Abstract
The MCM2-7 helicase complex is loaded on DNA replication origins during the G1 phase of the cell cycle to license the origins for replication in S phase. How the initiator primase–polymerase complex, DNA polymerase α (pol α), is brought to the origins is still unclear. We show that And-1/Ctf4 (Chromosome transmission fidelity 4) interacts with Mcm10, which associates with MCM2-7, and with the p180 subunit of DNA pol α. And-1 is essential for DNA synthesis and the stability of p180 in mammalian cells. In Xenopus egg extracts And-1 is loaded on the chromatin after Mcm10, concurrently with DNA pol α, and is required for efficient DNA synthesis. Mcm10 is required for chromatin loading of And-1 and an antibody that disrupts the Mcm10–And-1 interaction interferes with the loading of And-1 and of pol α, inhibiting DNA synthesis. And-1/Ctf4 is therefore a new replication initiation factor that brings together the MCM2-7 helicase and the DNA pol α–primase complex, analogous to the linker between helicase and primase or helicase and polymerase that is seen in the bacterial replication machinery. The discovery also adds to the connection between replication initiation and sister chromatid cohesion.
Keywords: And-1/CTF4, DNA replication, genome stability, cell cycle, DNA polymerase
In eukaryotic cells, DNA replication is dependent on the action of ORC, Cdc6, and Cdt1 to load the MCM2-7 complex onto replication origins, resulting in the formation of prereplicative complexes (pre-RC) (Bell and Dutta 2002). Following this, the activation of Cdc7/Dbf4 and Cdk2/Cyclin E protein kinases leads to the melting of origin DNA and initiation of DNA replication. MCM2-7 interacts with and is required to load Mcm10, which is itself critical for the loading of replication elongation factors like Cdc45 (Aparicio et al. 1997), GINS, and DNA polymerase α (pol α) (Mimura and Takisawa 1998; Takayama et al. 2003; Ricke and Bielinsky 2004; Gambus et al. 2006; Pacek et al. 2006). The loading of Mcm10 at replication origins precedes and is necessary for later loading of Replication Protein A (RPA) and DNA pol α onto replication origins. However, how the initiator primase–polymerase complex, DNA pol α, is brought to the origins is unclear.
Once DNA replication initiates, MCM2-7, Cdc45, Mcm10, and GINS proteins migrate with the replication fork along with DNA pol α and other replicative polymerases. In many bacteriophages the DNA helicase and the primase are part of the same molecule, while in bacteria, the helicase, DnaB and the primase, DnaG are in direct contact with each other (Soultanas 2005). In addition, a linker protein, the τ subunit, tethers the bacterial helicase DnaB to the catalytic subunits of the replicative polymerase (McHenry 2003; Bell 2006). Similarly in SV40, the helicase T antigen interacts directly with the replicative polymerase–primase, DNA pol α (Dornreiter et al. 1992; Huang et al. 1998). An analogous link is expected in the eukaryotic cellular replisome between the primase-containing DNA pol α and the MCM2-7 helicase, but such a linker has not yet been found.
Ctf4 (Chromosome transmission fidelity 4) in Saccharomyces cerevesiae was originally identified in a genetic screen for mutants affecting chromosome transmission fidelity (Kouprina et al. 1992). Later studies indicate that Ctf4 is required for sister chromatid cohesion (Hanna et al. 2001; Mayer et al. 2004; Petronczki et al. 2004), but the mechanism for this is unclear. Ctf4 was found to interact with DNA pol α in yeasts (Miles and Formosa 1992b; Zhou and Wang 2004). Although it is not essential for viability in budding yeast, mcl1, the homolog of CTF4 in Schizosaccharomyces pombe, is essential for viability, maintenance of genome integrity, DNA damage repair, and regulation of telomere replication (Williams and McIntosh 2002; Tsutsui et al. 2005). The Aspergillus nidulans homolog of CTF4, SepB, is also essential for viability (Harris and Hamer 1995). Mcl1/Ctf4 exhibits genetic interaction with genes involved in lagging strand DNA synthesis such as Rad2, Dna2, and Fen1 and physically interacts with DNA pol α (Formosa and Nittis 1999; Tsutsui et al. 2005), but its exact role in DNA replication has not yet been defined. The human homolog of Ctf4, And-1, was also identified and its function too is unknown (Kohler et al. 1997).
In this study we report that And-1/Ctf4/Mcl1 interacts with both Mcm10 and DNA pol α. The Mcm10–And-1 interaction is required for the loading of DNA pol α on chromatin and for DNA synthesis. DNA replication initiation factors were shown before to be linked to sister chromatid cohesion (Gillespie and Hirano 2004; Takahashi et al. 2004). Therefore, the implication of And-1/Ctf4 in replication initiation suggests a reason why it might be required for sister chromatid cohesion. The results explain how a crucial factor in the elongation machinery, the DNA polymerase–primase, is recruited to the initiation machinery at origins of replication, and begin to define the physical complex of proteins that is expected to tether the replicative helicase, MCM2-7 to the replicative polymerase–primase.
Results
Identification of And-1 as a protein that associates with Mcm10
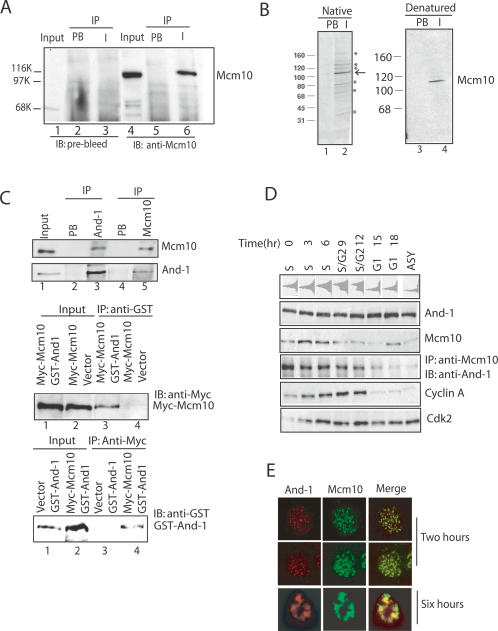
An antibody against full length of Mcm10 specifically immunoblotted and immunoprecipitated the 110-kDa Mcm10 protein from cell lysates (Fig. 1, lanes 4,6). Upon immunoprecipitation of Mcm10 from cells metabolically labeled with [35S]methionine, at least seven proteins of 30–170 kDa were detected in the Mcm10 precipitates (Fig. 1B, lane 2). Denaturation of cell lysates by boiling in 1% SDS resulted in only the 110-kDa Mcm10 protein being immunoprecipitated (Fig. 1B, lane 4), suggesting that the extra proteins in the native immunoprecipitate were brought down through noncovalent interactions with native Mcm10. Peptide sequences to Mcm10, Mcm6, and Mcm7 were identified when the protein bands in the nondenatured Mcm10 immunoprecipitate were excised and subjected to mass spectroscopy (Supplementary Table S1). Consistent with this, immunoblotting with a panMCM2-7 antibody detected five bands of 85–120 kDa in the Mcm10 immunoprecipitates (Supplementary Fig. S1; Supplementary Table S1), confirming previous reports that Mcm10 interacted with the MCM2-7 complex (Kawasaki et al. 2000; Lei et al. 2002).
Figure 1.
MCM10 is complexed to multiple proteins including And-1. (A) Characterization of the Mcm10 antibody. U2OS cells were lysed and immunoprecipitated with prebleed or anti-Mcm10. All immunoprecipitates were resolved on SDS-PAGE and immunoblotted with indicated antibodies. (IP) Immunoprecipitation; (PB) prebleed immunoprecipitates; (I) anti-Mcm10 immunoprecipitates. (B) SDS-PAGE and fluorography of immunoprecipitates. (Left panel) Lysates from [35S]methionine labeled 293T cells immunoprecipitated with prebleed (PB) or anti-Mcm10 (I) under native conditions. (Right panel) Lysates were denatured before immunoprecipitation. (*) Proteins coimmunoprecipitated with Mcm10 (arrow) under native conditions. (C) Mcm10 interacts with And-1. (Top) Cell lysates immunoprecipitated with prebleed, anti-And-1, or anti-Mcm10 and immunoblotted with anti-Mcm10 or anti-And-1 as indicated. (Middle) 293T cells transfected with plasmids expressing indicated proteins. Lysates were immunoprecipitated with anti-GST, followed by immunoblotting with anti-Myc antibody. (Bottom) Same as in the middle part, but the lysates were immunoprecipitated with anti-Myc and immunoblotted with anti-GST antibody. (D) Interaction of And-1 with Mcm10 is highest in S phase. (Top) Cells released for a thymidine/aphidicolin block were harvested for FACS at indicated time points. Immunoblot with indicated antibodies on cell lysates or on anti-Mcm10 immunoprecipitates. (E) And-1 and Mcm10 colocalize in S phase. 293T cells cotransfected with Flag-And-1 and Myc-Mcm10 were released from a thymidine/aphidicolin block and harvested at 2 or 6 h after release for immunofluorescence with anti-Flag for And-1 (red) and anti-Myc for Mcm10 (green). Mcm10 and And-1 colocalized in nuclei changing from a fine granular pattern seen by confocal microscopy in early S-phase cells (2 h after release) to a coarse clumpy pattern seen by indirect immunofluorescence in late S-phase cells (6 h after release).
