Abstract
Using fluorescent differential display, we identified, from ∼8000 displayed bands, a DNA fragment showing rapid induction in response to red light irradiation. This EARLY-PHYTOCHROME-RESPONSIVE1 gene (EPR1) encodes a novel nucleus-localized MYB protein harboring a single MYB domain that is highly similar to the circadian oscillator proteins CCA1 and LHY. EPR1 is regulated by both phytochrome A and phytochrome B, and the red-light induction of EPR1 is not inhibited by cycloheximide, demonstrating that EPR1 represents a primary phytochrome-responsive gene. Our results show that EPR1 overexpression results in enhanced far-red light–induced cotyledon opening and delayed flowering. In wild-type Arabidopsis plants grown in continuous light, the EPR1 transcript exhibits circadian rhythmicity similar to that of CCA1 and LHY. Moreover, EPR1 suppresses its own expression, suggesting that this protein is part of a regulatory feedback loop. Constitutive expression of CCA1 and LHY results in the loss of EPR1 rhythmicity, whereas increased levels of EPR1 have no effect on the central oscillator. We propose that EPR1 is a component of a slave oscillator that contributes to the refinement of output pathways, ultimately mediating the correct oscillatory behavior of target genes.
INTRODUCTION
Light is one of the most influential factors regulating various growth and developmental processes during the life cycle of plants. To cope with changes in the light environment, plants make effective use of several different photoreceptors, including phytochrome and cryptochrome. Phytochromes have been characterized extensively and consist of a small gene family of five distinct genes (PHYA to PHYE) in Arabidopsis (Sharrock and Quail, 1989; Clack et al., 1994) and three genes in rice (Kay et al., 1989; Dehesh et al., 1991; Tahir et al., 1998). Each phytochrome member executes specific but overlapping functions in a broad range of light-dependent responses in higher plants (Furuya and Kim, 2000; Fankhauser, 2001). Phytochrome A (phyA) mediates not only the far-red (FR) light–dependent high irradiance response (Nagatani et al., 1993; Parks and Quail, 1993; Whitelam et al., 1993) but also the photoirreversible, very low fluence response (Shinomura et al., 1996). By contrast, phytochrome B (phyB) regulates the red (R) light/FR light photoreversible response (Shinomura et al., 1996) in addition to continuous R light effects (Nagatani et al., 1991; Reed et al., 1993).
In addition to monitoring the light environment, plants also must anticipate light and dark periodic alterations. For this function, plants make use of their endogenous circadian clock to time molecular and physiological responses. Microarray analysis has shown that ∼6% of Arabidopsis genes are controlled by the clock at the transcriptional level (Harmer et al., 2000), demonstrating the complexity of the process.
There is an intimate relationship between light perception by photoreceptors and the circadian clock (Devlin and Kay, 2001). The circadian clock can be divided into three main elements: the input pathway, in which phytochrome and cryptochrome act as input photoreceptors (Somers et al., 1998), which entrains the clock to the correct phase; a central oscillator, which generates periods of ∼24 h; and the output pathway, which decodes this information into physiological rhythmicity (Young and Kay, 2001). Several protein components of the circadian central oscillator have been isolated, including TOC1, CCA1, and LHY (Millar et al., 1995; Schaffer et al., 1998; Wang and Tobin, 1998; Alabadi et al., 2001). CCA1 was identified initially as an early-phytochrome-responsive gene encoding a MYB-related transcription factor shown to bind to the promoter region of the Lhcb gene and by doing so regulating Lhcb expression (Wang et al., 1997). CCA1 and its homolog LHY were shown subsequently to be components of the central oscillator, acting redundantly in mediating the circadian rhythmicity of target genes (Schaffer et al., 1998; Wang and Tobin, 1998; Mas et al., 2003). In contrast to LHY and CCA1, which show peak levels at dawn, TOC1 peaks at dusk and is controlled by LHY and CCA1 by direct binding to the TOC1 promoter (Alabadi et al., 2001). Conversely, TOC1 has a positive effect on LHY and CCA1 gene expression, contributing to the dawn-induced expression peak. This reciprocal control mechanism forms the basis of the central oscillator. A more direct link between photoperception and circadian gene expression also has been demonstrated, by which CCA1 expression itself may be controlled by a direct binding of the PHYTOCHROME-INTERACTING FACTOR3/phytochrome complex to the CCA1 promoter region in a light-dependent manner (Ni et al., 1998, 1999; Martinez-Garcia et al., 2000). Recently, it was shown that the SRR1 gene mediates phyB signaling and is needed for circadian clock function, again emphasizing the close relationship between light perception and the circadian clock (Staiger et al., 2003a).
The identification of early-phytochrome-responsive genes, such as CCA1 and LHY, is important in dissecting the interaction between light perception and the circadian clock. Although several components of the Arabidopsis central oscillator have been identified, it is likely that the central circadian oscillator comprises several additional levels of regulation to fine-tune output pathways (Carre and Kim, 2002).
We applied the fluorescent differential display (FDD) technique to screen for early-phytochrome-responsive genes whose expression is altered rapidly after a short R light pulse in etiolated Arabidopsis seedlings. FDD is a highly sensitive mRNA fingerprinting technique that is suitable for detecting rare transcripts in addition to minute variations in transcript levels (Kuno et al., 2000). Here, we report the isolation and characterization of EPR1 (EARLY-PHYTOCHROME-RESPONSIVE1), which encodes a novel MYB protein that is highly similar to CCA1 and LHY (Wang et al., 1997; Schaffer et al., 1998). EPR1 regulates its own circadian expression pattern and is influenced by the central oscillator. Our studies suggest that EPR1 is part of a slave oscillator that contributes to and fine-tunes circadian output pathways.
RESULTS
FDD Screening for Early-Phytochrome-Responsive Genes in Etiolated Arabidopsis Seedlings
In an effort to identify early-phytochrome-responsive genes, we performed FDD analysis of Arabidopsis seedlings in response to short exposures of R light. Six-day-old etiolated wild-type seedlings were irradiated with 1 mmol m−2 R light for 30 s and then returned to darkness. Total RNA was isolated from these seedlings at 0, 30, and 60 min of dark incubation after R light irradiation, and arbitrary primed PCR-generated cDNA fingerprints were compared by FDD to identify differentially expressed bands. From ∼8000 displayed cDNA bands, we identified several fragments through repeated FDD analysis, and five of these were excised and sequenced (Table 1). For further analysis, we selected one of these fragments, a 490-bp cDNA fragment (CB18-490) that showed a gradual increase in transcript levels from 0 to 60 min of dark incubation after R light irradiation (Figure 1A). The reproducibility of the FDD banding pattern of this candidate band was confirmed by repeated FDD analysis using independently prepared RNA samples (data not shown). The excised CB18-490 FDD fragment was reamplified with the same primer set used for the FDD analysis, and the reamplified products were separated by electrophoresis using an agarose gel containing a base-specific DNA ligand to eliminate nonrelated contaminating cDNAs (Yoshikawa et al., 1998). The main band was recovered subsequently from the agarose gel, amplified by PCR, and cloned into an appropriate vector. This third-round amplification product also was subjected to direct DNA sequencing to verify that the cloned cDNA molecule represented the target transcript (CB18-490).
Table 1.
Characteristics of Identified Fragments in the Present FDD Screening
| Clone | Fragment Size (bp) | PCR Primers (5′ to 3′) | Best Homology with the Arabidopsis Genome (%) | Phy Regulation |
|---|---|---|---|---|
| CB18-490 | 490 | CCACAGCAGT,GT15C | CCA1/LHY (70) | Up |
| CX9-370 | 370 | GGTCTGGTTG,GT15C | CCA1/LHY | Up |
| CD10-440 | 440 | GGTCTACACC,GT15C | AtmybL2 (myb-related protein) (90) | Up |
| CD2-570 | 570 | GGACCCAACC,GT15C | XTR7 (xyloglucan endotransglycosylase) (95) | Down |
| CD9-530 | 530 | CTCTGGAGAC,GT15C | XTR7 | Down |
Figure 1.
