Abstract
Cryptochromes are photolyase-like blue/UV-A light receptors that regulate various light responses in animals and plants. Arabidopsis cryptochrome 1 (cry1) is the major photoreceptor mediating blue light inhibition of hypocotyl elongation. The initial photochemistry underlying cryptochrome function and regulation remain poorly understood. We report here a study of the blue light–dependent phosphorylation of Arabidopsis cry1. Cry1 is detected primarily as unphosphorylated protein in etiolated seedlings, but it is phosphorylated in plants exposed to blue light. Cry1 phosphorylation increases in response to increased fluence of blue light, whereas the phosphorylated cry1 disappears rapidly when plants are transferred from light to dark. Light-dependent cry1 phosphorylation appears specific to blue light, because little cry1 phosphorylation is detected in seedlings treated with red light or far-red light, and it is largely independent from phytochrome actions, because no phytochrome mutants tested significantly affect cry1 phosphorylation. The Arabidopsis cry1 protein expressed and purified from insect cells is phosphorylated in vitro in a blue light–dependent manner, consistent with cry1 undergoing autophosphorylation. To determine whether cry1 phosphorylation is associated with its function or regulation, we isolated and characterized missense cry1 mutants that express full-length CRY1 apoprotein. Mutant residues are found throughout the CRY1 coding sequence, but none of these inactive cry1 mutant proteins shows blue light–induced phosphorylation. These results demonstrate that blue light–dependent cry1 phosphorylation is closely associated with the function or regulation of the photoreceptor and that the overall structure of cry1 is critical to its phosphorylation.
INTRODUCTION
Plants rely on at least three types of photosensory receptors to regulate growth and development in response to the changing light environment. These photoreceptors include red/far-red light receptor phytochromes (Quail et al., 1995; Nagy and Schafer, 2002), blue/UV-A light receptor phototropins (Briggs and Huala, 1999; Briggs and Christie, 2002), and cryptochromes (Cashmore et al., 1999; Lin, 2002; Lin and Shalitin, 2003). Arabidopsis has at least two cryptochromes, cry1 and cry2, which mediate, among other responses, deetiolation and photoperiodic responses, respectively (Koornneef et al., 1980, 1991; Ahmad and Cashmore, 1993; Guo et al., 1998; Mockler et al., 1999, 2003; El-Din El-Assal et al., 2001). It has been found that cryptochromes interact with phyB and COP1 (Mas et al., 2000; Yang et al., 2000, 2001; Wang et al., 2001) and that cryptochromes mediate the light regulation of gene expression (Somers et al., 1998; Ma et al., 2001; Yanovsky and Kay, 2002).
Light-dependent protein phosphorylation plays important roles in the function of photoreceptors. For example, it has been shown that phytochromes and phototropins are light-regulated protein kinases that catalyze the phosphorylation of their respective photoreceptors and possibly other proteins (Huala et al., 1997; Christie et al., 1998; Yeh and Lagarias, 1998; Fankhauser et al., 1999). It also has been shown that cryptochromes are phosphoproteins in Arabidopsis and mammalian cells (Eide et al., 2002; Shalitin et al., 2002). Arabidopsis cry2 is phosphorylated in seedlings exposed to blue light but not in seedlings exposed to a similar range of light fluence of red or far-red light. Mutations in multiple phytochrome genes showed little effect on the blue light–dependent cry2 phosphorylation (Shalitin et al., 2002). On the other hand, it has been reported that Arabidopsis cry1 was phosphorylated by phyA in vitro and that cry1 phosphorylation in vivo could be induced under red light but suppressed by far-red light (Ahmad et al., 1998). Therefore, it remains unclear whether blue light–dependent phosphorylation is a common light response associated with plant cryptochromes and whether protein phosphorylation is associated with cryptochrome-mediated blue light responses in general. To address these questions, we investigated the light-dependent protein phosphorylation of Arabidopsis cry1.
RESULTS
Light-Dependent Phosphorylation of cry1
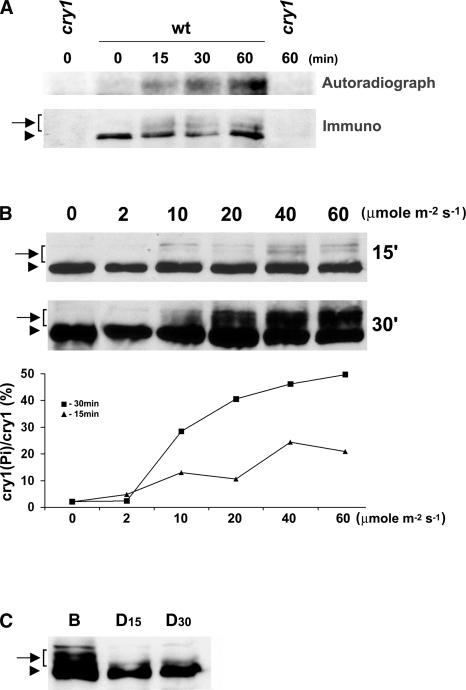
To determine whether Arabidopsis cry1, like cry2, is phosphorylated in response to blue light, we first tested whether cry1 might be metabolically labeled by 32P in dark-grown or light-treated plants. Etiolated seedlings were excised above the roots, placed in test tubes containing [32P]orthophosphate, and incubated in the dark for 3 h. The tissue aliquots then were exposed to blue light; the cry1 protein was immunoprecipitated before or after light treatment and examined using immunoblot analysis and autoradiography. As shown in Figure 1, cry1 was radioactively labeled by 32P in seedlings exposed to blue light for 15 min. By contrast, little 32P labeling of cry1 was detected in etiolated seedlings that were not treated with blue light (Figure 1A). Relatively more radioactively labeled cry1 was immunoprecipitated from seedlings exposed to blue light for an extended time (Figure 1A). These results clearly demonstrate that, like Arabidopsis cry2, cry1 also undergoes blue light–induced protein phosphorylation. As with many phosphoproteins, cry1 phosphorylation results in slow migration on a SDS-PAGE gel (Figure 1B). The immunoblot shows that in addition to a fast-migrating cry1 band detected in both etiolated and light-treated seedlings, at least two or three slow-migrating bands recognized by the anti-CRY1 antibodies were detected in plants treated with blue light (Figure 1B). The slow-migrating bands were sensitive to the phosphatase treatment (see below), indicating that they are phosphorylated forms of cry1. The multiple slow-migrating cry1 bands apparently represent differentially phosphorylated cry1 isoforms, suggesting that the phosphorylation of cry1 occurs at more than one residue.
