Abstract
The previously reported Arabidopsis dominant gain-of-function mutant accelerated cell death6-1 (acd6-1) shows spontaneous cell death and increased disease resistance. acd6-1 also confers increased responsiveness to the major defense signal salicylic acid (SA). To further explore the role of ACD6 in the defense response, we cloned and characterized the gene. ACD6 encodes a novel protein with putative ankyrin and transmembrane regions. It is a member of one of the largest uncharacterized gene families in higher plants. Steady state basal expression of ACD6 mRNA required light, SA, and an intact SA signaling pathway. Additionally, ACD6 mRNA levels were increased in the systemic, uninfected tissue of Pseudomonas syringae–infected plants as well as in plants treated with the SA agonist benzothiazole (BTH). A newly isolated ACD6 loss-of-function mutant was less responsive to BTH and upon P. syringae infection had reduced SA levels and increased susceptibility. Conversely, plants overexpressing ACD6 showed modestly increased SA levels, increased resistance to P. syringae, and BTH-inducible and/or a low level of spontaneous cell death. Thus, ACD6 is a necessary and dose-dependent activator of the defense response against virulent bacteria and can activate SA-dependent cell death.
INTRODUCTION
The small phenolic compound salicylic acid (SA) plays a central role in disease resistance in higher plants. Its synthesis is induced in response to many types of pathogens (Ryals et al., 1996). SA is both necessary and sufficient for general resistance to many pathogens. Plants carrying a nahG transgene whose product catabolizes SA or plants harboring a mutation in the SA biosynthetic pathway or signaling are more susceptible to many pathogens (Gaffney et al., 1993; Delaney et al., 1994; Wildermuth et al., 2001). Conversely, plants engineered to produce high SA levels constitutively and plants treated exogenously with SA or an SA agonist such as benzothiazole (BTH) have enhanced disease resistance (Friedrich et al., 1996; Verberne et al., 2000).
SA plays multiple roles in the regulation of plant defenses. It is required for the induction of broad-spectrum disease resistance in the systemic tissue of plants previously infected with a necrotizing pathogen (a phenomenon termed systemic acquired resistance) (Gaffney et al., 1993). Some plants also require SA to mount a strong resistance response during so called gene-for-gene resistance. In this response, plants have a resistance (R) gene allele that confers the ability to recognize specific pathogen proteins encoded by avr genes (Staskawicz, 2001). In some R-avr–mediated interactions, SA is required for the R gene–dependent host programmed cell death (called the hypersensitive response [HR]) and/or for disease resistance (Delaney et al., 1994; Brading et al., 2000; McDowell et al., 2000; Rate and Greenberg, 2001; Rairdan and Delaney, 2002). However, SA on its own is not sufficient to activate an HR and some defenses when produced at high levels in plants, suggesting that SA acts as a coactivator with another signal(s) to induce these responses (Rate et al., 1999). One such coactivator appears to be light, because the light receptors called phytochromes are important for some SA responses (Genoud et al., 2002).
The molecular basis of SA perception remains unclear, although several SA binding proteins have been identified (Chen et al., 1993; Klessig et al., 2000; Slaymaker et al., 2002). Several genes important for SA accumulation in response to pathogen attack and for its transduction have been found. Positive regulators of SA production during infection by some pathogens include NDR1 (Non-Race-Specific Disease Resistance1), a possible membrane protein (Century et al., 1997; Shapiro and Zhang, 2001), and EDS1 (Enhanced Disease Susceptibility1) and PAD4 (Phytoalexin Deficient4), which are interacting proteins that resemble lipases (Zhou et al., 1998; Falk et al., 1999; Jirage et al., 1999; Feys et al., 2001). Additionally, the Arabidopsis eds3 (Glazebrook et al., 2003), eds4 (Gupta et al., 2000), eds8 (Glazebrook et al., 2003), pad1 (Glazebrook et al., 1997), and pad2 (Glazebrook et al., 1997) mutants are impaired for SA production in response to the bacterial pathogen Pseudomonas syringae. NPR1/NIM1 (Nonexpressor of PR1/Noninducible Immunity1), an ankyrin repeat–containing protein, is important for transducing the SA signal. The ankyrin repeat is a motif containing ∼33 amino acids involved in protein–protein interactions (Sedgwick and Smerdon, 1999). npr1/nim1 Arabidopsis plants are hypersusceptible to many pathogens and fail to express SA-induced PR (pathogenesis-related) genes after treatment with SA or its agonists (Cao et al., 1994, 1997). Arabidopsis harboring a sni1 mutation (suppressor of npr1-1, inducible1) is potentiated for SA-induced PR gene expression. Thus, SNI1 is a negative regulator of the SA pathway (Li et al., 1999).
A number of Arabidopsis mutants constitutively accumulate high levels of SA. Generally, these mutants show increased disease resistance that requires SA, PAD4, EDS1, and/or NPR1, as well as other phenotypes, such as reduced size, altered morphology, and/or spontaneous cell death. Such mutants include accelerated cell death (acd) (Rate et al., 1999; Brodersen et al., 2002), constitutive expressor of PR genes (Bowling et al., 1997; Clarke et al., 2000), lesion-simulating disease (lsd) (Weymann et al., 1995; Rusterucci et al., 2001; Aviv et al., 2002), defense no death (dnd) (Yu et al., 1998), aberrant growth and death (agd) (Rate and Greenberg, 2001), and suppressor of salicylic acid insensitivity (Shirano et al., 2002). Most genes previously identified by this class of mutants are not known to be induced by SA; rather, their functions are influenced by the presence of SA or changes in SA signaling. One example is LSD1, a zinc-finger protein with similarity to GATA-type transcription factors. LSD1 is not controlled by SA signaling, at least with respect to its steady state transcript accumulation (Dietrich et al., 1997). However, lsd1 plants show uncontrolled cell death that is suppressed by the eds1 and pad4 mutations (Rusterucci et al., 2001). Another example is ACD11, a sphingosine transfer protein that may negatively regulate cell death and defenses in vivo by altering sphingolipid metabolism (Brodersen et al., 2002). Although the acd11 cell death phenotype requires SA, ACD11 transcript accumulation is not SA responsive (this work). These observations suggest that many ACD/LSD-type genes function by interacting with SA-controlled activities.
We previously reported the isolation of a dominant gain-of-function mutant in Arabidopsis, acd6-1, whose phenotypes all require SA. acd6-1 has reduced stature, increased SA, spontaneous cell death, high resistance to P. syringae, and constitutive defense responses. Treatment of SA-depleted acd6-1 plants with the SA agonist BTH results in the hyperactivation of defense-related genes (Rate et al., 1999). This finding suggests that acd6-1 is sensitized to SA signaling and that ACD6 could be involved in amplifying SA responses. We report here the cloning of ACD6, which encodes a novel protein containing putative ankyrin and transmembrane regions. Unlike most other LSD- and ACD-type genes characterized to date, the expression of ACD6 simultaneously requires both SA and light. Plants lacking ACD6 are less responsive to the SA agonist BTH, and upon infection they produce less SA and are more susceptible to P. syringae. Plants with extra genomic copies of ACD6 are more resistant to P. syringae, have modestly increased SA levels, and show BTH-inducible and/or spontaneous cell death. These facts suggest that ACD6 regulates SA and also is an effector of the SA pathway involved in disease resistance and cell death control.
RESULTS
The acd6-1 Mutant Has Altered Defense Regulation and Cell Wall Properties
acd6-1 plants have increased levels of SA (Vanacker et al., 2001), whereas SA-depleted acd6-1 plants are sensitized to the SA agonist BTH (Rate et al., 1999). These results indicate that the levels of SA signaling components may be altered in acd6-1 plants. To test this possibility, we examined the expression of the positive regulators of SA signaling, EDS1 and PAD4, and the SA-transducing component NPR1. Indeed, the abundance of EDS1, PAD4, and NPR1 mRNAs was increased in acd6-1 (Figure 1A). As a positive control for SA signaling activation, the level of PR1 mRNA also was increased in acd6-1, in agreement with our previous findings (Rate et al., 1999). Interestingly, the steady state mRNA levels of the SA-interacting and cell death–repressing genes ACD11 and LSD1 also were increased in acd6-1 (Figure 1A).
Figure 1.
