Abstract
The ability to logically engineer novel cellular functions promises a deeper understanding of biological systems. Here we demonstrate the rational design of cellular memory in yeast that employs autoregulatory transcriptional positive feedback. We built a set of transcriptional activators and quantitatively characterized their effects on gene expression in living cells. Modeling in conjunction with the quantitative characterization of the activator-promoter pairs accurately predicts the behavior of the memory network. This study demonstrates the power of taking advantage of components with measured quantitative parameters to specify eukaryotic regulatory networks with desired properties.
Keywords: Rational design, synthetic biology, positive feedback, memory, in vivo quantitation
The field of synthetic biology aims to design biological systems to perform tasks to better understand analogous natural systems and for direct applications in research and medicine (e.g., see Andrianantoandro et al. 2006; Drubin et al. 2007). Currently our ability to design biological systems is limited by the difficulty of predicting the behavior of even simple genetic circuits because often a given network topology will show qualitatively different behavior depending on the quantitative features of the underlying components. While advances have been made with networks composed of well-studied modules in bacteria (Guido et al. 2006), this challenge is acute in more complex eukaryotic systems where the components rarely have measured characteristics. Therefore, to make rapid progress in designing eukaryotic systems, synthetic biologists require both a pool of quantitatively annotated biological parts and the knowledge that these parts can be logically engineered into more complex networks with predictable function.
One such network, which carries intrinsic value and tests a bottom-up design approach, is a network that confers memory. Memory, which can be defined as a protracted response to a transient stimulus, is exemplified in differentiation where a precursor cell makes a permanent and heritable cell fate decision in response to transient signals. Two major feedback motifs characterized in natural systems that exhibit memory are mutual inhibition and autoregulatory positive feedback (e.g., see Xiong and Ferrell 2003; Ptashne 2004; Huang et al. 2006; Zordan et al. 2006). Initial work in building ectopic cellular memory has demonstrated a bistable transcriptional mutual repression switch, a “toggle switch,” in bacteria and mammalian cells (Gardner et al. 2000; Kramer et al. 2004). Others have executed autoregulatory positive feedback designs to varying levels of success (Becskei et al. 2001; Atkinson et al. 2003; Kramer and Fussenegger 2005; Vilaboa et al. 2005; Ingolia and Murray 2007). However, none of the eukaryotic synthetic network studies succeed at demonstrating predictable behavior of a system.
In this study we describe the rational design and construction of a high fidelity, modular memory device in yeast based on transcriptionally controlled autoregulatory positive feedback. This device heritably retains an induced state in individual cells in response to a transient stimulus. The rational design approach used here employs an extensive in vivo quantitative characterization of a set of synthetic transcription factors and the prediction of system behavior via network models incorporating these measured parameters. By successfully constructing this memory device, we established the essential parameters for maintaining an autoregulatory positive feedback loop in a dividing cellular system. Most importantly, we demonstrated predictability of system behavior in eukaryotes when the system is built from well-understood components.
Results and Discussion
Functional activators based on a modular architecture
To rationally engineer a memory device, we designed a set of fluorescently labeled synthetic transcription factors and their corresponding reporter genes to serve as candidate components. Each activator gene consists of a DNA-binding domain (DBD), two tandem copies of the monomeric red fluorescent protein (RFP) mCherry (Shaner et al. 2004), the viral activation domain VP64 (Beerli et al. 1998), and the SV40 nuclear localization sequence (NLS) (Kalderon et al. 1984; Lanford and Butel 1984), all under control of the GAL1/10 promoter (Fig. 1A). Each reporter gene has multiple copies of the DNA-binding sites corresponding to its given transcription factor upstream of the minimal CYC1 promoter, and its protein coding region encodes two tandem copies of the yellow fluorescent protein variant (YFP) Venus (Fig. 1A; Nagai et al. 2002). The DBDs used were the LexA DBD (Hurstel et al. 1986, 1988), an engineered variant of the murine zinc-finger Zif268 (ZifH) (Hurt et al. 2003), and three human zinc fingers (ERG2, Gli1, and YY1) (Chavrier et al. 1990; Kinzler and Vogelstein 1990; Shi et al. 1991).
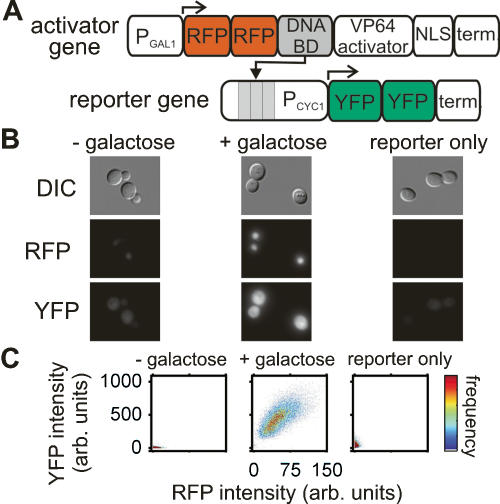
Figure 1.
