Abstract
The interface between cellular systems involving small noncoding RNAs and epigenetic change remains largely unexplored in metazoans. RNA-induced silencing systems have the potential to target particular regions of the genome for epigenetic change by locating specific sequences and recruiting chromatin modifiers. Noting that several genes encoding RNA silencing components have been implicated in epigenetic regulation in Drosophila, we sought a direct link between the RNA silencing system and heterochromatin components. Here we show that PIWI, an ARGONAUTE/PIWI protein family member that binds to Piwi-interacting RNAs (piRNAs), strongly and specifically interacts with heterochromatin protein 1a (HP1a), a central player in heterochromatic gene silencing. The HP1a dimer binds a PxVxL-type motif in the N-terminal domain of PIWI. This motif is required in fruit flies for normal silencing of transgenes embedded in heterochromatin. We also demonstrate that PIWI, like HP1a, is itself a chromatin-associated protein whose distribution in polytene chromosomes overlaps with HP1a and appears to be RNA dependent. These findings implicate a direct interaction between the PIWI-mediated small RNA mechanism and heterochromatin-forming pathways in determining the epigenetic state of the fly genome.
Keywords: PIWI, ARGONAUTE, HP1, heterochromatin, epigenetic, RNAi
RNA-induced gene silencing operates both post-transcriptionally and transcriptionally. Post-transcriptional gene silencing (PTGS) utilizes target gene-derived double-stranded RNA (dsRNA) or independently transcribed microRNAs (miRNAs) either to destroy a target mRNA or to modulate its translation (Tomari and Zamore 2005; Valencia-Sanchez et al. 2006). In contrast, transcriptional gene silencing (TGS) represents the interface between RNA silencing pathways and the chromatin modification system (Matzke and Birchler 2005). In TGS, small RNAs are incorporated into specialized effector complexes able to regulate chromatin modification, resulting in reduced access for the transcriptional machinery to chromatin. The TGS system that contributes to initiation and maintenance of heterochromatin formation in Schizosaccharomyces pombe is particularly well characterized (Verdel and Moazed 2005; Grewal and Jia 2007). In S. pombe, the RNA silencing system targets histone 3 Lys 9 (H3K9) methylation and recruits HP1 homolog Swi6 to the MAT locus and to repetitive elements in pericentric heterochromatin, setting up a heterochromatin spreading/maintenance loop that is dependent on the interaction of Swi6 with the Clr4 histone methyltransferase (HMT) (Grewal and Jia 2007). In plants, a specialized small RNA pathway utilizes heterochromatic RNAs to direct DNA methylation and presumably other chromatin modifications to effect silencing of target loci (for review, see Vaucheret 2007). In Caenorhabditis elegans, a single episode of RNA interference (RNAi) exposure can induce silencing inherited over many generations; mutations that abolish inheritance are all involved in chromatin structure, suggesting a chromatin-based mechanism (Vastenhouw et al. 2006). While the work in fission yeast and plants provides a possible paradigm for RNAi-dependent TGS in metazoans, a specialized TGS effector complex has not yet been characterized in an animal.
All RNA silencing complexes characterized to date contain a member of the ARGONAUTE (AGO)/PIWI protein family (Hall 2005). This protein family can be divided into AGO and PIWI subfamilies. The AGO subfamily proteins bind to ∼21-nucleotide (nt) small interfering RNAs (siRNAs) and miRNAs derived from long double-stranded transcripts and miRNA genes, respectively. In contrast, PIWI subfamily proteins bind to 24- to 30-nt Piwi-interacting RNAs (piRNAs) that are likely derived from long single-stranded precursors (Lin 2007). Five AGO/PIWI proteins are found in Drosophila: PIWI, AUBERGINE (AUB), AGO1, AGO2, and AGO3. Among them, PIWI binds to 24- to 26-nt piRNAs that are largely derived from repetitive sequences in the genome (Vagin et al. 2006; Brennecke et al. 2007) and has been broadly implicated in gene silencing (Pal-Bhadra et al. 2004; Saito et al. 2006; Vagin et al. 2006). Here we report that PIWI associates with chromatin and interacts directly with heterochromatin protein 1a (HP1a), a nonhistone chromosomal protein that plays diverse and critical roles in chromatin structure, transcription, DNA replication, chromosome segregation, and genomic stability (James et al. 1989; Hiragami and Festenstein 2005; Peng and Karpen 2007). Furthermore, a transgene with a mutation in PIWI that abrogates HP1 binding fails to rescue silencing in a PIWI-depleted line, demonstrating that this interaction has an important epigenetic function. The physical and functional interaction between HP1 and PIWI may represent a key link between small RNA pathways and TGS in Drosophila.