Identification and Isolation of EPR1.
(A) FDD screening for CB18-490. Total RNA was harvested from etiolated wild-type seedlings at 0, 30, and 60 min of dark incubation after 1 mmol m−2 R light irradiation.
(B) RNA gel blot analysis confirming the observed R light–induced expression of the isolated gene corresponding to the CB18-490 FDD fragment in (A). Total RNA (20 μg per lane) from etiolated wild-type seedlings at 0, 30, 60, 120, and 240 min of dark incubation after 1 mmol m−2 R light irradiation was hybridized with the CB18-490 probe. The transcript sizes were estimated using RNA size markers (0.16- to 1.77-kb RNA ladder). 18S rRNA was used for normalization.
(C) Nucleotide and deduced amino acid sequences of EPR1. Boldface letters indicate the arbitrary (OPB-18) and oligo(dT) primers that were used for FDD reactions. The amino acid sequence of the EPR1 MYB domain is underlined.
(D) Comparison of the MYB domain amino acid sequences of EPR1, LHY, and CCA1. Identical amino acid residues are boxed, asterisks represent the positions of conserved Trp and Ala residues in the MYB domain, and dashes indicate gaps.
To confirm the CB18-490 expression pattern, we used the isolated CB18-490 cDNA fragment as a probe for RNA gel blot analysis. Figure 1B shows that a transcript of ∼1.7 kb was induced rapidly and transiently by R light. CB18-490 showed peak transcript levels at 1 h of dark incubation after R light irradiation, followed by a rapid decline after another 2 h. This rapid and transient induction mimics the situation observed for several circadian clock components, in which light signals are thought to entrain and reset the clock (Devlin, 2002). These results demonstrate that the isolated CB18-490 FDD clone was the candidate clone representing an early-phytochrome-responsive gene.
CB18-490 Encodes a Novel Single MYB Domain Protein with Similarity to LHY and CCA1
The nucleotide sequence of the isolated CB18-490 FDD clone (497 bp) was analyzed by searching for similarities against the EMBL/GenBank databases using the BLASTN (Basic Local Alignment Search Tool) algorithm (Altschul et al., 1990). The database search revealed that a 280-bp region at the 5′ end of CB18-490 encodes a sequence with 70% similarity to the MYB DNA binding domain of CCA1 and LHY (Schaffer et al., 1998; Wang and Tobin, 1998), whereas sequences encoded by the remaining 210-bp region exhibited no significant similarity to CCA1 or LHY (Figure 1C).
By screening an Arabidopsis cDNA library, we isolated a full-length cDNA corresponding to the CB18-490 FDD clone (Figure 1C). Figure 1C shows the nucleotide sequence and the translated amino acid sequence of the longest clone (designated EPR1). The EPR1 cDNA represents a 1038-bp open reading frame (chromosome I) encoding a protein of 346 amino acids with a single MYB domain (Figure 1C). BLAST searches using the full-length EPR1 protein as the input query revealed that EPR1 has high overall similarity (70% identity) to Arabidopsis CCA1 and LHY (Wang et al., 1997; Schaffer et al., 1998) in the MYB domain region (Figure 1D). Further analysis also revealed that two of the three regularly spaced Trp residues, which are characteristic of MYB repeats, are present in the MYB domain of EPR1. At the predicted position of the third Trp residue, an Ala residue is present that is conserved among all three of these Arabidopsis single MYB repeat proteins (Figure 1D). Although CCA1 and LHY share extensive regions of similarity outside of their MYB domains, EPR1 shows similarity only to CCA1 and LHY in the MYB domain region. The predicted size of the EPR1 protein (346 amino acids) also is smaller than that of CCA1 (608 amino acids) and LHY (645 amino acids). EPR1 also shows remarkable similarity (87%) to a predicted gene on chromosome III (At3g10113) annotated as having similarity to MYB-related transcription factors. However, the extreme 5′ end this gene (55 nucleotides downstream of the predicted start codon) shows no significant similarity to EPR1. This gene probably originated from a gene duplication event and may represent an EPR1 homolog. Thus, the isolated EPR1 gene encodes a novel MYB protein containing a single MYB domain, showing high similarity to the Arabidopsis CCA1 and LHY proteins.
EPR1 Expression Is Mediated through the phyA Very Low Fluence Response and phyB
To investigate whether phyA can mediate EPR1 induction, the expression dynamics of EPR1 was analyzed in etiolated wild-type and phyA-deficient mutant (phyA) Arabidopsis seedlings with or without a brief pulse of FR light (10 mmol m−2). Figure 2A shows that EPR1 transcript levels in wild-type seedlings were approximately threefold higher in response to FR light irradiation (WT/FR60) compared with dark growth (WT/D). Furthermore, there were no significant differences in EPR1 transcript levels between FR light–treated (phyA/FR60) and dark control (phyA/D) phyA mutant seedlings, demonstrating that phyA regulates the induction of EPR1 expression by FR light through the very low fluence response.
Figure 2.
Expression and Localization Analysis of EPR1.
(A) RNA gel blot analysis confirming the phyA and phyB regulation of EPR1 expression in wild-type and phy mutant seedlings. Total RNA (10 μg) from etiolated wild-type Landsberg erecta seedlings (WT) and phyA mutant seedlings that were harvested after 60 min of dark incubation and 10 mmol m−2 FR light irradiation (FR60). Seedlings kept in total darkness (D) were used as a control.
(B) Total RNA (10 μg) from wild-type Landsberg erecta seedlings, phyA mutant seedlings, and phyA phyB double mutant seedlings that were exposed to R light (R; 1 mmol m−2), R light followed by FR light (R/FR; 1 mmol m−2 and 10 mmol m−2), or FR light only (FR; 10 mmol m−2) and then kept in darkness for 60 min. 18S rRNA was included for normalization purposes.
(C) Six-day-old etiolated wild-type Landsberg erecta seedlings were treated with 300 μM cycloheximide (CHX+) or without cycloheximide (CHX−) for 2 h before R light treatment (1 mmol m−2). Total RNA (25 ng) from 0, 30, 60, 120, and 240 min of dark incubation after R light treatment was analyzed by semiquantitative reverse transcriptase–mediated PCR. ACT8 and rbcS were used as controls.
(D) Subcellular localization of an EPR1:GFP fusion protein in onion epidermal cells. Epifluorescence analysis showing GFP fluorescence in the nucleus was confirmed by 4′,6-diamidino-2-phenylindole (DAPI) staining. Bars = 100 μm.
To examine the effect of other phytochrome members, EPR1 expression analysis was performed on total RNA extracted from etiolated wild-type, phyA, and phyA phyB double mutant Arabidopsis seedlings after various light treatments (1 mmol m−2 R light [R] or 1 mmol m−2 R light followed by 10 mmol m−2 FR light [R/F] or 10 mmol m−2 FR light [FR] and then darkness [D] for 60 min). In the wild type, EPR1 expression was induced in response to R and FR light (Figure 2B, WT/R and WT/FR). Interestingly, EPR1 expression was induced by 1 mmol m−2 R light in phyA seedlings (Figure 2B, phyA/D and phyA/R), and this R light induction was repressed by subsequent irradiation with FR light (Figure 2B, phyA/R/F). These results indicate that a type-II phytochrome, possibly phyB, photoreversibly regulates EPR1 expression.
The effect of the protein synthesis inhibitor cycloheximide (CHX) on EPR1 induction was examined to determine whether de novo protein synthesis was necessary for the observed light induction. Six-day-old etiolated wild-type seedlings were treated with CHX at 2 h before R light irradiation followed by semiquantitative reverse transcriptase–mediated PCR analysis (Figure 2C). As shown in Figure 2C, the R light induction of EPR1 expression was not inhibited by CHX. This result, in combination with the observed rapid light response, indicates that EPR1 represents a primary phytochrome-responsive gene in Arabidopsis.