Synthetic transcriptional activators in yeast. (A) Schematic diagram of an activator cascade composed of an activator gene and a reporter gene. (B) DIC and fluorescence images of live cells containing the ZifH activator cascade in the absence (−galactose) and presence (+galactose) of the inducer or only the ZifH reporter gene in the presence of the inducer. (C) Dot plots of YFP versus RFP fluorescence intensity for single cells grown as in B.
To evaluate the function of these constructs in yeast, we constructed two types of strains—one type carrying both the activator and the reporter gene, and the other carrying only the reporter gene. When activator expression was induced in the activator–reporter strains, we observed intense red nuclear fluorescence, and, in four of the five strains, a significant increase in cellular YFP intensity, indicating that the activators are correctly expressed and localized and the reporter genes are activated (Fig. 1B). To test whether reporter gene activation requires activator expression, we also examined fluorescence of the reporter-only strains under induction conditions. Three reporter-only strains showed no significant YFP fluorescence (Fig. 1B). These observations were confirmed over larger populations of cells by flow cytometry (Fig. 1C; Supplementary Table S1). We therefore conclude that the LexA, ZifH, and Gli1 activator–reporter strains each contain a functional transcriptional activation cascade.
Quantitative characterization of a subset of transcriptional activators
With this set of functional activators, we had components to construct a positive autofeedback loop. Modeling (see the next section, Creating a Cellular Memory Device) indicates that only feedback loops with particular quantitative features will result in systems that will switch between two stable states upon transient induction. Specifically, the production rate of a reporter as a function of the activator concentration constrains which systems will be bistable and maintain memory. Thus, we developed quantitative understanding of the in vivo behavior of the LexA and ZifH activator–reporter strains both at steady state and dynamically.
To probe their steady-state behavior, we measured by flow cytometry single-cell YFP and RFP intensities of the LexA and ZifH activator–reporter strains that were grown to steady state in the presence of various amounts of galactose. Variations in the transcriptional activator concentration for the LexA and ZifH activation cascades modulate to different degrees the transcriptional up-regulation of the reporter gene (Fig. 2A,B). To describe this relationship between activators and reporters, in which both concentrations are in the same units (i.e., molecules cell−1), we established calibration curves between mean fluorescence intensity per cell and mean number of proteins per cell as measured by quantitative immunoblotting for both RFP and YFP (Supplementary Fig. S1; Wu and Pollard 2005).
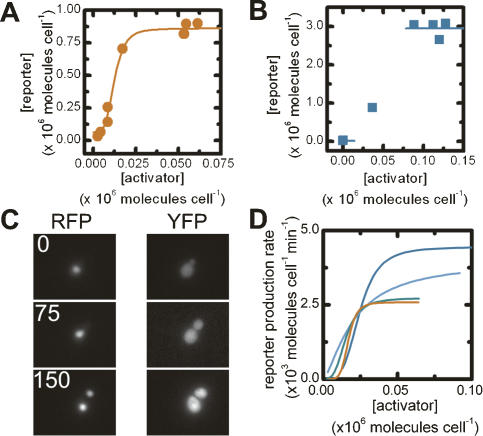
Figure 2.
Steady-state and dynamic behavior of activation cascades. The mean steady-state reporter versus activator concentrations for the ZifH (A) and LexA (B) activator–reporter strains. The solid lines show best fits to the data. Only s and β could be robustly determined in B. (C) Time-lapsed RFP and YFP fluorescence images of cells containing the ZifH activation cascade 0, 75, and 150 min after induction. (D) Rate of change of reporter concentration as a function of activator concentration for individual cells (red, blue, green, and light-blue lines).
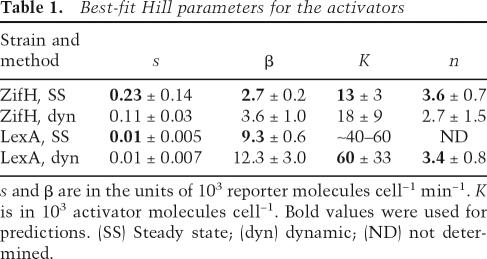
We chose to describe the relationship between the production rate of reporter protein and the activator concentration by a Hill-type relation. Cooperativity in this system is expected to arise from the inherent cooperativity of eukaryotic transcription (Polach and Widom 1996; Vashee et al. 1998; Wang et al. 1999). We fit the steady-state reporter concentration as a function of activator concentration (Eq. 4) to determine s (the basal expression rate of the reporter gene), β (the maximal production rate of the reporter protein), n (the Hill coefficient), and K (the activator concentration required to give half the maximal production rate) for multiple data sets for the two transcriptional activators. While we were able to obtain best-fit values for the ZifH activator, the lack of intermediate activator and reporter concentrations only permitted robust determination of s and β for the LexA activator strain (Fig. 2B; Table 1). The differences in the parameter values (Table 1) for two different DBDs show that different implementations of the same gene-circuit architecture can lead to quantitatively different behaviors.