Results
PIWI interacts specifically with HP1a
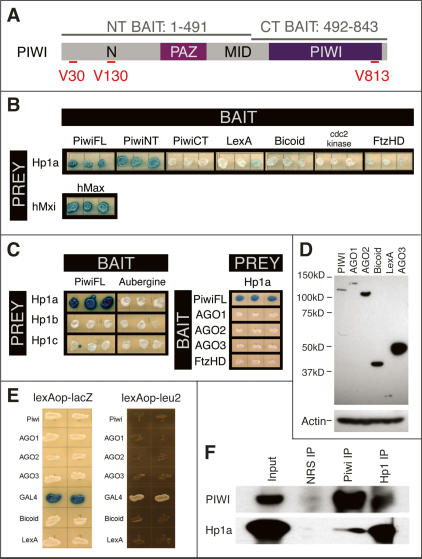
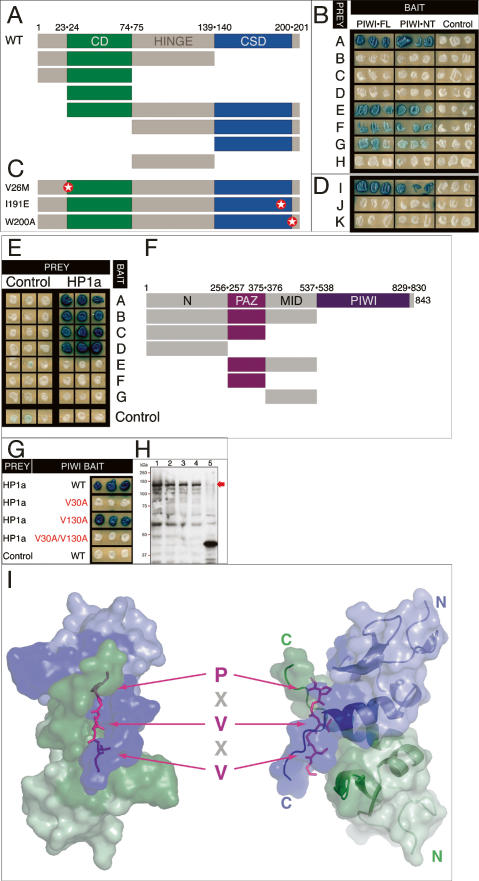
To search for PIWI interactors, we conducted yeast two-hybrid (Y2H) screens of a high-complexity ovarian cDNA library using three PIWI baits: full-length (PIWI-FL), N-terminal half (PIWI-NT), and C-terminal half (PIWI-CT) (Fig. 1A; see Supplemental Material). We screened 12.5, 7.5, and 2.5 million primary transformants with the three baits and recovered 102, 100, and 0 strong positives, respectively. Forty-four of 102 positives for PIWI-FL and 39 of 100 positives for PIWI-NT encoded cDNAs for Drosophila HP1a. HP1a binding to PIWI is strong and highly specific; HP1a does not interact with a panel of unrelated baits (Fig. 1B). The Drosophila melanogaster genome encodes two additional HP1-like chromatin proteins: HP1b and HP1c (Smothers and Henikoff 2001). However, a Y2H assay indicates that PIWI interacts specifically with HP1a, but not HP1b or HP1c (Fig. 1C). In turn, HP1a also fails to interact with two other PIWI subfamily proteins, AUB and AGO3, or the two AGO subfamily proteins, AGO1 and AGO2 (Fig. 1C). Note that AGO1 is expressed as strongly as PIWI, whereas AGO2 and AGO3 are expressed at much higher levels than PIWI in this yeast cell system (Fig. 1D). All of these proteins enter the budding yeast nucleus, as determined by two transcriptional blocking assays (Fig. 1E; see Materials and Methods)
Figure 1.
HP1a interacts specifically with PIWI. (A) Baits used in PIWI Y2H screens. PIWI contains N, PAZ, MID, and PIWI domains. Y2H baits are PIWI-FL, residues 1–843; PIWI-NT, residues 1–491; and and PIWI-CT, residues 492–843. The positions of the three “PxV” sequences—at V30, V130 and V813—are indicated. (B) HP1a is a strong PIWI interactor. Three independent transformants for each bait/prey combination were plated to assess LacZ activity of reporter plasmid pSH18-34. HP1a interacts strongly with PIWI-FL and PIWI-NT baits, but not with PIWI-CT or unrelated baits, including the LexA moiety of the bait plasmid pEG202, Drosophila BICOID, Drosophila Cdc2 kinase or Drosophila FUSHI-TARAZU homeodomain. The strongly interacting human Mxi and Max proteins serve as a positive control. (C) The PIWI–HP1a interaction is specific. Full-length PIWI interacts with HP1a, but not with closely related HP1b or HP1c. Similarly, HP1a shows no interaction with PIWI paralogs AUB, AGO1, AGO2, or AGO3. (D) AGO1 is expressed at a similar level to PIWI, whereas AGO2 and AGO3 are expressed much more abundantly than PIWI, as shown by the Western blot analysis. (E) AGO1, AGO2, and AGO3 proteins enter the nucleus, as shown by lexA–lacZ reporter and lexA–leu2 expression-blocking assays (see Materials and Methods). (F) PIWI and HP1a interact in vivo. Nuclear proteins from Drosophila embryos were incubated with antibodies specific for HP1a or PIWI and the resulting immunoprecipitates were analyzed for coprecipitation by Western blot analysis. For reference, HP1a and PIWI immunoprecipitates are compared with material generated using naive rabbit serum (NRS).
