 ± 0.1 | 2.7 ± 0.3 | 8.4 ± 1.7 | 2.0 ± 0.4 | 14.4 ± 0.5 | 48.5 ± 1.6 |
All values are given as mol % of fatty acids. The value for each line is the average of five samples ± sd. Values for 14:0 are not listed.
By contrast, the mol % of 18:1 in sfd1-1 ssi2 npr1 was 2.5-fold higher and that in sfd2-1 ssi2 npr1 was 2-fold higher than that in the ssi2 npr1 plant. The mol % of 18:1 in the sfd4 ssi2 npr1 plant was ∼18-fold higher than in the ssi2 npr1 plant and 6-fold higher than that in npr1. This increase in the mol % of 18:1 in sfd4 ssi2 npr1 was paralleled by a corresponding reduction in the mol % of 18:3. One striking and consistent alteration in the fatty acid compositions of all of the sfd ssi2 npr1 plants was the decrease in the mol % of hexadecatrienoic acid (16:3). 16:3 mol % in the sfd1-1 ssi2 npr1 mutant was 40% lower than that in the ssi2 npr1 plant, whereas 16:3 mol % in sfd2-1 ssi2 npr1 and sfd4 ssi2 npr1 were 82 and 90% lower, respectively, than that in ssi2 npr1. The decreased mol % of 16:3 in sfd4 ssi2 npr1 was paralleled by an increase in the mol % of 16:1.
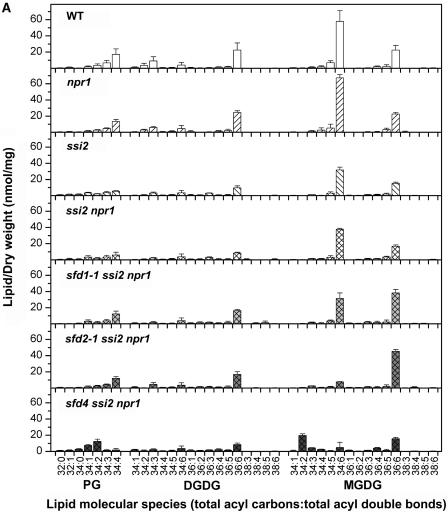
The lipid compositions were examined in more detail by comparing the molecular species of the membrane lipids in the leaves of ssi2, ssi2 npr1, sfd1-1 ssi2 npr1, sfd2-1 ssi2 npr1, and sfd4 ssi2 npr1 plants. Wild-type and npr1 plants served as controls for this experiment. Compared with the wild-type (214 nmol diacyl species/mg dry weight) and the npr1 mutant (213 nmol diacyl species/mg dry weight), the ssi2 and ssi2 npr1 plants contained lower amounts of lipid (147 and 155 nmol diacyl species/mg dry weight, respectively). As shown in Figure 5A, the lower lipid levels in ssi2 and ssi2 npr1 were attributable to a lower content of the lipids that are primarily plastid synthesized and localized; PG and MGDG species decreased by ∼40%, whereas DGDG levels decreased by ∼50%. The total lipid content was increased somewhat in the sfd1-1 ssi2 npr1 (191 nmol diacyl species/mg dry weight) and sfd2-1 ssi2 npr1 (166 nmol diacyl species/mg dry weight) plants, compared with 155 nmol diacyl species/mg dry weight for ssi2 npr1. This increase was primarily the result of an increase in the content of 36:6-MGDG (18:3-18:3 acyl combination) in sfd1-1 ssi2 npr1 and sfd2-1 ssi2 npr1 (Figure 5A). Total lipid in the sfd4 ssi2 npr1 mutant plant (153 nmol diacyl species/mg dry weight) was not increased. The large reduction in 16:3 levels in sfd2-1 ssi2 npr1 and sfd4 ssi2 npr1 were reflected in a >80% reduction in the level of 34:6-MGDG (16:3-18:3 acyl combination) (Figure 5A). 34:6 levels in sfd1-1 ssi2 npr1 were only 17% lower than those in ssi2 npr1. However, the ratio of 34:6-MGDG to 36:6-MGDG species was low in each of the sfd mutants, with ratios of 0.8 in sfd1-1 ssi2 npr1, 0.2 in sfd2-1 ssi2 npr1, and 0.3 in sfd4 ssi2 npr1, compared with 2.6 in the wild type and 2.3 in ssi2 npr1.
Figure 5.
Lipid Profiles of the sfd Mutants.
(A) Electrospray ionization–tandem mass spectrometry-generated profiles of the plastidic glycerolipids PG, DGDG, and MGDG in wild-type (WT), npr1, ssi2, ssi2 npr1, sfd1-1 ssi2 npr1, sfd2-1 ssi2 npr1, and sfd4 ssi2 npr1 plants.
(B) Electrospray ionization–tandem mass spectrometry-generated profiles of the extraplastidic glycerolipids PC, PE, and PI in wild-type (WT), npr1, ssi2, ssi2 npr1, sfd1-1 ssi2 npr1, sfd2-1 ssi2 npr1, and sfd4 ssi2 npr1 plants.
In addition, there were considerable differences in the molecular species within the phospholipid classes of ssi2 and ssi2 npr1 plants compared with those in wild-type and npr1 plants. For example, ssi2 and ssi2 npr1 plants contained higher proportions of PC, PE, and PI containing either a 36:2 or 36:3 acyl composition (Figure 5B). Analysis of the acyl fragments of these 36:2 and 36:3 molecular species in ssi2 npr1 plants indicated that they represent primarily the 18:0-18:2 and 18:0-18:3 combinations, respectively. The levels of 34:2-PC, 34:2-PE, and 34:2-PI were lower in the ssi2 and ssi2 npr1 plants than in wild-type or npr1 plants, whereas 34:3-PC, 34:3-PE, and 34:3-PI were similar or slightly higher in ssi2 and ssi2 npr1 (Figure 5B). Acyl fragment analysis showed that 34:2 species in npr1 and ssi2 npr1 represented mainly a combination of 18:2 and 16:0, whereas 34:3 species were mainly a 16:3-18:0 combination, but lower amounts of 16:1-18:2 and 18:0-16:3 combinations were detected. Interestingly, the trace amounts of 18:0-16:3 in phospholipids were higher in ssi2 npr1 plants than in npr1 plants. For example, in npr1 plants, 16:3-18:0 represents 0.3% of the 34:3-PE species, whereas in ssi2 npr1, 16:3-18:0 represents 1.4% of the 34:3-PE species. Levels of the 34:2-, 36:2-, and 36:3 PC, PE, and PI species in sfd1-1 ssi2 npr1, sfd2-1 ssi2 npr1, and sfd4 ssi2 npr1 were comparable to the levels of these species in ssi2 npr1 (Figure 5B). However, compared with the levels in wild-type, npr1, and ssi2 npr1 plants, the levels of 34:3-PC, 34:3-PE, and 34:3-PI species were lower in the sfd1-1 ssi2 npr1 and sfd4 ssi2 npr1 plants.