Defense Responses in the acd6-1 Mutant.
(A) RNA gel blot analysis of defense gene induction. EF1α was used as a loading control. This experiment was repeated three times with similar results.
(B) Staining of cell walls. Fourth and fifth leaves from the wild type (Columbia [Col]) and the acd6-1 mutant (a6) were examined for autofluorescence (top row) and callose (bottom row).
To determine if any of the SA regulatory genes were important for the acd6-1–conferred phenotypes, we constructed acd6-1 pad4 double mutants. The pad4 mutation partially suppressed the SA-dependent dwarfism of acd6-1, as measured by rosette diameter. Additionally, pad4 partially suppressed the increased SA levels and disease resistance of acd6-1 to P. syringae pv maculicola strain DG3 (Pma DG3) (Table 1). Thus, the SA regulatory component PAD4 was partially required for the acd6-1–conferred dwarfism and disease resistance phenotypes. acd6-1 plants also inhibit the extracellular bacterial pathogen P. syringae from delivering Avr proteins into host cells via the type-III secretion apparatus during infection, resulting in a reduced HR. However, acd6-1 plants retain the ability to respond to Avr proteins when they are expressed directly in acd6-1 cells (Rate et al., 1999). Thus, the cell walls of acd6-1 may have altered properties that prevent the penetration of the type-III apparatus. Indeed, callose, the primary cell wall component induced by wounding and pathogen infection (Adam and Somerville, 1996), was increased in the acd6-1 leaf cells (Figure 1B). Some acd6-1 leaf cells also showed strong autofluorescence, possibly resulting from the accumulation and cross-linking of phenolic compounds in and around the cells that had died spontaneously (Figure 1B). These changes in acd6-1 cell wall properties could reflect cell wall strengthening, a response reported in pathogen-infected plants (Hachler and Hohl, 1982; Kuc, 1990).
Table 1.
Role of PAD4 in acd6-1 Rosette Growth, Pathogenicity, and Defense Phenotypes
| Genotype | Rosette Diameter (mm ± se) |
Growth of Pma DG3 (cfu/leaf disc ± se) |
Free SA (μg/g Fresh Weight ± sd) |
Total SA (μg/g Fresh Weight ± sd) |
|---|---|---|---|---|
| acd6-1 | 6.6 ± 0.3a (n = 30) | 7.0 × 103 ± 4.2 × 103a (n = 6) | 6.8 ± 0.6a (n = 3) | 89.6 ± 5.6a (n = 3) |
| acd6-1 pad4 | 17.3 ± 0.4b (n = 30) | 1.1 × 105 ± 2.4 × 104b (n = 6) | 2.6 ± 0.2b (n = 3) | 38.7 ± 13.3b (n = 3) |
| pad4 | 47.9 ± 0.8c (n = 13) | 1.4 × 108 ± 1.4 × 107c (n = 6) | 0.8 ± 0.08c (n = 3) | 0.9 ± 0.01c (n = 3) |
| Wild type | 46.4 ± 0.5c (n = 14) | 4.3 × 106 ± 1.9 × 106d (n = 6) | 0.7 ± 0.06c (n = 3) | 1.1 ± 0.14c (n = 3) |
For rosette diameter measurements, 20-day-old plants were measured. Growth measurements were made 3 days after infection. The starting inoculum was 100 colony-forming units (cfu)/leaf disc. This experiment was repeated twice with similar results. Superscript letters indicate that values are statistically different (P < 0.0001 for rosette diameters; P < 0.008 for bacterial growth and SA levels).
ACD6 Is a Novel Ankyrin Protein
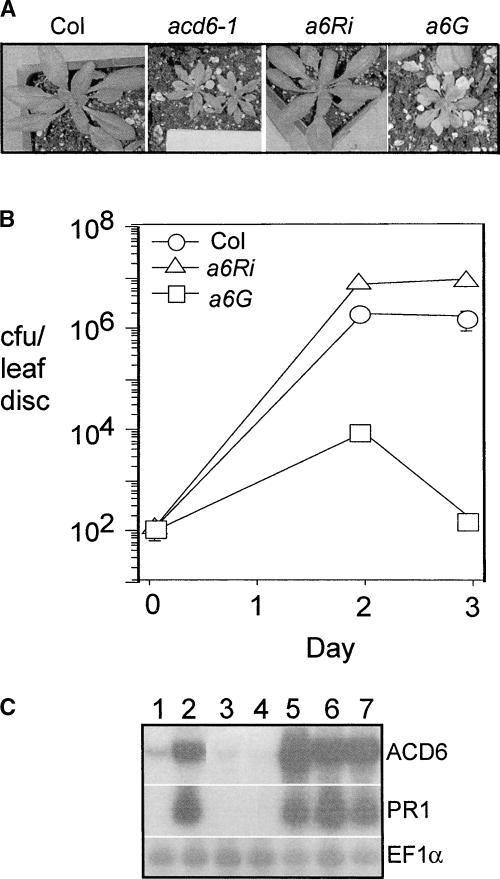
We fine-mapped ACD6 to a 93.1-kb region on chromosome IV. To identify the mutation, most of the coding sequences were amplified by PCR from both the wild type and the acd6-1 mutant and sequenced on both strands. Only one point mutation (a C-to-T change) was found in one of the open reading frames (At4g14400), which was confirmed to be ACD6. Three additional lines of evidence confirmed the cloning of ACD6. First, a PCR-based derived cleaved amplified polymorphic sequence marker based on this point mutation cosegregated with the acd6-1–conferred phenotype in the mapping population and in backcrosses (data not shown). Second, an ACD6 RNA interference (RNAi) construct introduced into the acd6-1 mutant (a6Ri) suppressed the dwarfism, cell death, and disease resistance phenotypes (Figures 2A and 2B). Finally, the ACD6-1 genomic clone introduced into the wild type (a6G) conferred the dwarfism, cell death, increased PR-1 transcript accumulation, and disease resistance phenotypes seen in the acd6-1 plants (Figures 2A to 2C).
Figure 2.
Suppression and Recapitulation of acd6-1 Mutant Phenotypes in Transgenic Plants.
(A) Three-week-old wild-type and transgenic plants. a6Ri, acd6-1 transformed with an ACD6RNAi construct; a6G, Col transformed with an ACD6-1 genomic clone. At least 25 independent a6Ri and a6G lines behaved similarly to the lines shown.
(B) P. syringae growth curve. Col, a6Ri, and a6G plants were infected with Pma DG3 (OD600 = 0.0001). Bars indicate standard errors; in some cases, the symbol obscures the error bars. The growth of bacteria in the three hosts was significantly different on days 2 and 3 (P < 0.001 [t test], n = 6). Similar results were obtained with several additional independent transformants (data not shown). This experiment was repeated three times with similar results. cfu, colony-forming units.
(C) RNA gel blot analysis of the steady state accumulation of ACD6 and PR1 mRNAs. EF1α was used as a loading control. Total RNA was extracted from Col (lane 1), acd6-1 (lane 2), two a6Ri lines (lanes 3 and 4), and three a6G lines (lanes 5 to 7).
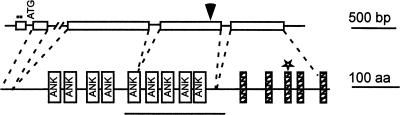
ACD6 encodes a novel protein containing ankyrin repeats and putative transmembrane regions (Figure 3). Isolation of the cDNA clone confirmed the size of the ACD6 mRNA detected by RNA gel blot analysis (2 kb). ACD6 is composed of five exons and is predicted to encode a protein with 670 amino acids. The first exon is untranslatable because it contains in-frame stop codons. Although not well described in plants, such untranslated exons are common in animals (for example, see Chen et al., 1996). The N-terminal region (∼70 amino acids) does not match any regions with known functions. The middle part contains nine ankyrin repeats, based on the SMART protein domain prediction program. Several transmembrane helices were predicted in the C-terminal region, although the exact number (usually between five and seven) varied depending on the algorithm. The missense mutation in acd6-1 caused a Leu-to-Phe substitution at position 591 of the protein in a predicted transmembrane helix (Figure 3). This change did not alter the predicted topology of the transmembrane-spanning region.
Figure 3.
ACD6 Encodes a Putative Protein with Ankyrin and Transmembrane Regions.