In order to determine whether the Y2H interaction observed between HP1a and PIWI occurs in vivo, coimmunoprecipitation assays were performed. Immunoprecipitates from 6- to 18-h Drosophila embryo nuclei generated with specific antibodies against HP1a are enriched for PIWI protein, by comparison with control immunoprecipitates with normal rabbit serum (Fig. 1F). The converse is also true: Anti-PIWI antibody immunoprecipitates also contain HP1a (Fig. 1F). The small amount of coprecipitating protein, as suggested by the relative band intensities displayed, implies that only a small fraction of the PIWI and HP1a pools are involved in this particular complex.
PIWI colocalizes with HP1a at many chromosomal sites
The presence of PIWI in Drosophila nuclei and in condensed regions of nurse cell polytene chromosomes has been reported (Cox et al. 2000; Lei and Corces 2006), but direct interaction with HP1a raises further questions about the global chromosomal distribution of PIWI. To more precisely locate PIWI within Drosophila nuclei, we utilized antibodies directed against a peptide sequence in the C terminus of the protein to stain polytene nuclei and chromosomes. This antibody recognizes a single 95-kDa (PIWI-sized) band in salivary gland and Kc cell extracts as shown by Western blot (Supplementary Fig. S1). Immunostaining of Drosophila embryos with this antibody further demonstrates the presence of PIWI in all nuclei of stage 5 embryos, where it colocalizes with HP1a (Fig. 2A). PIWI protein appears to be more abundant in prospective germline (pole cell and embryonic gonad) nuclei than in somatic nuclei, but is easily detected by immunostaining in all nuclei of embryos throughout embryogenesis (data not shown). Consistent with a role in tissues other than the germline, RT–PCR analysis indicates that PIWI is expressed in somatic tissues throughout development (Supplementary Fig. S2).
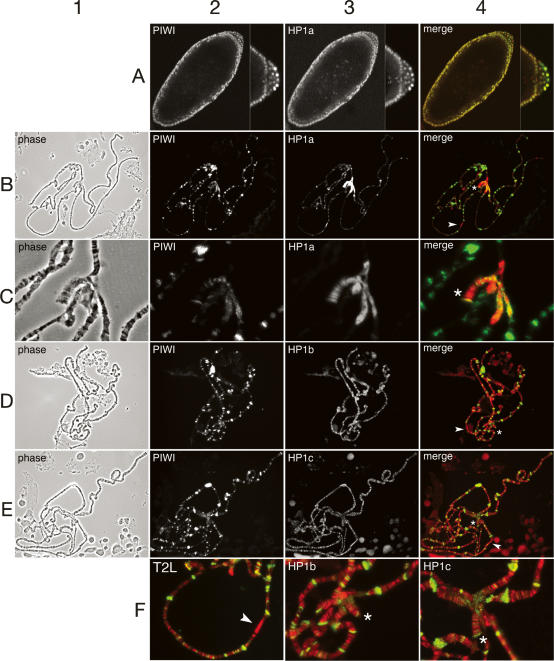
Figure 2.
PIWI is associated with chromatin, where it colocalizes with HP1a. All images are from wild-type (Oregon R) embryos or larval salivary glands. In merged images showing PIWI (green channel) and HP1 isoforms (red channel), the overlap is yellow. On merged images, arrowheads indicate cytological region 31; the fourth chromosome is denoted by an asterisk. (A) PIWI colocalizes with HP1 in all nuclei of stage 5 embryos; PIWI is more abundant in pole cell nuclei (posterior pole is shown at higher magnification at right of each embryo image). (B) PIWI shows extensive colocalization with HP1a to euchromatic bands along polytene chromosome arms, to telomeres, and to distinct regions of the chromocenter. HP1a is most concentrated in the chromocenter and along chromosome 4. (C) PIWI partially colocalizes with HP1a in the chromocenter and at distinct bands on the largely heterochromatic chromosome 4. (D–E) PIWI localization is distinct from and largely nonoverlapping with HP1b and HP1c. PIWI does not overlap with HP1b or HP1c in region 31. (F) Enlargements from merged images in C–E, emphasizing that PIWI colocalizes with HP1a at telomere 2L, is not concentrated in region 31A, and is distinct from both HP1b and HP1c in the chromocenter.