Table 1.
Best-fit Hill parameters for the activators
s and β are in the units of 103 reporter molecules cell−1 min−1. K is in 103 activator molecules cell−1. Bold values were used for predictions. (SS) Steady state; (dyn) dynamic; (ND) not determined.
Because the parameters derived from these steady-state experiments are based on a relatively few number of data points, we also measured them with a kinetic experiment. We grew each activator strain in noninduced conditions, switched the cells to galactose-containing media to induce activator synthesis, then used time-lapse microscopy to track activator and reporter concentrations via fluorescence in single cells as a function of time (Fig. 2C). The activator concentration gradually rises over several hours; this experiment thus provides measurements of the reporter protein production rate sampled over many different activator concentrations. We used Cell-ID (Gordon et al. 2007) to extract fluorescence intensities and volumes of single cells as a function of time. We calculated the activator concentration and reporter production rate for each time point (refer to Supplemental Material; Supplementary Figs. S2, S3), and for each cell, fit this relationship to the Hill equation (Eq. 1). Again, different activator strains showed distinctly different reporter response rates relative to the activator concentration, but the resulting best-fit Hill parameters for each activator–reporter pair are strikingly similar to the values from the steady-state experiments (Fig. 2D; Table 1). This similarity between two different types of measurements strongly suggests our parameters accurately describe the transcriptional activators’ in vivo behaviors. Such similarity also validates the use of the kinetic measurements as a new method of characterizing eukaryotic transcription factors in vivo.
Creating a cellular memory device
We next set out to construct a transcriptional positive feedback loop intended to confer “memory” of a stimulus to a yeast cell and its progeny (Fig. 3A). In our proposed memory device, a signal induces synthesis of a “sensor” transcription factor, which triggers the expression of an “autofeedback” transcription factor. This autofeedback activator binds to its own promoter and, under appropriate circumstances, will continue to activate its own expression even in the absence of stimulus, resulting in memory. This synthetic network can exist in three different steady states: never exposed to stimulus (“off”), stimulus present (“on”), and previously exposed to stimulus (“memory”). In our implementation, the sensor activator is labeled with RFP, the autofeedback activator is labeled with YFP, and the stimulus is galactose.
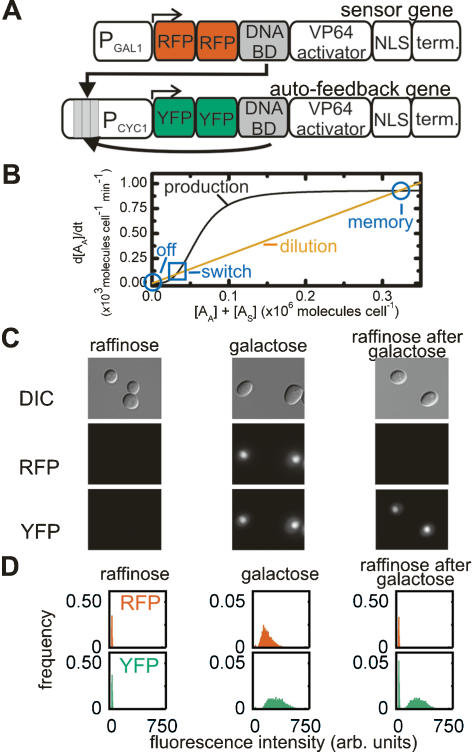
Figure 3.
A memory device in yeast. (A) Schematic diagram of the sensor and autofeedback genes of the memory device. (B) The predicted rate of change in the autofeedback activator concentration as a function of the total activator concentration for the LexA memory device. The decay rate (orange line) was calculated using Equation 2 and τ = 240 min, while the production rate (black line) was calculated using Equation 1; the values of s, n, and K for the LexA activator–promoter pair (Table 1, bold); and the estimated value of β for the LexA autofeedback activator. The high stable state (memory) and the unstable steady state (switch) of the autofeedback loop are indicated. (C) DIC and fluorescence images of live cells harboring the LexA memory device growing in the absence of stimulus (raffinose), in the presence of stimulus (galactose), and in the absence of stimulus after transient induction (raffinose after galactose). (D) The frequency distribution of RFP and YFP fluorescence intensities from single cells as in C.