Genetic Characterization of the sfd1 and sfd2 Mutants
The genetic basis of the sfd1 and sfd2 phenotypes was characterized in detail. F1 plants resulting from a cross between sfd1-1 ssi2 npr1 and the ssi2 npr1 parent exhibited the typical small-plant phenotype associated with the ssi2 mutant, suggesting that sfd1-1 is recessive to the wild-type SFD1 allele (Table 3). These F1 plants also developed lesions containing dead cells and expressed the PR1 gene at high levels (data not shown). The F2 progeny of these F1 plants segregated wild-type and ssi2-like plants in a nearly 1:3 ratio. Moreover, only the ssi2-like dwarf segregants expressed PR1, confirming that sfd1-1 is a recessive mutation at a single genetic locus. Likewise, the sfd1-2 phenotype also was confirmed to be a monogenic recessive trait (data not shown). The F1 plants generated by crossing the sfd1-1 ssi2 npr1 plant with the sfd1-2 ssi2 npr1 plant were large, devoid of lesions, and lacked PR1 expression. Moreover, none of the 72 F2 progeny derived from these F1 plants exhibited the dwarf and lesion-bearing phenotypes associated with ssi2, confirming that sfd1-1 and sfd1-2 are allelic (Table 3).
Table 3.
Genetic Analysis of the sfd1 and sfd2 Mutants
| F2 Progeny
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cross | F1 Phenotype a | Large | Intermediate | Dwarf | Mendelian Ratio |
Hypothesis | χ2 | Pb | Fit?/Remarks |
|
sfd1-1 ssi2 npr1 × ssi2 npr1 |
Dwarf | 11 | 0 | 32 | 1:2.9 | Monogenic recessive | 0.05 | 0.9 > P > 0.5 | Yes |
|
sfd1-1 ssi2 npr1 × sfd1-2 ssi2 npr1 |
Large | 72 | 0 | 0 | Allelic | ||||
|
sfd1-2 ssi2 npr1 × SFD1 SSI2 NPR1 |
Large | 144 | 0 | 2 | Linked to ssi2 | ||||
|
sfd2-1 ssi2 npr1 × ssi2 npr1 |
Intermediate | 9 | 17 | 10 | 1:1.9:1.1 | Monogenic semidominant | 0.17 | P > 0.9 | Yes |
Plants were scored for the presence/absence of the dwarf phenotype associated with ssi2.
Degrees of freedom used to calculate probability: sfd1-1 ssi2 npr1 × ssi2 npr1 (df = 1); sfd2-1 ssi2 npr1 × ssi2 npr1 (df = 2).
F1 plants (sfd2-1/+ ssi2 npr1) generated by crossing the sfd2-1 ssi2 npr1 plant with a ssi2 npr1 plant were intermediate in size compared with the parents (Figure 2A). In addition, the level of PR1 transcript in these sfd2-1/+ ssi2 npr1 plants was intermediate to levels of PR1 transcript in the ssi2 npr1 and homozygous sfd2-1 ssi2 npr1 plants (Figure 3D), suggesting that sfd2-1 is semidominant. Likewise, in contrast to the sfd2-1 ssi2 npr1 plant, the sfd2-1–mediated suppression of ssi2-conferred resistance to Psm was less pronounced in the sfd2-1/+ ssi2 npr1 plant (A. Nandi and J. Shah, unpublished data). The semidominance of sfd2-1 was confirmed in the F2 progeny, which segregated large, intermediate, and dwarf plants in a nearly 1:2:1 ratio (Table 3). Negligible levels of PR1 transcript were observed in the large F2 progeny plants, whereas moderate and high levels of constitutive PR1 expression were observed in the intermediate and dwarf plants, respectively (data not shown). F3 progeny of the intermediate F2 plants segregated large, intermediate, and dwarf plants, confirming that these F2 plants were heterozygous at the sfd2 locus. Similarly, the sfd2-2 and sfd2-3 alleles also exhibited a semidominant phenotype (data not shown). Crosses between sfd2-1 ssi2 npr1 and sfd2-2 ssi2 npr1, sfd2-1 ssi2 npr1 and sfd2-3 ssi2 npr1, and sfd2-2 ssi2 npr1 and sfd2-3 ssi2 npr1 suggested that sfd2-1, sfd2-2, and sfd2-3 are allelic. All F1 plants from these crosses retained the large sfd2 phenotype. Furthermore, none of them segregated dwarf ssi2-like F2 progeny plants. Further support for allelism among sfd2-1, sfd2-2, and sfd2-3 is provided by the identical lipid profiles of these mutants (A. Nandi, C. Buseman, M. Li, R. Welti, and J. Shah, unpublished data).
In our attempts to outcross sfd1 away from the ssi2 mutant allele, we discovered that sfd1 was linked to the ssi2 locus on chromosome 2. As shown in Table 3, of the 146 F2 progeny of an F1 plant obtained from a cross between sfd1-2 ssi2 npr1 and a wild-type plant, only two morphologically ssi2-like segregants were observed. Mapping populations composed of large and lesionless (sfd1/sfd1) F2 progeny of F1 plants derived from a cross between a sfd1-1 ssi2 npr1 plant in ecotype Nössen and the fab2 (allelic with ssi2) mutant in ecotype Columbia confirmed that sfd1 maps in the vicinity of SSI2. Likewise, mapping populations generated by crossing the sfd1-2 ssi2 npr1 plant with the fab2 mutant also localized sfd1-2 on chromosome 2, near SSI2. Cleaved amplified polymorphic sequence (Konieczny and Ausubel, 1993) and simple sequence length polymorphism (Bell and Ecker, 1994) analyses performed on 29 sfd1-1/sfd1-1 and 26 sfd1-2/sfd1-2 F2 progeny plants mapped the sfd1 locus between the SSLP markers F27D4 and the SSI2 gene, 20 centimorgan (cM) from F27D4 and 3 cM from SSI2. The size of the mapping population was increased subsequently to 301 F2 sfd1/sfd1 plants to map sfd1 between the SSLP marker nga168 (2.1 cM; 13 recombinant chromosomes) and the SSI2 gene (2.6 cM; 16 recombinant chromosomes).
Mapping populations generated by crossing sfd2-1 ssi2 npr1 with fab2 were used to map sfd2. Analysis of 20 large sfd2-1/sfd2-1 F2 progeny plants mapped the sfd2 locus to a 27-cM interval on chromosome 3, between the cleaved amplified polymorphic sequence marker T6H20 and the SSLP nga6, 20 cM from T6H20 and 7 cM from nga6.