Figure 1.
Blue Light–Dependent Phosphorylation of cry1.
(A) Five-day-old etiolated seedlings were cut above the roots and incubated with 300 μCi of 32P-H3PO4 in water for 3 h in the dark. The tissue aliquots were exposed to blue light (30 μmol·m−2·s−1) for the time indicated or kept in darkness. The cry1 protein was immunoprecipitated with anti-CRY1 antibodies, separated on a 10% SDS-PAGE gel, blotted to a membrane, and analyzed by autoradiography (Autoradiograph) and then by probing an immunoblot with anti-cry1 antibodies (Immuno).
(B) Immunoblots of samples prepared from wild-type seedlings exposed to blue light for 15 min (15′) or 30 min (30′) at the fluence rate indicated were probed with the anti-CRY1 antibody. The signals of the slow-migrating bands (indicated by the arrow-bracket) are normalized to the fast-migrating band in the same lane (arrowhead), represented as [cry1(Pi)/cry1 (%)], and plotted against the fluence rates (graph at bottom).
(C) Immunoblot showing dephosphorylation of cry1 in the dark. Etiolated seedlings were exposed to blue light (B; 30 μmol·m−2·s−1) for 1 h and then transferred to dark. Samples were prepared before or after seedlings were transferred to darkness for 15 or 30 min (D15 and D30).
Arrows with brackets indicate phosphorylated cry1; arrowheads represent nonphosphorylated cry1. wt, wild type.
We next analyzed the fluence response of cry1 phosphorylation. As shown in Figure 1B, cry1 phosphorylation increased in plants exposed to a higher fluence rate of blue light or exposed to blue light for a longer time. Cry1 phosphorylation was barely detectable in seedlings treated with the lowest fluence of blue light tested (2 μmol·m−2·s−1 for 30 min) (Figure 1B). At the highest fluence rate of blue light tested (60 μmol·m−2·s−1), the level of cry1 phosphorylation was approximately four times greater in seedlings exposed to light for 30 min than that in seedlings exposed to light for 15 min (Figure 1B). In seedlings treated with blue light for 30 min, the level of overall cry1 phosphorylation increased in response to the increased fluence rate of blue light (Figure 1B). These kinetics features of cry1 phosphorylation are in contrast to the bell-shaped fluence response curve of cry2 phosphorylation, for which the relative phosphorylation increases in seedlings exposed to relatively low fluence but decreases at higher fluence (Shalitin et al., 2002). The different fluence responses of cry1 and cry2 phosphorylation are consistent with the fact that cry1 and cry2 are light-stable and light-labile proteins, respectively, and that the phosphorylation of cry2 triggers its degradation (Shalitin et al., 2002).
We found that the total fluence used in the cry1 phosphorylation assay was significantly lower than the total fluence needed to show the gross morphological change resulting from the blue light inhibition of hypocotyl growth. Such a morphological change usually requires exposure of the seedlings to comparable fluence rates of blue light for days instead of minutes. This finding is consistent with cryptochrome phosphorylation being required for its function. We also noted that cry1 was not phosphorylated to completion under the conditions tested. For example, at the highest fluence tested (60 μmol·m−2·s−1 blue light for 30 min), the slow-migrating isoforms that represent phosphorylated cry1 accounted for ∼50% of the total cry1 pool (Figure 1B). It is less likely that the nonphosphorylated cry1 represents an artifact of nonspecific phosphatase activity during homogenization, because a phosphatase inhibitor (NaF) was included in the homogenization buffer (see Methods). The response of cry1 phosphorylation to changing light conditions was examined further in seedlings transferred from blue light to dark (Figure 1C). In this experiment, the phosphorylated cry1 disappeared almost completely within 15 min after plants were transferred to dark. Because cry1 protein is relatively stable in both dark and light conditions, this result indicates that cry1 is likely dephosphorylated by a protein phosphatase.
A recent study showed the involvement of a protein phosphatase, PP7, in the blue light inhibition of hypocotyl elongation (Moller et al., 2003). It would be interesting to determine whether PP7 is involved in cryptochrome dephosphorylation. Because cry1 function depends specifically on blue light, the observation that cry1 phosphorylation is blue light dependent would be consistent with the hypothesis that the unphosphorylated cry1 is inactive but the phosphorylated cry1 is the active photoreceptor. However, alternative interpretations cannot be excluded at present. For example, blue light might activate cry1 via a change of conformation rather than by phosphorylation of the protein, and the active cry1 might trigger phosphorylation and inactivation of the photoreceptor as a negative feedback regulation. Provided that phosphorylated and unphosphorylated cryptochrome molecules have different physiological activities, the presence of both isoforms in plants grown in light would allow a rapid adjustment of photoreceptor activity in response to the changing light environment.