Immunostaining of larval polytene chromosomes with the above anti-PIWI antibody reveals a complex banded distribution on polytenes that is partially coincident with HP1a (Fig. 2B). Confirming its specificity for endogenous PIWI protein, the pattern generated by this antibody on chromosomes and in Western blots is completely blocked by the peptide antigen used to generate the antiserum (Supplementary Figs. S1, S3) and is virtually absent in chromosomes from larvae homozygous for the protein-null piwi2 allele (Supplementary Fig. S3).
While HP1a prominently localizes to the chromocenter (an agglomeration of pericentric heterochromatin) and to the largely heterochromatic fourth chromosome, it is also found in all telomeres and in >200 bands along polytene chromosome arms (James et al. 1989; Fanti et al. 2003). A prominent site of HP1a binding on the chromosome arms is cytological region 31A, wherein HP1a localizes to eight discrete bands and functions in a manner distinct in many ways from its action in the chromocenter (Fanti et al. 2003; Cryderman et al. 2005). PIWI colocalizes with HP1a in a complex pattern at multiple classes of chromatin loci (Fig. 2B,C,F2), but is completely absent from region 31A (Fig. 2F2). While HP1a is uniformly distributed within the chromocenter, PIWI has an irregular and particulate distribution within the pericentric heterochromatin of each chromosome (Fig. 2C). The uniform distribution of HP1a reflects its role as a foundational building block in pericentric chromatin, whereas the focal distribution of PIWI may indicate a localized function at a subset of sequences. PIWI deposition also overlaps with HP1a in polytene telomeres (Fig. 2B,F2; Supplementary Fig. S4). Furthermore, there is significant overlap between HP1a-positive and PIWI-positive euchromatic bands along the arms of chromosomes 1–3 (Fig. 2B). In chromosome 4, PIWI is restricted to a subset of distinct bands relative to HP1a (Fig. 2B,C). Chromosome 4 is a mosaic of alternating heterochromatin and euchromatin with a high density of repetitive sequence elements. The mechanism whereby interspersed heterochromatin domains are established in chromosome 4 is unknown, but there are indications that certain repetitive elements participate in this process, possibly by an RNA-induced silencing mechanism (Sun et al. 2004; Haynes et al. 2006).
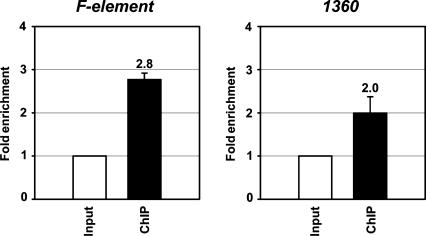
To further verify the colocalization of PIWI and HP1a on chromosomes, we conducted chromatin immunoprecipitation (ChIP) assays to isolate MYC-PIWI-associated chromatin from a lysate of adult flies that carry a fully functional myc-piwi transgene (Cox et al. 2000; see Materials and Methods). Chromatin precipitated by MYC-PIWI antibodies was recovered and the DNA was assayed by real-time PCR to quantify the association of MYC-PIWI with the F and 1360 elements—two transposable elements known to be preferentially bound by HP1a (De Lucia et al. 2005). MYC-PIWI is enriched at F and 1360 elements 2.8- and 2.0-fold, respectively, over the housekeeping gene rpL32 (Fig. 3). These results further support the colocalization of PIWI and HP1a at specific genomic sites.
Figure 3.
PIWI is enriched in the HP1a-rich F and 1360 elements. Shown are the ratios of the abundance of PIWI-associated DNA of F and 1360 elements versus rpL32 DNA obtained by MYC-PIWI immunoprecipitation of cross-linked chromatin. The results represent the average values of six independent samples.
PIWI does not colocalize with HP1b or HP1c on chromosomes
Given the specificity of the Y2H interaction observed between HP1a and PIWI, we hypothesized that HP1b and HP1c would not share the same chromosomal colocalization. Upon immunostaining with antibodies specific for HP1b or HP1c, we found that unlike HP1a, neither HP1b nor HP1c displayed significant colocalization with PIWI on chromosomes. HP1b and HP1c each localizes to many bands along the euchromatin arms, none of which appears to overlap with strong PIWI signals (Fig. 2D–F). This is consistent with a previous report indicating that neither HP1b nor HP1c shows significant overlap with HP1a in the chromocenter or euchromatic polytene bands (Smothers and Henikoff 2001), and is in agreement with our Y2H results, which indicate that PIWI association is HP1a specific.