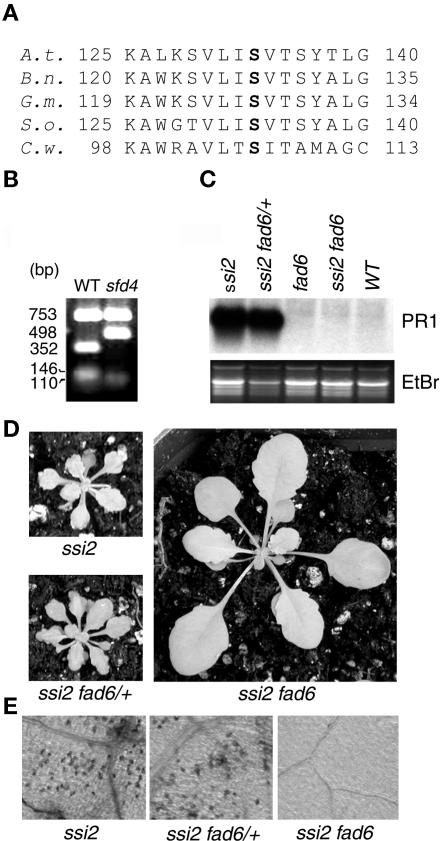
sfd4 Contains a Mutation in the FAD6 Gene
The fatty acid composition of polar lipids in sfd4 ssi2 npr1 showed a decrease in content of the trienoic acids 16:3 and 18:3 and a corresponding increase in the concentrations of 16:1 and 18:1 (Table 2), suggesting that it is blocked in the desaturation of 16:1 and 18:1. This is similar to the defect in the fad6 mutant, which is deficient in a plastidic 16:1/18:1 ω6 desaturase activity (Browse et al., 1989). Therefore, we sequenced the FAD6 locus from wild-type Nössen and sfd4 ssi2 npr1 plants. Indeed, the sfd4 ssi2 npr1 plant contained a single nucleotide change (C→T transition) in the FAD6 gene (Falcone et al., 1994), which is expected to result in a Ser-133→Phe-133 change in the corresponding protein. Ser-133 is highly conserved among FAD6 homologs from various plant and cyanobacterial species (Figure 6A). This C→T transition mutation in the sfd4 ssi2 npr1 plant resulted in an EcoRV restriction polymorphism (Figure 6B), thus confirming the presence of a mutation in the FAD6 gene. To confirm that FAD6 is required for the manifestation of the ssi2-conferred development and defense phenotypes, we generated the ssi2 fad6 double mutant. As shown in Figures 6D and 6E, the fad6 mutant allele suppressed the ssi2-conferred dwarfing and spontaneous cell death phenotypes. In addition, the ssi2-conferred constitutive PR1 expression also was suppressed by the fad6 allele (Figure 6C). These effects of the fad6 allele on the ssi2 phenotype were recessive to the wild-type FAD6 allele, and the ssi2 fad6/+ plant retained all of the ssi2-conferred phenotypes.
Figure 6.
sfd4 Contains a Mutation in the FAD6 Gene.
(A) Alignment of amino acid sequences in a conserved region of plastidic ω6 desaturases from Arabidopsis thaliana (A.t.), rape (Brassica napus; B.n.), soybean (Glycine max; G.m.), spinach (Spinacia oleracea; S.o.), and the green alga Chlamydomonas sp W80 (C.w.). The Ser-133 that is mutated to Phe-133 in sfd4 is shown in boldface. Numbers at left and right indicate amino acid positions.
(B) EcoRV restriction polymorphism generated by a C→T mutation at the FAD6 locus in the sfd4 ssi2 npr1 plant. PCR-amplified products from the wild type (WT) and sfd4 ssi2 npr1 were digested with EcoRV and resolved on an agarose gel. The size of each band in base pairs is listed at left.
(C) Comparison of PR1 expression in the leaves of 4-week-old soil-grown ssi2 plants that are homozygous for the wild-type FAD6 allele (ssi2), heterozygous for the fad6-1 mutant allele (ssi2 fad6/+), and homozygous for the fad6-1 mutant allele (ssi2 fad6). All RNAs were resolved on denaturing gels, transferred to Nytran Plus membranes, and probed for the indicated genes. Gel loading was monitored by photographing the ethidium bromide–stained gel (EtBr) before transferring the RNA to a Nytran Plus membrane.
(D) Morphological phenotype of 4-week-old soil-grown ssi2 plants that are homozygous for the wild-type FAD6 allele (ssi2), heterozygous for the fad6-1 mutant allele (ssi2 fad6/+), and homozygous for the fad6-1 mutant allele (ssi2 fad6). All plants were photographed from the same distance.
(E) Light microscopy of trypan blue–stained leaves of 4-week-old soil-grown ssi2 plants that are homozygous for the wild-type FAD6 allele (ssi2), heterozygous for the fad6-1 mutant allele (ssi2 fad6/+), and homozygous for the fad6-1 mutant allele (ssi2 fad6). All photographs were taken at the same magnification.
DISCUSSION
The sfd1, sfd2, and sfd4 mutations suppress key phenotypes of the ssi2 mutation, including dwarfing, lesion development, the NPR1-independent expression of the PR1 gene, and resistance to Psm. The high mol % of 18:0 in leaves of sfd1-1 ssi2 npr1, sfd2-1 ssi2 npr1, and sfd4 ssi2 npr1 plants imply that increased 18:0 levels are not sufficient to cause the ssi2 phenotypes. The increases in mol % of 18:1 in the leaves of the sfd1-1 ssi2 npr1, sfd2-1 ssi2 npr1, and sfd4 ssi2 npr1 plants could account for the suppression of some ssi2-conferred phenotypes. However, pharmacological experiments argue against this increase in 18:1 mol % suppressing the ssi2-conferred PR1 expression. To the contrary, the application of 18:1 free fatty acid to leaves of the wild-type plant activates PR1 expression (A. Nandi and J. Shah, unpublished data). It is likely that other changes in the sfd plants contribute to the suppression phenotype.
A striking change in the fatty acid composition of each of the sfd mutants, but especially sfd2-1 ssi2 npr1 and sfd4 ssi2 npr1 plants, was the decreased content of 16:3 (Table 2). This decrease in 16:3 content was reflected by a reduction in the content of the major plastid-localized lipid species, 34:6-MGDG, in the sfd1-1 ssi2 npr1, sfd2-1 ssi2 npr1, and sfd4 ssi2 npr1 plants (Figure 5A). 16:3 is the precursor for the synthesis of C-16 fatty acid–derived oxylipin compounds (e.g., dinor-12-oxo-phytodienoic acid), which has been postulated to fine-tune defense signaling in plants (Weber et al., 1997; Stinzi et al., 2001). In addition, dinor-12-oxo-phytodienoic acid may modulate crosstalk between C-16 and C-18 fatty acid–derived oxylipins (Stinzi et al., 2001). A 16:3-derived molecule(s) that is depressed in the sfd mutants may contribute to some of the ssi2-conferred phenotypes. Indeed, compared with the npr1 plant, increased numbers of species with 16:3-18:0 acyl combinations were found in the phospholipids of the ssi2 npr1 plant. 16:3 usually is not found at more than trace levels outside of the plastids. This incorporation of 16:3 into new molecular species and potentially into nonplastidic compartment(s) in the ssi2 npr1 plant, coupled with the decrease in 16:3-containing complex lipid levels in all of the sfd mutants, suggests the potential importance of a complex lipid(s) containing 16:3, and perhaps a signal derived from this species, in the ssi2-conferred phenotypes.