Mutations of Phytochromes Have Little Effect on cry1 Phosphorylation
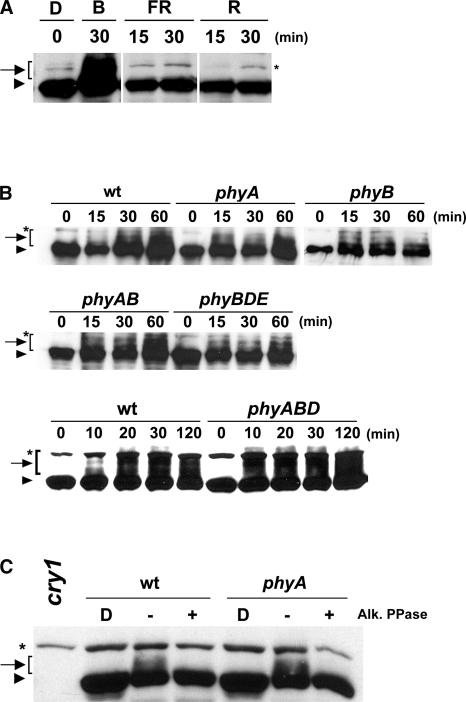
To further investigate whether cry1 phosphorylation is specific to blue light, we analyzed cry1 protein in seedlings exposed to different wavelengths of light. Figure 2 shows that cry1 phosphorylation was detected in seedlings exposed to blue light for 30 min, but little phosphorylation of cry1 was detected in seedlings exposed to a similar fluence range of red light, far-red light, or dark (Figure 2A). To determine whether phytochromes affect blue light–dependent cry1 phosphorylation, we analyzed cry1 phosphorylation in phytochrome mutants. As shown in Figure 2B, cry1 phosphorylation clearly was detected in the phyA and phyB monogenic mutants as well as in those mutant lines that are impaired in multiple phytochrome genes, including phyAB, phyBDE, and phyABD (Franklin et al., 2003). The slow-migrating cry1 isoforms in wild-type and phyA mutant seedlings disappeared completely after phosphatase treatment (Figure 2C). This result confirmed that the slow-migrating bands recognized by the anti-CRY1 antibody were phosphorylated cry1 and that the phyA mutation did not abolish the blue light–induced cry1 phosphorylation.
Figure 2.
Blue Light Specificity and the Effect of Phytochrome Mutations on cry1 Phosphorylation.
(A) Immunoblot showing cry1 phosphorylation in seedlings exposed to blue light (B; 30 μmol·m−2·s−1), far-red light (FR; 50 μmol·m−2·s−1), or red light (R; 50 μmol·m−2·s−1) for the time indicated or kept in the dark (D).
(B) Immunoblot showing cry1 phosphorylation in samples prepared from the wild type or from phyA, phyB, phyA phyB, phyBDE, and phyABD mutants. Seedlings were grown in the dark and then exposed to blue light (20 μmol·m−2·s−1) for the time indicated. wt, wild type.
(C) Immunoblot showing cry1 phosphorylation in samples prepared from wild-type or phyA seedlings grown in the dark and then exposed to blue light (30 μmol·m−2·s−1) for 1 h. Protein extracts from light-treated seedlings were incubated with alkaline phosphatase (Alk. PPase) at 30°C for 30 min (+) or analyzed without phosphatase treatment (−).
Asterisks indicated a band nonspecifically recognized by the antibody.
The findings that cryptochromes are phosphorylated in blue light but not in red or far-red light (Figure 2A) and that none of the phytochrome mutants tested showed an easily discernible effect in the phosphorylation of cry1 (Figures 2B and 2C) suggest that no single phytochrome tested is solely responsible for the blue light–induced phosphorylation of cry1. However, because it is difficult to precisely quantify each of the multiple bands of phosphorylated cry1, a minor or quantitative effect of phytochrome mutations on the blue light–dependent phosphorylation of cry1 cannot be excluded. For example, it appears that cry1 phosphorylation may occur slightly faster in the phytochrome mutants tested than in the wild-type controls (Figure 2B), although this remains to be examined more carefully.
Cry1 Phosphorylation Is Closely Associated with Its Function or Regulation
The structure-function relationship is critical to our understanding of the molecular mechanisms underlying cryptochrome phosphorylation and function. Although we previously reported cry2 phosphorylation in Arabidopsis (Shalitin et al., 2002), a direct test of the structure-function relationship of cry2 phosphorylation has been difficult, because few missense cry2 mutants are available (Koornneef et al., 1991; Guo et al., 1998). Isolation of a large number of cry2 mutations has been difficult (H. Yang and C. Lin, unpublished data), because the late-flowering phenotype is associated not only with cry2 but also with mutations in other flowering-time genes and the long-hypocotyl phenotype of cry2 is not robust enough for efficient genetic screening (Guo et al., 1998; Lin et al., 1998). By contrast, a large number of cry1 mutants in Arabidopsis were isolated that showed long hypocotyls when grown in continuous blue light (Koornneef et al., 1980; Liscum and Hangarter, 1991; Ahmad et al., 1995; Bagnall et al., 1996; Bruggemann et al., 1996).