The 125-kDa interactor was identified by mass spectrometry as the acidic nucleoplasmic DNA-binding protein, human And-1 (Supplementary Table S1; Kohler et al. 1997), indicating that And-1 may play a role in the DNA replication. To test this interaction, we raised an antibody against fragment (776–927) of And-1. Mcm10 was detected in the And-1 immunoprecipitates (Fig. 1C, top, lane 3) and And-1 was detected in the Mcm10 immunoprecipitate (Fig. 1C, top, lane 5). When Myc-Mcm10 and GST-And-1 were expressed by transient transfection of human 293T cells, Myc-Mcm10 coimmunoprecipitated with GST-And-1 and vice-versa (Fig. 1C, bottom). Thus, endogenous or transfected Mcm10 physically associates with endogenous or transfected And-1.
To determine when in the cell cycle And-1 interacts with Mcm10, U2OS osteosarcoma cells were synchronized in early S phase by thymidine/aphidicolin block and released (Fig. 1D). And-1 levels remained stable throughout the cell cycle, while Mcm10 protein levels fluctuated with an increase in S phase (Fig. 1D), as has been previously described (Izumi et al. 2001, 2004). The And-1 content of Mcm10 immunoprecipitates closely parallels the cellular levels of Mcm10, with the most abundant coimmunoprecipitation occurring in S phase when Mcm10 is most abundant (Fig. 1D, cf. lanes 1–4 and 7). We also examined the localization of both Mcm10 and And-1 in the S phase by immunostaining. Mcm10 and And-1 colocalized in nuclei changing from a fine granular immunofluorescence pattern in early S-phase cells to a coarse clumpy pattern in late S-phase cells (Fig. 1E). In summary, mammalian And-1 and Mcm10 associate with each other and colocalize in S phase.
Both Mcm10 and And-1 interact with DNA pol α p180
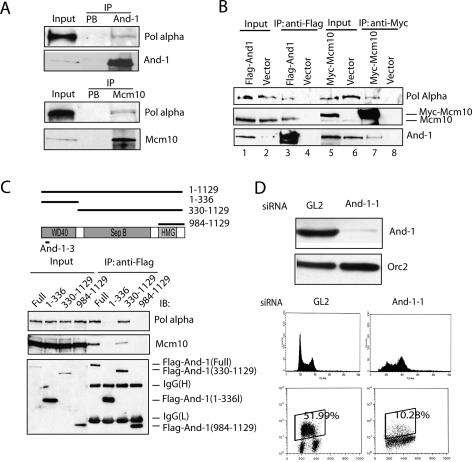
The budding yeast homolog of And-1, Ctf4, and Mcm10 interact with DNA pol α (Miles and Formosa 1992a, b; Ricke and Bielinsky 2004). To test whether the same was true in human cells, endogenous And-1 and Mcm10 were immunoprecipitated from cell lysates. Endogenous p180 can be detected in both And-1 and Mcm10 immunoprecipitates (Fig. 2A). From the analysis of the immunoblots, a very small percent of human pol α p180 coimmunoprecipitates with human And-1 and Mcm10, respectively, most likely reflecting the lability of these interactions in solution and post-lysis. Upon transient expression of Flag-And-1 in 293T cells, the p180 subunit of endogenous DNA pol α was detected in Flag-And-1 immunoprecipitates (Fig. 2B, lane 3). Endogenous p180 was also detected in Myc-Mcm10 immunoprecipitates (Fig. 2B, lane 7). Therefore, human And-1 forms a complex with Mcm10 and Pol α. The coimmunoprecipitation persisted in ethidium bromide or after DNase treatment, suggesting that DNA bridges were not involved in the coimmunoprecipitation. Whether And-1–Mcm10 and pol α form a ternary complex needs to be further investigated.
Figure 2.
And-1 interacts with p180 (Pol α) and is required for S phase. (A) Endogenous And-1 and Mcm10 associate with endogenous DNA Pol α. Immunoprecipitates with indicated antibodies immunoblotted for p180. (PB) Prebleed immunoprecipitates. Five percent of input p180 is loaded for comparison. (B) And-1, Mcm10, and Pol α coexist in a complex. 293T cells transfected with indicated plasmids were lysed and immunoprecipitated with anti-Flag or anti-Myc antibodies, followed by immunoblotting with indicated antibodies. (C) The WD40 domain is dispensable and the Sep B domain is required for interaction between And-1 and Pol α or Mcm10. (Top) Schematic of deletions and target site of And1-3. (Bottom) 293T cells transfected with indicated Flag-And-1 mutants were lysed and immunoprecipitated with anti-Flag, resolved on SDS-PAGE, and immunoblotted with indicated antibodies. (D) And-1 is required for S-phase progression. HCT116 cells were transfected twice with indicated siRNAs and harvested at 72 h of first transfection. Immunoblots with anti-And-1 and anti-Orc2 is shown. Cells were labeled with BrdU for 1 h before harvest. Propidium-iodide and two-color FACS shown: BrdU staining on Y-axis and propidium-iodide staining on X-axis. BrdU-labeled S-phase fractions are indicated.
We next made a series of truncation mutants of And-1 to determine whether a discrete part of And-1 is required for the association with Mcm10 and Pol α p180. And-1 contains three major domains: a WD40 repeat-containing domain at the N terminus, a SepB homology domain in the middle, and an HMG domain at the C terminus (Fig. 2C, top). Full-length And-1 and And-1 (330–1129) interact with p180 and Mcm10, and And-1 (1–336) does not, indicating that the N-terminal WD40 domain is dispensable for the interaction with p180 or Mcm10 (Fig. 2C). The extreme C terminus of And-1 (984–1129), however, is not sufficient for interaction with p180 or Mcm10.
And-1 is required for efficient DNA replication
Given that And-1 complexes with Mcm10 and Pol α, we next tested whether And-1, like Mcm10 and Pol α, is involved in DNA replication. Two separate small interfering RNA (siRNA) duplexes were designed to silence the expression of And-1 in HCT116 colon cancer cells (Figs. 2D, 3A, lanes 2,3). Propidium-iodide staining showed that depletion of And-1 increased the percent of cells between G1 and G2 peaks in the S-phase fraction (Fig. 2D, middle panel). To establish whether the And-1-depleted cells were in active DNA synthesis, they were labeled with Bromodeoxyuridine (BrdU) for 1 h before harvest. The incorporation of BrdU was monitored by two-color flow cytometry analysis of propidium-iodide-stained cells that were immunostained by anti-BrdU-FITC antibodies. As shown in Figure 2D (bottom panel), depletion of And-1 caused a fivefold decrease in the percentage of cells incorporating BrdU. Intriguingly, the And-1-depleted cells were not arrested in G1 or G2, but accumulated with arrested BrdU incorporation in early, middle, and late S phase (as judged from the propidium-iodide stain intensity), suggesting that And-1 may be critical for initiation events not only at early firing origins, but also initiation at late firing origins and/or for the elongation phase of DNA replication.
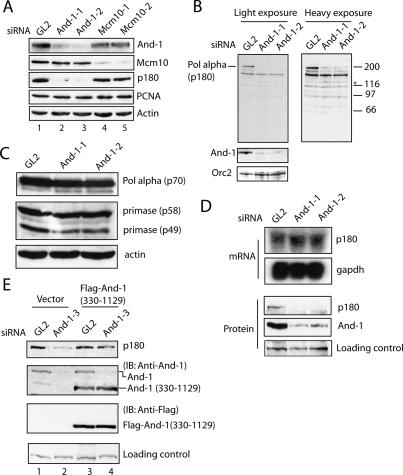
Figure 3.
And-1 is required for stability of DNA pol α (p180). (A) And-1 is required for the maintenance of p180 pol α levels. HCT116 cells were transfected twice with indicated siRNAs and harvested at 72 h after first siRNA transfection. Cell lysates were resolved by SDS-PAGE and immunoblotted with indicated antibodies. Actin and PCNA serve as loading controls. (B) A 125-kDa band related to p180 becomes visible after And-1 depletion. HCT116 cells treated as in A were immunoblotted for pol α (p180) (top panel), And-1 (middle panel), and Orc2 (bottom panel). (*) A 125-kDa band. The 150-kDa band visible in all lanes in the top left panel is a cross-reacting protein unrelated to p180: It does not decrease after siRNA to p180. (C) DNA pol α (p70), primase (p58), and primase (p49) protein levels are not affected in And-1-depleted cells. The same lysates as in B were immunoblotted for indicated proteins. (D) mRNA level of p180 is not affected in And-1-depleted cells. (Top panel) HCT116 cells were treated as in A and mRNA levels of p180 analyzed by Northern blotting. The blot was also probed for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a loading control. Northern blotting was performed as described elsewhere (Lee et al. 2005). (Bottom panel) Same cells treated as in the top panel were lysed and immunoblotted for indicated proteins. (E) Rescue of p180 level by exogenous And-1. 293T cells transfected with either empty vector or plasmid expressing Flag-And-1 (330–1129) were transfected with indicated siRNAs. Cell lysates were resolved on SDS-PAGE and immunoblotted with indicated antibodies.