EPR1 Shows Constitutive and Uniform Nuclear Localization
The amino acid sequence similarity between EPR1 and other MYB transcription factors, such as CCA1 and LHY, indicated that EPR1 might localize to the nucleus. To test this possibility, we constructed a CaMV35S-EPR1:green fluorescent protein (GFP) fusion cassette followed by transient transfection assays in onion epidermal cells. Epifluorescence microscopy revealed that the EPR1:GFP fusion protein was localized exclusively to nuclei (Figure 2D). More detailed studies involving optical sectioning of whole nuclei in planta and three-dimensional rendering algorithms (Volocity II, Improvision, Coventry, UK) revealed that the EPR1:GFP signal was distributed uniformly within each nucleus (data not shown), in sharp contrast to that for LAF1, a MYB transcription activator showing subnuclear localization to nuclear bodies (Ballesteros et al., 2001). We also tested whether EPR1 localization was affected by different light conditions in transgenic Arabidopsis plants harboring the EPR1:GFP fusion protein. From these studies, we observed no difference in localization patterns in darkness or in response to R, FR, or blue (B) light irradiation (data not shown), indicating that EPR1 is localized constitutively to the nucleus.
EPR1 Overexpression Results in a Variety of Light-Dependent Phenotypes
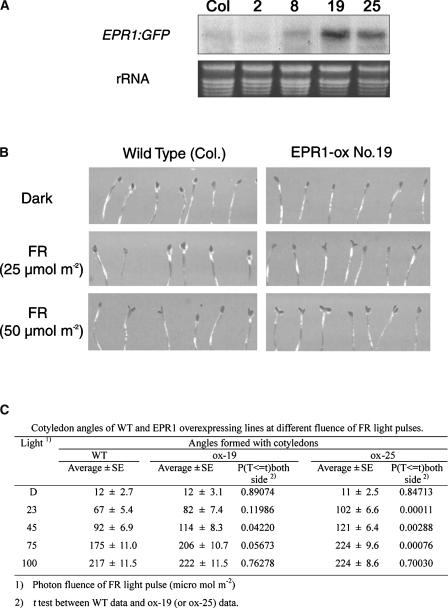
To investigate the physiological role of EPR1, we made use of transgenic Arabidopsis plants overexpressing EPR1. From RNA gel blot analysis, we selected two independent transgenic lines showing increased EPR1 levels (EPR1 ox-19 and EPR1 ox-25) and two transgenic lines showing nearly wild-type expression levels (EPR1 ox-2 and EPR1 ox-8) as controls (Figure 3A). We subjected both wild-type and transgenic seedlings to various light regimes and recorded phenotypic differences. In terms of hypocotyl elongation, we observed no significant differences between wild-type seedlings and seedlings with increased EPR1 levels upon R, FR, or B light irradiation (data not shown). However, cotyledon-opening characteristics were altered in transgenic seedlings compared with wild-type seedlings (Figure 3B). At low fluence of intermittent FR light irradiation (25 μmol m−2), wild-type cotyledons remain closed; however, both EPR1 ox-19 and EPR1 ox-25 transgenic seedlings with increased EPR1 levels exhibited clear cotyledon opening. EPR1 ox-19 is shown in Figure 3B. To verify that the differences in cotyledon opening between wild-type and EPR1-overexpressing seedlings were statistically significant, we measured the cotyledon angles of wild-type, EPR1 ox-19, and EPR1 ox-25 seedlings in response to increasing fluences of FR light and performed a t test (Figure 3C). As can be seen from the P values (Figure 3C), the measured angle differences are clearly significant at 23 to 75 μmol m−2 FR light. Furthermore, the FR light–induced cotyledon-opening response in Arabidopsis seedlings clearly was dependent on phyA, because this response was abolished in phyA-deficient seedlings (data not shown). Together, our results suggest that EPR1 is involved in phyA-mediated cotyledon opening in Arabidopsis.
Figure 3.
FR Light–Induced Cotyledon Opening of Transgenic Arabidopsis Overexpressing EPR1.
(A) RNA gel blot analysis (20 μg of total RNA) of wild-type Columbia (Col) and four transgenic lines overexpressing EPR1.
(B) Cotyledon opening of wild-type Columbia and EPR1-overexpressing line 19 (EPR1 ox-19) was induced by intermittent FR light irradiation.
(C) Cotyledon angles of the wild type (WT) and EPR1-overexpressing lines 19 and 25 (EPR1 ox-19 and EPR1 ox-25) at different fluences of FR light pulses. Accompanying statistical analysis (t test) showed that the differences in cotyledon angles were significant. D, darkness.
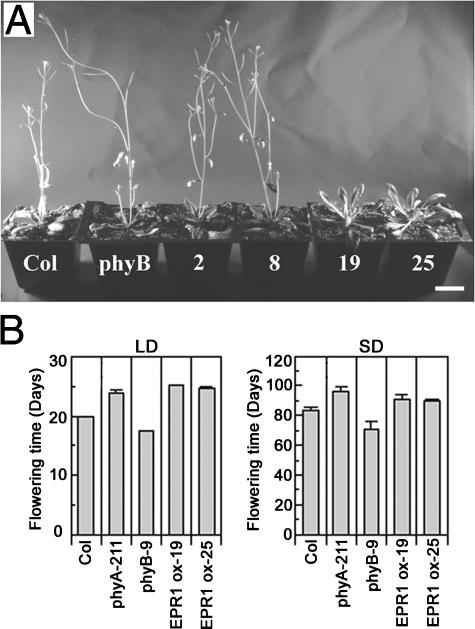
We also analyzed the effect of EPR1 overexpression in adult plants. When wild-type and EPR1-overexpressing plants (EPR1 ox-19 and EPR1 ox-25) were grown under long-day (LD) conditions (16 h of light/8 h of dark), we found that EPR1 overexpression resulted in a delay in flowering (Figure 4A), and quantitative analysis showed that EPR1-overexpressing plants flowered 5 to 6 days later than wild-type plants (Figure 4B, LD). As an internal control, we determined the flowering time of transgenic EPR1 ox-2 and EPR1 ox-8 plants and showed that the time of flowering was not different from that of wild-type plants (Figure 4A). In addition, EPR1 overexpression also resulted in a slight delay in flowering under short-day (SD) conditions (8 h of light/16 h of dark), as shown in Figure 4B. The delay in flowering under SD conditions was less than that observed under LD conditions (almost overlapping error bars). Although the delay in flowering was not as dramatic as that seen in plants overexpressing CCA1 and LHY (Schaffer et al., 1998; Wang and Tobin, 1998), these results indicate that EPR1 overexpression causes a slightly altered photoperiod flowering response.
Figure 4.
Late-Flowering Phenotype of Transgenic Arabidopsis Overexpressing EPR1.
(A) Transgenic Arabidopsis plants showing wild-type levels of EPR1 (EPR1 ox-2 and EPR1 ox-8) and increased EPR1 levels (EPR1 ox-19 and EPR1 ox-25) were grown under LD conditions (16 h of light/8 h of dark) for 24 days. Wild-type Columbia (Col) and phyB mutant plants were used as controls. Bar = 1 cm.
(B) Quantitative analysis of flowering time under LD and SD conditions of two EPR1-overexpressing lines (EPR1 ox-19 and EPR1 ox-25). The time of bolting was the day when the plant had a bolt of 1 cm. Wild-type (Col), phyA-211, and phyB-9 were used as controls. The error bars represent standard errors.