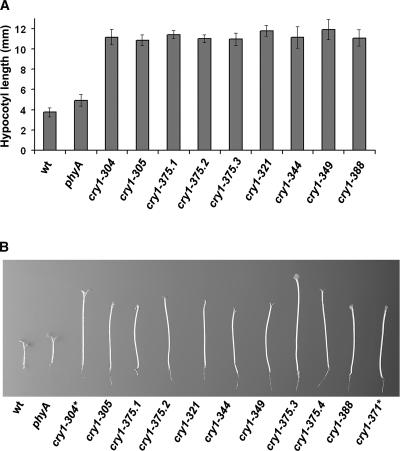
To investigate the role of cry1 phosphorylation, we isolated and characterized missense cry1 mutants that showed little blue light inhibition of hypocotyl elongation but still expressed apparently full-length CRY1 apoprotein. It was expected that these missense mutations would allow the identification of the amino acid residues critical for cry1 function and/or phosphorylation. Ethyl methanesulfonate–mutagenized seeds of a phyA mutant line were screened to isolate individuals with long hypocotyls when grown in continuous blue light (Figure 3). We then used immunoblot analysis with anti-CRY1 antibodies to select those mutants that expressed apparently full-length CRY1 apoprotein. Most cry1 mutations isolated expressed little or truncated CRY1 protein (data not shown). However, nine lines were found to express apparently full-length CRY1 apoprotein at a level indistinguishable from that of the wild type, and they were studied further (Figures 3 and 4). As shown in Figure 3, the parental phyA mutant line was slightly taller than the wild type when grown under continuous blue light, but the newly isolated cry1 lines that express full-length CRY1 apoprotein grew significantly taller than the phyA parent or the wild type. These newly isolated cry1 mutants showed very similar phenotypes to the reference cry1-304 allele or a cry1 phyA double mutant that expresses no CRY1 apoprotein (Mockler et al., 1999) (Figure 3B, cry1-371), and they all contained mutations in the CRY1 gene (see below).
Figure 3.
Long-Hypocotyl Phenotype of the Newly Isolated cry1 Mutants.
(A) Seedlings were grown in continuous blue light (25 μmol·m−2·s−1) for 5 days, and hypocotyl lengths were measured. wt, wild type.
(B) Representative seedlings of parental control (phyA), a cry1 reference allele (cry1-304), and nine newly isolated cry1 alleles are shown. cry1-304 and cry1-371 express no CRY1 apoprotein, whereas the other alleles express full-length CRY1 apoprotein.
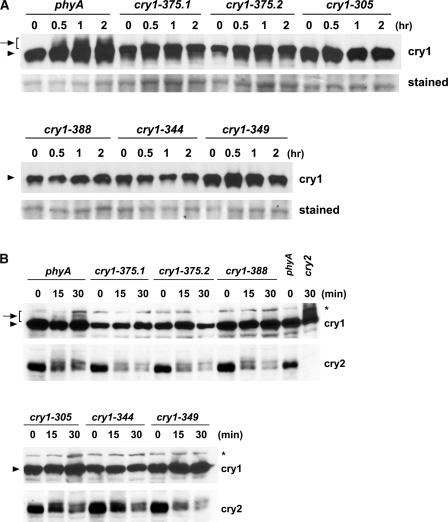
Figure 4.
Immunoblots Showing the Lack of cry1 Phosphorylation and Normal cry2 Phosphorylation in Different cry1 Mutant Alleles.
(A) Samples were prepared from etiolated seedlings exposed to 25 μmol·m−2·s−1 blue light for the time indicated or from samples kept in the dark (0 h), and immunoblots were probed with anti-CRY1 antibodies (cry1). For a loading control, the membrane was stained with Ponceau red, and a portion of the stained blot showing unspecified proteins is included (stained).
(B) Samples were prepared from etiolated seedlings exposed to 7 μmol·m−2·s−1 blue light for the time indicated or from samples kept in the dark (0 min). The immunoblot was probed first with anti-CRY2 antibody (cry2), stripped, and reprobed with anti-CRY1 antibody (cry1).
We tested whether these cry1 mutations affected cry1 phosphorylation. In contrast to the phyA mutant that exhibited apparently normal cry1 phosphorylation in response to blue light of both high fluence (Figure 4A) and low fluence (Figure 4B), none of the nine cry1 alleles showed easily detectable cry1 phosphorylation (Figure 4). These results suggest a positive correlation between the light-dependent phosphorylation of cry1 and its activity in mediating the blue light inhibition of hypocotyl elongation. Because both cry1 and cry2 are phosphorylated in response to blue light and their functions are partially redundant (Mockler et al., 1999), we tested if the two cryptochromes would affect the phosphorylation of each other. Figure 4B shows that the cry2 mutation had no apparent effect on cry1 phosphorylation. Similarly, cry2 phosphorylation clearly was detectable in all of the cry1 mutant alleles tested at the relatively low fluence range that is optimal to show cry2 phosphorylation (Shalitin et al., 2002) (Figure 4B). It appears that cry1 and cry2 are phosphorylated largely independently in response to blue light.
Mutations That Impair cry1 Phosphorylation Are Found throughout the CRY1 Apoprotein
CRY1 is a 681-residue protein comprising an N-terminal photolyase-related domain (∼500 amino acids) and a C-terminal domain (∼180 amino acids). The CRY1 C-terminal domain is unrelated to the DNA photolyase, but it contains a DAS motif that is rich in Ser and is well conserved in cryptochromes throughout the plant kingdom (Lin, 2002). To determine which residues of CRY1 might be important for its phosphorylation and function, we sequenced the CRY1 gene in all nine of the apparently missense cry1 mutants. As expected, mutations were detected in the CRY1 coding sequence of every cry1 allele tested (Figure 5). The mutated residues are distributed throughout the sequence of CRY1 apoprotein (Figure 5). Among the nine independent cry1 lines sequenced, seven contain mutations in the photolyase-related domain and two have mutations in the C-terminal domain (cry1-321 and cry1-349); four contain the same mutation, G347R (cry1-375.1, -375.2, -375.3, and -375.4). The sequence corresponding to Gly-347 apparently is prone to mutagenesis, because G347R also was reported in two previously isolated cry1 alleles, hy4-15 and hy4-16 (Ahmad et al., 1995). One mutation results in the conservative change R536K in the C-terminal domain (cry1-321). Some of the cry1 mutations (cry1-375 and cry1-344) may affect the functionality common to photolyase and cryptochromes, such as chromophore binding or energy transfer, because these mutant residues are within or adjacent to a motif (WRWG) that is well conserved among photolyases and cryptochromes (Ahmad and Cashmore, 1993; Kanai et al., 1997).