And-1 is required for stabilization of DNA pol α (p180) in human cells
An intriguing result came to light when we silenced And-1 in HCT116 cells and examined the protein levels of Mcm10, And-1, or the p180 subunit of DNA pol α. p180 was markedly reduced in cells when And-1 was depleted (Fig. 3A [lanes 2,3], B). We noticed that a 120-kDa polypeptide reacting to the anti-p180 increased as p180 decreased, and may be a transient product of proteolysis of p180 (Fig. 3B). Pol α p180 normally forms a pol α–primase complex with three subunits, p70, p58, and p49. Unlike p180, the other subunits of pol α were not markedly decreased by And-1 depletion, although p49 is decreased a little (Fig. 3C). Decreased p180 protein levels in And-1-depleted cells suggest that And-1 either stabilizes p180 by forming a complex with it or regulates expression of pol α at the mRNA level. To distinguish between these two possibilities, we examined the mRNA level of pol α (p180) in And-1-depleted cells. As shown in Figure 3D, the mRNA level of p180 was not affected by the absence of And-1, suggesting that p180 is down-regulated at a post-transcriptional level. In budding yeast, p180 is degraded in cells when Mcm10 is depleted (Ricke and Bielinsky 2004). It appears that the function of stabilizing p180 has evolved in mammalian cells from Mcm10 to a partner of Mcm10, And-1 (Fig. 3A, lanes 2–5). Collectively, these results suggest that And-1 is required to stabilize the p180 subunit of pol α in human somatic cells.
To rule out off-target activities of the And-1 siRNA duplexes, we designed a third siRNA oligo And-1–3 targeting the N-terminal part of And-1. And-1–3 knocks down endogenous full-length And-1, but not a transiently expressed exogenous And-1 (330–1129), which interacts with pol α p180 (Figs. 2C, 3E). Again, p180 protein was degraded in And-1-depleted and vector-transfected cells (Fig. 3E, lane 2). However, the decrease of p180 after transfection of And-1–3 was rescued by the expression of exogenous And-1 (330–1129) (Fig. 3E, lane 4), indicating that the destabilization of p180 was specifically due to loss of And-1.
And-1 is required for efficient DNA replication in Xenopus egg extracts
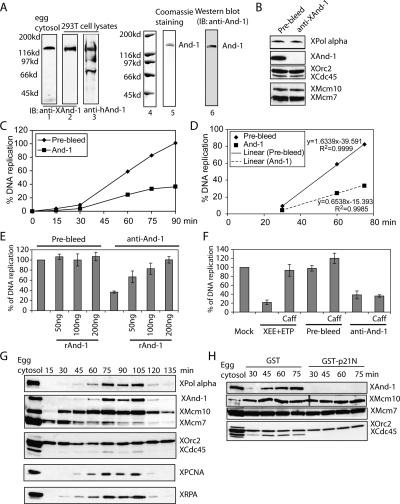
The requirement of And-1 for stabilizing p180 in mammalian cells complicated efforts to test whether And-1 recruits p180 to the chromatin. We therefore switched to the Xenopus egg system to test the functional relationship between XAnd-1 and pol α. An antibody against the C-terminal part of Xenopus And-1 specifically recognized XAnd-1 in Xenopus egg extracts and human And-1 in 293T cell lysates (Fig. 4A, left panel). The anti-XAnd-1 serum depleted XAnd-1 protein from egg extracts without depleting Mcm10 or p180 (Fig. 4B). Extracts depleted of XAnd-1 exhibited a significant decrease in DNA replication compared with mock-depleted extracts (Fig. 4C). And-1 depletion inhibited the initial velocity of DNA synthesis to 40% of that in prebleed-depleted extracts (Fig. 4D). To confirm that this deficit was solely due to the removal of XAnd-1, we expressed recombinant human His-And-1 from a baculovirus in insect Sf9 cells. The purified recombinant human And-1 protein was stained by Coomassie and its identity confirmed by immunoblotting (Fig. 4A right panel). The DNA replication defect of And-1-depleted extracts was completely reversed by addition of recombinant human And-1 (Fig. 4E). Therefore, And-1 itself is required for DNA replication in egg extracts and the effect is not due to codepletion of And-1-associated proteins (Fig. 4B).
Figure 4.
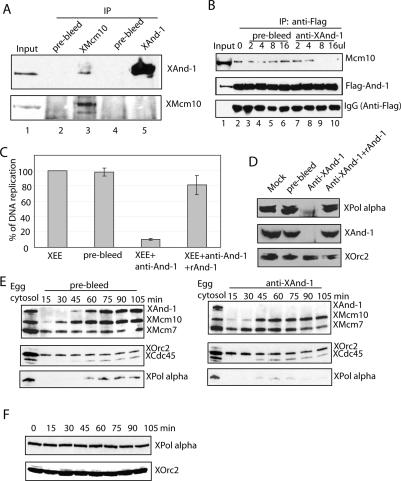
Xenopus And-1 is required for efficient DNA replication. (A) Characterization of the anti-XAnd-1 antibody and purification of recombinant hAnd-1. (Lanes 1–3) Xenopus egg cytosol or cell lysates from 293T cells were resolved on SDS-PAGE and immunoblotted with anti-XAnd-1 or anti-hAnd-1 antibody as indicated. (Lanes 5,6) Full-length His-tagged human And-1 was expressed in SF9 insect cells by a recombinant baculovirus and purified. Coomassie staining and Western blotting of purified And-1. (B) Immunodepletion of XAnd-1 from egg extracts. Egg cytosols depleted with prebleed or with anti-XAnd-1 were immunoblotted with indicated antibodies. (C) XAnd-1 is required for efficient DNA synthesis. Control or XAnd-1-depleted egg cytosols were used for replication. Percentage of DNA replication represented at indicated time points. Mean ± SD of three experiments. (D) Depletion of And-1 inhibits the initial velocity of DNA replication to 40%. Linear correlation plots to show initial maximal velocity of DNA replication in control or XAnd-1-depleted extracts. The data points are derived from the experiment in C. (E) Inhibition of DNA replication in XAnd-1-depleted extracts can be rescued by addition of recombinant hAnd-1 protein. Indicated amount of recombinant hAnd-1 was added to egg cytosol depleted with prebleed or with anti-XAnd-1. Percentage of DNA replication after 90 min is shown. Mean ± SD of two experiments. (F) Inhibition of DNA replication in XAnd-1-depleted extracts is not due to activation of checkpoint. DNA replication after 90 min is shown. Mean ± SD of two experiments. (Mock) Replication in egg extract without any additions; (XEE + ETP) etoposide (30 μM) added to activate checkpoint; (Prebleed and anti-And-1) extracts depleted by indicated antibodies, no etoposide added. Caffeine (5 mM) was added as indicated. (G) XAnd-1 is loaded onto chromatin after pre-RC and Mcm10. Xenopus egg cytosol incubated with sperm chromatin at 23°C and chromatin-bound proteins analyzed by Western blot using indicated antibodies at indicated times. (H) Cdk2 kinase activity is required for chromatin loading of XAnd-1. Xenopus egg cytosol supplemented with 3 μM GST or GST-p21N were treated as in G.
We will show later (Fig. 6A, below) that XAnd-1 indeed associates with XMcm10, but the antibody used here selectively immunoprecipitates free XAnd-1, accounting for the lack of codepletion of the And-1-associated proteins. To rule out the possibility that depletion of XAnd-1 leads to DNA damage and activates a checkpoint, the DNA replication assay was repeated in the presence of caffeine, which is known to inhibit ATM/ATR kinases and bypass the DNA damage checkpoint. Addition of caffeine did not relieve the DNA replication inhibition seen after And-1 depletion, in contrast to the situation after topoisomerase inhibition by etoposide (Fig. 4F), suggesting that inhibition in DNA replication after depletion of XAnd-1 is not due to the activation of a checkpoint.
Figure 6.