For this device to exhibit memory, the autofeedback loop must contain two stable states. In terms of our mathematical model, the production rate of the autofeedback activator must be balanced by its decay rate at three activator concentrations (Eq. 5; Fig. 3B). The lowest and highest of these concentrations correspond to the “off” and “memory” stable states, while the intermediate concentration is an unstable steady state (“switch”) where the system switches between the stable states. In the off stable state, the basal levels of activator stimulate low levels of autoactivator synthesis that decrease rapidly due to the comparatively high rate of autoactivator decay. In contrast, in the memory state, the high concentration of autoactivator catalyzes a fast rate of its own synthesis that balances the decay rate and perpetuates its own production. Under this model the growth rate of the yeast cells becomes important, since the effective dilution of the activator during cell growth dictates its decay rate (Supplementary Fig. S3). Bistability of the autofeedback circuit can only be achieved if the synthesis of the autoactivator is a cooperative process (n > 1). For the memory state to respond specifically to a transient stimulus two more conditions must be met. First, the sum of the noninduced level of sensor activator and autoactivator must be less than the switch concentration; otherwise, the system will always be in the memory state. Second, in the presence of the stimulus, the sensor activator concentration must be greater than the switching concentration in order to jump start autoactivator production and shift the system into the memory state (Fig. 3B).
We used the measured Hill parameters, together with several assumptions, to predict the behavior of an autofeedback loop in which either LexA or ZifH were used as the DBD. First, we assume that the transcriptional activator–promoter pair dictates K and n such that these parameters are comparable in an activator–reporter strain and its corresponding memory device strain. Since β depends on the protein being synthesized, we also assume that s and β for autoactivator synthesis can be scaled from s and β for reporter synthesis by a proportionality constant. This constant is set to the ratio of the RFP intensity from a strain carrying a tandem set of mCherry modules under control of the GAL1/10 promoter to the RFP fluorescence from the LexA or ZifH activator strain (0.10 and 0.043, respectively). Our model, which has no free parameters with these assumptions, indicates that the LexA activator can be used to make a switchable bistable system at certain growth rates (Fig. 3B). However, this model predicts that the basal levels of ZifH activator will be greater than the “switch” concentration for the ZifH system, and thus cause a switch into the memory state independently of exposure to a stimulus (data not shown). This effect is predicted for the ZifH memory loop because of the higher leakiness of the ZifH promoter (i.e., its higher value of s), coupled with the more potent effect of ZifH activator on gene expression (i.e., its lower value of K) (Table 1). We tested the model predictions by building both networks and characterizing their responses.
The LexA memory network indeed responds specifically to induction and shows bistability, creating a high-fidelity in vivo three-state memory device. Cells with the LexA network never exposed to the induction stimulus show no significant RFP or YFP fluorescence (Fig. 3C,D); i.e., are in the “off” state. Cells grown to steady-state in the presence of the induction stimulus are in the “on” state, showing significant fluorescence for both RFP and YFP. After the stimulus is removed and cells re-establish a steady-state, the sensing gene is no longer expressed, but the feedback activator continues to be present at the same level (Fig. 3C,D). These cells are in the “memory” state. Observation over approximately eight cell divisions by microscopy shows that ∼90% of cells retain the memory state continuously (data not shown). Bistability of the loop was verified by flow cytometry, which shows that in all three states, individual cells fall into two well-separated populations based on their fluorescence intensity (Fig. 3D). In contrast, the ZifH memory network is, as predicted, fixed in the “memory” state before induction (Supplementary Table S1).