Figure 5.
Lesions of the cry1 Mutants.
Sequences of the cry1 mutants were aligned with the deduced amino acid sequence of CRY1 of the Columbia accession (WT; At4g08920). Residue numbers of CRY1 are indicated above the sequence. White and gray boxes highlight mutant residues of the different alleles.
Blue Light–Dependent cry1 Phosphorylation in Vitro
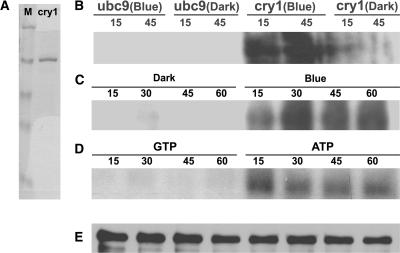
In an attempt to establish an in vitro cryptochrome phosphorylation assay, we found that cry1 was phosphorylated in vitro without the addition of a protein kinase. In these experiments, the His-tagged cry1 protein was expressed using the baculovirus expression system in Sf9 insect cells and purified using nickel affinity chromatography (Lin et al., 1995) (Figure 6A). The cry1 protein was incubated with γ-32P-ATP in vitro in the presence or absence of blue light, fractionated by SDS-PAGE, and examined using autoradiography (Figures 6B and 6D) or immunoblot analysis (Figure 6E). Figure 6 shows that although little cry1 phosphorylation was detected in the dark, cry1 phosphorylation clearly was detected in the reactions exposed to blue light (Figure 6B). A control protein (ubc9) expressed similarly in the baculovirus system and purified using nickel affinity chromatography was not phosphorylated, regardless of the blue light treatment (Figure 6B). The cry1 phosphorylation was dependent on ATP, because cry1 was readily labeled by γ-32P-ATP but not by γ-32P-GTP (Figure 6D). Light-dependent in vitro phosphorylation also has been reported for both Arabidopsis cry1 and human cry1 (Bouly et al., 2003). These results are consistent with the hypothesis that cryptochromes may possess blue light–dependent autophosphorylation activity. However, this hypothesis needs to be tested further.
Figure 6.
Blue Light–Dependent Phosphorylation of cry1 in Vitro.
(A) The cry1 protein purified from insect cells (cry1; 1 μg) and the molecular mass marker proteins (M; 1 μg per band) were fractionated on a SDS-PAGE gel and stained with Coomassie blue.
(B) to (D) Autoradiographs showing cry1 phosphorylation in vitro. Arabidopsis cry1 ([B] and [C]) and the control protein ubc9 (B) were incubated with γ-32P-ATP, and the reaction products were kept in the dark or exposed to blue light (33 μmol·m−2·s−1) for the time indicated (15 to 60 min). cry1 phosphorylation reaction products incubated with γ-32P-ATP or γ-32P-GTP under blue light (33 μmol·m−2·s−1) for the time indicated (15 to 60 min) are shown in (D).
(E) The immunoblot of the same membrane used in (C) was probed with the anti-CRY1 antibody.
DISCUSSION
We have shown here that Arabidopsis cry1 is phosphorylated in response to blue light under both in vivo and in vitro conditions. In Arabidopsis seedlings exposed to blue light, multiple isoforms of phosphorylated cry1 were detected, suggesting that cry1 is phosphorylated at multiple residues. The relative amount of all of the phosphorylated cry1 isoforms increased in response to increased fluence of blue light, whereas cry1 was dephosphorylated rapidly in the absence of light. The cry1 phosphorylation seems specific to blue light because little cry1 phosphorylation was detected in seedlings exposed to similar fluence ranges of red light or far-red light. Moreover, the blue light–dependent phosphorylation of cry1 was largely independent of phytochromes, and the phosphorylation of cry1 appears not to be cry2 dependent and vice versa. Together, the previous study of cry2 phosphorylation (Shalitin et al., 2002) and these results demonstrate that blue light–dependent phosphorylation is a general light reaction associated with plant cryptochromes.
A question raised by the blue light–dependent cry1 phosphorylation concerns the role(s) of this protein modification. To address this question, we isolated and characterized missense cry1 mutants that showed little blue light inhibition of hypocotyl elongation but still expressed the full-length CRY1 apoproteins. Missense mutations were found in both the N-terminal chromophore binding domain and the C-terminal domain of these cry1 mutant proteins. Importantly, none of the cry1 mutant proteins showed the blue light–dependent phosphorylation. It also is interesting that none except one of the missense cry1 mutations identified affect phosphorylatable residues, such as Ser, Thr, or Tyr. Similarly, no mutation affecting phosphorylatable residues of CRY1 was isolated from a previous screen (Ahmad et al., 1995). These results are unlikely to be attributable to the method of ethyl methanesulfonate mutagenesis, which often results in C/G to A/T transitions affecting several codons for Ser or Thr (data not shown). Moreover, it appears that the mutagenesis is at least partially saturated for cry1, because several cry1 mutants isolated in this study and in a previous study (Ahmad et al., 1995) contain the identical mutation (G347R). The observations that none of the cry1 mutations isolated showed phosphorylation but few affected phosphorylatable residues may be explained by the notions that cry1 is phosphorylated at multiple residues and that cry1 phosphorylation may be required for its function. This is because if cry1 activity is dependent on its phosphorylation at multiple residues, a mutation at only one of the phosphorylatable residues may not eliminate its activity and therefore may not be isolated as a strong allele from our function-based genetic screen. On the other hand, a missense mutation that completely abolishes cry1 phosphorylation would inevitably eliminate its activity, regardless of the mutation occurs at a phosphorylatable residue or not.