Interaction between Mcm10 and And-1 is important for loading of And-1 and pol α onto chromatin. (A) Immunoprecipitate of Xenopus egg lysates immunoblotted for And-1 and for Mcm10. Anti-Mcm10 IP coimmunoprecipitates XAnd1, but the anti-XAnd-1 antibody does not coimmunoprecipitate XMcm10. (B) Anti-XAnd-1 antibody disrupts Mcm10–And-1 interaction in human cell lysates. Human cell lysates containing Flag-And-1 were preincubated with prebleed or anti-XAnd-1, immunoprecipitated with anti-Flag, and the precipitates immunoblotted for human Mcm10. (C) Anti-XAnd-1 antibody inhibits DNA replication in Xenopus egg extracts. Either anti-XAnd-1 or control (prebleed) was added to DNA replication assay and recombinant hAnd-1 protein was added where indicated. Percentage DNA replication after 90 min is shown. Mean ± SD of two experiments. (D) Addition of recombinant hAnd-1 rescues loading of DNA pol α in reactions neutralized by anti-XAnd-1. Chromatin-associated proteins were isolated from reactions containing antibodies indicated at top and immunoblotted for proteins indicated on the right. The remainder are as in C. (E) Anti-XAnd-1 antibody blocks chromatin loading of And-1 and of pol α. Xenopus egg cytosols supplemented with prebleed or anti-XAnd-1 antibody were treated as in Figure 4G to examine chromatin loading of various proteins. (F) Pol α (p180) protein levels are stable in the egg cytosol supplemented with anti-XAnd-1 antibody (5% v/v). Equal amounts of egg cytosol were collected for immunoblotting at indicated time points after addition of anti-XAnd-1 antibody.
XAnd-1 binds to chromatin after pre-RC assembly in a Cdk2 activity-dependent manner
To gain mechanistic insight into how And-1 is involved in DNA replication, we examined the chromatin loading of And-1 during DNA replication. In an unperturbed Xenopus egg extract, Orc2 and Mcm7 are already bound to sperm chromatin by 15 min (Fig. 4G), consistent with their role in pre-RC assembly. Mcm10 binds soon after, while And-1, Cdc45, and pol α bind to chromatin simultaneously and with slower kinetics than Mcm10. RPA and PCNA also bind to chromatin with kinetics similar to that of And-1. These results suggest that And-1, unlike MCM2-7 and ORC, is not a component of the pre-RC for licensing. Instead, And-1 is loaded at a step after Mcm10 binding at about the time that the pre-RC changes to an active replication fork. Consistent with this, inhibition of cdk2 kinase by addition of the cdk inhibitor GST-p21N (Chen et al. 1995) to the egg extracts blocked the loading of And-1 and Cdc45 to chromatin (Fig. 4H). Under these conditions, the nuclear envelope formation was not affected (Supplementary Figs. S3, S4). Thus, And-1 loads onto the chromatin after pre-RC formation, after Mcm10 loading, and after cdk2 activation, at or soon after the origin-unwinding step.
And-1 is not a cofactor of DNA polymerase
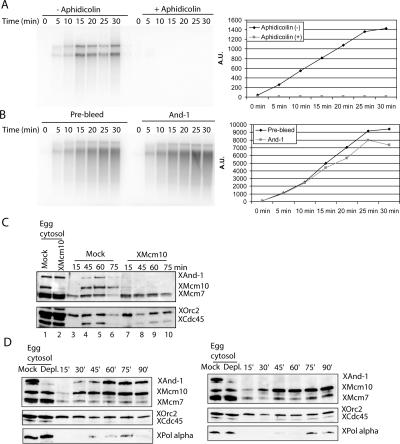
The inhibition in DNA replication after depletion of XAnd-1 from the Xenopus egg extracts could be explained if And-1 is a cofactor of DNA polymerase complex. To test this, origin-independent DNA synthesis was assessed in Xenopus egg extracts using M13 single-strand DNA as substrate. Within 30 min, M13 DNA was replicated in the extracts, and this was completely inhibited after addition of aphidicolin, an inhibitor of DNA pol α (Fig. 5A). The depletion of XAnd-1 from the extracts did not have any effect on M13 single-strand DNA replication as compared with control (Fig. 5B, Prebleed) depletion, suggesting that And-1 is not a cofactor of DNA pol α complex.
Figure 5.
XAnd-1 is not a cofactor of DNA polymerase, and the roles of Mcm10 and And-1 in loading of DNA replication factors on chromatin. (A) DNA polymerase-dependent single-strand DNA replication. Xenopus egg extract with or without aphidicolin (50 mg/mL) was used for single-stand DNA (M13) replication assay. Aliquot of the DNA replication reaction at indicated time point was resolved on 1% agarose gel and autoradiographed. Image and quantitation of the radioactive products are shown. (B) Depletion of XAnd-1 does not affect single-strand DNA replication. Control or XAnd-1-depleted egg extracts was used. The remainder are as in A. (C) XMcm10 is required for chromatin loading of XAnd-1. Xenopus egg cytosol was immunodepleted by anti-Mcm10 or normal serum (Mock) and treated as described in Figure 4G to examine the chromatin association of proteins. (D) Depletion of XAnd-1 delays loading of DNA pol α onto chromatin. Control (left panel) or XAnd-1-depleted egg extracts (right panel) incubated with sperm chromatin at 23°C and chromatin-bound proteins analyzed by Western blot using indicated antibodies at indicated times. Mock-depleted and And-1-depleted cytosol are shown in both panels on the extreme left.
Mcm10 is required for chromatin loading of XAnd-1
Given that XMcm10 binds to chromatin before XAnd-1, and XMcm10 interacts with XAnd-1, we hypothesized that XMcm10 is required for loading of XAnd-1 onto origins for DNA replication initiation. XMcm10 was immunodepleted from the egg extracts to test its requirement for the chromatin loading of various factors during the replication reaction (Fig. 5C). Although XAnd-1 was not codepleted from the egg extracts (Fig. 5C, lane 2), the depletion of XMcm10 blocked the loading of And-1 and Cdc45 on chromatin (Fig. 5C, lanes 7–10) and not that of the pre-RC components. Thus, XMcm10 is required to load XAnd-1 on chromatin. It is worth noting here that although XMcm10 coimmunoprecipitates with XAnd-1 (Fig. 6A, lane 3), the interaction is labile and egg extracts are highly enriched in replication factors, so that it is not surprising that there is an excess of free XAnd-1 in the extract after XMcm10 depletion.
Interruption of Mcm10–And-1 interaction blocks loading of And-1 and of p180
Next, we looked at loading of various DNA replication proteins in XAnd-1-depleted extracts and found that as compared with control depleted extracts, there was a significant delay in loading of DNA pol α (p180) (Fig. 5D). Given that XMcm10 is required for loading of XAnd-1 onto chromatin, we hypothesized that the interaction between XAnd-1 and XMcm10 is important for loading of XAnd-1 and p180 onto chromatin. We noted that, although XAnd-1 can be detected in XMcm10 immunoprecipitates, immunoprecipitation of XAnd-1 by the antibody to the C-terminal part of And-1 fails to coimmunoprecipitate Mcm10 (Fig. 6A, lane 5). This result can be explained if the epitope recognized by the anti-XAnd-1 antibody is necessary for And-1–Mcm10 interaction. To confirm this, we tested whether the anti-XAnd-1 disrupts the interaction of human And-1 with Mcm10. Anti-XAnd-1 antibody was added to lysates from 293T cells in which Flag-And-1 is transiently transfected. Anti-XAnd-1 antibody inhibited the coimmunoprecipitation of Mcm10 with Flag-And-1 at 1.6% v/v (Fig. 6B, lanes 9,10), suggesting that the C-terminal epitope on And-1 recognized by the anti-And-1 antibody is indeed important for interaction with Mcm10.
We could therefore use the anti-XAnd-1 to interrupt the And-1–Mcm10 interaction and thus test the importance of the interaction in DNA replication. Addition of XAnd-1 antibody to the egg extract (5% v/v) suppressed DNA replication (Fig. 6C), while adding back recombinant His-hAnd-1 together with anti-XAnd-1 rescued both the DNA replication in egg extracts and the loading of DNA polymerase on to chromatin (Fig. 6C,D). To ascertain the step in the replication initiation process that is disrupted by the anti-XAnd-1 antibody, the chromatin association of replication factors was studied (Fig. 6E). Just as with depletion of Mcm10, addition of the antibody that disrupts Mcm10–And-1 interaction impairs the loading of And-1 on chromatin (Fig. 6E, right panel). Chromatin loading of Orc2, Mcm7, Mcm10, and Cdc45 is not impaired (Fig. 6E), consistent with the possibility that the antibody disrupts And-1 loading at a step after Mcm10 loading. Most important, the anti-XAnd-1 antibody also inhibits chromatin loading of p180 (Fig. 6E) without affecting the level of p180 in the extracts (Fig. 6F), suggesting that And-1 on the chromatin facilitates the stable association of p180 with chromatin. Consistent with this, immunodepletion of XAnd-1 significantly delayed the loading of p180 without decreasing or changing the kinetics of the association of Cdc45 or Mcm10 (Fig. 5D). The partial effect of And-1 immunodepletion (Figs. 4C, 5C) compared with the more dramatic effect of And-1 immunoneutralization (Fig. 6C,E) is most likely due to the partial depletion of XAnd-1 with the antibody (see Materials and Methods). These results are consistent with the proposal that the Mcm10–And-1 interaction is necessary for the loading of And-1 on chromatin, with the latter being required for the loading of DNA pol α on chromatin.