Barring the ssi2-conferred increased 18:0 levels, the fatty acid composition and lipid species profile of sfd1-1 ssi2 npr1 are strikingly similar to those in the gly1 mutant (Miquel et al., 1998). gly1 and sfd1 map to the same region on chromosome II, in the vicinity of SSI2, and may contain mutations in the same gene. Glycerol application restores the lipid composition of the gly1 mutant, suggesting that the defect in gly1 is caused by the limited availability of glycerol-3-phosphate for complex lipid biosynthesis in the plastids (Miquel et al., 1998). Similarly, glycerol application restores ssi2-conferred PR1 expression in the sfd1-1 ssi2 npr1 and sfd1-2 ssi2 npr1 plants (A. Nandi and J. Shah, unpublished data), further suggesting that gly1 and sfd1 contain mutations in the same gene.
The sfd4 mutation is predicted to alter the highly conserved Ser-133 to Phe-133 in the FAD6 protein. FAD6 encodes a 16:1/18:1 ω6 desaturase that catalyzes the desaturation of plastid-localized 16:1 and 18:1 to 16:2 and 18:2, respectively (Browse et al., 1989; Falcone et al., 1994). In wild-type plants, 16:2 and 18:2 can be converted further to 16:3 and 18:3, respectively, by other desaturases. Like the fad6 mutant, sfd4 ssi2 npr1 contains higher levels of the monounsaturated 16:1 and 18:1 fatty acids at the expense of the trienoic fatty acids 16:3 and 18:3, the two predominant PUFAs in Arabidopsis (Wallis and Browse, 2002). Mutations in the FAD5 gene also affect the synthesis of 16:3 (Kunst et al., 1989). However, unlike the fad6 mutation, mutations in fad5 do not affect 18:3 levels. FAD5 encodes a plastidic palmitoyl desaturase (Mekhedov et al., 2000). Compared with ssi2 npr1 and npr1, 16:3 levels are very low in leaves of the sfd2 ssi2 npr1 plant, and no decrease in the level of 18:3 was observed (Table 2). These data suggest that SFD2, like FAD5, might affect the synthesis or accumulation of 16:3. However, sfd2 does not map to the FAD5 locus; thus, it identifies a novel gene that affects 16:3 biosynthesis or accumulation. Therefore, our results suggest an important role for 16:3, or complex lipids containing it, in the manifestation of one or more ssi2-conferred phenotypes.
SA Signaling in the sfd Mutants
The activation of JA signaling can antagonize SA signaling in Arabidopsis (Kloek et al., 2001). It is possible that the ssi2-conferred PR1 expression is the result of the loss of this inhibitory effect of JA. Indeed, the loss of ssi2-conferred PR1 expression in sfd1 ssi2 npr1 plants correlates with the restoration of JA-activated PDF1.2 expression. However, JA responses are not restored in the sfd2 ssi2 npr1 and sfd4 ssi2 npr1 mutants, yet ssi2-conferred NPR1-independent PR1 expression is suppressed in these plants, arguing against JA signaling per se affecting PR1 expression in the ssi2 mutant. However, we cannot exclude the possibility that the sfd2 and sfd4 mutants affect a step after the point of this crosstalk between the JA and SA pathways.
The simultaneous absence of ssi2-conferred cell death, dwarfism, SA and PR1 transcript accumulation, and enhanced resistance to Psm in the sfd1 ssi2 npr1 and sfd4 ssi2 npr1 plants suggests that SFD1 and SFD4 target a step common to the expression of these ssi2 phenotypes (Figure 7). The sfd1-1 and sfd4/fad6 mutants blocked the ssi2-conferred PR1 expression equally well in wild-type NPR1 and npr1 mutant backgrounds (Figures 3C and 6C). The lack of an increased SA level could account for the absence of ssi2-conferred PR1 expression in the sfd1-1 ssi2 and sfd4/fad6 ssi2 plants. However, unlike the sfd1 ssi2 npr1 and sfd4 ssi2 npr1 plants, the sfd2-1 ssi2 npr1 plant accumulates increased SA levels. Furthermore, in contrast to the sfd2-1 ssi2 npr1 plant, PR1 was expressed constitutively in sfd2-1 ssi2, which contains the wild-type NPR1 gene. Thus, SFD2 is required only for the ssi2-conferred cell death, dwarfing, and for the functioning of the NPR1-independent defense pathway. The fatty acid composition in sfd2-1 ssi2 was comparable to that in sfd2-1 ssi2 npr1 (Table 2). Hence, the inability of sfd2-1 to suppress the NPR1-dependent PR1 expression is not caused by differences in lipid composition between the sfd2-1 ssi2 and sfd2-1 ssi2 npr1 plants. Instead, the high SA content may suffice to activate PR1 expression through the NPR1-regulated pathway in the sfd2-1 ssi2 plant.
Figure 7.
Working Model of the Interplay of ssi2, sfd1, sfd2, and sfd4 in Defense Signaling in Arabidopsis.
This model is a refinement of Figure 1. The sfd1, sfd2, and sfd4 mutant alleles suppress (−) the ssi2-conferred dwarfing, spontaneous development of lesions containing dead cells, NPR1-independent expression of PR1, and enhanced resistance to Psm. In addition, sfd1 and sfd4 also suppress the ssi2-conferred accumulation of high SA levels. However, SA application is ineffective in restoring PR1 expression in sfd1 ssi2 npr1, sfd2 ssi2 npr1, and sfd4 ssi2 npr1 plants, implicating the involvement of another ssi2-contributed factor in the activation of the NPR1-independent pathway leading to the expression of PR1 and enhanced resistance to Psm. The absence of SA accumulation in the ssi2 nahG and ssi2 eds5 plants does not ameliorate the ssi2-conferred cell death phenotype, suggesting that high levels of SA do not have a causal role in the cell death phenotype. The sfd2 ssi2 npr1 plants accumulate increased SA levels despite the lack of spontaneous cell death, suggesting that cell death is not the primary factor that promotes SA accumulation in the ssi2 mutant. Hence, ssi2-conferred cell death and SA accumulation are shown to be independent of each other. The sfd1 mutant alleles restore JA-inducible PDF1.2 expression in sfd1 ssi2 npr1 plants. sfd1 is shown to impinge on a step that is common to the activation of cell death, SA accumulation, the activation of the NPR1-independent defense pathway, and the repression of JA signaling in the ssi2 mutant. However, because sfd1 does not restore resistance to B. cinerea, despite restoring PDF1.2 expression in the sfd1 ssi2 npr1 plants, an additional ssi2-modulated mechanism is shown to suppress (−) the defense against B. cinerea. sfd4 does not restore JA-inducible PDF1.2 expression in sfd4 ssi2 npr1. Hence, sfd4 is shown to suppress (−) a step that is common to the activation of cell death and dwarfing, SA accumulation, and the activation of the NPR1-independent defense pathway. sfd2 is shown to interfere with (−) a step common to the activation of ssi2-conferred cell death and the activation of NPR1-independent signaling.