Which enzyme catalyzes the cryptochrome phosphorylation is another interesting question. The observation that purified cry1 can be phosphorylated in vitro suggests an autophosphorylation activity of the photoreceptor, although the activities of an insect kinase copurified with cry1 cannot be excluded completely. Given that plant and animal cryptochromes share relatively low sequence similarity (Todo, 1999) and the light-dependent nature of the in vitro cry1 phosphorylation (Figure 6) (Bouly et al., 2003), the “contamination” hypothesis has to make an intriguing, albeit not impossible, assumption that the presumed insect kinase copurified with cry1 not only would have recognized Arabidopsis cry1 but also would have distinguished its conformational change in response to light. On the other hand, an obvious difficulty with the “autophosphorylation” hypothesis is that cryptochromes generally lack a well-recognized protein kinase motif. However, a careful examination of the CRY1 sequence revealed a number of putative nucleotide binding P-loop (Saraste et al., 1990; Sinha et al., 1999) motifs (data not shown), and at least one of the putative P-loops is well conserved between Arabidopsis and fern cryptochromes (Imaizumi et al., 2000). Additional investigations are needed to resolve these competing hypotheses and to fully understand the mechanism and physiological role(s) of blue light–dependent cryptochrome phosphorylation.
METHODS
Plant Materials
Arabidopsis thaliana accession Columbia was used throughout this study. Lights and filters used were as described (Mockler et al., 2003). Mutant lines, except for the newly isolated cry1, were as described previously (Reed et al., 1994; Devlin et al., 1999; Mockler et al., 1999, 2003; Shalitin et al., 2002; Franklin et al., 2003; Halliday et al., 2003). The two cry2 mutant alleles in Col background, which were originally called cry2-1 and cry2-2 (Guo et al., 1998), are renamed as cry2-4 and cry2-5, respectively. Seeds were sown on compound soil, kept in the dark at 4°C for 3 days, treated with white light for 8 h to promote germination, and grown in dark or light conditions as indicated in the figure legends. The cry1 mutants were isolated from the phyA (phyA-211) mutant background (Reed et al., 1994). The phyA seeds were treated with ethyl methanesulfonate (0.3%) for 11 h, rinsed thoroughly, and grown as 30 subpopulations to prepare independent pools of M2 seeds. M2 seeds (∼1,000,000) were sown and grown in continuous blue light (25 μmol·m−2·s−1) for 5 days, and individuals that showed long hypocotyls were isolated from independent M2 pools. Putative strong cry1 mutants that expressed CRY1 apoprotein with a molecular mass similar to that of wild-type CRY1 were selected for further study. The cry1 mutations were confirmed and the mutant residues identified by direct DNA sequencing of the cry1 cDNAs amplified from individual lines using reverse transcriptase–mediated PCR. Four independently isolated lines, cry1-315, -320, -375, and -376, later were found to contain the same mutations and were renamed cry1-375.1, -375.2, -375.3, and -375.4, respectively.
Immunoblot Analysis and Immunoprecipitation Assays
Immunoprecipitation was performed as described previously with minor modifications (Shalitin et al., 2002). Etiolated seedlings were excised from the hypocotyl base under a dim red safelight and homogenized in ice-cold immunoprecipitation buffer (20 mM Tris-HCl, pH 8, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.2% Triton X-100, 10 μM NaF, and 1 mM phenylmethylsulfonyl fluoride) and complete protease inhibitor cocktail (Roche, Mannheim, Germany). Extracts were passed through 0.2-mm filters to remove tissue debris. The filtrates were incubated with anti-CRY1 antiserum (1:1000) on ice for 1 h. Protein A–Sepharose beads (Sigma) were added (1:60) to the mixture, which then was incubated on ice for 1 h. Immunoprecipitated samples were washed three times with ice-cold washing buffer (50 mM Tris-HCl, pH 8, 140 mM NaCl, 1 mM EDTA, and 0.1% Triton X-100). The bound proteins were denatured by mixing with SDS-PAGE sample buffer and boiling for 5 min.
Samples were fractionated on a 10% SDS-PAGE minigel for 4 h at constant current (18 mA, up to 110 V), and the protein bands were blotted to a nitrocellulose membrane using a semidry electrophoresis apparatus (Amersham Bioscience, Piscataway, NJ) at a constant voltage (6 V) for 12 h. Immunoblots were probed with the anti-CRY1 antiserum (1:5000 dilution in PBST [7 mM Na2HPO4, 3 mM NaH2PO4, 130 mM NaCl, and 0.3% Tween 20]), washed in PBST three times, reacted with goat anti-rabbit IgG conjugated with horseradish peroxidase (1:20,000; Amersham), washed, and exposed to x-ray film using the enhanced chemiluminescence (ECL) method according to the manufacturer's instructions (Amersham). The same blot sometimes was stripped and reprobed with different antibodies (Lin et al., 1998). Protein signals were digitized by scanning the ECL films and quantified using NIH Image software. The exposure time for ECL of different immunoblots was not controlled precisely, so the signal intensities from different immunoblots are not directly comparable. Anti-CRY1 and anti-CRY2 antibodies were described previously (Lin et al., 1996, 1998).