Structures of the ACD6 genomic DNA (top) and the predicted ACD6 protein (bottom). In the DNA structure, the boxes indicate exons, lines indicate untranslated regions and introns, and dots indicate the in-frame stop codons in exon 1. ATG is the putative translation start site. The arrowhead indicates the T-DNA insertion site in acd6-T. In the protein structure, boxes labeled ANK indicate ankyrin repeats, hatched boxes indicate transmembrane helices, and the star indicates the Leu-to-Phe mutation in acd6-1. The line below the structure indicates the fragment used for the ACD6 RNAi construct. The broken lines between the two structures connect the exons to their encoded protein regions. aa, amino acids.
Many sequences from higher plants in GenBank shared significant similarity with ACD6, both at the amino acid level and in the organization of the predicted functional regions. With an expectation value of <10−4, there were 34 genes encoding ACD6-like proteins in the Arabidopsis genome that contained ankyrin and transmembrane regions. The overall similarity between ACD6 and the ACD6-like proteins varied from 9 to 64%.
ACD6 Expression Is SA Dependent
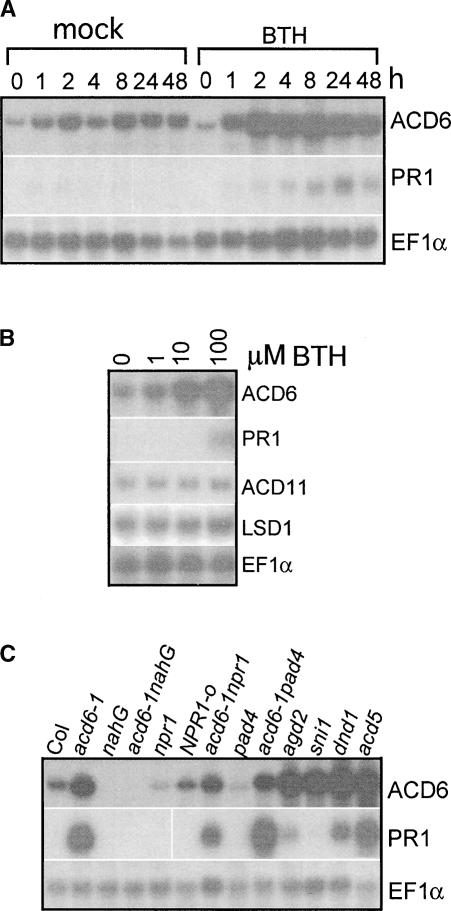
acd6-1 and wild-type plants harboring the ACD6-1 transgene both showed increased steady state ACD6-1 and/or ACD6 mRNA accumulation (Figure 2C). Because SA levels in acd6-1 are increased (Vanacker et al., 2001), it seemed possible that ACD6 could be regulated by SA. Indeed, ACD6 mRNA abundance increased after BTH treatment in both time-course and dose-response experiments (Figures 4A and 4B). Compared with ACD6, the PR1 gene was much less responsive to BTH (Figures 4A and 4B). Unlike ACD6, the ACD11 and LSD1 genes associated with defense and cell death were not induced by BTH (Figure 4B).
Figure 4.
SA-Dependent ACD6 Gene Expression.
(A) Time-course induction by 100 μM BTH. This experiment was repeated three times with similar results.
(B) BTH dose-dependent gene induction. Col leaves were collected 24 h after treatment with BTH at the indicated concentrations. This experiment was repeated three times with similar results.
(C) Gene expression in different genotypes in the Col background. RNA samples were extracted from 20-day-old plants. This experiment was repeated twice with similar results.
Consistent with the BTH treatment results, mutants with high SA levels, such as agd2, dnd1, and acd5 (Yu et al., 1998; Greenberg et al., 2000; Rate and Greenberg, 2001), had high steady state ACD6 mRNA levels (Figure 4C). Conversely, plants with low SA levels (nahG), impaired SA regulation (pad4), or impaired signaling (npr1) had greatly reduced accumulation of ACD6 mRNA. In addition, a mutation in a negative regulator of systemic acquired resistance (SNI1) potentiated the expression of ACD6 mRNA. Consistent with the findings that acd6-1 acts partially through NPR1 and PAD4 (Rate et al., 1999) (Figure 1A, Table 1), the acd6-1 npr1 and acd6-1 pad4 double mutants had reduced ACD6-1 mRNA levels relative to that seen in acd6-1 alone. The pattern of PR1 mRNA accumulation was similar to that of ACD6, with some variations in the level of induction (Figure 4C). These data suggest that a threshold level of SA or SA signaling is required for the basal ACD6 mRNA accumulation, which can be induced further when SA levels are increased.
ACD6 Expression Is Induced Systemically but Not Locally during Pathogen Infection
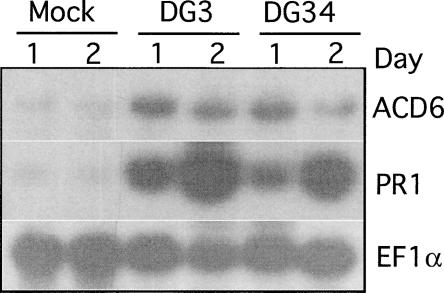
Because the steady state ACD6 mRNA level was under SA regulation, we further examined the expression of the ACD6 gene during P. syringae infection, a condition known to induce SA accumulation (Zhou et al., 1998). In a 48-h time course of wild-type plants infected with Pma DG3 (virulent) and Pma DG34 (avirulent, carrying avrRpm1), no changes in the steady state level of ACD6 transcript were observed. However, PR1 transcript levels were increased upon infection (data not shown). Because SA levels also increase in systemic tissue of P. syringae–infected plants (Delaney et al., 1995; Summermatter et al., 1995), we tested the steady state ACD6 mRNA level in the uninfected leaves adjacent to the infected leaves. The abundance of the ACD6 mRNA was higher in the systemic tissue of infected plants than in the tissue from mock-treated plants. As expected, the level of the PR1 mRNA also was induced systemically (Figure 5). Thus, ACD6 is a systemically induced gene.
Figure 5.
ACD6 Gene Expression during Pathogen Infection.
Col leaves were inoculated with 10 mM MgSO4 (mock treatment), Pma DG3, and Pma DG34 (carrying avrRpm1) at OD600 = 0.01. RNA was extracted at the indicated times from the uninfected leaves of inoculated plants. This experiment was repeated three times with similar results.
ACD6 Expression Requires Light
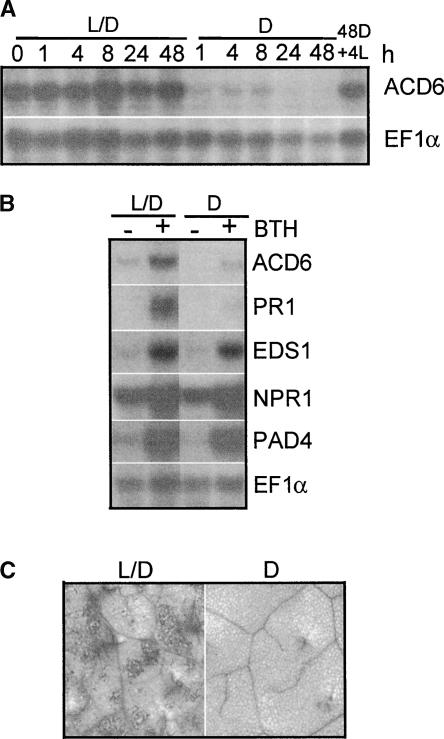
Light is important for at least some aspects of SA signal transduction (Genoud and Metraux, 1999; Genoud et al., 2002). To test the effect of light on the expression of ACD6, wild-type plants maintained in the 16-h-light/8-h-dark cycle were moved to constant dark 4 h after the lights came on in the morning. Within 1 h, the abundance of the ACD6 mRNA was reduced significantly, and the decrease continued for up to 48 h. Interestingly, when the 48-h dark-treated plants were exposed to light again for only 4 h, the steady state level of ACD6 mRNA was restored back to the level seen in the control plants maintained in the normal light/dark cycle (Figure 6A). In agreement with these findings, the normal dark period also resulted in the downregulation of ACD6 (data not shown).
Figure 6.