PIWI is localized at genomic sites enriched for methylated H3K9
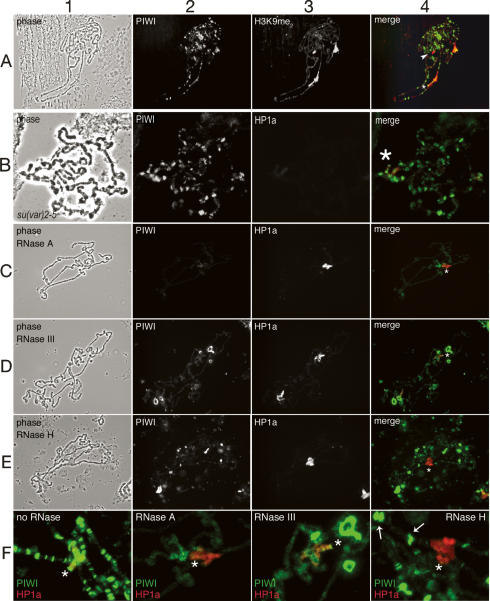
The ability to visualize PIWI on chromosomes permits us to investigate the relationship between PIWI, HP1a, and the epigenetic mark—methylated H3K9 (H3K9me). In Drosophila and other organisms, much (but not all) stable localization of HP1a depends on the H3K9me2/3 mark provided by the SUVAR3-9 family of HMTs. The H3K9me2 staining pattern of Drosophila polytene chromosomes and proper targeting of HP1a have a weak dependence on piwi gene function, seen in piwi2 homozygous larvae (Pal-Bhadra et al. 2004). As would be expected given its colocalization with HP1a, PIWI is present at sites enriched for H3K9me2 (Fig. 4A). A simple explanation is that initial HP1a deposition depends on RNA-induced, PIWI-directed targeting; subsequent interaction of HP1a with H3K9 HMTs results in H3K9 methylation, perhaps as a nucleation step of heterochromatin formation. This hypothetical mechanism would predict that PIWI localization should not depend on the presence of HP1a. This prediction is borne out in Figure 4B, which demonstrates that PIWI localization in chromatin is not globally altered in Su(var)2-5 animals that are largely depleted of HP1a at this stage (Fig. 4B). However, it should be noted that because of maternal loading of chromosomal proteins into the zygote, both HP1a and PIWI are presumably present in the embryos of the respective homozygous or heteroallelic mutant animals (recovered by crossing heterozygous mutant parents) (Megosh et al. 2006), and so are present when heterochromatin is first established.
Figure 4.
PIWI chromatin binding overlaps H3K9 methylation and depends on RNA hybrids but not HP1a. Images from indirect immunofluorescent detection of PIWI and HP1a. Labels and color channels are as in Figure 3. All images are of wild-type (Oregon R) chromosomes, except B, which shows Su(var)2-5 mutant chromosomes. (A) PIWI shows significant overlap with dimethyl-H3K9 in the chromocenter and fourth chromosome but not region 31 (arrowhead). (B) PIWI localization to polytene chromosomes is not globally perturbed in the absence of HP1a. Su(var)2-5 chromosomes are shown at higher magnification because of their small size by comparison with wild-type chromosomes. (C–F) Patterns of PIWI and HP1a (determined by indirect immunofluorescence) on chromosomes treated with different RNase activities. In each case, the HP1a-binding pattern remains essentially the same as on untreated chromosomes. On all merged images the fourth chromosome is indicated by an asterisk. (C) Mild treatment with RNase A eliminates the PIWI-binding pattern seen on untreated chromosomes without perturbing HP1a. (D) RNase III treatment results in loss of PIWI binding to bands in the euchromatic arms without abolishing PIWI bound in pericentric chromatin and in bands on chromosome 4. Delocalized PIWI is concentrated in ring-like bodies that may represent nucleolar fragments in the polytene preparation. (E) PIWI binding in pericentric chromatin and chromosome 4 is abolished by RNase H digestion; binding along the euchromatic arms is diminished at some sites more than others, but not abolished. (F) Enlargement of chromocenters from merged images in C–E with a representative image from untreated chromosomes for comparison. Arrows point to the brightly staining PIWI bands in euchromatic arms, not affected by RNase H treatment.