Phosphorylation and Dephosphorylation Assays
In vivo labeling analysis was performed as described previously (Shalitin et al., 2002) with minor modifications. Five-day-old etiolated seedlings (∼50) were cut above the hypocotyl base and incubated with 32P (300 μCi of H3PO4 [ICN, Costa Mesa, CA] in deionized water) for 3 h in the dark at room temperature. Excess 32P was removed by rinsing with water. The tissue aliquots were either exposed to light or kept in the dark before harvesting for immunoprecipitation or immunoblot analyses. For alkaline phosphatase treatment, tissues were homogenized in calf intestinal alkaline phosphatase buffer (100 mM NaCl, 50 mM Tris-HCl, pH 7.9, 10 mM MgCl2, and 1 mM DTT) and divided into two aliquots. Alkaline phosphatase (New England Biolabs, Beverly, MA) was added to one of the two aliquots (0.25 unit/μL) and incubated at 30°C for 30 min. The dephosphorylation reactions were stopped by adding SDS-PAGE sample buffer and boiling for 5 min. The samples were examined by immunoblot analysis as described above.
The in vitro protein phosphorylation experiments were performed essentially as reported previously (Yeh and Lagarias, 1998) with minor modifications. Arabidopsis cry1 was expressed and purified using the baculovirus expression system as described (Lin et al., 1995). The control protein ubc9 (yeast E2 ubiquitin-like conjugating protein) was expressed and purified similarly. Cry1 or ubc9 (1 μg/reaction) was incubated with 100 μM γ-32P-ATP or γ-32P-GTP (2000 to 5000 cpm/pmol) in the phosphorylation buffer (25 mM Tris-HCl, pH 7.5, 0.2 mM EDTA, 5 mM MgCl2, and 4 mM 2-mercaptoethanol) for 15 to 60 min. The phosphorylation reactions were prepared under dim red light, moved to dark or exposed to blue light, and stopped by adding 4× SDS-PAGE sample buffer and boiling for 5 min. The proteins were separated on 10% SDS-PAGE gels and transferred to nitrocellulose membranes for autoradiography and immunoblot analyses.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Chentao Lin, clin@mcdb.ucla.edu.
Acknowledgments
We thank Garry Whitelam and Keara Franklin for providing the multiple phytochrome mutant lines before publication. This work was supported in part by research grants from the National Institutes of Health (GM56265 to C.L.), the National Science Foundation (MCB-0091391 to C.L.), and the University of California, Los Angeles-FGP. T.M. was supported in part by the National Science Foundation Integrative Graduate Education and Research Traineeship (DGE-9987641).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.013011.
References
- Ahmad, M., and Cashmore, A.R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366, 162–166. [DOI] [PubMed] [Google Scholar]
- Ahmad, M., Jarillo, J.A., Smirnova, O., and Cashmore, A.R. (1998). The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol. Cell 1, 939–948. [DOI] [PubMed] [Google Scholar]
- Ahmad, M., Lin, C., and Cashmore, A.R. (1995). Mutations throughout an Arabidopsis blue-light photoreceptor impair blue-light-responsive anthocyanin accumulation and inhibition of hypocotyl elongation. Plant J. 8, 653–658. [DOI] [PubMed] [Google Scholar]
- Bagnall, D.J., King, R.W., and Hangarter, R.P. (1996). Blue-light promotion of flowering is absent in hy4 mutants of Arabidopsis. Planta 200, 278–280. [DOI] [PubMed] [Google Scholar]
- Bouly, J.P., Giovani, B., Djamei, A., Mueller, M., Zeugner, A., Dudkin, E.A., Batschauer, A., and Ahmad, M. (2003). Novel ATP-binding and autophosphorylation activity associated with Arabidopsis and human cryptochrome-1. Eur. J. Biochem. 270, 2921–2928. [DOI] [PubMed] [Google Scholar]
- Briggs, W.R., and Christie, J.M. (2002). Phototropins 1 and 2: Versatile plant blue-light receptors. Trends Plant Sci. 7, 204–210. [DOI] [PubMed] [Google Scholar]
- Briggs, W.R., and Huala, E. (1999). Blue-light photoreceptors in higher plants. Annu. Rev. Cell Dev. Biol. 15, 33–62. [DOI] [PubMed] [Google Scholar]
- Bruggemann, E., Handwerger, K., Essex, C., and Storz, G. (1996). Analysis of fast neutron-generated mutants at the Arabidopsis thaliana locus. Plant J. 10, 755–760. [DOI] [PubMed] [Google Scholar]
- Cashmore, A.R., Jarillo, J.A., Wu, Y.J., and Liu, D. (1999). Cryptochromes: Blue light receptors for plants and animals. Science 284, 760–765. [DOI] [PubMed] [Google Scholar]
- Christie, J.M., Reymond, P., Powell, G.K., Bernasconi, P., Raibekas, A.A., Liscum, E., and Briggs, W.R. (1998). Arabidopsis NPH1: A flavoprotein with the properties of a photoreceptor for phototropism. Science 282, 1698–1701. [DOI] [PubMed] [Google Scholar]
- Devlin, P.F., Robson, P.R., Patel, S.R., Goosey, L., Sharrock, R.A., and Whitelam, G.C. (1999). Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation growth and flowering time. Plant Physiol. 119, 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide, E.J., Vielhaber, E.L., Hinz, W.A., and Virshup, D.M. (2002). The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Iɛ. J. Biol. Chem. 277, 17248–17254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Din El-Assal, S., Alonso-Blanco, C., Peeters, A.J., Raz, V., and Koornneef, M. (2001). A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat. Genet. 29, 435–440. [DOI] [PubMed] [Google Scholar]
- Fankhauser, C., Yeh, K.C., Lagarias, J.C., Zhang, H., Elich, T.D., and Chory, J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284, 1539–1541. [DOI] [PubMed] [Google Scholar]
- Franklin, K.A., Praekelt, U., Stoddart, W.M., Billingham, O.E., Halliday, K.J., and Whitelam, G.C. (2003). Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol. 131, 1340–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Yang, H., Mockler, T.C., and Lin, C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279, 1360–1363. [DOI] [PubMed] [Google Scholar]