Light Is Required for ACD6 Expression and the acd6-1 Phenotype.
(A) A time course of steady state ACD6 mRNA accumulation. Twenty-day-old Col plants grown in a 16-h-light/8-h-dark cycle were kept in these conditions (L/D) or shifted to continuous dark (D) 4 h after the lights normally came on (time 0) for the indicated times and then switched back to light for 4 h (4L).
(B) Effect of light on BTH-induced defense gene expression. Col plants were treated with 100 μM BTH and subjected to the normal 16-h-light/8-h-dark cycle (L/D) or dark treatment (D) for 24 h.
(C) Cell death staining. acd6-1-nahG plants were treated with 100 μM BTH or water and grown in the normal 16-h-light/8-h-dark (L/D; left) or 24-h dark (D; right) condition for 1 day. The fourth leaves were stained with trypan blue to detect cell death. Wild-type control and water-treated acd6-1-nahG tissue showed no cell death under the conditions used here (data not shown). These experiments were repeated three times with similar results.
BTH-induced ACD6 mRNA accumulation in wild-type plants was reduced greatly in the dark (Figure 6B). A similar suppression by dark was found with the PR1 transcript. However, for EDS1 and PAD4 mRNAs, only the steady state basal but not the BTH-induced transcripts were reduced slightly during the 24-h dark treatment (Figure 6B). In addition, the level of NPR1 mRNA under basal or BTH induction conditions was unaffected by light. Thus, light differentially regulated defense gene expression.
Consistent with the requirement of light for ACD6 transcript accumulation, it also was required for the acd6-1–conferred cell death phenotypes. Under normal light conditions (16 h of light/8 h of dark), SA-depleted acd6-1-nahG plants that lacked cell death showed cell death upon BTH treatment (Rate et al., 1999). However, in constant dark, cell death was suppressed completely (Figure 6C).
ACD6 Loss-of-Function Plants Are More Susceptible to Disease and Have Attenuated Defenses
In a6Ri plants, suppression of the steady state ACD6 mRNA levels correlated well with a modest increase in the susceptibility of the plants to Pma DG3 (Figures 2B and 2C) as well as to the congenic avirulent strains Pma DG6 and Pma DG34 carrying avrRpt2 and avrRpm1, respectively (data not shown). Wild-type plants harboring the ACD6 RNAi construct behaved similarly to a6Ri plants (data not shown). The expression of genes with the closest similarity to ACD6 (At4g14390 [ACL1] and At4g05040 [ACL2]) was not affected by the RNAi transgene (data not shown). These genes had 86 and 79% similarity, respectively, at the DNA level with ACD6 in the region of the RNAi. Other genes related to ACD6 showed <55% similarity at the DNA level in this region, making it less likely that the ACD6 RNAi would affect their expression.
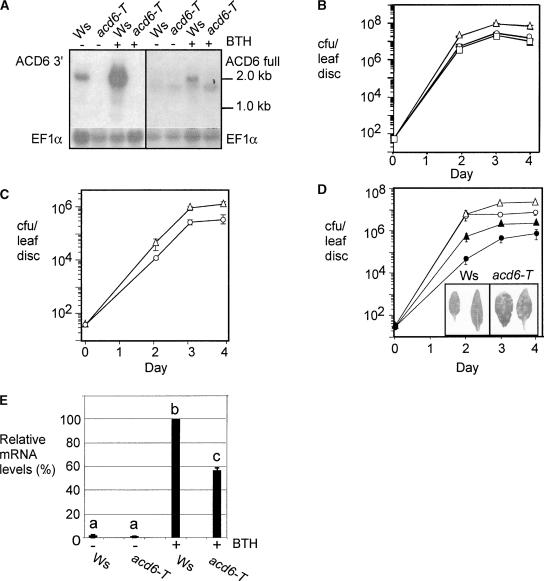
However, to be certain that the RNAi results are attributable to the specific downregulation of ACD6, we isolated and characterized a loss-of-function acd6 mutant with a T-DNA insertion in the fourth exon of ACD6 (acd6-T) in the Wassilewskija (Ws) ecotype background (Figure 3). Using an ACD6-specific probe complementary to the 3′ region of ACD6, no ACD6 transcript was detectable by RNA gel blot analysis in acd6-T, whereas a 2.0-kb band was visible in Ws (Figure 7A, left blot). It seemed possible that the T-DNA insertion could lead to the accumulation of a truncated ACD6 transcript predicted to be 1.2 kb, shorter than any of the predicted transcripts of ACD6-like genes. However, with the full-length ACD6 cDNA as a probe on a duplicate blot, neither the 1.2-kb truncated transcript nor the full-length ACD6 mRNA was observed in acd6-T plants (Figure 7A, right blot). A transcript of ∼1.8 kb, which also was detected with an ACL2 (At4g05040)-specific probe (data not shown), was visible in both wild-type and the acd6-T plants. This band was not induced significantly by treatments that induced ACD6 transcript accumulation, such as with the SA agonist BTH (Figure 7A). Thus, acd6-T appears to be a null mutant.
Figure 7.
Defense Response and Disease Susceptibility of an acd6-T Loss-of-Function Mutant.
(A) Steady state accumulation of ACD6 mRNA using an ACD6-specific probe (ACD6 3′) (left) and the full-length ACD6 cDNA probe (ACD6 full) (right). EF1α served as a loading control. Wild-type Ws and the acd6-T mutant were treated with 300 μM BTH or water for 1 day.
(B) and (C) Disease susceptibility and complementation of acd6-T plants. Wild-type Ws (circles), acd6-T (triangles), and/or acd6-T complemented with genomic ACD6 (squares) were infected with Pto DC3000 (B) or Pto DC3000 carrying avrRpt2 (C) at an OD600 = 0.0001. The growth of Pto DC3000 in acd6-T was significantly different from that in the wild type and complemented on days 2, 3, and 4 (P < 0.02 [t test], n = 6). The growth of Pto DC3000 carrying avrRpt2 in acd6-T was significantly different from that in the wild type on days 3 and 4 (P < 0.007 [t test], n = 6). These experiments were repeated twice with similar results. cfu, colony-forming units.
(D) Reduced disease resistance of acd6-T treated with BTH. Wild-type Ws and acd6-T were pretreated with 300 μM BTH or water for 2 days and then subjected to infection by Pto DC3000 (OD600 = 0.0001). Open circles, water-treated Ws; closed circles, BTH-treated Ws; open triangles, water-treated acd6-T; closed triangles, BTH-treated acd6-T. Bars indicate standard errors; in some cases, the error bars are obscured by the symbols. For water-treated plants, the growth of bacteria in acd6-T was significantly different from that in the wild type on days 3 and 4 (P < 0.001 [t test], n = 6). For BTH-treated plants, the growth of bacteria in acd6-T was significantly different from that in the wild type on days 2, 3, and 4 (P < 0.001 [t test]). The inset shows BTH-treated and infected leaves taken 3 days after the infection. Note the increased lesion numbers on the acd6-T plants. This experiment was repeated three times with similar results.
(E) Quantitation of the relative abundance of PR1 transcript in Ws and acd6-T plants after BTH treatment. Plants were treated as in (A). PR1 mRNA levels were normalized to the level of EF1α mRNA. Different letters indicate that the values are significantly different from each other (P < 0.01). The data were averaged from four independent experiments.
The acd6-T mutant was morphologically normal and displayed modestly increased susceptibility to P. syringae pv tomato strain DC3000 (Pto DC3000) as well as to Pto DC3000 carrying the avrRpt2 avirulence gene (Figures 7B to 7D) (Pma DG3 was not used because it does not cause disease on the parental Ws ecotype). acd6-T/ACD6 heterozygotes showed wild-type disease susceptibility, suggesting that the acd6-T mutation was recessive (data not shown). Furthermore, the susceptibility of acd6-T plants to Pto DC3000 was complemented by the ACD6 genomic clone (Figure 7B).