PIWI association with chromatin is RNA dependent
This hypothetical mechanism, in which PIWI acts as the core of a TGS effector complex, also predicts that PIWI binding to chromosomes is RNA dependent. We tested this prediction by assessing the impact of ribonuclease (RNase) treatment of chromosomes on PIWI and HP1a distribution; we found that mild treatment with the single-stranded RNA-specific RNase A markedly reduced PIWI localization on chromosomes without significantly altering the distinctive HP1a-binding pattern at the chromocenter, chromosome 4, telomeres, or region 31A (Fig. 4C,F2), in keeping with previously published analyses of HP1a (Piacentini et al. 2003). In mammalian cells, HP1 binding to pericentric chromatin was disrupted by digestion with RNase A (Maison et al. 2002). However, those experiments were performed under more extensive digestion conditions, using 100-fold more RNase A and longer digestion times than were used here.
The sensitivity of PIWI chromatin binding to RNase A digestion raises questions regarding the nucleic acid targets of the PIWI:RNA complexes presumed to dictate the PIWI pattern on chromosomes. Does PIWI utilize its guide RNA to identify sequences in genomic DNA, or to identify associated RNA transcripts? We used RNases with specificity for dsRNA (RNase III) and RNA:DNA heteroduplex (RNase H) to address this question and found that the PIWI pattern is partially sensitive to each of these nucleases. While PIWI binding in the polytene arms and at telomeres is most sensitive to RNase III digestion (Fig. 4D,F3), PIWI protein in heterochromatic domains is most sensitive to RNase H digestion (Fig. 4E,F4). These results suggest that RNA-dependent PIWI chromatin association occurs by at least two distinct mechanisms. In the chromosome arms and at telomeres, PIWI appears to bind via hybridization with RNA targets, probably nascent transcripts. In heterochromatin, where active transcription is rare, PIWI may bind instead through hybridization with genomic DNA. We note that, like RNase A, neither of these nucleases perturbs binding of HP1a in its characteristic locations, presumably reflecting the redundant modes of binding that stabilize HP1a in chromatin.
HP1a binds to a PxVxV motif in the N-terminal domain of PIWI
To gain more insight into the molecular mechanism of PIWI–HP1a interaction we mapped the intermolecular contacts between the two proteins. We pinpointed PIWI–HP1a interaction requirements using two approaches: Y2H and NMR. HP1 family proteins contain three domains (Fig. 5A): an N-terminal chromodomain (CD) that mediates binding to H3K9me2/3, a flexible hinge region, and a C-terminal chromoshadow domain (CSD) that mediates dimerization and interaction with many HP1 target proteins (Hiragami and Festenstein 2005). Deletion analysis demonstrates that the HP1a CSD alone is necessary and sufficient for binding to NT-PIWI and FL-PIWI baits (Fig. 5B). To investigate further, we utilized two Drosophila HP1a point mutations analogous to the well-characterized mouse HP1-β (mHP1β) mutations that disrupt CSD dimerization or target interaction (Brasher et al. 2000; Thiru et al. 2004; Hines 2006). In mHP1β, two CSDs symmetrically dimerize to form the interface for the asymmetrical binding of a single target peptide (Thiru et al. 2004). The Drosophila W200A mutant is equivalent to mHP1β W170A, which does not affect dimerization but abolishes HP1 binding to the PXVXL motif in targets, whereas the I191E mutant is equivalent to mHP1β W161E, which disrupts the dimerization interface (Fig. 5C; Brasher et al. 2000; Thiru et al. 2004; Hines 2006). The inability of the Drosophila I191E HP1a to dimerize has been validated by in vitro studies (Hines 2006). PIWI binding is lost in the presence of these HP1a mutations (Fig. 5D). In contrast, PIWI interaction in the Y2H assay is maintained in the HP1a CD mutation V26M (Fig. 5D), which abolishes H3K9me2/3 binding (Jacobs et al. 2001). Thus, HP1a requires an intact CSD dimer interface for binding to PIWI.
Figure 5.