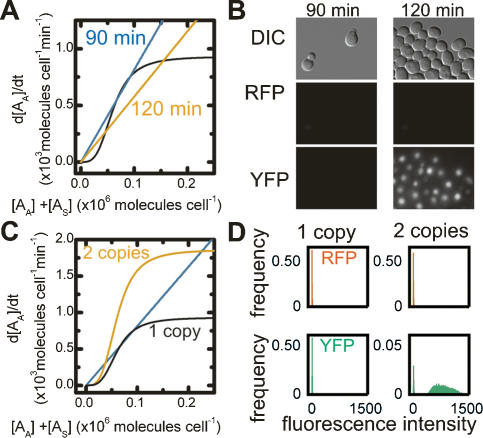
The above results for the LexA system were obtained in raffinose with a doubling rate of 240 min. As discussed, the effective degradation rate of the activator is determined by the doubling rate of the cells. In glucose liquid culture the doubling time of the culture decreases to ∼90 min. Under these growth conditions, our model suggests that the LexA loop is just on the border between a mono- and bistable system (Fig. 4A), and would likely lose the memory state: The production of the autoactivator cannot keep up with the increased dilution of the autoactivator, so in the absence of the “sensor” activator, the system relaxes to the “off” state. In fact, we observed no RFP or YFP fluorescence from the LexA memory strain after transient induction, indicating it cannot hold the memory state when grown in glucose liquid culture (Fig. 4B). Our model also suggests that a modest change in growth rate—i.e., from a doubling time of 90 min to 120 min—would return the autofeedback loop to a distinctly bistable region of phase space (Fig. 4A); such a change would sufficiently slow the decay of the autoactivator so that the production rate can keep pace with autoactivator dilution. This change in growth rate was achieved by growing the cells in glucose on solid media rather than in liquid culture, and indeed, the cells with a slower doubling time retain memory; i.e., maintain YFP fluorescence after transient induction (Fig. 4B). Thus, we can reliably tune the functionality of our memory device by controlling growth rate.
Figure 4.
Regulating bistability of the memory device. (A) The calculated rate of change of the autofeedback activator concentration versus the total activator concentration for the LexA memory device. Two decay rates, calculated using Equation 2 with τ = 90 min (blue line) and 120 min (orange line), predict the effect of decreasing the growth rate on bistability of the autofeedback loop. The production rate versus activator concentration (black line) is as in Figure 3B. (B) DIC and fluorescence images of cells carrying the LexA memory device that were grown in inducer, then to steady state in glucose with a doubling time of 90 or 120 min. (C) The predicted rate of change of autofeedback activator concentration as a function of the total activator concentration for the LexA memory device and the dual autofeedback memory device. The production rates were calculated using Equation 1, the values of n and K for the LexA activator–reporter strain, and either β (black line) or 2*β (orange line) for the LexA autofeedback activator corresponding to one and two copies of the autofeedback gene, respectively. The decay rate (blue line) corresponds to a doubling time of 90 min. (D) The frequency distribution of RFP and YFP intensities from the LexA memory device strain with one and two copies of the autofeedback gene grown in glucose after induction.
As a further test of our model, we attempted to modify the memory system to hold the memory state even under rapid growth conditions. Possible alterations include increasing the Hill coefficient (n), decreasing the concentration of activator required for half maximal activation (K), or increasing the maximum production rate (β). Increasing n or decreasing K would require alterations to the transcriptional activator or promoter and would require recharacterization of the system. We therefore modified β by simply adding an identical autofeedback gene at a second locus. Assuming the two copies act identically, this change doubles s and β of autoactivator, without affecting n or K. We predicted that the basal expression rate of two autofeedback genes would still be low enough to maintain the “off” state in the absence of inducer, but the increased production rate would maintain the memory state at high growth rate (Fig. 4C). Indeed, the dual autofeedback memory device switches specifically and remains in the memory state even in liquid glucose culture (Fig. 4D).
Here we demonstrated a memory device composed of synthetic transcription factors in an autoregulatory positive feedback motif that retains an induced state in a heritable fashion in response to a transient stimulus. This result supports previous studies showing that autoregulatory positive feedback loops are capable of a switch-like output in both yeast and mammalian cells (Becskei et al. 2001; Kramer and Fussenegger 2005; Vilaboa et al. 2005; Ingolia and Murray 2007). However, the device presented here integrates high-fidelity memory, with both potential interchangeability with regard to stimulus and a quantitative, single-cell description of the system. Such a defined memory module can be exploited in research applications to identify cells that experience specific events and determine whether this correlates with later behavior, or could be incorporated into a more complex network that causes a cell to “differentiate” in a certain fashion after experiencing a defined event. The memory module also potentially fills a direct need in industrial biotechnology: It permits high induction of recombinant proteins without the high cost of large quantities of inducer.
Furthermore, as far as we know, this is the first demonstration in a eukaryotic system that quantitatively characterized components can be used to build a functional circuit with predictable behavior. This success opens the door for rapid construction of eukaryotic devices using rational design with these or other well-characterized parts, and it suggests that a limited set of important control parameters may govern the behavior of naturally occurring autofeedback loops. Specifically, our results highlight the rate of synthesis and decay of the autofeedback elements as potential regulation points for naturally occurring autofeedback circuits that govern cell fate decisions, and suggest that the mechanisms that generate cooperativity in bacteria are not required for native eukaryotic systems. In general, these studies reinforce the idea that output of a system is not dictated simply by the network motif, rather the quantitative characteristics of system components are key to obtaining desired behavior. With this work, we demonstrate that we can predictably engineer in vivo eukaryotic networks, paving the way for complex synthetic devices that can tackle sophisticated technical and scientific challenges.