- Halliday, K.J., Salter, M.G., Thingnaes, E., and Whitelam, G.C. (2003). Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J. 33, 875–885. [DOI] [PubMed] [Google Scholar]
- Huala, E., Oeller, P.W., Liscum, E., Han, I.S., Larsen, E., and Briggs, W.R. (1997). Arabidopsis NPH1: A protein kinase with a putative redox-sensing domain. Science 278, 2120–2123. [DOI] [PubMed] [Google Scholar]
- Imaizumi, T., Kanegae, T., and Wada, M. (2000). Cryptochrome nucleocytoplasmic distribution and gene expression are regulated by light quality in the fern Adiantum capillus-veneris. Plant Cell 12, 81–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai, S., Kikuno, R., Toh, H., Ryo, H., and Todo, T. (1997). Molecular evolution of the photolyase-blue-light photoreceptor family. J. Mol. Evol. 45, 535–548. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Hanhart, C.J., and van der Veen, J.H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229, 57–66. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Rolff, E., and Spruit, C.J.P. (1980). Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z. Pflanzenphysiol. Bd. 100, 147–160. [Google Scholar]
- Lin, C. (2002). Blue light receptors and signal transduction. Plant Cell 14 (suppl.), S207.–S225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C., Ahmad, M., and Cashmore, A.R. (1996). Arabidopsis cryptochrome 1 is a soluble protein mediating blue light-dependent regulation of plant growth and development. Plant J. 10, 893–902. [DOI] [PubMed] [Google Scholar]
- Lin, C., Robertson, D.E., Ahmad, M., Raibekas, A.A., Jorns, M.S., Dutton, P.L., and Cashmore, A.R. (1995). Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269, 968–970. [DOI] [PubMed] [Google Scholar]
- Lin, C., and Shalitin, D. (2003). Cryptochrome structure and signal transduction. Annu. Rev. Plant Biol. 54, 469–496. [DOI] [PubMed] [Google Scholar]
- Lin, C., Yang, H., Guo, H., Mockler, T., Chen, J., and Cashmore, A.R. (1998). Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc. Natl. Acad. Sci. USA 95, 2686–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum, E., and Hangarter, R. (1991). Arabidopsis mutants lacking blue light-dependent inhibition of hypocotyl elongation. Plant Cell 3, 685–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., Li, J., Qu, L., Hager, J., Chen, Z., Zhao, H., and Deng, X.W. (2001). Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13, 2589–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas, P., Devlin, P.F., Panda, S., and Kay, S.A. (2000). Functional interaction of phytochrome B and cryptochrome 2. Nature 408, 207–211. [DOI] [PubMed] [Google Scholar]
- Mockler, T., Yang, H., Yu, X., Parikh, D., Cheng, Y.C., Dolan, S., and Lin, C. (2003). Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc. Natl. Acad. Sci. USA 100, 2140–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler, T.C., Guo, H., Yang, H., Duong, H., and Lin, C. (1999). Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126, 2073–2082. [DOI] [PubMed] [Google Scholar]
- Moller, S.G., Kim, Y.S., Kunkel, T., and Chua, N.H. (2003). PP7 is a positive regulator of blue light signaling in Arabidopsis. Plant Cell 15, 1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, F., and Schafer, E. (2002). Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 53, 329–355. [DOI] [PubMed] [Google Scholar]
- Quail, P.H., Boylan, M.T., Parks, B.M., Short, T.W., Xu, Y., and Wagner, D. (1995). Phytochromes: Photosensory perception and signal transduction. Science 268, 675–680. [DOI] [PubMed] [Google Scholar]
- Reed, J.W., Nagatani, A., Elich, T.D., Fagan, M., and Chory, J. (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104, 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste, M., Sibbald, P.R., and Wittinghofer, A. (1990). The P-loop: A common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15, 430–434. [DOI] [PubMed] [Google Scholar]
- Shalitin, D., Yang, H., Mockler, T.C., Maymon, M., Guo, H., Whitelam, G.C., and Lin, C. (2002). Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417, 763–767. [DOI] [PubMed] [Google Scholar]
- Sinha, K.M., Ghosh, M., Das, I., and Datta, A.K. (1999). Molecular cloning and expression of adenosine kinase from Leishmania donovani: Identification of unconventional P-loop motif. Biochem. J. 339, 667–673. [PMC free article] [PubMed] [Google Scholar]
- Somers, D.E., Devlin, P.F., and Kay, S.A. (1998). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282, 1488–1490. [DOI] [PubMed] [Google Scholar]
- Todo, T. (1999). Functional diversity of the DNA photolyase/blue light receptor family. Mutat. Res. 434, 89–97. [DOI] [PubMed] [Google Scholar]
- Wang, H., Ma, L.G., Li, J.M., Zhao, H.Y., and Deng, X.W. (2001). Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294, 154–158. [DOI] [PubMed] [Google Scholar]
- Yang, H.Q., Tang, R.H., and Cashmore, A.R. (2001). The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13, 2573–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H.-Q., Wu, Y.-J., Tang, R.-H., Liu, D., Liu, Y., and Cashmore, A.R. (2000). The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103, 815–827. [DOI] [PubMed] [Google Scholar]
- Yanovsky, M.J., and Kay, S.A. (2002). Molecular basis of seasonal time measurement in Arabidopsis. Nature 419, 308–312. [DOI] [PubMed] [Google Scholar]
- Yeh, K.C., and Lagarias, J.C. (1998). Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase an-cestry. Proc. Natl. Acad. Sci. USA 95, 13976–13981. [DOI] [PMC free article] [PubMed] [Google Scholar]