HP1a requires an intact CSD dimer interface to bind PIWI. (A) PIWI requires an intact CSD dimer interface to bind HP1a. Cartoon of the HP1a deletion series used to map PIWI-binding requirements. The 206-residue HP1a contains a CD, a hinge domain, and a CSD. Domain junction residue numbers are indicated (WT, full-length, wild type). (B) Only prey possessing the CSD (prey A, E, F, and G) interact with PIWI-FL and PIWI-NT baits. (C,D) PIWI interaction is lost with HP1a mutations predicted to disrupt either its target binding interface within the intact CSD dimer (W200A mutant) or dimerization (191E mutant), but not with a mutation in the CD (V26M). Positions of the respective point mutations are indicated by stars. (E) Only baits containing the N domain of PIWI (baits A–D) interact with HP1a-FL. (F) Cartoon of the PIWI deletion series used to map HP1a binding requirements. Domain junction residue numbers are indicated. (G) V30 of PIWI but not V130 is required for HP1a interaction. Wild type and V130A PIWI mutant produce comparably strong LacZ signals, whereas the signal in V30A or the V30A/V130A double mutant is undetectable. (H) Western blot of protein extracts of yeast strains in G. Each bait is expressed at comparable levels, as detected by polyclonal antibody to LexA, the DNA-binding moiety common to each bait. (Lane 1) Wild-type PIWI. (Lane 2) V30A. (Lane 3) V130A. (Lane 4) V30A/V130A. (Lane 5) BICOID homeodomain. The LexA-PIWI baits (red arrow) are predicted to be 120.2 kDa. (I) Model of the HP1a CSD dimer in complex with the PIWI peptide (TSRGSGDGPRVKVFRGSSSGD). Top and side views are shown in left and right panels, respectively. Each monomer of the HP1a CSD dimer is color-coded (green or blue). The CSD-binding motif (PxVxV) in the PIWI peptide is shown in stick models (magenta). Side chains of the conserved residues (P,V,V) are displayed. Chemical shift perturbations are calculated as δCS = [δH2 + (0.2δN)2]1/2. Residues of HP1a that experience significant resonance perturbation (δCS > 0.06 ppm) during PIWI peptide titration are colored in dark green and dark blue and are mapped on the CSD surface and the ribbon diagram. The homology model of the complex was built using the XLOOK program (Lee 1993), and the figure was generated by PyMOL (Delano Scientific).
We then used a PIWI deletion series to map the region of PIWI that interacts with HP1a by the Y2H assay (Fig. 5E). Each AGO/PIWI protein contains an N-terminal domain, a Mid domain, and highly conserved PAZ and PIWI domains (Fig. 5F). Deletion analyses indicate that HP1a requires only the N-terminal domain of PIWI for interaction (Fig. 5E). This domain contains two sequences similar to a known vertebrate HP1-binding motif, the monomeric PxVxL motif (Lechner et al. 2005) that binds the dimerized CSD. By phage display, Drosophila HP1a binds a similar consensus, PxVx [M or L or V] (Smothers and Henikoff 2000). The PIWI N-terminal domain contains two PxV sequences: PRVKV, centered on V30, and PRVRM centered on V130 (Fig. 1A). Mutating V30 to alanine (Fig. 5G) abolishes HP1a binding. In contrast, mutating V130, which lies in an equally good consensus sequence, to alanine does not impact the interaction (Fig. 5G). Each PIWI mutant bait is expressed at levels comparable with the wild type, ruling out differences attributable to differential bait stability (Fig. 5H).
To confirm the V30 interaction, we synthesized a 21mer PIWI peptide centered on V30 of the HP1a-binding sequence for NMR titration with the HP1a CSD dimer (Fig. 5I; Supplementary Fig. S5). The 1H and 15N chemical shifts of a number of residues of the HP1a CSD showed significant perturbations after peptide addition, suggesting that these residues are involved in PIWI binding. The exchange rate was slow on the NMR time scale, consistent with tight binding between the HP1a CSD dimer and the PIWI peptide. A structural model of Drosophila HP1a CSD dimer complex was built based on its high degree of sequence homology with the mHP1β CSD, which binds the CAF-1 peptide (Fig. 5I; Thiru et al. 2004). Consistent with such a structural model, where a single PIWI peptide binds across the HP1a CSD dimer, CSD residues that experience significant resonance perturbation during NMR titration are primarily distributed along the β-strands close to the PIWI peptide, the C-terminal portion of the central helices, and the C-terminal extended loop.
The HP1a-interacting PRVKV motif is required for the silencing function of PIWI
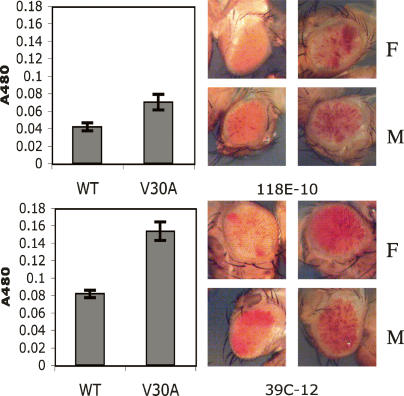
The direct interaction between dimerized HP1a CSD and the PIWI V30-binding motif potentially represents a very direct means by which PIWI protein could act to recruit heterochromatin-forming components to specific sites in the Drosophila genome. In order to test the functional significance of the PIWI–HP1a interface defined here for gene silencing, we introduced transgenes encoding either the wild-type or V30A mutant form of PIWI under the native piwi promoter in flies. The endogenous function of the V30A mutant product was compromised; when introduced into a (lethal) piwi-null genetic background, only the wild-type transgene was able to rescue viability (data not shown). Previous work has shown that hypomorphic alleles of piwi act as dominant suppressors of heterochromatic gene silencing (Pal-Bhadra et al. 2004; Haynes et al. 2006). We tested the capacity of either transgene to rescue dominant silencing defects produced in flies heterozygous for the protein-null piwi2 allele. In the haplo-insufficient piwi background, the presence of one or two copies of the wild-type transgene supported greater silencing of variegating white reporters than did the presence of the V30A transgene (Fig. 6). These data strongly suggest that the V30 motif is directly involved in silencing observed at white reporters embedded in constitutive heterochromatin.