Discussion
In this study, we show that And-1 is required for efficient DNA replication in both human cells and Xenopus egg extracts. In human cells, And-1 is required for the stabilization of DNA pol α, a function that, in budding yeast, is carried out by Mcm10 itself. Fortunately, And-1 is not required for the stabilization of p180 in Xenopus egg extracts, allowing us to test whether And-1 is also required for the loading of p180 on chromatin. Such small regulatory differences between Xenopus egg extracts and human somatic cells have been seen before (for example, in the degradation of Cdt1) (Arias and Walter 2006; Senga et al. 2006) and are often very useful for mechanistic studies.
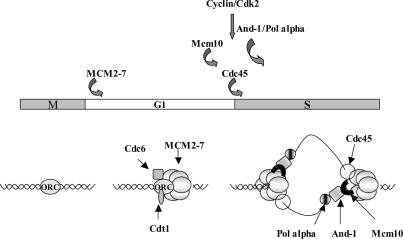
We found that XMcm10 is required for chromatin loading of XAnd-1, which itself facilitates the loading of DNA pol α onto chromatin (Fig. 7). Mcm1 0 interacts with MCM2-7, and here we show that Mcm10 interacts with And-1 and DNA pol α. And-1 and Mcm10 could therefore recruit Pol α to the MCM2-7 loaded at origins of replication (Fig. 7), although it is not clear that MCM2-7–Mcm10–And-1–p180 form a ternary complex. Cdc45 has been shown to play a role in recruiting pol α (Mimura and Takisawa 1998), but is not sufficient to recruit p180 when And-1 is absent from the chromatin (Fig. 6E). Like And-1, the loading of Cdc45 on chromatin requires cdk2 kinase activity and the presence of Mcm10. Together, these results suggest that Cdc45, Mcm10, and And-1 cooperate in the loading of DNA pol α on chromatin.
Figure 7.
And-1 and Mcm10 cooperate to load pol α. A model for the recruitment of elongation factors like pol α being dependent on the prior recruitment of MCM2-7, Mcm10, and And-1. The And-1–Mcm10 complex is part of a protein complex linking the MCM2-7 helicase with the primase-containing pol α complex. Other proteins that are not indicated here, like GINS, may also be part of the linking complex.
Ctf4/And-1 was reported before to physically and genetically interact with DNA pol α, but such a critical role of Ctf4 in DNA replication was unsuspected. Our results demonstrate that Ctf4/And-1 is required for maintaining the stability of pol α (in mammalian cells) and facilitates the loading of the polymerase at origins of replication. An additional role of Ctf4 in replication forks is possible based on the following evidence: (1) Deletion of CTF4 in cdc17-1 ts mutants decreases the maximum permissive temperature (Wittmeyer and Formosa 1997). CDC17 codes for the p180 homolog in S. cerevisiae. (2) As an essential gene in S. pombe, mcl1 ts mutants exhibit double-stranded breaks at the restrictive temperature (Tsutsui et al. 2005). (3) Ctf4 is part of the complex of proteins at a replication fork (Gambus et al. 2006; Lengronne et al. 2006). Thus, although we specifically examined the initiation stage of DNA replication, it is entirely possible that And-1 also helps maintain pol α on chromatin near the elongation complex at the replication fork. Such a role in elongation would be consistent with the observations that And-1 is required late in S phase (Fig. 2D) and that Mcm10, p180, and Ctf4 are components of the replication fork. The fate of elongating vertebrate replication complexes after rapid inactivation of And-1/Ctf4 is necessary to critically evaluate this model.
The replicative polymerase in Escherichia coli, Pol III holoenzyme, is composed of many subunits individually required for DNA replication. Proteins executing similar functions as the Pol III subunits also exist in eukaryotes, but are rearranged as individual proteins or as complexes that are purified independently of the eukaryotic polymerases. Thus, the β2 subunits of E. coli are ring-shaped clamps that retain the polymerase on the DNA template, but the function of the β subunit is taken over by PCNA in eukaryotes (Kong et al. 1992; Krishna et al. 1994). Similarly, the γ–δ complex in E. coli is the ATP-dependent clamp loader whose function is carried out in eukaryotes by RF-C (Jeruzalmi et al. 2001; Bowman et al. 2004). The τ subunit of the Pol III holoenzyme has one part that is closely related to the γ subunit of the γ–δ complex and another part that is unique. The unique part gives the τ subunit several critical functions (McHenry 2003). The τ subunit interacts with the dnaB helicase, stimulates its processivity, and links the helicase to the polymerase (Kim et al. 1996). In addition, direct physical contact is required between the dnaB helicase and the dnaG primase for DNA replication (Soultanas 2005). Similar protein links between the eukaryotic MCM2-7 helicase and the DNA α polymerase–primase complex have not yet been discovered. We speculate that Mcm10 and And-1 form part of the complex that interacts both with MCM2-7 and with DNA pol α, most likely in cooperation with GINS and Cdc45. The GINS complex in yeast interacts with Ctf4 (Gambus et al. 2006). Xenopus Mcm10, GINS, and Cdc45 are present in the replisome with the helicase and the polymerase, but experimental separation of the unwindosome from the polymerase reveals that GINS and Cdc45 partition with the MCM2-7 helicase in the unwindosome, while Mcm10 is lost from the unwindosome (Pacek et al. 2006). Thus, the bridge between the helicase MCM2-7 and the polymerase at the replication fork is likely to contain Cdc45 and GINS on the helicase side and Mcm10 and And-1 on the polymerase side, with multipartite protein–protein interactions stabilizing the complex. Physical interaction between dnaB and dnaG in bacteria keeps the lagging strand polymerase at the replication fork poised to synthesize the next Okazaki fragment (Soultanas 2005). The other possibility, therefore, is that And-1–Mcm10, Cdc45, and GINS serve to keep the lagging strand polymerase–primase complex, DNA pol α, at the replication fork so that it can cycle from one Okazaki fragment to the next.
CTF4 was initially identified in a genetic screen for yeast mutants affecting chromosome transmission fidelity (Kouprina et al. 1992). The essential role of And-1 in pol α function in mammals and Xenopus reported here agrees with the essential functions of Mcl1 and SepB (Harris and Hamer 1995; Williams and McIntosh 2002), but contradicts with the viability of S. cerevisiae lacking Ctf4. We speculate that a more stable Mcm10–p180 interaction in S. cerevisiae and the existence of other interactions involving Mcm10, Cdc45, and GINS in the bridge between the helicase and the polymerase explains why S. cerevisiae CTF4 is nonessential. S. cerevisiae Mcm10 protein is known to directly interact with p180 of DNA pol α and is required for the stability of the p180 (Ricke and Bielinsky 2004). A similar role of Mcm10 in human cells could not be seen in our experiments, suggesting that vertebrate cells use the Mcm10 partner, And-1, to maintain the stability of DNA pol α. Since Ctf4 moves with the replication fork in budding yeast (Gambus et al. 2006), it could, however, play a role in optimizing the association of the polymerase with initiator and elongation proteins in S. cerevisiae without being essential.
Chromosome missegregation and defects in sister-chromatid cohesion are noted in S. cerevisiae with mutations in CTF4. In vertebrates, the loading of cohesins on chromatin is dependent on the loading of MCM2-7 proteins in late mitosis/early G1(Gillespie and Hirano 2004; Takahashi et al. 2004), well before the step when Ctf4/And-1 is loaded on chromatin. In yeast, preRC components are not even required for loading cohesins. There is a second step, however, where the already loaded cohesin rings are activated to grasp the sister chromatids as the replication fork passes through a chromosomal segment (Blow and Tanaka 2005; Lengronne et al. 2006). Thus, a likely explanation of the cohesion defect in ctf4 yeast may lie in a defective function of DNA polymerase at a yeast replication fork lacking Ctf4, leading to a defect in the activation of cohesion.
The role of And-1 in DNA replication initiation ties together several disparate phenomena noted in the field for more than a decade: (1) Ctf4 is involved in chromosome transmission fidelity. (2) Ctf4 physically and genetically interacts with DNA pol α. (3) Ctf4/Mcl1 shows genetic interaction with proteins involved in lagging stand synthesis. (4) Mcm10 is required at origins of replication for the loading of DNA pol α. Our results suggest that And-1 acts at a critical step downstream from Mcm10: It interacts with Mcm10 and is required for the proper loading of DNA pol α and replication initiation. Most likely, the interaction persists as MCM2-7, Mcm10, And-1, and DNA pol α leave the origin of replication as part of the replication fork. The results also shed light on an important and unexplained question: How does the initiation machinery ending with the loading of Mcm2-7 succeed in bringing components of the elongation machinery (starting with the DNA polymerase–primase) to origins of replication?
Materials and methods
Cell culture and flow cytometry
293T and U2OS were grown in DMEM medium supplemented with 10% heat-inactivated donor calf serum and antibiotics. HCT116 cells were grown in McCoy’s medium supplemented with 10% heat-inactivated donor calf serum. Flow cytometry analysis was performed as described previously (Zhu et al. 2004).