EPR1 Expression follows a Circadian Rhythm and Regulates Its Own Expression in Continuous Light
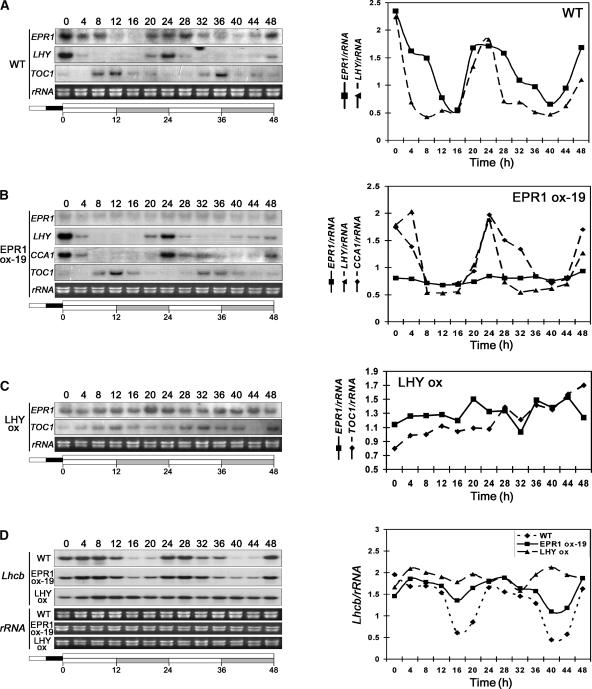
The disruption of circadian expression patterns in Arabidopsis has been shown to affect photoperiodism. For example, disruption of circadian rhythms by CCA1 and LHY overexpression in Arabidopsis causes a late-flowering phenotype under LD conditions (Schaffer et al., 1998; Wang and Tobin, 1998). The late-flowering phenotype of transgenic plants exhibiting increased EPR1 levels (Figure 4) suggested that EPR1 may be involved in circadian regulation. To test this notion, EPR1 RNA expression dynamics was monitored in wild-type Arabidopsis seedlings. Plants were grown for 14 days under 12-h-light/12-h-dark cycles and then transferred to continuous light (LL) conditions. Plants were harvested every 4 h for 48 h followed by RNA gel blot analysis. This analysis demonstrated that EPR1 exhibited a circadian pattern of expression under LL conditions in wild-type seedlings (Figure 5A) but with a peak of expression slightly broader than that of LHY and CCA1 (Schaffer et al., 1998; Wang and Tobin, 1998). As expected, LHY showed defined peaks of expression at subjective dawn (e.g., at 24 h), whereas EPR1 showed a broader peak of expression up to 4 h after subjective dawn (Figure 5A, 24 and 28 h). This finding may indicate a distinct phase of expression. Similar circadian rhythmic expression patterns of EPR1 also were observed in continuous darkness (data not shown).
Figure 5.
Circadian Expression Analysis of EPR1 and Multiple Circadian Rhythm–Regulated Genes.
EPR1, LHY, CCA1, and TOC1 levels of expression in wild-type (WT) plants (A), in EPR1-overexpressing plants (EPR1 ox-19) (B), and in LHY-overexpressing plants (C). Lhcb expression levels were analyzed in wild-type, EPR1-overexpressing (EPR1 ox-19), and LHY-overexpressing (LHY ox) plants (D). All plants were grown under 12-h-light/12-h-dark cycles for 2 weeks and then transferred to LL conditions (time 0). Plants were harvested every 4 h for 48 h, and total RNA isolated and expression levels were determined by RNA gel blot analysis and quantification. rRNA was used as a loading control.
Because oscillatory autoregulatory feedback loops have been found in all circadian clock mechanisms examined to date (Dunlap, 1999), we examined whether EPR1 regulates its own expression. RNA gel blot analysis of EPR1-overexpressing plants was performed as described above in LL conditions but using the EPR1 3′ untranslated region as a probe. As can be seen in Figure 5B, EPR1 overexpression resulted in repression of the endogenous EPR1 transcript and loss of rhythmicity. These results demonstrate that EPR1 expression is regulated by the circadian clock in a manner similar to LHY and CCA1 and that EPR1 is part of a regulatory feedback loop.
Constitutive Expression of LHY and CCA1 Causes the Arrhythmic Expression of EPR1, Whereas LHY, CCA1, and TOC1 Are Not Affected by EPR1 Overexpression in Continuous Light
To determine whether the overexpression of known components of the central circadian oscillator affects EPR1 rhythmicity, we analyzed EPR1 expression in LHY-overexpressing plants. Figure 5C shows that EPR1 rhythmic expression was disrupted severely in plants with increased LHY levels in LL conditions. Similarly, EPR1 circadian rhythmicity was disrupted in CCA1-overexpressing plants (data not shown). These findings indicate that EPR1 expression was controlled by the central oscillator mediated by LHY and CCA1.
We further tested whether EPR1 overexpression had any affect on CCA1 and LHY expression in LL conditions. In EPR1-overexpressing plants, both CCA1 and LHY showed wild-type–like circadian rhythmicity (Figure 5B), indicating that EPR1 does not influence LHY or CCA1 cycling and that EPR1 probably is not part of the central oscillator. However, to further verify this finding, we analyzed the effect of EPR1 overexpression on TOC1 rhythmicity. In wild-type plants, TOC1 showed circadian oscillations, with peak expression levels coinciding with periods of minimal expression of CCA1, LHY (Strayer et al., 2000) (Figure 5A), and EPR1 (Figure 5A), whereas in LHY-overexpressing plants, TOC1 rhythmicity was abolished, as expected (Figure 5C). In EPR1-overexpressing plants, TOC1 exhibited wild-type–like expression patterns in LL conditions (Figure 5B), corroborating our notion that EPR1 is not part of the central oscillator.
Constitutive Expression of EPR1 Inhibits the Circadian Rhythm of Lhcb
Our data suggest that EPR1 is part of a regulatory circadian feedback loop peripheral to the central oscillator in Arabidopsis. Thus, we analyzed whether the constitutive expression of EPR1 had any effect on known target genes of the clock, such as Lhcb. In wild-type plants, Lhcb showed robust rhythmicity, with peak expression levels delayed by ∼4 h compared with LHY and CCA1 and delayed only slightly compared with EPR1 peak levels (Figure 5D). By contrast, Lhcb rhythmicity was abolished in plants overexpressing LHY (Figure 5D). In EPR1-overexpressing plants, Lhcb rhythmicity was inhibited (Figure 5D), indicating that the circadian rhythm of Lhcb is partly controlled by the oscillatory behavior of EPR1. Based on these data, EPR1 is a likely component of an oscillator peripheral to the central oscillator in Arabidopsis that is involved in controlling and fine-tuning the circadian rhythms of target genes.
DISCUSSION
There is a close relationship between photoreceptors and the circadian clock (Devlin and Kay, 2001), and both R and FR light have been shown to be dominant signals involved in the rapid resetting of the clock through the action of phytochromes (Nagy et al., 1993). Given the observations that several key phytochrome signaling events involve rapid light-dependent nuclear translocation events that ultimately regulate the expression of light-regulated target genes (Møller et al., 2002; Quail, 2002), it is becoming increasingly important to identify early-phytochrome-responsive genes. The use of FDD for the isolation of genes that show rapid and minute changes in response to environmental cues has advantages that complement current methods, such as microarray technology, including high sensitivity and the random nature of the technique. Here, we report the successful use of FDD for the isolation of a new R light–induced gene, EPR1, that encodes a component of the circadian clock in Arabidopsis.