The inability of applied SA to restore PR1 expression in the sfd1-1 ssi2 npr1, sfd2-1 ssi2 npr1, and sfd4 ssi2 npr1 plants (Figure 3B and data not shown) suggests the requirement for an additional factor, in addition to SA, for the activation of NPR1-independent PR1 expression. A 16:3-derived product and/or regulated activity could be this second factor required for the activation of SA signaling. Lipid signaling was shown by Anderson et al. (1998) to be an important factor in modulating the SA-activated expression of the PR1 gene in tobacco. Alternatively, this second factor could be made available by the cell death and/or dwarfing that occurs in the ssi2 mutant.
JA Signaling in sfd Mutants
Lack of an 18:1-derived signal/factor was suggested to be responsible for the inability of JA to activate PDF1.2 expression in the ssi2 mutant (Kachroo et al., 2001). The restoration of wild-type levels of 18:1 (Table 2) could account for the establishment of JA-activated PDF1.2 expression in the sfd1-1 ssi2 npr1 plant (Figure 3E). However, comparable levels of 18:1 in the sfd2-1 ssi2 npr1 plant and the eightfold higher levels of 18:1 in sfd4 ssi2 npr1 were not sufficient to restore JA-activated PDF1.2 expression, suggesting that factors in addition to 18:1 are involved. Because JA is ineffective at activating PDF1.2 expression in SA-deficient ssi2 nahG plants (Figure 3E) (Kachroo et al., 2001) and in sfd4 ssi2 npr1 (Figure 3E), it is highly unlikely that the increased SA levels in ssi2 npr1 and sfd2-1 ssi2 npr1 suppress JA-activated PDF1.2 expression. The restoration of JA-activated PDF1.2 expression in the sfd1-1 ssi2 npr1 plant suggests that an SFD1-dependent signal may be an inhibitor of PDF1.2 expression in the ssi2 plant (Figure 7). In wild-type plants, the 18:1-derived signal may antagonize this inhibitory activity, allowing JA to activate PDF1.2 expression.
JA application activates PDF1.2 expression and confers protection against the necrotrophic pathogen B. cinerea (Penninckx et al., 1996, 1998; Thomma et al., 1999). In addition, enhancing PDF1.2 expression by the overexpression of ETHYLENE RESPONSE FACTOR1, a regulator of PDF1.2 expression in Arabidopsis, enhances resistance to B. cinerea (Berrocal-Lobo et al., 2002). An increased susceptibility to B. cinerea parallels the inability of the ssi2 mutant to express the PDF1.2 gene in response to JA application (Kachroo et al., 2001). However, the restoration of MeJA-activated PDF1.2 expression in the sfd1-1 ssi2 npr1 plant (Figure 3E) was not sufficient to restore resistance to B. cinerea (Figure 4B). However, we cannot exclude the possibility that the sfd1-1 ssi2 npr1 plant, although responsive to MeJA, may not be very sensitive to it or to other factors that activate PDF1.2. For example, although exogenously applied MeJA activated PDF1.2 expression in sfd1-1 ssi2 npr1 plants, the PDF1.2 gene transcript accumulated to lower levels than in the MeJA-treated control npr1 plant (Figure 3E). In addition, application of 1-aminocyclopropane-1-carboxylic acid was ineffective at activating PDF1.2 expression in sfd1-1 ssi2 npr1 plants (A. Nandi and J. Shah, unpublished data).
Govrin and Levine (2000) have shown that hypersensitive response–associated cell death in Arabidopsis enhances susceptibility to B. cinerea. It is likely that cell death in ssi2 predisposes the mutant plant to infection by B. cinerea. However, the prevalence of the ssi2-conferred enhanced susceptibility to B. cinerea in the sfd1-1 ssi2 npr1, sfd2-1 ssi2 npr1, and sfd4 ssi2 npr1 plants (Figure 4B), despite the absence of ssi2-conferred cell death (Figure 2B), argues against cell death being an important factor in the susceptibility of the ssi2 mutant to B. cinerea. The loss of ssi2-activated high-level SA accumulation in the sfd1-1 ssi2 npr1 and sfd4 ssi2 npr1 plants also excludes any involvement of SA in the susceptibility of the ssi2 mutant to B. cinerea.
In conclusion, we show here that the mere increase in 18:0 level resulting from the deficiency of a plastidic fatty acid desaturase activity is not sufficient to cause the development defects and the altered defense phenotypes observed in the ssi2 mutant. Instead, the signals/factors derived from the PUFA 16:3 may be involved in the activation of one or more of the ssi2-conferred phenotypes. Although the exact identity of these signaling molecules and the mechanism by which they exert their effects on plant development and defense responses are unclear, the genetic dissection of these signaling mechanisms in concert with lipid compositional analysis in mutants should facilitate our understanding of how plants generate and decipher complex information involving lipids.
METHODS
Cultivation of Plants and Pathogens
Arabidopsis thaliana seeds were germinated at 22°C in a tissue culture chamber exposed to a 14-h-light (80 μE·m−2·s−1)/10-h-dark cycle on Murashige and Skoog (1962) (Sigma, St. Louis, MO) agar supplemented with 1% sucrose. Nine-day-old seedlings were transferred to soil and grown at 22°C in growth chambers programmed for a 14-h-light (90 μE·m−2·s−1)/10-h-dark cycle.
Pseudomonas syringae pv maculicola (Psm) ES4326 was propagated at 28°C on King's B medium (King et al., 1954) containing streptomycin (100 μg/mL). An overnight culture was used to infect plants. Botrytis cinerea IMI169558 was cultivated on maltose medium at 22°C in a growth chamber programmed for a 14-h-light (80 μE·m−2·s−1)/10-h-dark cycle. Spores were harvested and infections performed as described previously (Nandi et al., 2003).
Infection of Plants with Bacterial and Fungal Pathogens
Infections with Psm ES4326 were performed on 4-week-old soil-grown plants as described previously (Nandi et al., 2003). For every set of infections, 25 (five replications of five leaves in each sample) leaf discs (0.15 cm2) were harvested at 3 days after infection and placed in preweighed tubes. After the weight of each sample was determined, bacterial counts were determined as described previously (Shah et al., 1997). Bacterial counts were expressed as colony-forming units per milligram of leaf tissue.
B. cinerea infections were performed on 4-week-old soil-grown plants as described previously (Nandi et al., 2003). Three leaves per plant were pricked with a needle. Infection was initiated by placing a 10-μL drop of freshly harvested spore suspension (5 × 105 spores/mL) on each needle prick. The inoculum was allowed to air dry. Thereafter, plants were covered with a transparent plastic dome and cultivated at 22°C in a growth chamber programmed for a 14-h-light (90 μE·m−2·s−1)/10-h-dark cycle. The number of leaves showing varied levels of necrosis were scored at 4 days after inoculation.