Interestingly, the acd6-T plants were less responsive to the SA agonist BTH. Thus, acd6-T plants treated with BTH showed impaired disease resistance early during infection (on day 2) with Pto DC3000, relative to the wild-type parent Ws (Figure 7D). This impaired resistance of BTH-treated acd6-T plants was accompanied by an increase in the severity of disease symptoms elicited by Pto DC3000 (Figure 7D, inset). Consistent with these observations, acd6-T plants showed compromised induction of PR1 mRNA accumulation upon BTH treatment relative to that seen in Ws (Figure 7E). In addition to reduced responsiveness to BTH, acd6-T plants had transiently reduced SA production and PR1 expression after P. syringae infection (Figure 8; note levels at 12 h). In summary, ACD6 is important for disease resistance and the timely activation of defenses against virulent P. syringae.
Figure 8.
Reduced Defenses in acd6-T Plants.
(A) Reduced SA levels in P. syringae–infected acd6-T plants. Plants were inoculated with 10 mM MgSO4 or Pto DC3000 at OD600 = 0.01. Samples were analyzed in triplicate. Asterisks indicate P < 0.05 at 12 h. FW, fresh weight.
(B) Reduced PR1 expression in P. syringae–infected acd6-T plants. Plants treated as in (A) were used for RNA isolation. This experiment was repeated twice with similar results.
Plants Overexpressing ACD6 Are More Resistant to Disease and Have Increased Defenses
To determine whether ACD6 was sufficient to confer disease resistance, we introduced extra copies of ACD6 genomic clones into wild-type Col plants. Strikingly, 13 independent transformants carrying at least one extra copy of ACD6 were more resistant to Pto DC3000. Examples of the growth of Pto DC3000 in two such lines are shown in Figure 9. Plants with extra copies of ACD6 also showed increased steady state ACD6 mRNA (Table 2). These plants all had modestly increased SA levels, although only some lines had statistically significant increases in SA (Table 2). Additionally, 2 of the 13 independent transgenic lines showed spontaneous microscopic cell death, whereas the rest showed cell death after treatment with BTH (Table 2 and data not shown). All of the transgenic plants were similar in size to wild-type plants. Collectively, these data support a role for ACD6 as a necessary and dose-dependent component of the defense response against virulent P. syringae and suggest a role for ACD6 in activating SA-dependent cell death.
Figure 9.
Growth of P. syringae in Plants with Extra Copies of ACD6.
Wild-type (Col) and T2 plants from two independent transformants with extra copies of ACD6 (lines 12 and 14) were infected with Pto DC3000 at OD600 = 0.0001. Bars indicate standard errors (n = 6); in some cases, the error bars are obscured by the symbols. Bacteria grew significantly more in the wild type than in all of the lines carrying extra copies of ACD6 (P < 0.01). This experiment was repeated twice with similar results. cfu, colony-forming units.
Table 2.
Relative ACD6 mRNA Accumulation, Cell Death, and SA Levels in Plants with Extra Copies of ACD6
| Line | Transgene Copy Number |
Relative ACD6 mRNA Level (n = 2) |
BTH-Induced Cell Death |
Free SA (μg/g Fresh Weight; n = 3) |
Total SAa (μg/g Fresh Weight; n = 3) |
|---|---|---|---|---|---|
| Untransformed | 0 | 1.0 | − | 0.8 ± 0.4 | 1.1 ± 0.6 |
| 3 | 1 | 3.2 ± 0.5 | + | 1.0 ± 0.2 | 2.2 ± 0.7 (P = 0.114) |
| 4 | 1 | 5.4 ± 0.3 | + | 1.3 ± 0.05 | 2.7 ± 0.6 (P = 0.030) |
| 12 | 2 | 2.4 ± 0.1 | + | 1.2 ± 0.2 | 2.9 ± 0.6 (P = 0.020) |
| 14 | ≥3 | 5.8 ± 0.1 | +b | 0.7 ± 0.1 | 2.5 ± 0.7 (P = 0.054) |
The copy number of the representative homozygotic ACD6 overexpression lines was determined by germinating the T1 seeds on agar medium containing Murashige and Skoog (1962) salts and 40 μg/mL BASTA. The relative ACD6 mRNA level was normalized to the level of EF1α mRNA. To score cell death on leaf tissue, plants were treated with 100 μM BTH or water for 4 days and stained with trypan blue.
t test results shown are for the wild type compared with each overexpression line. This experiment was repeated twice with similar results.
This line also showed some cell death before BTH treatment.
DISCUSSION
SA has been known for many years to be a key signal molecule that mediates plant disease resistance. However, the mechanism by which SA is regulated, perceived, and transduced is understood only in outline. In particular, only a few SA-regulated signaling components are known to be important for disease resistance. Here, we showed that the Arabidopsis ACD6 gene encodes a novel component of the light-dependent branch of the SA signaling pathway. The abundance of the ACD6 mRNA is regulated by both light and SA. Furthermore, ACD6 transcript is induced in the systemic tissue of P. syringae–infected plants. Finally, we showed that plants lacking ACD6 transiently produce less SA, are less responsive to the SA agonist BTH, and are more susceptible to virulent P. syringae. By contrast, plants with an extra copy(s) of ACD6 have modestly increased SA levels, are more resistant to virulent P. syringae, and show BTH-inducible and/or spontaneous cell death. These observations are consistent with our previous findings that SA-depleted acd6-1 gain-of-function plants are more responsive to the SA agonist BTH and that acd6-1 plants show SA-dependent disease resistance and defenses (Rate et al., 1999). Thus, ACD6 appears to be a regulator and an effector of the SA pathway.
ACD6 encodes a novel protein with ankyrin repeats and transmembrane regions, which are hallmarks of some signal transduction proteins. Ankyrin repeats are involved in diverse processes but share the property that they often are involved in protein–protein interactions (Sedgwick and Smerdon, 1999). The best characterized ankyrin protein from plants is NPR1/NIM1, which is involved in SA-dependent disease resistance and in an SA-independent resistance response elicited by certain root-associated bacteria (Pieterse et al., 1998). NPR1 localizes to the nucleus during SA signaling (Kinkema et al., 2000) and functions as a transcriptional coactivator to regulate the defense response (Zhang et al., 1999; Fan and Dong, 2002). ACD6 does not show significant similarity to NPR1, although both proteins have repeat regions in the ankyrin family. The possible transmembrane helices in ACD6 suggest that ACD6 may act at the membrane to activate defenses, as has been shown or suggested for other defense signaling components (Century et al., 1997; Boyes et al., 1998; Falk et al., 1999; Jirage et al., 1999; Mackey et al., 2002). Although the mutant ACD6-1 protein is not predicted to have altered membrane topological properties, it may have an altered conformation that switches it to a constitutively active form to amplify SA signaling.
The positive activators of SA accumulation, EDS1 and PAD4, have been proposed to form a signal-amplification loop with SA (Falk et al., 1999; Jirage et al., 1999). Like eds1 and pad4, acd6-T is compromised for SA production upon infection. However, the effect of acd6-T on SA production after virulent P. syringae infection was more transient than that reported for pad4 and eds1 (Jirage et al., 1999; Feys et al., 2001). Consistent with a role for ACD6 in the regulation of SA production, ACD6-overexpressing plants have modestly increased SA levels in uninfected plants. Unlike ACD6, the regulation of EDS1 and PAD4 appears to be largely independent of light. The expression of PAD4 and EDS1 is much higher in the acd6-1 gain-of-function mutant, whereas the loss of PAD4 function results in lower expression of ACD6 (Figures 1A and 4C). In addition, pad4 partially suppresses the acd6-1 disease resistance and dwarf phenotypes shown previously to be SA dependent (Table 1). Thus, this suppression is likely the result of reduced SA levels in acd6-1 pad4 plants relative to acd6-1 alone. These results collectively suggest an interaction of the signaling pathways mediated by EDS1, PAD4, and ACD6, possibly by regulating SA and SA signaling, the key common molecule in the Arabidopsis defense response.
The regulation of ACD6 by SA and light is likely to occur at the level of transcriptional activation, because the promoter region of ACD6 has many predicted SA and light cis-regulatory elements (data not shown). Other possible modes of regulation, such as those that affect mRNA and/or protein stability, also are possible. Although we have not explored the molecular basis of the light requirement for ACD6 transcript accumulation, there is precedent for a role for the photoreceptor phytochrome in amplifying SA signaling to induce the transcription of some genes and to control cell death during HR (Genoud et al., 2002). The regulation of ACD6 by both light and SA suggests one way that plants can integrate external and internal signals to effect the amplification of the defense response.