EPR1 Represents an Early-Phytochrome-Responsive Gene That Encodes a Novel Member of the MYB Protein Family
EPR1 was predicted to encode a DNA binding protein that is highly similar to the MYB class of transcription factors. This prediction was based on the high similarity with several other reported MYB DNA binding proteins showing the presence of a MYB domain within the N-terminal region (Figure 1D). Although most MYB proteins identified in plants contain two or three MYB domain repeats, EPR1 contains only one. In R2R3 two-repeat MYB proteins, both repeats are required for DNA binding, suggesting that single MYB domain proteins, such as EPR1, bind DNA in a unique manner. Although representing a smaller family than the R2R3s, a number of single MYB domain proteins have been identified in plants (Feldbrugge et al., 1997; Wang et al., 1997). EPR1 is highly similar (70%) to LHY and CCA1, which are both single MYB domain proteins (Wang et al., 1997; Schaffer et al., 1998). CCA1 has been shown to bind to the promoter element of at least two Lhcb genes in Arabidopsis, and it has been demonstrated that this binding is responsible for its phytochrome responsiveness (Wang et al., 1997). Furthermore, CCA1 and LHY are components of the circadian central oscillator in Arabidopsis that modulates the circadian rhythms of genes such as Lhcb (Schaffer et al., 1998; Wang and Tobin, 1998). Recently, through studies with the lhy cca1 double mutant, it was shown that CCA1 and LHY are partially redundant, acting as negative regulators required for the central circadian oscillator function (Mizoguchi et al., 2002), indicating that single-domain MYB factors do show overlapping functionality.
EPR1 was identified initially based on its rapid induction by a brief pulse of R light (Figure 1A), and we have shown that this induction does not require de novo protein synthesis (Figure 2C). Although the induction of EPR1 gene expression was very rapid and peaked at 1 h of dark incubation after R light irradiation, the transcript declined rapidly, and after an additional 3 h of dark incubation, the transcript levels were similar to those before light induction (Figure 1B). Further analysis also revealed that EPR1 expression was induced rapidly by FR light irradiation and that this induction was abolished in phyA-deficient seedlings (Figure 2A), indicating that EPR1 gene expression is regulated by phyA. Interestingly, EPR1 expression was induced by low-fluence R light in the absence of phyA, and this induction was repressed by subsequent FR light irradiation (Figure 2B), suggesting that EPR1 is regulated photoreversibly by a phytochrome other than phyA, probably phyB. The rapid induction kinetics of EPR1 is reminiscent of LHY and CCA1 expression profiles showing rapid but transient induction by R and FR light irradiation (Schaffer et al., 1998; Wang and Tobin, 1998). CCA1 induction precedes Lhcb induction by ∼3 h, consistent with the notion that CCA1 is a mediator of Lhcb gene expression (Wang et al., 1997). EPR1 induction also precedes Lhcb induction, and it is possible that EPR1 plays a role similar to that of CCA1 in directly regulating the expression of light-regulated genes.
Increased EPR1 Levels Affect Multiple Phytochrome- and Circadian Rhythm–Regulated Processes
In addition to its direct transcriptional role on Lhcb gene expression, CCA1 and probably LHY clearly are not limited to the regulation of Lhcb genes in Arabidopsis. It is evident that both CCA1 and LHY play direct roles in a multitude of physiological and molecular processes that are under the control of the circadian clock, such as hypocotyl growth and photoperiodic responses (Schaffer et al., 1998; Wang and Tobin, 1998). Although EPR1 is highly similar to CCA1, constitutive overexpression of EPR1 (Figure 3A) does not result in the loss of hypocotyl growth inhibition, as observed upon CCA1 overexpression (Wang and Tobin, 1998). This finding indicates that the mode of action of EPR1 is different, at least in part, from that of CCA1 and possibly LHY. In contrast to CCA1- and LHY-overexpressing plants, an increase in EPR1 levels results in accelerated cotyledon opening at low fluences (23 to 75 μmol m−2) of FR light irradiation (Figures 3B and 3C). This finding is in marked contrast to the effect in wild-type seedlings, which remain fully closed at these fluence levels, showing only partial expansion at considerably higher fluences of FR light (50 μmol m−2) (Figures 3B and 3C). As expected, this FR light–induced cotyledon-opening phenotype is dependent on phyA (data not shown). Cotyledon opening in wild-type Arabidopsis is controlled by multiple photoreceptors, as shown when grown under white light (Neff and Chory, 1998), demonstrating the complex nature of this response. It is interesting that increased levels of a single MYB protein can affect such a complex pathway, which may indicate that EPR1 is involved in the regulation of several genes, as observed for CCA1 and LHY (Schaffer et al., 1998; Wang and Tobin, 1998).
Physiological studies have demonstrated that the circadian clock is involved in photoperiodism (Thomas and Vince-Pruce, 1997), and several circadian rhythm mutants have been shown to affect photoperiod perception (Hayama and Coupland, 2003). Transgenic plants with constitutive overexpression of CCA1 or LHY display a severe late-flowering phenotype under LD conditions (Schaffer et al., 1998; Wang and Tobin, 1998) but only a partial late-flowering phenotype under SD conditions (Green et al., 2002), suggesting a disruption in the circadian rhythm involved in photoperiodism. Our studies reveal that EPR1 overexpression results in a delay in flowering under LD (16 h of light/8 h of dark) and SD (8 h of light/16 h of dark) conditions (Figure 4A), although the flowering delay under SD conditions was less pronounced. The apparent late-flowering phenotype observed under SD conditions was similar to that observed for transgenic plants overexpressing CCA1 under SD conditions (Green et al., 2002). However, as a result of the weaker flowering phenotype of EPR1-overexpressing plants under LD conditions compared with CCA1-overexpressing plants, the SD flowering delay in Figure 4B appears more marked. The less severe flowering phenotype of EPR1-overexpressing plants under LD conditions suggests that the EPR1 slave oscillator probably controls a subset of circadian rhythm–regulated processes to varying degrees, as suggested for AtGRP7 (Heintzen et al., 1997). Our results suggest that as with CCA1 and LHY, EPR1 may be involved in regulating photoperiod responses, although to a lesser extent. The control of flowering time in Arabidopsis is dependent on sensing photoperiods, and it has been shown to be dependent on several photoreceptors: phyA-deficient plants flower late under LD conditions (Reed and Chory, 1994), phyB-deficient plants flower early (Reed et al., 1996), and cry2-deficient plants flower late under LD conditions (Koornneef, 1991).
EPR1 Is a Component of a Slave Oscillator in Arabidopsis
The rapid R and FR induction of EPR1 coupled with the late-flowering phenotype of EPR1-overexpressing plants suggested that EPR1 might be regulated by and function as part of the circadian clock. In wild-type plants, EPR1 exhibited clear circadian rhythmicity under continuous light conditions, with peak expression levels occurring at subjective dawn (Figure 5A). Although similar to CCA1 and LHY, EPR1 showed a slightly longer peak duration (up to 4 h after subjective dawn), suggesting that EPR1 cycles with a slightly different phase than CCA1 and LHY (Figure 5A). Multiplex reverse transcriptase–mediated PCR analysis further corroborated these findings, showing a slight shift in the cycling phase (data not shown). This out-of-phase relationship and the existence of three CCA1 recognition motifs (AAAATCT) in the 5′ regulatory region of EPR1 suggested that CCA1 (and/or LHY) might be involved in mediating EPR1 oscillations (Figure 6). Because light induction of EPR1 does not require de novo protein synthesis (Figure 2C), it is probable that light induction and the circadian regulation of EPR1 involve different transcription factors: LHY and CCA1 may mediate the rhythmic expression of EPR1 but may not be required for its induction by light.
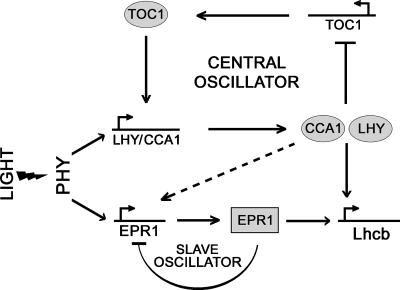
Figure 6.
Model Showing the Possible Position of EPR1 in the Circadian Clock.
Light induces CCA1 and LHY expression at dawn, which in turn leads to TOC1 repression by the direct action of CCA1 and LHY. During the course of the day, CCA1 and LHY relieve TOC1 repression, leading to TOC1 accumulation during the night phase, which ultimately results in the activation of CCA1 and LHY at dawn. EPR1 is induced by light and EPR1 rhythmicity is regulated by the central oscillator, possibly through the action of CCA1/LHY (dashed arrow). EPR1 is capable of regulating its own expression, forming a slave oscillator. EPR1 also mediates Lhcb rhythmicity and may be involved in the fine-tuning of specific output rhythms.