Chemical Treatment of Plants
For methyl jasmonate treatment, leaves from 4-week-old plants, excised at the base of the petioles, were floated on 5 mL of a methyl jasmonate solution (5 μM in 0.1% ethanol; Bedoukian Research, Danbury, CT) on tissue culture plates. As the control, leaves from the same plants were floated simultaneously on 5 mL of 0.1% ethanol solution. Leaves were harvested 48 h later, quick frozen in liquid nitrogen, and stored at −80°C until RNA extraction.
Salicylic acid (SA; 500 μM) treatment of 4-week-old soil-grown plants was performed as described previously (Shah et al., 1997). As controls, plants were treated similarly with water. Leaves were harvested 48 h after treatment and quick-frozen in liquid nitrogen. Leaf samples were stored at −80°C until RNA extraction.
RNA Extraction and RNA Gel Blot Analyses
Leaf tissue was ground under liquid nitrogen, and RNA was extracted using acid guanidinium thiocyanate-phenol-chloroform as described by Chomczynski and Sacchi (1987). RNA gel blot analysis and the synthesis of random primed probes for PR1, BGL2, and PDF1.2 were performed as described previously (Shah et al., 1997, 1999).
DNA Extraction and PCR Analyses
Arabidopsis genomic DNA from leaf tissue was isolated by the method of Konieczny and Ausubel (1993). PCR analyses to distinguish between the wild-type NPR1 and the npr1-5 mutant alleles were performed as described previously (Shah et al., 1999). A cleaved amplified polymorphic sequence (CAPS) marker was used to distinguish between the ssi2-1 and SSI2 alleles (Kachroo et al., 2001). For studies with sfd4, the primers FAD6-F1 (5′-GTCGCTTCTTCTGCATTTTC-3′) and FAD6-R1 (5′-TGATGACTCAAACTCCTCTG-3′) were used for PCR. Thirty cycles of PCR were performed. Each cycle consisted of incubation at 95°C for 45 s followed by 56°C for 30 s and 72°C for 2 min. The PCR products were digested with EcoRV to distinguish the sfd4 mutant allele from the wild-type SFD4 allele. PCR amplification of the other CAPS and simple sequence length polymorphism markers used in this study were performed as described at http://www.arabidopsis.org.
Histochemistry and Microscopy
Leaves from 4-week-old soil-grown plants were used for trypan blue staining of dead cells. Samples were processed and analyzed as described by Rate et al. (1999).
SA Quantitation
Total SA (SA plus SA-glucoside) in 0.25 to 0.5 g (fresh weight) of leaf tissue was extracted in methanol. The methanol extracts were dried and resuspended in 1.25 mL of 100 mM sodium acetate buffer, pH 5.5, containing 20 units of β-glucosidase (EC 3.2.1.21; almond). After 1.5 h of incubation at 37°C, extracts were acidified to pH 1.0 with 10% (w/v) trichloroacetic acid and subjected to SA extraction and quantification by spectrofluorescence HPLC as described previously (Enyedi, 1999).
Fatty Acid and Lipid Profiling
For fatty acid analysis, leaves from two to three plants were cut and immediately transferred to 3 mL of isopropanol containing 0.01% butylated hydroxytoluene at 75°C. After 15 min, 1.5 mL of chloroform plus 0.6 mL of water were added. The tubes were shaken for 1 h, followed by removal of the extract. The leaves were reextracted five times with chloroform:methanol (2:1) containing 0.01% butylated hydroxytoluene. During each extraction, the tubes were agitated for 30 min. The extracted leaf tissue was heated overnight at 105°C and weighed. The weight of the dried and extracted tissue was the “dry weight” of the sample. Dry weights ranged from 14 to 78 mg. The combined extracts were washed once with 1 mL of 1 M KCl and once with 2 mL of water. The solvent was evaporated under nitrogen, and the lipid extract was dissolved in 1 mL of chloroform.
For analysis of the fatty acyl species in complex lipids, 200 μL of the sample was applied to a silicic acid (0.3 g) column in chloroform. Neutral lipids were eluted with 10 mL of chloroform, and complex lipids were eluted with 10 mL of methanol. The solvent was evaporated from the methanol fraction, and fatty acid methyl esters were formed from 4% of each sample in duplicate by derivatization in 1.5 M methanolic HCl. After heating at 78°C for 90 min, 2 volumes of water was added to the reaction mixture. Each mixture was extracted twice with pentane and dried with sodium sulfate. The solvent was evaporated, and each sample was dissolved in carbon disulfide and analyzed by gas-liquid chromatography with a Supelco 30-meter Omegawax 250 capillary column (Sigma-Aldrich) at 155°C. The temperature of the injector was 220°C, and the flame ionization detector was also at 220°C. The fatty acyl composition for each sample was an average of the two analyses. The fatty acyl composition of each Arabidopsis line is the average ± sd of five samples.
An automated electrospray ionization–tandem mass spectrometry approach was used to profile lipid composition in Arabidopsis leaves. Sample preparation, processing, data acquisition and analysis, and acyl group identification were as described previously (Welti et al., 2002).
Mutagenesis and Selection of the sfd Mutants
Four thousand ssi2 npr1-5 seeds (ecotype Nössen) were mutagenized with 0.3% ethyl methanesulfonate (Sigma-Aldrich) as described previously (Shah et al., 1997). M2 seeds were harvested as pools; each pool contained M2 seeds derived from ∼150 ethyl methanesulfonate–mutagenized M1 seeds. The M2 seeds were germinated in soil, in parallel with seeds of ssi2 npr1 and wild-type plants. Large plants were identified, and RNA was extracted from the leaves. RNA was analyzed by RNA gel blot analysis for PR1 expression. RNA extracted from wild-type and ssi2 npr1 plants served as negative and positive controls, respectively. Individual plants were allowed to set seeds. sfd phenotypes were confirmed in M3 progeny.
Genetic Analysis
Backcrosses were performed by pollinating flowers of the sfd mutant lines with pollen from the ssi2 npr1 parental line (Shah et al., 2001). This line contains the npr1-5 allele (Shah et al., 1997, 1999). A backcrossed homozygous line was used for all other genetic analyses. To generate the sfd2-1 ssi2 line, flowers of a sfd2-1 ssi2 npr1 plant were crossed with pollen from a wild-type plant. The success of the cross was confirmed by CAPS analysis of F1 plants for heterozygosity at the npr1 and ssi2 loci. The genotype of the F1 generation is sfd2-1/+ ssi2/+ npr1/+. Large plants segregating in the F2 generation were screened using PCR to identify plants that were homozygous for the ssi2 mutant allele. Because of their large stature, these plants were presumed to be homozygous for the sfd2-1 allele. These plants then were tested by PCR to identify those that were homozygous for the wild-type NPR1 allele. The F2 lines that did not segregate any dwarf plants were confirmed to be sfd2-1 ssi2.