Materials and methods
Plasmid cloning and yeast strains
Constructs were made via a BioBrick assembly method (Knight 2003; Phillips and Silver 2006). Genes were cloned into yeast integrating shuttle vectors (Sikorski and Hieter 1989). Yeast strains are single-site integrations of the strain PSY580A (MATa, ura3-52, trp1Δ63, leu2Δ1). Activator and reporter genes were integrated into the LEU2 and URA3 loci, respectively. The autofeedback gene was inserted in the URA3 locus, the second at TRP1. Yeast were grown for at least 24 h at 30°C in media for induction (2% galactose, 2% raffinose), noninduction (2% raffinose), and repression (2% glucose). For measurements, cells were grown to mid-log phase (approximate OD600 of 0.3–0.6).
Flow cytometry and fluorescence microscopy
For flow cytometry analysis, we used a Dako MoFlo (Glostrup, Denmark) with 488- and 568-nm tunable lasers.
For live-cell imaging, a drop of cell suspension was placed on an agarose pad with media, and placed cell-side down on a glass-bottom dish. Images were acquired on an Eclipse TE2000-E equipped with a 60× objective (Nikon), Orca 285 CCD camera, Metamorph 6.3r7 software, and JP2 (YFP) and HcRed (RFP) filter sets (Chroma).
YFP and RFP intensity values were normalized such that a control strain had the same intensity between experiments. Reported intensity values are normalized, background-corrected averages.
Modeling of reporter and autoactivator synthesis
We use a single mathematical framework to describe the relationship between the activator concentration and the production rate of either the reporter or autofeedback activator. We assume that the rate of production of either the reporter or autoactivator species, (dX/dt)produce, can be described by the sum of a basal rate and a Hill function:
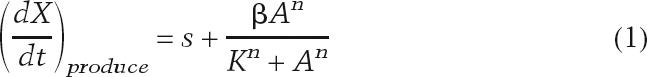 |
where s is the basal rate of production of activator, n is the Hill cooperativity, K is the concentration of activator that yields half the maximal production rate, β is the maximal rate of protein production, and A is the activator concentration (Alon 2006).
The decay rate of either the reporter or autoactivator concentration is affected by both dilution due to cell growth and protein degradation. However, since the reporter protein and the autoactivator are degraded at a rate that is many times slower than their dilution rate (Supplementary Fig. S3), the decay in the species concentration can be simplified to
 |
where τ is the time required for the number of cells to double by growth. The overall change in the concentration of the reporter or autofeedback activator can then be written as
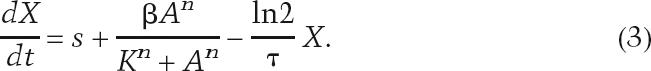 |
At steady state, the production rate is balanced by the decay rate, so the relationship between the steady-state activator concentration, A, and steady-state reporter concentration, R, can be written as
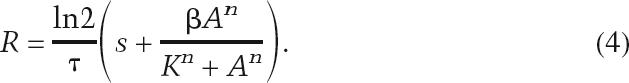 |
In the case of the positive feedback loop, we assume that the autofeedback gene is activated identically by the sensor-derived activator and the autofeedback activator, so we rewrite Equation 3 as
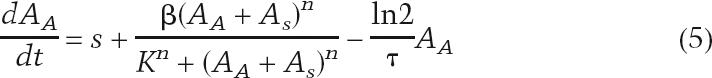 |
where AA is the autoactivator concentration and AS is the sensor-derived activator concentration.
All mathematical modeling was performed using MATLAB (The MathWorks).