Cloning and protein expression
GST-tagged And-1 was constructed as follows: The cDNA encoding the entire ORF from amino acids 1–1129 was PCR-amplified using primers containing BamHI and NotI sites, digested with BamHI and NotI, and ligated to pEBG digested with BamHI and NotI. The same sequence of And-1 was also cloned into pEFF vector digested with BamHI and NotI to produce Flag-tagged And-1. Fragments of And-1 encoding amino acids 1–336, 330–1129, and 984–1129 were cloned into pEFF for the expression of truncated Flag-tagged And-1 mutants using the same digestion sites. All of the truncated mutants have either an added NLS (nuclear localization sequence) or a native NLS. Nuclear localization of the truncation mutants was confirmed by immunofluorescence.
Full-length human And-1 was cloned into baculovirus by using a Bac-To-Bac Baculovirus expression system (Invitrogen). His-tagged And-1 protein was expressed in SF9 insect cells and lysed in 50 mM Tris (pH 7.7), 150 mM KCl, 0.1% Triton-X 100, and protease inhibitor. The soluble lysate was mixed with Ni-NTA agarose for 3 h. Beads were washed three times with 50 mM Tris (pH 7.7) and 300 mM KCl. His-And-1 was eluted from the beads with elution buffer 50 mM Tris (pH 7.7), 300 mM KCl, and 250 mM imidazole. The elution buffer was exchanged for the protein storage buffer (50 mM Tris at pH 7.7, 100 mM KCl, 10% glycerol) using the QuickChange PD10 column.
GST and GST-p21N were expressed and purified as described (Chen et al. 1995).
Antibodies and siRNA oligonucleotides
Mcm10, hAnd-1, and XAnd-1 antibodies were generated as follows: Full-length hMcm10 or a fragment of representing amino acids 776–927 of hAnd-1 (450-base-pair EcoR1 fragment of clone 4523768 from Invitrogen) were subcloned into pET 28a vector. The recombinant his-tagged products were purified by nickel affinity isolation and used as immunogens to generate antibodies in rabbits. Anti-XAnd-1 antibody was raised against a synthetic peptide (KPLGQSANNKLSAFAFKKE) of XAnd-1 (Open Biosystems).
Antibodies to Cdk2 (M2), pol α (G-16), GST (z-5), and myc (9E10) were obtained from Santa Cruz Biotechnology. Anti-Flag antibody was obtained from Sigma. Pan MCM2-7 antibody AS.1 was from Dr. Stephen Bell (Massachussetts Institute of Technology, Cambridge, MA); anti-XOrc2, anti-XCdc45, anti-XMcm7, and anti-XMcm10 were from Dr. Johannes Walter (Harvard Medical School, Boston, MA); and antibodies to the subunits of pol α were from Dr. Teresa Wang (Stanford University, Stanford, CA). Anti-BrdU antibody was from Pharmingen.
siRNA oligonucleotides were made to the following target sequences (sense): And-1-1 (AAGATGGTCAAGAAGGCA GCA), And-1-2 (AAGCAGGCATCTGCAGCATCC), and And-1-3 (TGACTCGCTTCACTACAAA). Mcm10-1 (AAGCCTT CTCTGGTCTGCGGC), Mcm10-2 (AAGCCCATAAGCTTT GCCTGC), and control GL2 were as described earlier (Zhu et al. 2004). siRNA transfections were performed twice with 100 nM siRNA oligonucleotide duplexes using Oligofectamine (Invitrogen) according to the manufacturer’s instructions. Cells were harvested for analysis 72 h after first transfection.
Xenopus DNA replication and chromatin-binding assays
Xenopus replication assay was performed as described elsewhere (Saxena et al. 2004) with slight modification. Briefly, BrdU was used instead of [αP32]ATP and the amounts of BrdU incorporated were determined by ELISA. For replication assay with addition of antibody, Xenopus egg cytosols (10 μL) were incubated with 0.5 μL of anti-XAnd-1 antibody (and 400 ng of recombinant h-And-1 where indicated), followed by the addition of sperm chromatin and BrdU. The reaction mixtures were incubated for 90 min at 23°C before adding stop buffer. Chromatin-binding assay was performed following the method as described (Pacek and Walter 2004), with one minor modification. The extracts containing sperm chromatin (up to 15 μL) were diluted with 60 μL of cold ELB/NP-40 buffer (0.25 M sucrose, 2.5 mM MgCl2, 50 mM KCl, 10 mM HEPES at pH 7.5, 0.1% NP-40), layered onto a 180-μL sucrose cushion (ELB containing 0.5 M sucrose) in Beckman 5 × 44-mm microcentrifuge tubes and centrifuged as described (Pacek and Walter 2004).
Immunodepletion
To deplete protein in egg cytosol, 0.2 vol of antibody-bound protein A-sepharose (Pharmacia) was incubated with egg extracts for 1 h (three times) at 4°C for XAnd-1, and for 1 h (two times) at 4°C for XMcm10. The sensitivity of the anti-XAnd-1 antibody in immunoblots was such that we can say that XAnd-1 was depleted to <10% wild-type levels, but cannot say that we had depleted to <1% wild-type level, which is often necessary to see inhibition of DNA replication after replication protein depletion in Xenopus egg extracts.
Immunoprecipitation and Western blot
The immunoprecipitations were usually performed as follows: Cells were lysed in lysis buffer (25 mM HEPES-KOH at pH 7.6, 150 mM KAc, 5 mM MgCl2, 1 mM Na2 EGTA, 10% glycerol, 0.1% Nonidet P-40, protease inhibitor) for 20 min on ice, followed by sonication. After centrifugation, the resulting supernatants were mixed with antibody or prebleed for immunoprecipitation for 2 h, followed by incubation with protein A beads for 1 h. Beads were then washed three times with lysis buffer. Associated proteins were eluted by incubating beads with SDS loading buffer for immunoblotting. For immunoprecipitation of Mcm10 in Figure 1, native lysis and wash buffer contained 150 mM NaCl, 50 mM Tris-HCl (pH 7.4), 0.2% NP40, 2 mM EDTA, and protease inhibitor cocktail. For denaturing washes, the solution used was as above with the addition of 1% NP-40, 1% Deoxycholic acid, and 0.5% SDS. For And-1 immunoprecipitates in Figure 1B, HCT116 cells were treated with MG132 (10 μM) for 5 h, lysed in 500 μL of lysis buffer as above. After sonication and centrifugation, supernatants (∼1000 μg) were mixed with 2 μL of antibody or prebleed for immunoprecipitation as above.
For results shown in Figure 6B, 293T cells overexpressing Flag-And-1 were lysed, and indicated amounts of anti-XAnd-1 antibodies were added to 500 μL lysates and mixed for 1 h at 4°C. Anti-Flag agarose was then used to immunoprecipitate Flag-And-1 and associated proteins. All immunoprecipitates were resolved by SDS-PAGE and immunoblotted for Flag-And-1 and for associated Mcm10.
For Western blots, cells were harvested by trypsinization in cold trypsin solution (∼2–3 min), followed by one quick wash in cold PBS solution. SDS loading buffer was then added to cell pellets to lyse cells, followed by sonication and boiling. Proteins were resolved by SDS-PAGE. To ensure that the decrease in p180 protein level after And-1 depletion was not due to post-lysis proteolysis, cells were also lysed directly into SDS-PAGE sample buffer to inactivate all cellular proteases (Supplementary Fig. S2).
Immunofluorescence
Cells grown on cover slips were extracted with CSK buffer (100 mM NaCl, 300 mM sucrose, 3mM MgCl2, 10 mM PIPES at pH 7.0) containing 0.5% Triton X-100 at room temperature before fixation in 4% paraformaldehyde in PBS for 15 min at room temperature. Cells were then blocked with 3% BSA in PBS-T (0.02% Tween-20) solution for 1 h. Primary antibodies were added at 1:500 dilution and incubated for 1 h at room temperature. After washing in 3% BSA PBS-T solution, tetramethylrhodamine isothiocyante- or fluorescein isothiocyanate-conjugated secondary antibody was then added and incubated at room temperature for 1 h. Finally, cells were washed and fixed in mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories) for confocal microscopy (2-h time point) immunofluorescence microscopy (6-h time point).
Cell synchronization
Cells were treated with 2 mM thymidine for 12 h, washed, and then released into thymidine-free medium for 12 h. Aphidicolin was then added to a final concentration of 1 μg/mL, and cells were incubated for an additional 12 h. Cells were washed four times with PBS and incubated in serum-containing medium. Samples for FACS analysis, immunostaining, and lysis were collected at the times indicated in Figure 1D.