The constitutive overexpression of EPR1 results in a complete suppression of the endogenous EPR1 transcript, suggesting that EPR1 can feedback repress its own expression, which could be responsible for the trough levels during normal EPR1 circadian oscillations in wild-type plants (Figure 5B). This feedback repression probably is mediated by direct transcriptional regulation; however, it is possible that this occurs post-transcriptionally, as observed for AtGRP7 (Staiger et al., 2003b).
The findings that EPR1 shows circadian expression patterns and regulates its own expression suggested that EPR1 may be part of an oscillator function. It has been established that LHY and CCA1 are central oscillator components (Figure 6) and that they act, at least in part, redundantly in mediating circadian oscillations (Schaffer et al., 1998; Wang and Tobin, 1998; Mizoguchi et al., 2002). We have shown that EPR1 rhythmicity is abolished in plants that overexpress LHY (Figure 5C), demonstrating that EPR1 rhythmicity is mediated by the central oscillator. LHY and CCA1 also have been shown to negatively regulate TOC1 in the circadian central loop (Alabadi et al., 2001); however, EPR1 does not seem to be part of the central oscillator, because EPR1 overexpression does not disrupt the circadian rhythmicity of LHY or CCA1 (Figure 5B). TOC1 acts as a positive regulator of LHY and CCA1 expression to ensure high levels at dawn (Alabadi et al., 2001), and it is possible that TOC1 regulates EPR1 levels. This situation is similar to that observed for AtGRP7 in Arabidopsis, in which it has been proposed that AtGRP7 receives information from the central oscillator through TOC1 (Millar et al., 1995) and acts as part of a slave oscillator to regulate a subset of clock-controlled transcripts (Heintzen et al., 1997). Our finding that EPR1 overexpression does not disrupt TOC1 rhythmicity (Figure 5B) further verified the idea that EPR1 is not part of the central oscillator loop in Arabidopsis but probably is part of a slave oscillator (Figure 6).
LHY and CCA1 ultimately mediate the circadian oscillation of target genes such as Lhcb (Wang and Tobin, 1998), and we predicted that EPR1 overexpression would result in changes in Lhcb expression profiles. We have shown that constitutive expression of EPR1 results in the inhibition of Lhcb rhythmicity (Figure 5D) but that the effect is not as dramatic as that observed upon LHY overexpression. This result is consistent with our hypothesis that EPR1 is part of a slave oscillator that mediates the oscillation of target genes (Figure 6).
Concluding Remarks
The current circadian clock model in plants still needs refining. Although a convincing relationship between the input and output pathways and the central oscillator has been proposed, the interaction between these different components remains ill-defined. It has been demonstrated that the output pathway can feed back into the central oscillator and moreover that components involved in the clock input also oscillate (Harmer et al., 2000). In addition, core components of the central oscillator have been shown to have added levels of complexity. For example, CCA1 loss-of-function plants show Lhcb oscillation with a shorter period, indicating that additional factors are involved in mediating rhythmicity (Green and Tobin, 1999). These observations suggest that the central circadian clock loop has additional levels of regulation (Figure 6). This notion is emphasized by data suggesting that TOC1 has a dual role in darkness and in controlling clock outputs (Mas et al., 2003). It is becoming clear that that the central oscillator framework in Arabidopsis probably has multiple positive and negative feedback loops that contribute to the circadian oscillations derived from the central oscillator. Indeed, interconnected circadian feedback loops clearly are present in Drosophila and mammals (Glossop et al., 1999; Shearman et al., 2000). The results presented here suggest that the nucleus-localized phytochrome-induced EPR1 MYB protein represents one component of such a slave oscillator in Arabidopsis involved in the fine-tuning of rhythmic behavior in plants.
METHODS
Plant Material and Growth Conditions for Fluorescent Differential Display Screening
Arabidopsis thaliana (Landsberg erecta) wild type together with phyA-201 (Nagatani et al., 1993), phyB-1 (Reed et al., 1996), and the phyA-201 phyB-1 double mutant (Hamazato et al., 1997) were used for all experiments described unless stated otherwise. Seeds were plated onto filter paper placed on top of 20% MS solid medium (Murashige and Skoog, 1962) supplemented with 0.6% sucrose and then exposed to far-red (FR) light (25 μmol m−2) for 10 min to inhibit dark germination. Wild-type, phyA-201, and phyB-1 seeds were transferred subsequently to 23 ± 1°C in darkness for 16 h, exposed to red (R) light (30 μmol m−2) for 8 h to induce germination, and then kept in total darkness at 23 ± 1°C for 5 days. Seeds of the phyA-201 phyB-1 double mutant were sown and incubated under continuous white light (12 W m−2) for 60 h to induce germination. The germinated seedlings were kept in total darkness at 23 ± 1°C for 3.5 days.
Light Treatment
Etiolated 6-day-old seedlings were irradiated with a pulse of R light and then returned immediately to darkness. At 0, 30, and 60 min after R light irradiation, seedlings were harvested and quickly frozen in liquid nitrogen. R light was obtained by filtering light from white fluorescent tubes [FL20SSW/18(G); Hitachi, Tokyo, Japan] through a 3-mm-thick red acrylic plate (Shinkolite A102; Mitsubishi Rayon, Tokyo, Japan). FR light was obtained by filtering light from far-red fluorescent tubes (FL20S.FR-74; Toshiba, Tokyo, Japan) through a 3-mm-thick far-red acrylic plate (Deraglass 102; Asahikasei, Tokyo, Japan). Fluence rates were measured with an Optical Power Meter (1830-C; Newport Co., Irvine, CA).
Fluorescent Differential Display
Total RNA was extracted from etiolated seedlings by the phenol/SDS/LiCl method (Verwoerd et al., 1989) and then treated with RNase-free DNase (Ambion, Austin, TX) for 30 min. Fluorescent differential display (FDD) was performed as described previously (Kuno et al., 2000). First-strand cDNAs were synthesized from each total RNA preparation (2.5 μg) using three different Texas Red–labeled 3′-anchored oligo(dT) primers (5′-Texas Red-GT15G-3; Nissibo, Tokyo, Japan) and a SuperScript Preamplification System (Gibco BRL, Rockville, MD). cDNAs produced from 25 ng of total RNA were amplified by PCR using combinations of Texas Red–labeled anchored oligo(dT) and arbitrary 10-mer primers (Kits B, D, F, and X; Operon Technologies, Alameda, CA). PCR was performed as follows: 94°C for 3 min, 40°C for 5 min, and 72°C for 5 min followed by 24 cycles of 94°C for 15 s, 40°C for 2 min, and 72°C for 1 min, with an additional extension step at 72°C for 5 min. Electrophoresis and detection of the PCR products were performed with an automated fluorescent DNA sequencer (SQ5500; Hitachi).
For the isolation of cDNAs of interest, preparative electrophoresis was performed and the FDD patterns visualized by a fluorescent image analyzer (FMBIO II Multi-View; Takara, Shiga, Japan). The gel on the glass plate was laid over the printed FDD image, and the bands of interest were excised. The cDNAs were eluted into distilled water and then amplified by PCR using the appropriate pairs of primers. Reamplification products were purified with a PCR purification kit (Qiagen, Chatsworth, CA) and subcloned into the pGEM-T vector (Promega, Madison, WI).
Measurement of Transcript Levels
RNA gel blot analysis was performed as described previously (Kuno et al., 2000). To detect EPR1 mRNA, a 338-bp PCR fragment, corresponding to the region of the EPR1 gene not containing the MYB domain, was used to prepare high-activity 32P-labeled probes synthesized by linear PCR amplification. Transcript sizes were determined by comparing electrophoretic mobility with those of RNA standards (0.16- to 1.77-kb RNA ladder; Gibco BRL).