Acknowledgments
We thank R. Ward and J. Wintermute for helpful comments on the manuscript. C.M.A. was supported by NIH Training Grant #T32CA09361, D.A.D. was supported through an NIH NRSA, and E.PS.G. was supported by an NIH Training Grant. This work was supported in part by NIH grants to P.A.S.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1586107
References
- Alon U. An introduction to systems biology: Design principles of biological circuits. Chapman & Hall/CRC Press; Boca Raton, FL: 2006. [Google Scholar]
- Andrianantoandro E., Basu S., Karig D.K., Weiss R., Basu S., Karig D.K., Weiss R., Karig D.K., Weiss R., Weiss R. Synthetic biology: New engineering rules for an emerging discipline. Mol. Syst. Biol. 2006;2:2006.0028. doi: 10.1038/msb410007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson M.R., Savageau M.A., Myers J.T., Ninfa A.J., Savageau M.A., Myers J.T., Ninfa A.J., Myers J.T., Ninfa A.J., Ninfa A.J. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell. 2003;113:597–607. doi: 10.1016/s0092-8674(03)00346-5. [DOI] [PubMed] [Google Scholar]
- Becskei A., Seraphin B., Serrano L., Seraphin B., Serrano L., Serrano L. Positive feedback in eukaryotic gene networks: Cell differentiation by graded to binary response conversion. EMBO J. 2001;20:2528–2535. doi: 10.1093/emboj/20.10.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli R.R., Segal D.J., Dreier B., Barbas C.F., Segal D.J., Dreier B., Barbas C.F., Dreier B., Barbas C.F., Barbas C.F. Toward controlling gene expression at will: Specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl. Acad. Sci. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavrier P., Vesque C., Galliot B., Vigneron M., Dolle P., Duboule D., Charnay P., Vesque C., Galliot B., Vigneron M., Dolle P., Duboule D., Charnay P., Galliot B., Vigneron M., Dolle P., Duboule D., Charnay P., Vigneron M., Dolle P., Duboule D., Charnay P., Dolle P., Duboule D., Charnay P., Duboule D., Charnay P., Charnay P. The segment-specific gene Krox-20 encodes a transcription factor with binding sites in the promoter region of the Hox-1.4 gene. EMBO J. 1990;9:1209–1218. doi: 10.1002/j.1460-2075.1990.tb08228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin D.A., Way J.C., Silver P.A., Way J.C., Silver P.A., Silver P.A. Designing biological systems. Genes & Dev. 2007;21:242–254. doi: 10.1101/gad.1507207. [DOI] [PubMed] [Google Scholar]
- Gardner T.S., Cantor C.R., Collins J.J., Cantor C.R., Collins J.J., Collins J.J. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- Gordon A., Colman-Lerner A., Chin T.E., Benjamin K.R., Yu R.C., Brent R., Colman-Lerner A., Chin T.E., Benjamin K.R., Yu R.C., Brent R., Chin T.E., Benjamin K.R., Yu R.C., Brent R., Benjamin K.R., Yu R.C., Brent R., Yu R.C., Brent R., Brent R. Single-cell quantification of molecules and rates using open-source microscope-based cytometry. Nat. Methods. 2007;4:175–181. doi: 10.1038/nmeth1008. [DOI] [PubMed] [Google Scholar]
- Guido N.J., Wang X., Adalsteinsson D., McMillen D., Hasty J., Cantor C.R., Elston T.C., Collins J.J., Wang X., Adalsteinsson D., McMillen D., Hasty J., Cantor C.R., Elston T.C., Collins J.J., Adalsteinsson D., McMillen D., Hasty J., Cantor C.R., Elston T.C., Collins J.J., McMillen D., Hasty J., Cantor C.R., Elston T.C., Collins J.J., Hasty J., Cantor C.R., Elston T.C., Collins J.J., Cantor C.R., Elston T.C., Collins J.J., Elston T.C., Collins J.J., Collins J.J. A bottom-up approach to gene regulation. Nature. 2006;439:856–860. doi: 10.1038/nature04473. [DOI] [PubMed] [Google Scholar]
- Huang G., Wang H., Chou S., Nie X., Chen J., Liu H., Wang H., Chou S., Nie X., Chen J., Liu H., Chou S., Nie X., Chen J., Liu H., Nie X., Chen J., Liu H., Chen J., Liu H., Liu H. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc. Natl. Acad. Sci. 2006;103:12813–12818. doi: 10.1073/pnas.0605270103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurstel S., Granger-Schnarr M., Daune M., Schnarr M., Granger-Schnarr M., Daune M., Schnarr M., Daune M., Schnarr M., Schnarr M. In vitro binding of LexA repressor to DNA: Evidence for the involvement of the amino-terminal domain. EMBO J. 1986;5:793–798. doi: 10.1002/j.1460-2075.1986.tb04283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurstel S., Granger-Schnarr M., Schnarr M., Granger-Schnarr M., Schnarr M., Schnarr M. Contacts between the LexA repressor—or its DNA-binding domain—and the backbone of the recA operator DNA. EMBO J. 1988;7:269–275. doi: 10.1002/j.1460-2075.1988.tb02809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt J.A., Thibodeau S.A., Hirsh A.S., Pabo C.O., Joung J.K., Thibodeau S.A., Hirsh A.S., Pabo C.O., Joung J.K., Hirsh A.S., Pabo C.O., Joung J.K., Pabo C.O., Joung J.K., Joung J.K. Highly specific zinc finger proteins obtained by directed domain shuffling and cell-based selection. Proc. Natl. Acad. Sci. 2003;100:12271–12276. doi: 10.1073/pnas.2135381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N.T., Murray A.W., Murray A.W. Positive-feedback loops as a flexible biological module. Curr. Biol. 2007;17:668–677. doi: 10.1016/j.cub.