ACD6 is a member of a large plant-specific gene family. Of the 158 Arabidopsis proteins with ankyrin repeat regions, 34 have the same overall organization as ACD6 and also are similar to ACD6 at the amino acid level in both the putative ankyrin repeat regions and in the transmembrane regions. None of the ACD6-like proteins have been assigned a function in Arabidopsis. A number of the ACD6-like genes are located in a large cluster on chromosome IV, which may make them susceptible to intergenic recombination, similar to some disease resistance (R) loci containing clustered genes (Ellis et al., 2000). It is intriguing that some ecotypes of Arabidopsis also have a duplicated copy of ACD6 lacking introns, although whether the additional copies have functional promoters is unclear (Rate, 2000). This observation could indicate that ACD6 is under adaptive selection. Because ACD6 and its related proteins represent one of the largest protein families in Arabidopsis, it is striking that no other loss-of-function mutants in the ACD6 family have been reported. Possibly, ACD6 and some of the ACD6-like genes have overlapping and/or redundant functions that have been difficult to detect in loss-of-function mutant screens. It also is possible that some of the eds mutants that could not be mapped, such as eds4 (Gupta et al., 2000), have defects in ACD6 or ACD6-like genes. This possibility highlights the utility of obtaining gain-of-function mutants to identify the components of signaling pathways.
In some host–pathogen interactions, SA is important for cell death during the HR (Rate et al., 1999) and may play a role in micro-HR formation during systemic acquired resistance (Alvarez et al., 1998). Does ACD6 play a role in cell death control? The acd6-1 gain-of-function mutant shows spontaneous SA- and light-dependent cell death (Rate et al., 1999). This cell death is localized, possibly as a result of the action of the cell death–suppressor genes ACD11 and LSD1, whose transcripts are upregulated in acd6-1. The loss-of-function mutant (acd6-T), although showing enhanced disease susceptibility and increased disease symptoms (especially in BTH-treated plants; Figure 7D), did not show an obvious difference in the HR in dose-response experiments with two different avirulent pathogens (data not shown). However, other genes in the ACD6 family may act redundantly to control SA-dependent cell death. Indeed, a number of ACD6 family members were found in stress-induced cDNA libraries (H. Lu and J.T. Greenberg, unpublished data). Interestingly, BTH treatment of ACD6-overexpressing plants elicits localized cell death, suggesting that ACD6 does play a role in cell death control. The isolation of additional loss- and gain-of-function mutants in ACD6 family members should be valuable for discerning their possible functions in mediating both disease resistance and/or cell death.
METHODS
Plant Materials, Treatments, and Pathogen Infection
Seeds of Arabidopsis thaliana acd6-1, npr1-1, acd6-1 npr1, acd6-1-nahG, nahG, NPR1-o (an NPR1-overexpressing line), acd5, agd2, pad4-1, sni1, and dnd1 were in the Columbia (Col) background. Isolation and/or construction of acd6-1, acd6-1 npr1, acd6-1-nahG, acd5, and agd2 were described previously (Rate et al., 1999; Greenberg et al., 2000; Rate and Greenberg, 2001). npr1-1, NPR1-o, and sni1 (Cao et al., 1994, 1998; Li et al., 1999) were from Xinnian Dong (Duke University, Durham, NC). nahG B15 (Delaney et al., 1995) seeds were from Syngenta (Research Triangle Park, NC). dnd1 (Yu et al., 1998) was from Andrew Bent (University of Wisconsin, Madison). pad4-1 (Glazebrook et al., 1997) was from Jane Glazebrook (University of Minnesota, St. Paul). acd6-1 pad4 was constructed by crossing acd6-1 with pad4-1. Double homozygous mutants were identified using cleaved amplified polymorphic sequence markers in the F2 generation. An acd6 T-DNA insertion mutant (acd6-T) in the Wassilewskija (Ws) background was obtained by screening the T-DNA insertion library at the University of Wisconsin (www.biotech.wisc.edu).
Arabidopsis plants were grown as described (Rate et al., 1999) in a 16-h-light (6 am to 10 pm)/8-h-dark cycle. Unless specified otherwise, each treatment of 20-day-old Arabidopsis plants was started at ∼10 am. Benzo(1,2,3)thiadiazole-7-carbothioic acid (benzothiazole [BTH]) was a gift from Robert Dietrich (Syngenta). For BTH treatment, plants were sprayed with BTH at the concentrations indicated in the legends to Figures 4, 6, and 7 and Table 2 until all of the leaves were wet. BTH treatment of Ws and acd6-T was performed on 16- to 18-day-old plants. During dark treatments, plants were covered with an aluminum foil–wrapped plastic dome. The top of the dome was cut off to minimize the increase of humidity. Pseudomonas syringae pv maculicola strain DG3 (a recA derivative of ES4326), P. syringae pv maculicola strain DG6 (an ES4326 recA derivative expressing the type-III effector avrRpt2), and P. syringae pv maculicola strain DG34 (an ES4326 recA derivative expressing the type-III effector avrRpm1) were described previously (Guttman and Greenberg, 2001). P. syringae pv tomato strain DC3000 (Pto DC3000) was obtained from F.M. Ausubel (Massachusetts General Hospital and Harvard University, Boston, MA). Pto DC3000 carrying avrRpt2 was constructed by transforming Pto DC3000 with pMMXR1 (Dong et al., 1991). Bacterial culturing, infection, and growth curve analysis were performed as described previously (Greenberg et al., 1994), except that Pto DC3000 was plated on Luria-Bertani agar (Sambrook et al., 1989) containing 100 μg/mL rifampicin.
RNA Gel Blot Analysis
The fourth and fifth leaves from treated and control plants were used for total RNA extraction as described (Rate et al., 1999). For probes, each DNA fragment shown in Table 3 was amplified by PCR from Col genomic DNA or ACD6 cDNA as indicated in the table, confirmed by sequencing, and labeled with α-32P-dCTP by primer extension using a corresponding reverse primer. Unless specified otherwise, the expression of ACD6 was detected with ACD6 3′, an ACD6-specific probe. The blots were hybridized as described (Rate et al., 1999). Each experiment was repeated at least twice.
Table 3.
DNA Fragments Used in This Report
| Fragment Name | Primer Names | Primer Sequence |
|---|---|---|
| ACD6-full* | ACD6-full (rev); ACD6-full (for) | 5′-TTCGGAACACGCCACACAACCA-3′; 5′-GCGATGGACAGTTCTGGAGCAG-3′ |
| ACD6 3′ (specific)* | ACD6 3′ (rev); ACD6 3′ (for) | 5′-CTTGTGAAGTATCGTCTATATTGCTCTTG-3′; 5′-AAGCTTGTTTTGGTTGTGTGG-3′ |
| ACD6-RNAi* | ACD6-RNAi (rev); ACD6-RNAi (for) | 5′-AAACGGATCCATTTAAATAAGAGCCGCTACCACGAGAAGAG-3′; 5′-ATCTACTAGTGGCGCGCCTGAAGGCCAAAAGTATAGGTGTC-3′ |
| ACD6-screen | ACD6-screen (rev); ACD6-screen (for) | 5′-TCTCTTCGAATGATATACTTTTCACGCTT-3′; 5′-CAATCGTAAGTGATACAAAATAGTTAGAAGTG-3′ |
| ACD11 | ACD11 (rev); ACD11 (for) | 5′-TTTTTCAGGCTCTTCACCAATCA-3′; 5′-CATTAGCCGTGCCAGTTTGTAGGATG-3′ |
| ACL1-specific | ACL1-specific (rev); ACL1-specific (for) | 5′-TGTCATCATCATCAAAGGAAATACGAAAC-3′; 5′-AGTACGCTGGTGACTTTCTCG-3′ |
| ACL2-specific | ACL2-specific (rev); ACL2-specific (for) | 5′-GTCAATGATACCAAGAAGAGGGGG-3′; 5′-AAGTGAGTTTCTTCCCGAATCCCT-3′ |
| EF1α | EF1α (for); EF1α (rev) | 5′-GCTGTCCTTATCATTGACTCCACC-3′; 5′-TCATACCAGTCTCAACACGTCC-3′ |
| LSD1 | LSD1 (rev); LSD1 (for) | 5′-CAACGGACATGGGGTTTTCTAC-3′; 5′-GGTTGCCCATGCTCCTTCCAG-3′ |
| NPR1 | NPR1 (rev); NPR1 (for) | 5′-CACAATTGATACTATCTCTATTGG-3′; 5′-TTTCGGCGATCTCCATTGCAGC-3′ |
| PAD4 | PAD4 (rev); PAD4 (for) | 5′-GCGATGCATCAGAAGAG-3′; 5′-TTAGCCCAAAAGCAAGTATC-3′ |
| PG1-2 | PG1-2 (for); PG1-2 (rev) | 5′-GCCCAAGGAAAGCGTCGT-3′; 5′-GCAGACTCATCACCCCCATACTTA-3′ |
| PG19 | PG19 (for); PG19 (rev) | 5′-CGGCTTTGGCTTGCTTTCACC-3′; 5′-CCCGTTGTCTCGCATATTCCATTG-3′ |
| PR1 | PR1 (rev); PR1 (for) | 5′-CACATAATTCCCACGAGGATC-3′; 5′-GTAGGTGCTCTTGTTCTTCCC-3′ |
Asterisks indicate fragments amplified from an ACD6 cDNA by PCR. The rest of the fragments were amplified from the Col genome. for, forward; rev, reverse.