The sfd1-1 ssi2 line was generated from a cross involving the sfd1-1 ssi2 npr1 and ssi2 plants. The genotype of these dwarf and lesion-bearing F1 plants is sfd1-1+ ssi2/ssi2 npr1/+. Large plants segregating in the F2 generation were analyzed using PCR for plants that are homozygous for the NPR1 allele. F3 seeds from these sfd1-1 ssi2 plants were used in experiments.
Mapping populations for sfd1 were generated by crossing the sfd1-1 ssi2 npr1 plant in ecotype Nössen with the fab2 mutant in ecotype Columbia. The resulting F1 plants were dwarf and expressed the PR1 gene at increased levels. The large F2 progeny plants were used to map sfd1-1. Similarly, F2 progeny plants from a cross between sfd1-2 ssi2 npr1 and fab2 were used to map sfd1-2. F2 progeny from a cross of a sfd2-1 ssi2 npr1 plant (ecotype Nössen) and the fab2 mutant (ecotype Columbia) were used to map sfd2-1. F2 progeny homozygous for sfd2-1, identified as large plants, were used for mapping.
A cross between the ssi2 and fad6-1 (ecotype Columbia) plants was used to generate the ssi2 fad6 double mutant. F2 progeny plants from this cross were analyzed for the presence of the ssi2 mutant allele by PCR (Kachroo et al., 2001). One-quarter of the plants that were homozygous for the ssi2 mutant allele lacked the ssi2-conferred dwarf phenotype, suggesting recessive epistasis, with fad6-1 epistatic to ssi2. The presence of the fad6 or wild-type FAD6 allele was monitored by PCR for the AGa and AthDET1 markers, which flank the FAD6 locus and exhibit polymorphism between ecotypes Columbia and Nössen. All of the ssi2/ssi2 F2 plants that did not exhibit the ssi2-conferred dwarf phenotype were homozygous for the Columbia pattern at these two markers, confirming that they were homozygous for the fad6-1 mutant allele.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Jyoti Shah, shah@ksu.edu.
Acknowledgments
We thank Ethan Baughman for his help in interpreting the lipid molecular species acyl fragmentation data and William Broekaert for providing a culture of B. cinerea IMI169558. We are grateful to Todd Williams for acquiring MS data and to Xuemin Wang for his suggestions and comments on this work. This material is based on work supported by the Cooperative State Research, Education, and Extension Service, U.S. Department of Agriculture, under Agreement 2002-35319-11655 and Grant MCB-0110979 from the National Science Foundation (to J.S. and R.W.). A Research Experience for Undergraduates supplement to National Science Foundation Grant MCB-0110979 and a fellowship from the Kansas Biomedical Research Infrastructure Network provided support (for C.M.B.). J.S. also acknowledges the National Science Foundation for Major Research Instrumentation Grant DBI-0079539, which provided for plant growth chambers to perform this work. This is Kansas Agricultural Experimental Station contribution 03-132-J.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.015529.
References
- Anderson, M.D., Chen, Z., and Klessig, D.F. (1998). Possible involvement of lipid peroxidation in salicylic acid-mediated induction of PR1 gene expression. Phytochemistry 47, 555–566. [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo, M., Molina, A., and Solano, R. (2002). Constitutive expression of ETHYLENE RESPONSE FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29, 23–32. [DOI] [PubMed] [Google Scholar]
- Browse, J., Kunst, L., Anderson, S., Hugly, S., and Somerville, C. (1989). A mutant of Arabidopsis deficient in the chloroplast 16:1/18:1 desaturase. Plant Physiol. 90, 522–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse, J., Warwick, N., Somerville, C.R., and Slack, C.R. (1986). Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the ‘16:3’ plant Arabidopsis thaliana. Biochem. J. 235, 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, A.S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski, P., and Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159. [DOI] [PubMed] [Google Scholar]
- Clarke, J.D., Volko, S.M., Ledford, H., Ausubel, F.M., and Dong, X. (2000). Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12, 2175–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P., Friedrich, L., and Ryals, J.A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 92, 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P., Uknes, S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D., Gaffney, T., Gut-Rella, M., Kessmann, H., Ward, E., and Ryals, J. (1994). A central role of salicylic acid in plant disease resistance. Science 266, 1247–1250. [DOI] [PubMed] [Google Scholar]
- Dewdney, J., Rueber, T.L., Wildermuth, M.C., Devoto, A., Cui, J., Stutius, L.M., Drummond, E.P., and Ausubel, F.M. (2000). Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J. 24, 205–218. [DOI] [PubMed] [Google Scholar]
- Durner, J., Shah, J., and Klessig, D.F. (1997). Salicylic acid and disease resistance in plants. Trends Plant Sci. 2, 266–274. [Google Scholar]
- Enyedi, A.J. (1999). Induction of salicylic acid synthesis and systemic acquired resistance using the active oxygen species generator rose bengal. J. Plant Physiol. 154, 106–112. [Google Scholar]
- Falcone, D.L., Gibson, S., Lemieux, B., and Somerville, C. (1994). Identification of a gene that complements an Arabidopsis mutant deficient in chloroplast ω6 desaturase activity. Plant Physiol. 106, 1453–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J.F., Beneditti, C.E., Penfold, C.N., and Turner, J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., Ward, E., Kessmann, H., and Ryals, J. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261, 754–756. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J., Rogers, E.E., and Ausubel, F.M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143, 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govrin, E.M., and Levine, A. (2000). The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 10, 751–757. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., and Jones, J.D.G. (1996). Resistance gene-dependent plant defense responses. Plant Cell 8, 1773–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, M., and Ryals, J. (1996). Systemic acquired resistance signal transduction. Crit. Rev. Plant Sci. 15, 583–606. [DOI] [PubMed] [Google Scholar]
- Kachroo, P., Shanklin, J., Shah, J., Whittle, E.J., and Klessig, D.F. (2001). A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc. Natl. Acad. Sci. USA 98, 9448–9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, E.O., Ward, M.K., and Raney, D.E. (1954). Two simple media for the demonstration of phycocyanin and fluorescein. J. Lab. Clin. Med. 44, 301–307. [PubMed] [Google Scholar]
- Klessig, D.F., and Malamy, J. (1994). The salicylic acid signal in plants. Plant Mol. Biol. 26, 1439–1458. [DOI] [PubMed] [Google Scholar]
- Kloek, A.P., Verbsky, M.L., Sharma, S.B., Schoelz, J.E., Vogel, J., Klessig, D.F., and Kunkel, B.N. (2001). Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 26, 509–522. [DOI] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Kunkel, B.N., and Brooks, D.M. (2002). Cross talk between signaling pathways in plant defense. Curr. Opin. Plant Biol. 5, 325–331. [DOI] [PubMed] [Google Scholar]