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D., Richardson W.D., Markham A.F., Smith A.E., Richardson W.D., Markham A.F., Smith A.E., Markham A.F., Smith A.E., Smith A.E. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- Kinzler K.W., Vogelstein B., Vogelstein B. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol. Cell. Biol. 1990;10:634–642. doi: 10.1128/mcb.10.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight T. 2003. [Google Scholar]
- Kramer B.P., Fussenegger M., Fussenegger M. Hysteresis in a synthetic mammalian gene network. Proc. Natl. Acad. Sci. 2005;102:9517–9522. doi: 10.1073/pnas.0500345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer B.P., Viretta A.U., Daoud-El-Baba M., Aubel D., Weber W., Fussenegger M., Viretta A.U., Daoud-El-Baba M., Aubel D., Weber W., Fussenegger M., Daoud-El-Baba M., Aubel D., Weber W., Fussenegger M., Aubel D., Weber W., Fussenegger M., Weber W., Fussenegger M., Fussenegger M. An engineered epigenetic transgene switch in mammalian cells. Nat. Biotechnol. 2004;22:867–870. doi: 10.1038/nbt980. [DOI] [PubMed] [Google Scholar]
- Lanford R.E., Butel J.S., Butel J.S. Construction and characterization of an SV40 mutant defective in nuclear transport of T antigen. Cell. 1984;37:801–813. doi: 10.1016/0092-8674(84)90415-x. [DOI] [PubMed] [Google Scholar]
- Nagai T., Ibata K., Park E.S., Kubota M., Mikoshiba K., Miyawaki A., Ibata K., Park E.S., Kubota M., Mikoshiba K., Miyawaki A., Park E.S., Kubota M., Mikoshiba K., Miyawaki A., Kubota M., Mikoshiba K., Miyawaki A., Mikoshiba K., Miyawaki A., Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Phillips I., Silver P.A., Silver P.A. 2006. [Google Scholar]
- Polach K.J., Widom J., Widom J. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J. Mol. Biol. 1996;258:800–812. doi: 10.1006/jmbi.1996.0288. [DOI] [PubMed] [Google Scholar]
- Ptashne M. A genetic switch: Phage lambda revisited. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2004. [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y., Giepmans B.N., Palmer A.E., Tsien R.Y., Palmer A.E., Tsien R.Y., Tsien R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosomasp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shi Y., Seto E., Chang L.S., Shenk T., Seto E., Chang L.S., Shenk T., Chang L.S., Shenk T., Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashee S., Melcher K., Ding W.V., Johnston S.A., Kodadek T., Melcher K., Ding W.V., Johnston S.A., Kodadek T., Ding W.V., Johnston S.A., Kodadek T., Johnston S.A., Kodadek T., Kodadek T. Evidence for two modes of cooperative DNA binding in vivo that do not involve direct protein–protein interactions. Curr. Biol. 1998;8:452–458. doi: 10.1016/s0960-9822(98)70179-4. [DOI] [PubMed] [Google Scholar]
- Vilaboa N., Fenna M., Munson J., Roberts S.M., Voellmy R., Fenna M., Munson J., Roberts S.M., Voellmy R., Munson J., Roberts S.M., Voellmy R., Roberts S.M., Voellmy R., Voellmy R. Novel gene switches for targeted and timed expression of proteins of interest. Mol. Ther. 2005;12:290–298. doi: 10.1016/j.ymthe.2005.03.029. [DOI] [PubMed] [Google Scholar]
- Wang J., Ellwood K., Lehman A., Carey M.F., She Z.S., Ellwood K., Lehman A., Carey M.F., She Z.S., Lehman A., Carey M.F., She Z.S., Carey M.F., She Z.S., She Z.S. A mathematical model for synergistic eukaryotic gene activation. J. Mol. Biol. 1999;286:315–325. doi: 10.1006/jmbi.1998.2489. [DOI] [PubMed] [Google Scholar]
- Wu J.Q., Pollard T.D., Pollard T.D. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- Xiong W., Ferrell J.E., Ferrell J.E. A positive-feedback-based bistable ‘memory module’ that governs a cell fate decision. Nature. 2003;426:460–465. doi: 10.1038/nature02089. [DOI] [PubMed] [Google Scholar]
- Zordan R.E., Galgoczy D.J., Johnson A.D., Galgoczy D.J., Johnson A.D., Johnson A.D. Epigenetic properties of white–opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc. Natl. Acad. Sci. 2006;103:12807–12812. doi: 10.1073/pnas.0605138103. [DOI] [PMC free article] [PubMed] [Google Scholar]