For circadian expression experiments, wild-type, LHY-ox, CCA1-ox, and EPR1-ox plants were grown under 12-h-light/12-h-dark cycles for 2 weeks, transferred to continuous light (LL) conditions, and then subjected to total RNA extraction every 4 h for 48 h. RNA gel blots containing 15 μg of total RNA were hybridized with 32P-labeled probes amplified by PCR from first-strand cDNA using the following primer combinations: EPR1, EPR1-forward(F) (5′-CGCGGAAGAATCTCACAAACCAT-3′) and EPR1-reverse(R) (5′-CATCTGAAGCTACCAGCGTCAATGA-3′); CCA1, CCA1-F (5′-TTCACGGGAGGGAAGTCAGA-3′) and CCA1-R (5′-CGGGAGGCCAAAATGATGAGC-3′); LHY, LHY-F (5′-CGCGGTTCAAGATGTTCCCAAGA-3′) and LHY-R (5′-CTGTAGCAGCGGCAATGGCAG-TT-3′); EPR 3′ untranslated region, EPR1-3′-L (5′-TTTAGCCATTGTACAGTTTGGAGTC-3′) and EPR1-3′-R (5′-TCTTCTTAACGTGAGACAGCAGG-3′); TOC1, TOCF (5′-TGATTCCACGAGTTTGGGAGA-3′) and TOCB (5′-TGTGGGTTGGTGGACCTAAGA-3′); and Lhcb, CAB5 (5′-CTCTCACTCACAAGTTAGTCATA-3′) and CAB3 (5′-GCAACAGTCTTCCTCATTGTCA-3′). Hybridized membranes were exposed to x-ray film, stripped, and reprobed. All circadian experiments were performed in duplicate. Quantification of band intensities was performed using ImageQuant Software (Molecular Dynamics, Sunnyvale, CA)
Construction and Screening of a cDNA Library
cDNA was prepared from poly(A) RNA isolated from young Arabidopsis seedlings grown under continuous white light conditions for 7 days. The plasmid cDNA library was constructed using the SuperScript plasmid system (Gibco BRL) according to the manufacturer's protocol. A full-length cDNA clone corresponding to the CB18-490 fragment was screened using the CloneCapture cDNA Selection Kit (Clontech, Palo Alto, CA) and the FDD fragment as a probe according to the manufacturer's protocol.
Nuclear Localization Analysis
Full-length EPR1 cDNA was amplified by PCR from pSPORT1-EPR1 using Pwo polymerase (Boehringer Mannheim) incorporating a unique XhoI restriction site immediately upstream from the translational start codon (5′-ATCTCGAGATGGCCGCTGAGGATCGAAGTGAGG-3′) and a unique KpnI restriction site immediately upstream from the termination codon (5′-ATGGTACCGCATATACGTGCTCTTTGG-3′). The amplified fragment was cloned into pPCR-SCRIPT (Stratagene), subjected to DNA sequencing, and subcloned into pGFP2 (Kost et al., 1998) to generate an EPR1:GFP translational fusion cassette under the control of the 35S promoter of Cauliflower mosaic virus. The resulting construct, pGFP-EPR1, was bombarded into onion epidermal cells, and GFP fluorescence was visualized using epifluorescence and a Zeiss LSM410 inverted confocal microscope (Jena, Germany). Nuclei were visualized using 4′,6-diamidino-2-phenylindole and UV light illumination.
Plant Transformation and Analysis of Transgenic Plants
For in planta nuclear localization and overexpression analysis, the CaMV35S-EPR1:GFP cassette was cloned into the promoterless binary vector pBA002a (Kost et al., 1998) and transformed into Arabidopsis ecotype Columbia by vacuum infiltration (Clough and Bent, 1998). Primary transformants were selected on MS solid medium containing 10 mg/L phosphinothricin (Reidel-de Haen AG, Seelze, Germany) and self-fertilized. T2 seeds were collected and used for further analysis.
Cotyledon Opening and Flowering Time Experiments
Seeds of the wild type, transgenic lines, and phy mutants were plated on agar plates (10% MS medium with 0.7% [w/v] agar) and kept at 4°C for 3 days. The plates were transferred to 23°C and irradiated with intermittent FR light (10 s of FR light and 170 s of darkness) for 7 days with a light-emitting diode irradiation system (Shinomura et al., 2000). The photon fluence of FR light for each pulse was 23 to 100 μmol m−2. Cotyledon opening of ∼100 independent seedlings was observed, and the cotyledon angles were measured using a custom-made imaging system and analysis software.
For the determination of flowering time, Arabidopsis plants were sown on soil and grown under long-day (16 h of light and 8 h of dark) and short-day (8 h of light and 16 h of dark) conditions. Flowering time was measured by the duration from germination to bolting when the plant had a bolt of 1 cm.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Simon Geir Møller, sgm5@le.ac.uk.
Accession Number
The accession number for the EPR1 cDNA is AB115696.
Acknowledgments
We thank Isabelle Carre for supplying CCA1-ox and LHY-ox seeds and for useful discussions and Kimie Mase, Hisayo Shimizu, and Hee-Jung Chung for technical assistance. This work was partly supported by grants from the Hitachi Advanced Laboratory and the Program for the Promotion of Basic Research Activities for Innovative Biosciences to M.F., from the Ministry of Agriculture, Forestry, and Fisheries (Japan) Rice Genome Project (Grants SY1108 and IP1006) to T.S., and from the National Institutes of Health (Grant GM-44640) to N.-H.C. and by a North Atlantic Treaty Organization Science Fellowship, support from The Royal Society, and Biotechnology and Biological Science Research Council Grants 91/P16510 and 91/REI18421 to S.G.M.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.014217.