Salicylic Acid Measurements
The fourth and fifth leaves from treated and control plants were used for salicylic acid extractions and analysis as described (Vanacker et al., 2001).
Histochemical Staining
Autofluorescence and callose staining with 0.01% aniline blue of leaf tissue was performed as described (Adam and Somerville, 1996) and examined with epifluorescence illumination from an Axioskop microscope (Zeiss, Jena, Germany). All tissues were photographed with the same exposure time. Trypan blue staining for cell death detection was described previously (Rate et al., 1999).
Positional Cloning of ACD6
Recombinant plants were created by crossing the homozygous acd6-1 mutant with ecotype Landsberg. The F2 plants showing the acd6-1 homozygous phenotype (dwarfed and harboring cell death patches on the leaves) in 25% of the population were used for mapping. After scoring 1384 homozygous acd6-1 recombinant plants with 42 cleaved amplified polymorphic sequence or simple sequence length polymorphism markers, acd6-1 was placed between two markers, PG1-2 and PG19 (Table 3), on chromosome IV, spanning 93.1 kb of sequence and containing 22 putative genes. The coding regions of 15 of the 22 genes (∼40 kb) from both the wild type and the mutant were amplified by PCR and sequenced. The genes not sequenced comprised retrotransposons and reverse transcriptase or were at the borders of the region defined by recombination.
To obtain an ACD6-1 genomic clone, 15 μg of genomic DNA from the acd6-1 mutant was digested with BamHI and SacI and separated on a 10 to 40% sucrose gradient. Fractions with 10 to 15 kb of DNA were pooled and ligated into binary vector pBIN19 by electroporating into Escherichia coli strain DH5α. The resulting library was plated and screened by colony hybridization using an α-32P-dCTP–labeled 2663-bp DNA fragment called ACD6-screen as a probe (Table 3). One positive clone was found and was confirmed to contain the ACD6-1 genomic region by sequencing and restriction map analysis. A full-length ACD6 cDNA was isolated by screening a Col cDNA library (a kind gift from Fumiaki Katagiri, University of Minnesota, St. Paul) using colony hybridization.
DNA Construction and Plant Transformation
The 7670-bp EcoRV-SacI fragment of the ACD6-1 gene was further subcloned into pBIN19 and used to transform Col using the dipping method (Clough and Bent, 1998). To construct an ACD6 RNA interference clone, a 691-bp DNA fragment from ACD6 cDNA was amplified by PCR (Table 3) and cloned as two inverted repeats into the binary vector pFGC1008 (http://ag.arizona.edu/chromatin/fgc1008.html). This construct was used to transform Col and acd6-1 plants. A wild-type genomic clone of ACD6 was subcloned as a 7670-bp EcoRV-SacI fragment from BAC clone F14B19 (ABRC Stock Center, Ohio State University, Columbus) into the pBin19 vector. The transformants were selected on agar plates containing Murashige and Skoog (1962) salts and appropriate antibiotics or on soil by spraying with BASTA at a dilution of 1:2000 (AgrEvo USA, Wilmington, DE).
Sequence Analysis
The putative functional regions of ACD6 were predicted by SMART (http://smart.embl-heidelberg.de/). The prediction of the transmembrane domain was made using several programs found at the following World Wide Web sites: http://www.ch.embnet.org/software/TMPRED_form.html, http://www.cbs.dtu.dk/services/TMHMM, and http://kr.expasy.org. CLUSTAL W (version 1.81) (http://www.ebi.ac.uk/clustalw/) was used for multiple sequence alignments.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Jean T. Greenberg, jgreenbe@midway.uchicago.edu.
Accession Number
The GenBank accession number for ACD6 is AY344843.
Acknowledgments
We thank Joanna Jelenska, Boris Vinatzer, Nan Yao, Jocelyn Malamy, Mark Johnson, and Ravishankar Palanivelu for valuable advice and critical reading of the manuscript. We thank Brian Traw and Joy Bergelson for their assistance with SA quantitation. We are grateful to the ABRC at Ohio State University for BAC clones and to Carolyn Napoli at the University of Arizona for pFGC1008 vector. This work was supported by National Institutes of Health Grant 5R01 GM54292 to J.T.G.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.015412.
References
- Adam, L., and Somerville, S.C. (1996). Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana. Plant J. 9, 341–356. [DOI] [PubMed] [Google Scholar]
- Alvarez, M.E., Pennell, R.I., Meijer, P.J., Ishikawa, A., Dixon, R.A., and Lamb, C. (1998). Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92, 773–784. [DOI] [PubMed] [Google Scholar]
- Aviv, D.H., Rusterucci, C., Iii, B.F., Dietrich, R.A., Parker, J.E., and Dangl, J.L. (2002). Runaway cell death, but not basal disease resistance, in lsd1 is SA- and NIM1/NPR1-dependent. Plant J. 29, 381–391. [DOI] [PubMed] [Google Scholar]
- Bowling, S.A., Clarke, J.D., Liu, Y., Klessig, D.F., and Dong, X. (1997). The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9, 1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes, D., Nam, J., and Dangl, J. (1998). The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc. Natl. Acad. Sci. USA 95, 15849–15854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brading, P.A., Hammond-Kosack, K.E., Parr, A., and Jones, J.D. (2000). Salicylic acid is not required for Cf-2- and Cf-9-dependent resistance of tomato to Cladosporium fulvum. Plant J. 23, 305–318. [DOI] [PubMed] [Google Scholar]
- Brodersen, P., Petersen, M., Pike, H.M., Olszak, B., Skov, S., Odum, N., Jorgensen, L.B., Brown, R.E., and Mundy, J. (2002). Knockout of Arabidopsis accelerated-cell-death11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev. 16, 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, A.S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. [DOI] [PubMed] [Google Scholar]
- Cao, H., Li, X., and Dong, X. (1998). Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc. Natl. Acad. Sci. USA 95, 6531–6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century, K.S., Shapiro, A.D., Repetti, P.P., Dahlbeck, D., Holub, E., and Staskawicz, B.J. (1997). NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278, 1963–1965. [DOI] [PubMed] [Google Scholar]
- Chen, Y., Duhl, D.M.J., and Barsh, G.S. (1996). Opposite orientations of an inverted duplication and allelic variation at the mouse agouti locus. Genetics 144, 265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z., Silva, H., and Klessig, D.F. (1993). Active oxygen species in the induction of plant systemic acquired resistance by salicylic acid. Science 262, 1883–1886. [DOI] [PubMed] [Google Scholar]
- Clarke, J.D., Volko, S.M., Ledford, H., Ausubel, F.M., and Dong, X. (2000). Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12, 2175–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Delaney, T.P., Friedrich, L., and Ryals, J.A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 92, 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P., Uknes, S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D., Gaffney, T., Gut-Rella, M., Kessmann, H., Ward, E., and Ryals, J. (1994). A central role of salicylic acid in plant disease resistance. Science 266, 1247–1250. [DOI] [PubMed] [Google Scholar]
- Dietrich, R.A., Richberg, M.H., Schmidt, R., Dean, C., and Dangl, J.L. (1997). A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell 88, 685–694. [DOI] [PubMed] [Google Scholar]
- Dong, X., Mindrinos, M., Davis, K.R., and Ausubel, F.M. (1991). Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell 3, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J., Dodds, P., and Pryor, T. (2000). Structure, function and evolution of plant disease resistance genes. Curr. Opin. Plant Biol. 3, 278–284. [DOI] [PubMed] [Google Scholar]
- Falk, A., Feys, B.J., Frost, L.N., Jones, J.D., Daniels, M.J., and Parker, J.E. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 96, 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, W., and Dong, X. (2002). In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell 14, 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J., Moisan, L.J., Newman, M.A., and Parker, J.E. (2001). Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20, 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich, L., et al. (1996). A benzothiadiazole derivative induces systemic acquired resistance. Plant J. 10, 61–70. [Google Scholar]
- Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., Ward, E., Kessmann, H., and Ryals, J. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261, 754–756. [DOI] [PubMed] [Google Scholar]
- Genoud, T., Buchala, A.J., Chua, N.H., and Metraux, J.P. (2002). Phytochrome signalling modulates the SA-perceptive pathway in Arabidopsis. Plant J. 31, 87–95. [DOI] [PubMed] [Google Scholar]
- Genoud, T., and Metraux, J.P. (1999). Crosstalk in plant cell signaling: Structure and function of the genetic network. Trends Plant Sci. 4, 503–507. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J., Chen, W., Estes, B., Chang, H.S., Nawrath, C., Metraux, J.P., Zhu, T., and Katagiri, F. (2003). Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 34, 217–228. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J., Zook, M., Mert, F., Kagan, I., Rogers, E.E., Crute, I.R., Holub, E.B., Hammerschmidt, R., and Ausubel, F.M. (1997). Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics 146, 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, J.T., Guo, A., Klessig, D.F., and Ausubel, F.M. (1994). Programmed cell death in plants: A pathogen-triggered response activated coordinately with multiple defense functions. Cell 77, 551–563. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T., Silverman, F.P., and Liang, H. (2000). Uncoupling salicylic acid-dependent cell death and defense-related responses from disease resistance in the Arabidopsis mutant acd5. Genetics 156, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, V., Willits, M.G., and Glazebrook, J. (2000). Arabidopsis thaliana EDS4 contributes to salicylic acid (SA)-dependent expression of defense responses: Evidence for inhibition of jasmonic acid signaling by SA. Mol. Plant-Microbe Interact. 13, 503–511. [DOI] [PubMed] [Google Scholar]
- Guttman, D.S., and Greenberg, J.T. (2001). Functional analysis of type III effectors AvrRpt2 and AvrRpm1 of P. syringae using a single copy genomic integration system. Mol. Plant-Microbe Interact. 14, 145–155. [DOI] [PubMed] [Google Scholar]
- Hachler, H., and Hohl, H.R. (1982). Histochemistry of papillae in potato tuber tissue infected with Phytophthora infestans. Bot. Helv. 92, 23–31. [Google Scholar]
- Jirage, D., Tootle, T.L., Reuber, T.L., Frost, L.N., Feys, B.J., Parker, J.E., Ausubel, F.M., and Glazebrook, J. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 96, 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkema, M., Fan, W., and Dong, X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12, 2339–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klessig, D.F., et al. (2000). Nitric oxide and salicylic acid signaling in plant defense. Proc. Natl. Acad. Sci. USA 97, 8849–8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuc, J. (1990). Compounds from plants that regulate or participate in disease resistance. Ciba Found. Symp. 154, 213–224. [DOI] [PubMed] [Google Scholar]
- Li, X., Zhang, Y., Clarke, J.D., Li, Y., and Dong, X. (1999). Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98, 329–339. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Holt, B.F., Wiig, A., and Dangl, J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108, 743–754. [DOI] [PubMed] [Google Scholar]
- McDowell, J.M., Cuzick, A., Can, C., Benyon, J., Dangl, J.L., and Holub, E.B. (2000). Downy mildew (Peronospora parasitica) resistance genes in Arabidopsis vary in functional requirements for NDR1, EDS1, NPR1 and salicylic acid accumulation. Plant J. 22, 523–529. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Pieterse, C.M., van Wees, S.C., van Pelt, J.A., Knoester, M., Laan, R., Gerrits, H., Weisbeek, P.J., and van Loon, L.C. (1998). A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10, 1571–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan, G.J., and Delaney, T.P. (2002). Role of salicylic acid and NIM1/NPR1 in race-specific resistance in Arabidopsis. Genetics 161, 803–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate, D.N. (2000). Novel Genes of the Plant Defense Response Pathway in Arabidopsis thaliana. PhD dissertation (Boulder: University of Colorado).
- Rate, D.N., Cuenca, J.V., Bowman, G.R., Guttman, D.S., and Greenberg, J.T. (1999). The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11, 1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate, D.N., and Greenberg, J.T. (2001). The Arabidopsis aberrant growth and death2 mutant shows resistance to Pseudomonas syringae and reveals a role for NPR1 in suppressing hypersensitive cell death. Plant J. 27, 203–211. [DOI] [PubMed] [Google Scholar]
- Rusterucci, C., Aviv, D.H., Holt, B.F., III, Dangl, J.L., and Parker, J.E. (2001). The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13, 2211–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J.A., Neuenschwander, U.H., Willits, M.G., Molina, A., Steiner, H.Y., and Hunt, M.D. (1996). Systemic acquired resistance. Plant Cell 8, 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sedgwick, S.G., and Smerdon, S.J. (1999). The ankyrin repeat: A diversity of interactions on a common structural framework. Trends Biochem. Sci. 24, 311–316. [DOI] [PubMed] [Google Scholar]
- Shapiro, A.D., and Zhang, C. (2001). The role of NDR1 in avirulence gene-directed signaling and control of programmed cell death in Arabidopsis. Plant Physiol. 127, 1089–1101. [PMC free article] [PubMed] [Google Scholar]
- Shirano, Y., Kachroo, P., Shah, J., and Klessig, D.F. (2002). A gain-of-function mutation in an Arabidopsis Toll Interleukin1 Receptor–Nucleotide Binding Site–Leucine-Rich Repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14, 3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker, D.H., Navarre, D.A., Clark, D., del Pozo, O., Martin, G.B., and Klessig, D.F. (2002). The tobacco salicylic acid-binding protein 3 (SABP3) is the chloroplast carbonic anhydrase, which exhibits antioxidant activity and plays a role in the hypersensitive defense response. Proc. Natl. Acad. Sci. USA 99, 11640–11645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz, B.J. (2001). Genetics of plant-pathogen interactions specifying plant disease resistance. Plant Physiol. 125, 73–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summermatter, K., Sticher, L., and Metraux, J.P. (1995). Systemic responses in Arabidopsis thaliana infected and challenged with Pseudomonas syringae pv. syringae. Plant Physiol. 108, 1379–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacker, H., Lu, H., Rate, D.N., and Greenberg, J.T. (2001). A role for salicylic acid and NPR1 in regulating cell growth in Arabidopsis. Plant J. 28, 209–216. [DOI] [PubMed] [Google Scholar]
- Verberne, M.C., Verpoorte, R., Bol, J.F., Mercado-Blanco, J., and Linthorst, H.J. (2000). Overproduction of salicylic acid in plants by bacterial transgenes enhances pathogen resistance. Nat. Biotechnol. 18, 779–783. [DOI] [PubMed] [Google Scholar]
- Weymann, K., Hunt, M., Uknes, S., Neuenschwander, U., Lawton, K., Steiner, H.Y., and Ryals, J. (1995). Suppression and restoration of lesion formation in Arabidopsis lsd mutants. Plant Cell 7, 2013–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth, M.C., Dewdney, J., Wu, G., and Ausubel, F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. [DOI] [PubMed] [Google Scholar]
- Yu, I.C., Parker, J., and Bent, A.F. (1998). Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc. Natl. Acad. Sci. USA 95, 7819–7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Fan, W., Kinkema, M., Li, X., and Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 96, 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, N., Tootle, T.L., Tsui, F., Klessig, D.F., and Glazebrook, J. (1998). PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10, 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]