- Kunst, L., Browse, J., and Somerville, C. (1989). A mutant of Arabidopsis deficient in desaturation of palmitic acid in leaf lipids. Plant Physiol. 90, 943–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxalt, A.M., and Munnik, T. (2002). Phospholipid signaling in plant defense. Curr. Opin. Plant Biol. 5, 332–338. [DOI] [PubMed] [Google Scholar]
- Lightner, J., James, D.W., Jr., Dooner, H.K., and Browse, J. (1994. a). Altered body morphology is caused by increased stearate levels in a mutant of Arabidopsis. Plant J. 6, 401–412. [Google Scholar]
- Lightner, J., Wu, J., and Browse, J. (1994. b). A mutant of Arabidopsis with increased levels of stearic acid. Plant Physiol. 106, 1443–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn, M., and Browse, J. (1996). The critical requirement for linolenic acid in pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon, T.A., and Stumpf, P.K. (1982). Purification and characterization of the stearoyl-acyl carrier protein desaturase and the acyl-carrier protein thioesterase from maturing seeds of safflower. J. Biol. Chem. 267, 1502–1509. [PubMed] [Google Scholar]
- Mekhedov, S., de Ilárduya, O.M., and Ohlrogge, J. (2000). Toward a functional catalog of the plant genome: A survey of genes for lipid biosynthesis. Plant Physiol. 122, 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel, M., Cassagne, C., and Browse, J. (1998). A new class of Arabidopsis mutants with reduced hexadecatrienoic acid fatty acid levels. Plant Physiol. 117, 923–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongrand, S., Cassagne, C., and Bessoule, J.J. (2000). Import of lyso-phosphatidylcholine into chloroplasts likely at the origin of eukaryotic plastidial lipids. Plant Physiol. 22, 845–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik, T., Irvine, R.F., and Musgrave, A.P. (1998). Phospholipid signaling in plants. Biochim. Biophys. Acta 1389, 222–272. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Nandi, A., Kachroo, P., Fukushige, H., Hildebrand, D.F., Klessig, D.F., and Shah, J. (2003). Ethylene and jasmonic acid signaling pathways affect NPR1-independent expression of defense genes without impacting resistance to Pseudomonas syringae and Peronospora parasitica in the Arabidopsis ssi1 mutant. Mol. Plant-Microbe Interact. 16, 588–599. [DOI] [PubMed] [Google Scholar]
- Nawrath, C., and Metraux, J.P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11, 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge, J., Browse, J., and Somerville, C.R. (1991). The genetics of plant lipids. Biochim. Biophys. Acta 1082, 1–26. [DOI] [PubMed] [Google Scholar]
- Penninckx, I.A.M.A., Eggermont, K., Terras, F.R.G., Thomma, B.P.H.J., De Samblanz, G.W., Buchala, A., Métraux, J.-P., Manners, J.M., and Broekaert, W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8, 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A.M.A., Thomma, B.P.H.J., Buchala, A., Metraux, J.P., and Broekaert, W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate, D.N., Cuenca, J.V., Bowman, G.R., Guttman, D.S., and Greenberg, J.T. (1999). The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defense, and cell growth. Plant Cell 11, 1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond, P., and Farmer, E.E. (1998). Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1, 404–411. [DOI] [PubMed] [Google Scholar]
- Ryals, J.A., Neuenschwander, U.H., Willits, M.G., Molina, A., Steiner, H.-Y., and Hunt, M.D. (1996). Systemic acquired resistance. Plant Cell 8, 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J., Kachroo, P., and Klessig, D.F. (1999). The Arabidopsis ssi1 mutation restores pathogenesis-related gene expression in npr1 plants and renders defensin gene expression salicylic acid dependent. Plant Cell 11, 191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J., Kachroo, P.K., Nandi, A., and Klessig, D.F. (2001). A recessive mutation in the Arabidopsis SSI2 gene confers SA- and NPR1-independent expression of PR genes and resistance against bacterial and oomycete pathogens. Plant J. 25, 563–574. [DOI] [PubMed] [Google Scholar]
- Shah, J., and Klessig, D.F. (1999). Salicylic acid: Signal perception and transduction. In Biochemistry and Molecular Biology of Plant Hormones, Vol. 33, K. Libbenga, M. Hall, and P.J.J. Hooykaas, eds (London: Elsevier), pp. 513–541.
- Shah, J., Tsui, F., and Klessig, D.F. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant-Microbe Interact. 10, 69–78. [DOI] [PubMed] [Google Scholar]
- Shanklin, J., and Cahoon, E.B. (1998). Desaturation and related modifications of fatty acids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 611–641. [DOI] [PubMed] [Google Scholar]
- Shanklin, J., and Somerville, C.R. (1991). The cDNA clones for stearoyl-ACP desaturase from higher plants are not homologous to yeast or mammalian genes encoding stearoyl-CoA desaturase. Proc. Natl. Acad. Sci. USA 88, 2510–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirano, Y., Kachroo, P., Shah, J., and Klessig, D.F. (2002). A gain-of-function mutation in an Arabidopsis Toll Interleukin1 Receptor–Nucleotide Binding Site–Leucine-Rich Repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14, 3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville, C., Browse, J., Jaworski, J.G., and Ohlrogge, J.B. (2000). Lipids. In Biochemistry and Molecular Biology of Plants, B. Buchanan, W. Gruissem, and R. Jones, eds (Rockville, MD: American Society of Plant Physiologists), pp. 456–527.
- Staswick, P.E., Yuen, G., and Lehman, C.C. (1998). Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J. 15, 747–754. [DOI] [PubMed] [Google Scholar]
- Stinzi, A., and Browse, J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97, 10625–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinzi, A., Weber, H., Reymond, P., Browse, J., and Farmer, E.E. (2001). Plant defense in the absence of jasmonic acid: The role of cyclopentenones. Proc. Natl. Acad. Sci. USA 98, 12837–12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P.H.J., Eggermont, K., Tierens, F.M.-J., and Broekaert, W.F. (1999). Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol. 121, 1093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan, P., Shockey, J., Levesque, C.A., Cook, R.J., and Browse, J. (1998). A role for jasmonate in pathogen defense of Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 7209–7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis, J.G., and Browse, J. (2002). Mutants of Arabidopsis reveal many roles for membrane lipids. Prog. Lipid Res. 41, 254–278. [DOI] [PubMed] [Google Scholar]
- Weber, H. (2002). Fatty acid-derived signals in plants. Trends Plant Sci. 7, 217–224. [DOI] [PubMed] [Google Scholar]
- Weber, H., Vick, B.A., and Farmer, E.E. (1997). Dinor-oxo-phytodienoic acid: A new hexadecanoid signal in the jasmonate family. Proc. Natl. Acad. Sci. USA 94, 10473–10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welti, R., Li, W., Li, M., Sang, Y., Biesiada, H., Zhou, H.-E., Rajashekar, C.B., Williams, T.D., and Wang, X. (2002). Profiling membrane lipids in plant stress response. J. Biol. Chem. 277, 31994–32002. [DOI] [PubMed] [Google Scholar]