Silver staining and protein extraction
Following electrophoresis, the gel was fixed in 50% methanol, 5% acetic acid for 20 min, and then washed in 50% methanol and deionized water for 10 min each. The gel was sensitized for 5 min by incubation in 0.02% sodium thiosulfate. Following two 1-min rinses in water, the gel was immersed in 0.1% silver-nitrate solution and incubated for 30 min at 4°C. After rinsing twice with water, the staining was developed in 0.04% formalin containing 2% sodium carbonate and then rinsed in 5% acetic acid. The gel was stored in 1% acetic acid until bands were excised for mass spectroscopy.
Acknowledgments
We thank Johannes Walter and Teresa Wang for the generous gift of critical antibodies, Todd Stukenberg for advice on the experiments with Xenopus egg extracts, and Dutta laboratory members for helpful discussions. This work was supported by RO1 CA60499 to A.D., RO1 GM67225 to S.K., and K08 DK067240 to C.U.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1585607
References
- Aparicio O.M., Weinstein D.M., Bell S.P., Weinstein D.M., Bell S.P., Bell S.P. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- Arias E.E., Walter J.C., Walter J.C. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat. Cell Biol. 2006;8:84–90. doi: 10.1038/ncb1346. [DOI] [PubMed] [Google Scholar]
- Bell S.D. Molecular biology: Prime-time progress. Nature. 2006;439:542–543. doi: 10.1038/439542a. [DOI] [PubMed] [Google Scholar]
- Bell S.P., Dutta A., Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Blow J.J., Tanaka T.U., Tanaka T.U. The chromosome cycle: Coordinating replication and segregation. Second in the cycles review series. EMBO Rep. 2005;6:1028–1034. doi: 10.1038/sj.embor.7400557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman G.D., O’Donnell M., Kuriyan J., O’Donnell M., Kuriyan J., Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp–clamp loader complex. Nature. 2004;429:724–730. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- Chen J., Jackson P.K., Kirschner M.W., Dutta A., Jackson P.K., Kirschner M.W., Dutta A., Kirschner M.W., Dutta A., Dutta A. Separate domains of p21 involved in the inhibition of Cdk kinase and PCNA. Nature. 1995;374:386–388. doi: 10.1038/374386a0. [DOI] [PubMed] [Google Scholar]
- Dornreiter I., Erdile L.F., Gilbert I.U., von Winkler D., Kelly T.J., Fanning E., Erdile L.F., Gilbert I.U., von Winkler D., Kelly T.J., Fanning E., Gilbert I.U., von Winkler D., Kelly T.J., Fanning E., von Winkler D., Kelly T.J., Fanning E., Kelly T.J., Fanning E., Fanning E. Interaction of DNA polymerase α-primase with cellular replication protein A and SV40 T antigen. EMBO J. 1992;11:769–776. doi: 10.1002/j.1460-2075.1992.tb05110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T., Nittis T., Nittis T. Dna2 mutants reveal interactions with DNA polymerase α and Ctf4, a Pol α accessory factor, and show that full Dna2 helicase activity is not essential for growth. Genetics. 1999;151:1459–1470. doi: 10.1093/genetics/151.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A., Jones R.C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R.D., Labib K., Jones R.C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R.D., Labib K., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R.D., Labib K., Kanemaki M., van Deursen F., Edmondson R.D., Labib K., van Deursen F., Edmondson R.D., Labib K., Edmondson R.D., Labib K., Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- Gillespie P.J., Hirano T., Hirano T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr. Biol. 2004;14:1598–1603. doi: 10.1016/j.cub.2004.07.053. [DOI] [PubMed] [Google Scholar]
- Hanna J.S., Kroll E.S., Lundblad V., Spencer F.A., Kroll E.S., Lundblad V., Spencer F.A., Lundblad V., Spencer F.A., Spencer F.A. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell. Biol. 2001;21:3144–3158. doi: 10.1128/MCB.21.9.3144-3158.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S.D., Hamer J.E., Hamer J.E. sepB: An Aspergillus nidulans gene involved in chromosome segregation and the initiation of cytokinesis. EMBO J. 1995;14:5244–5257. doi: 10.1002/j.1460-2075.1995.tb00209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.G., Weisshart K., Gilbert I., Fanning E., Weisshart K., Gilbert I., Fanning E., Gilbert I., Fanning E., Fanning E. Stoichiometry and mechanism of assembly of SV40 T antigen complexes with the viral origin of DNA replication and DNA polymerase α-primase. Biochemistry. 1998;37:15345–15352. doi: 10.1021/bi9810959. [DOI] [PubMed] [Google Scholar]
- Izumi M., Yatagai F., Hanaoka F., Yatagai F., Hanaoka F., Hanaoka F. Cell cycle-dependent proteolysis and phosphorylation of human Mcm10. J. Biol. Chem. 2001;276:48526–48531. doi: 10.1074/jbc.M107190200. [DOI] [PubMed] [Google Scholar]
- Izumi M., Yatagai F., Hanaoka F., Yatagai F., Hanaoka F., Hanaoka F. Localization of human Mcm10 is spatially and temporally regulated during the S phase. J. Biol. Chem. 2004;279:32569–32577. doi: 10.1074/jbc.M314017200. [DOI] [PubMed] [Google Scholar]
- Jeruzalmi D., O’Donnell M., Kuriyan J., O’Donnell M., Kuriyan J., Kuriyan J. Crystal structure of the processivity clamp loader γ (γ) complex of E. coli DNA polymerase III. Cell. 2001;106:429–441. doi: 10.1016/s0092-8674(01)00463-9. [DOI] [PubMed] [Google Scholar]
- Kawasaki Y., Hiraga S., Sugino A., Hiraga S., Sugino A., Sugino A. Interactions between Mcm10p and other replication factors are required for proper initiation and elongation of chromosomal DNA replication in Saccharomyces cerevisiae. Genes Cells. 2000;5:975–989. doi: 10.1046/j.1365-2443.2000.00387.x. [DOI] [PubMed] [Google Scholar]
- Kim S., Dallmann H.G., McHenry C.S., Marians K.J., Dallmann H.G., McHenry C.S., Marians K.J., McHenry C.S., Marians K.J., Marians K.J. τ couples the leading- and lagging-strand polymerases at the Escherichia coli DNA replication fork. J. Biol. Chem. 1996;271:21406–21412. doi: 10.1074/jbc.271.35.21406. [DOI] [PubMed] [Google Scholar]
- Kohler A., Schmidt-Zachmann M.S., Franke W.W., Schmidt-Zachmann M.S., Franke W.W., Franke W.W. AND-1, a natural chimeric DNA-binding protein, combines an HMG-box with regulatory WD-repeats. J. Cell Sci. 1997;110:1051–1062. doi: 10.1242/jcs.110.9.1051. [DOI] [PubMed] [Google Scholar]
- Kong X.P., Onrust R., O’Donnell M., Kuriyan J., Onrust R., O’Donnell M., Kuriyan J., O’Donnell M., Kuriyan J., Kuriyan J. Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: A sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- Kouprina N., Kroll E., Bannikov V., Bliskovsky V., Gizatullin R., Kirillov A., Shestopalov B., Zakharyev V., Hieter P., Spencer F., Kroll E., Bannikov V., Bliskovsky V., Gizatullin R., Kirillov A., Shestopalov B., Zakharyev V., Hieter P., Spencer F., Bannikov V., Bliskovsky V., Gizatullin R., Kirillov A., Shestopalov B., Zakharyev V., Hieter P., Spencer F., Bliskovsky V., Gizatullin R., Kirillov A., Shestopalov B., Zakharyev V., Hieter P., Spencer F., Gizatullin R., Kirillov A., Shestopalov B., Zakharyev V., Hieter P., Spencer F., Kirillov A., Shestopalov B., Zakharyev V., Hieter P., Spencer F., Shestopalov B., Zakharyev V., Hieter P., Spencer F., Zakharyev V., Hieter P., Spencer F., Hieter P., Spencer F., Spencer F. CTF4 (CHL15) mutants exhibit defective DNA metabolism in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1992;12:5736–5747. doi: 10.1128/mcb.12.12.5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna T.S., Kong X.P., Gary S., Burgers P.M., Kuriyan J., Kong X.P., Gary S., Burgers P.M., Kuriyan J., Gary S., Burgers P.M., Kuriyan J., Burgers P.M., Kuriyan J., Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Kim H.K., Chung S., Kim K.S., Dutta A., Kim H.K., Chung S., Kim K.S., Dutta A., Chung S., Kim K.S., Dutta A., Kim K.S., Dutta A., Dutta A. Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J. Biol. Chem. 2005;280:16635–16641. doi: 10.1074/jbc.M412247200. [DOI] [PubMed] [Google Scholar]