References
- Alabadi, D., Oyama, T., Yanovsky, M.J., Harmon, F.G., Mas, P., and Kay, S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293, 880–883. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Ballesteros, M.L., Bolle, C., Lois, L.M., Moore, J.M., Vielle-Calzada, J.P., Grossniklaus, U., and Chua, N.-H. (2001). LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev. 15, 2613–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carre, I.A., and Kim, J.Y. (2002). MYB transcription factors in the Arabidopsis circadian clock. J. Exp. Bot. 53, 1551–1557. [DOI] [PubMed] [Google Scholar]
- Clack, T., Mathews, S., and Sharrock, R.A. (1994). The phytochrome apoprotein family in Arabidopsis is encoded by five genes: The sequences and expression of PHYD and PHYE. Plant Mol. Biol. 25, 413–427. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Dehesh, K., Tepperman, J., Christensen, A.H., and Quail, P.H. (1991). phyB is evolutionarily conserved and constitutively expressed in rice seedling shoots. Mol. Gen. Genet. 225, 305–313. [DOI] [PubMed] [Google Scholar]
- Devlin, P.F. (2002). Signs of the time: Environmental input to the circadian clock. J. Exp. Bot. 53, 1535–1550. [DOI] [PubMed] [Google Scholar]
- Devlin, P.F., and Kay, S.A. (2001). Circadian photoperception. Annu. Rev. Physiol. 63, 677–694. [DOI] [PubMed] [Google Scholar]
- Dunlap, J.C. (1999). Molecular bases for circadian clocks. Cell 96, 271–290. [DOI] [PubMed] [Google Scholar]
- Fankhauser, C. (2001). The phytochromes, a family of red/far-red absorbing photoreceptors. J. Biol. Chem. 276, 11453–11456. [DOI] [PubMed] [Google Scholar]
- Feldbrugge, M., Sprenger, M., Hahlbrock, K., and Weisshaar, B. (1997). PcMYB1, a novel plant protein containing a DNA-binding domain with one MYB repeat, interacts in vivo with a light-regulatory promoter unit. Plant J. 5, 1079–1093. [DOI] [PubMed] [Google Scholar]
- Furuya, M., and Kim, B.-C. (2000). Do phytochromes interact with diverse partners? Trends Plant Sci. 5, 87–89. [DOI] [PubMed] [Google Scholar]
- Glossop, N.R.J., Lyons, L.C., and Hardin, P.E. (1999). Interlocked feedback loops within the Drosophila circadian oscillator. Science 286, 766–771. [DOI] [PubMed] [Google Scholar]
- Green, R.M., Tingay, S., Wang, Z.Y., and Tobin, E. (2002). Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 129, 576–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, R.M., and Tobin, E.M. (1999). Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc. Natl. Acad. Sci. USA 96, 4176–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamazato, F., Shinomura, T., Hanzawa, H., Chory, J., and Furuya, M. (1997). Fluence and wavelength requirements for Arabidopsis CAB gene induction by different phytochromes. Plant Physiol. 115, 1533–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer, S.L., Hogenesch, J.B., Straume, M., Chang, H.S., Han, B., Zhu, T., Wang, X., Kreps, J.A., and Kay, S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113. [DOI] [PubMed] [Google Scholar]
- Hayama, R., and Coupland, G. (2003). Shedding light on the circadian clock and the photoperiodic control of flowering. Curr. Opin. Plant Biol. 6, 13–19. [DOI] [PubMed] [Google Scholar]
- Heintzen, C., Nater, M., Apel, K., and Staiger, D. (1997). AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94, 8515–8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay, S.A., Keith, B., Shinozaki, K., Chye, M.L., and Chua, N.-H. (1989). The rice phytochrome gene: Structure, autoregulated expression, and binding of GT-1 to a conserved site in the 5′ upstream region. Plant Cell 1, 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M. (1991). Isolation of higher plant developmental mutants. Symp. Soc. Exp. Biol. 45, 1–19. [PubMed] [Google Scholar]
- Kost, B., Spielhofer, P., and Chua, N.H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualises the actin cytoskeleton in growing pollen tubes. Plant J. 16, 393–401. [DOI] [PubMed] [Google Scholar]
- Kuno, N., Muramatsu, T., Hamazato, F., and Furuya, M. (2000). Identification by large-scale screening of phytochrome-regulated genes in etiolated seedlings of Arabidopsis using a fluorescent differential display technique. Plant Physiol. 122, 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia, J.F., Huq, E., and Quail, P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288, 859–863. [DOI] [PubMed] [Google Scholar]
- Mas, P., Alabadi, D., Yanovsky, M.J., Oyama, T., and Kay, S.A. (2003). Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15, 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.J., Carre, I.A., Strayer, C.A., Chua, N.-H., and Kay, S.A. (1995). Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267, 1161–1163. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, T., Wheatley, K., Hanzawa, Y., Wright, L., Mizoguchi, M., Song, H.R., Carre, I.A., and Coupland, G. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 5, 629–641. [DOI] [PubMed] [Google Scholar]
- Møller, S.G., Ingles, P.J., and Whitelam, G.C. (2002). The cell biology of phytochrome signaling. New Phytol. 154, 553–590. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nagatani, A., Kay, S.A., Deak, M., Chua, N.-H., and Furuya, M. (1991). Rice type I phytochrome regulates hypocotyl elongation in transgenic tobacco seedlings. Proc. Natl. Acad. Sci. USA 88, 5207–5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani, A., Reed, J.W., and Chory, J. (1993). Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol. 102, 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, F., Fejes, E., Wehmeyer, B., Dallman, G., and Schäfer, E. (1993). The circadian oscillator is regulated by a very low fluence response of phytochrome in wheat. Proc. Natl. Acad. Sci. USA 90, 6290–6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M.M., and Chory, J. (1998). Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 118, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95, 657–667. [DOI] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1999). Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400, 781–784. [DOI] [PubMed] [Google Scholar]
- Parks, B.M., and Quail, P.H. (1993). hy8, a new class of Arabidopsis long hypocotyl mutants deficient in functional phytochrome A. Plant Cell 5, 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail, P.H. (2002). Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 3, 85–93. [DOI] [PubMed] [Google Scholar]
- Reed, J.W., and Chory, J. (1994). Mutational analyses of light-controlled seedling development in Arabidopsis. Semin. Cell Biol. 5, 327–334. [DOI] [PubMed] [Google Scholar]
- Reed, J.W., Foster, K.R., Morgan, P.W., and Chory, J. (1996). Phytochrome B affects responsiveness to gibberellins in Arabidopsis. Plant Physiol. 112, 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J.W., Nagpal, P., Poole, D.S., Furuya, M., and Chory, J. (1993). Mutations in the gene for red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5, 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carre, I.A., and Coupland, G. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93, 1219–1229. [DOI] [PubMed] [Google Scholar]
- Sharrock, R.A., and Quail, P.H. (1989). Novel phytochrome sequences in Arabidopsis thaliana: Structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev. 3, 1745–1757. [DOI] [PubMed] [Google Scholar]
- Shearman, L.P., Sriram, S., Weaver, D.R., Maywood, E.S., Chaves, I., Zheng, B., Kume, K., Lee, C.C., van der Horst, G.T., Hastings, M.H., and Reppert, S.M. (2000). Interacting molecular loops in the mammalian circadian clock. Science 288, 1013–1019. [DOI] [PubMed] [Google Scholar]
- Shinomura, T., Nagatani, A., Hanzawa, H., Kubota, M., Watanabe, M., and Furuya, M. (1996). Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 93, 8129–8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura, T., Uchida, K., and Furuya, M. (2000). Elementary processes of photoperception by phytochrome A for high-irradiance response of hypocotyl elongation in Arabidopsis. Plant Physiol. 112, 147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers, D.E., Webb, A.A.R., Pearson, M., and Kay, S.A. (1998). The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125, 485–494. [DOI] [PubMed] [Google Scholar]
- Staiger, D., Allenbach, L., Salathia, N., Fiechter, V., Davis, S.J., Millar, A.J., Chory, J., and Fankhauser, C. (2003. a). The Arabidopsis SRR1 gene mediates phyB signaling and is required for normal circadian clock function. Genes Dev. 17, 256–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger, D., Zecca, L., Kirk, D.A., Apel, K., and Eckstein, L. (2003. b). The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J. 33, 361–371. [DOI] [PubMed] [Google Scholar]
- Strayer, C., Oyama, T., Schultz, T.F., Raman, R., Somers, D.E., Mas, P., Panda, S., Kreps, J.A., and Kay, S.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289, 768–771. [DOI] [PubMed] [Google Scholar]
- Tahir, M., Kanegae, H., and Takano, M. (1998). PHYC (phytochrome C) gene in rice: Isolation and characterization of a complete coding sequence (accession No. AB018442). Plant Physiol. 118, 1555. [Google Scholar]
- Thomas, B., and Vince-Pruce, D. (1997). Photoperiodism in Plants. (San Diego, CA: Academic Press).
- Verwoerd, T.C., Dekker, B.M., and Hoekema, A. (1989). A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 17, 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.Y., Kenigsbuch, D., Sun, L., Harel, E., Ong, M.S., and Tobin, E.M. (1997). A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9, 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.Y., and Tobin, E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217. [DOI] [PubMed] [Google Scholar]
- Whitelam, G.C., Johnson, E., Peng, J., Carol, P., Anderson, M.L., Cowl, J.S., and Harberd, N.P. (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5, 757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa, Y., Mukai, H., Asada, K., Hino, F., and Kato, I. (1998). Differential display with carboxy-X-rhodamine-labeled primers and the selection of differentially amplified cDNA fragments without cloning. Anal. Biochem. 256, 82–91. [DOI] [PubMed] [Google Scholar]
- Young, M.W., and Kay, S.A. (2001). Time zones: A comparative genetics of circadian clocks. Nat. Rev. Genet 2, 702–715. [DOI] [PubMed] [Google Scholar]