- Lei M., Cheng I.H., Roberts L.A., McAlear M.A., Tye B.K., Cheng I.H., Roberts L.A., McAlear M.A., Tye B.K., Roberts L.A., McAlear M.A., Tye B.K., McAlear M.A., Tye B.K., Tye B.K. Two mcm3 mutations affect different steps in the initiation of DNA replication. J. Biol. Chem. 2002;277:30824–30831. doi: 10.1074/jbc.M201816200. [DOI] [PubMed] [Google Scholar]
- Lengronne A., McIntyre J., Katou Y., Kanoh Y., Hopfner K.P., Shirahige K., Uhlmann F., McIntyre J., Katou Y., Kanoh Y., Hopfner K.P., Shirahige K., Uhlmann F., Katou Y., Kanoh Y., Hopfner K.P., Shirahige K., Uhlmann F., Kanoh Y., Hopfner K.P., Shirahige K., Uhlmann F., Hopfner K.P., Shirahige K., Uhlmann F., Shirahige K., Uhlmann F., Uhlmann F. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol. Cell. 2006;23:787–799. doi: 10.1016/j.molcel.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Mayer M.L., Pot I., Chang M., Xu H., Aneliunas V., Kwok T., Newitt R., Aebersold R., Boone C., Brown G.W., Pot I., Chang M., Xu H., Aneliunas V., Kwok T., Newitt R., Aebersold R., Boone C., Brown G.W., Chang M., Xu H., Aneliunas V., Kwok T., Newitt R., Aebersold R., Boone C., Brown G.W., Xu H., Aneliunas V., Kwok T., Newitt R., Aebersold R., Boone C., Brown G.W., Aneliunas V., Kwok T., Newitt R., Aebersold R., Boone C., Brown G.W., Kwok T., Newitt R., Aebersold R., Boone C., Brown G.W., Newitt R., Aebersold R., Boone C., Brown G.W., Aebersold R., Boone C., Brown G.W., Boone C., Brown G.W., Brown G.W., et al. Identification of protein complexes required for efficient sister chromatid cohesion. Mol. Biol. Cell. 2004;15:1736–1745. doi: 10.1091/mbc.E03-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHenry C.S. Chromosomal replicases as asymmetric dimers: Studies of subunit arrangement and functional consequences. Mol. Microbiol. 2003;49:1157–1165. doi: 10.1046/j.1365-2958.2003.03645.x. [DOI] [PubMed] [Google Scholar]
- Miles J., Formosa T., Formosa T. Evidence that POB1, a Saccharomyces cerevisiae protein that binds to DNA polymerase α, acts in DNA metabolism in vivo. Mol. Cell. Biol. 1992a;12:5724–5735. doi: 10.1128/mcb.12.12.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles J., Formosa T., Formosa T. Protein affinity chromatography with purified yeast DNA polymerase α detects proteins that bind to DNA polymerase. Proc. Natl. Acad. Sci. 1992b;89:1276–1280. doi: 10.1073/pnas.89.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura S., Takisawa H., Takisawa H. Xenopus Cdc45-dependent loading of DNA polymerase α onto chromatin under the control of S-phase Cdk. EMBO J. 1998;17:5699–5707. doi: 10.1093/emboj/17.19.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M., Walter J.C., Walter J.C. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 2004;23:3667–3676. doi: 10.1038/sj.emboj.7600369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M., Tutter A.V., Kubota Y., Takisawa H., Walter J.C., Tutter A.V., Kubota Y., Takisawa H., Walter J.C., Kubota Y., Takisawa H., Walter J.C., Takisawa H., Walter J.C., Walter J.C. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell. 2006;21:581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- Petronczki M., Chwalla B., Siomos M.F., Yokobayashi S., Helmhart W., Deutschbauer A.M., Davis R.W., Watanabe Y., Nasmyth K., Chwalla B., Siomos M.F., Yokobayashi S., Helmhart W., Deutschbauer A.M., Davis R.W., Watanabe Y., Nasmyth K., Siomos M.F., Yokobayashi S., Helmhart W., Deutschbauer A.M., Davis R.W., Watanabe Y., Nasmyth K., Yokobayashi S., Helmhart W., Deutschbauer A.M., Davis R.W., Watanabe Y., Nasmyth K., Helmhart W., Deutschbauer A.M., Davis R.W., Watanabe Y., Nasmyth K., Deutschbauer A.M., Davis R.W., Watanabe Y., Nasmyth K., Davis R.W., Watanabe Y., Nasmyth K., Watanabe Y., Nasmyth K., Nasmyth K. Sister-chromatid cohesion mediated by the alternative RF-CCtf18/Dcc1/Ctf8, the helicase Chl1 and the polymerase-α-associated protein Ctf4 is essential for chromatid disjunction during meiosis II. J. Cell Sci. 2004;117:3547–3559. doi: 10.1242/jcs.01231. [DOI] [PubMed] [Google Scholar]
- Ricke R.M., Bielinsky A.K., Bielinsky A.K. Mcm10 regulates the stability and chromatin association of DNA polymerase-α. Mol. Cell. 2004;16:173–185. doi: 10.1016/j.molcel.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Saxena S., Yuan P., Dhar S.K., Senga T., Takeda D., Robinson H., Kornbluth S., Swaminathan K., Dutta A., Yuan P., Dhar S.K., Senga T., Takeda D., Robinson H., Kornbluth S., Swaminathan K., Dutta A., Dhar S.K., Senga T., Takeda D., Robinson H., Kornbluth S., Swaminathan K., Dutta A., Senga T., Takeda D., Robinson H., Kornbluth S., Swaminathan K., Dutta A., Takeda D., Robinson H., Kornbluth S., Swaminathan K., Dutta A., Robinson H., Kornbluth S., Swaminathan K., Dutta A., Kornbluth S., Swaminathan K., Dutta A., Swaminathan K., Dutta A., Dutta A. A dimerized coiled-coil domain and an adjoining part of geminin interact with two sites on Cdt1 for replication inhibition. Mol. Cell. 2004;15:245–258. doi: 10.1016/j.molcel.2004.06.045. [DOI] [PubMed] [Google Scholar]
- Senga T., Sivaprasad U., Zhu W., Park J.H., Arias E.E., Walter J.C., Dutta A., Sivaprasad U., Zhu W., Park J.H., Arias E.E., Walter J.C., Dutta A., Zhu W., Park J.H., Arias E.E., Walter J.C., Dutta A., Park J.H., Arias E.E., Walter J.C., Dutta A., Arias E.E., Walter J.C., Dutta A., Walter J.C., Dutta A., Dutta A. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J. Biol. Chem. 2006;281:6246–6251. doi: 10.1074/jbc.M512705200. [DOI] [PubMed] [Google Scholar]
- Soultanas P. The bacterial helicase–primase interaction: A common structural/functional module. Structure. 2005;13:839–844. doi: 10.1016/j.str.2005.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T.S., Yiu P., Chou M.F., Gygi S., Walter J.C., Yiu P., Chou M.F., Gygi S., Walter J.C., Chou M.F., Gygi S., Walter J.C., Gygi S., Walter J.C., Walter J.C. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat. Cell Biol. 2004;6:991–996. doi: 10.1038/ncb1177. [DOI] [PubMed] [Google Scholar]
- Takayama Y., Kamimura Y., Okawa M., Muramatsu S., Sugino A., Araki H., Kamimura Y., Okawa M., Muramatsu S., Sugino A., Araki H., Okawa M., Muramatsu S., Sugino A., Araki H., Muramatsu S., Sugino A., Araki H., Sugino A., Araki H., Araki H. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes & Dev. 2003;17:1153–1165. doi: 10.1101/gad.1065903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui Y., Morishita T., Natsume T., Yamashita K., Iwasaki H., Yamao F., Shinagawa H., Morishita T., Natsume T., Yamashita K., Iwasaki H., Yamao F., Shinagawa H., Natsume T., Yamashita K., Iwasaki H., Yamao F., Shinagawa H., Yamashita K., Iwasaki H., Yamao F., Shinagawa H., Iwasaki H., Yamao F., Shinagawa H., Yamao F., Shinagawa H., Shinagawa H. Genetic and physical interactions between Schizosaccharomyces pombe Mcl1 and Rad2, Dna2 and DNA polymerase α: Evidence for a multifunctional role of Mcl1 in DNA replication and repair. Curr. Genet. 2005;48:34–43. doi: 10.1007/s00294-005-0584-2. [DOI] [PubMed] [Google Scholar]
- Williams D.R., McIntosh J.R., McIntosh J.R. mcl1+, the Schizosaccharomyces pombe homologue of CTF4, is important for chromosome replication, cohesion, and segregation. Eukaryot. Cell. 2002;1:758–773. doi: 10.1128/EC.1.5.758-773.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmeyer J., Formosa T., Formosa T. The Saccharomyces cerevisiae DNA polymerase α catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol. Cell. Biol. 1997;17:4178–4190. doi: 10.1128/mcb.17.7.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Wang T.S., Wang T.S. A coordinated temporal interplay of nucleosome reorganization factor, sister chromatin cohesion factor, and DNA polymerase α facilitates DNA replication. Mol. Cell. Biol. 2004;24:9568–9579. doi: 10.1128/MCB.24.21.9568-9579.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Chen Y., Dutta A., Chen Y., Dutta A., Dutta A. Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol. Cell. Biol. 2004;24:7140–7150. doi: 10.1128/MCB.24.16.7140-7150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]