Figure 6.
A PIWI transgene bearing the V30A mutation is deficient in rescuing dominant defects in white reporter silencing produced by the piwi2 mutation. Eye pigmentation of PIWIWT; piwi2/CyO;P[wvar] flies are compared with pigmentation in PIWIV30A;piwi2/CyO;P[wvar] flies. Variegating white reporters (P[wvar]) analyzed are embedded in pericentric (118E-10) or fourth chromosome (39C-12) heterochromatin. (F) Female; (M) male.
Discussion
We showed that Drosophila PIWI interacts directly with HP1a and is distributed on chromosomes in a pattern overlapping HP1a. Association of PIWI with constitutive heterochromatin domains including pericentric heterochromatin, telomeres, and part of the banded portion of chromosome 4 is consistent with the profile of PIWI-associated small RNAs, which includes sequences homologous to telomeric and centromeric repetitive elements as well as some that are overrepresented in chromosome 4 (Saito et al. 2006; Brennecke et al. 2007; Gunawardane et al. 2007). We determined that the PIWI–HP1a interaction is mediated through binding of a PxVxL-type motif by dimerized CSDs of HP1a. Furthermore, we showed that the intact PxVxL motif is required in vivo for effective heterochromatic silencing. This is consistent with the wealth of data implicating PIWI at the interface of RNA silencing mechanisms with epigenetic phenomena, in particular heterochromatin-induced silencing. PIWI is a nuclear protein (Cox et al. 2000) required for silencing of transposons and retroviruses (Sarot et al. 2004; Vagin et al. 2006). PIWI is required for effective silencing of multiple copies of dispersed transgenes (Pal-Bhadra et al. 2002), of tandem repeats of the white gene at euchromatic insertion sites, and of white reporter genes in the pericentric heterochromatin or fourth chromosome (Pal-Bhadra et al. 2004; Haynes et al. 2006). Silencing in the latter two cases is dependent on HP1a (Wallrath and Elgin 1995; Fanti et al. 1998; Haynes et al. 2006). Here we showed that PIWI interacts directly with HP1a. Some PIWI signal overlaps with HP1a in polytene chromosomes, suggesting that PIWI and HP1a together regulate the epigenetic state of diverse regions in the genome. This overlap is spatially complex and could therefore represent numerous varied roles for PIWI, HP1a, and the PIWI–HP1a interaction. Heterochromatin is first assembled early, while the Drosophila embryo is still a syncytium (nuclear division cycles 10–14). This is critical for the silencing observed in PEV, which may be compromised later in development (Lu et al. 1998). While the HP1a–PIWI association observed on polytene chromosomes might be similar to that inferred in the embryo, it might also represent a different function (e.g., see Piacentini et al. 2003).
Recently, PIWI has been shown to bind in the germline to piRNAs, many of which are from heterochromatic regions (Saito et al. 2006; Brennecke et al. 2007). The colocalization of PIWI and HP1a in pericentric heterochromatin may indeed reflect a simple relationship between the piRNA pathway, histone modification, and heterochromatin formation similar to the S. pombe system wherein AGO1-mediated TGS locally targets H3K9 methylation to create a binding site for the HP1 homolog Swi6 (Volpe et al. 2002; Noma et al. 2004). In Drosophila, our data on the specific interaction between PIWI and HP1a raise the possibility of an alternate pathway to HP1a-mediated heterochromatinization: A PIWI–piRNA complex might directly recruit HP1a to piRNA-corresponding genomic sequences, which could then recruit HMTs such as SU(VAR)3-9 to effect nucleation/spreading. This would represent an H3K9me-independent mode for initial HP1 localization, an alternative but potentially equally effective means for triggering local formation of heterochromatin. Conversely, if heterochromatin formation is targeted by a different mechanism, the presence of HP1a could allow stable binding of PIWI to heterochromatin for PTGS, a process that might be necessary to maintain silencing throughout the lifetime of the fly.
Materials and methods
Y2H screens and assays
The PIWI Y2H screens assays were conducted using the Clontech Matchmaker LexA Two-Hybrid System according to the manufacturer’s protocols (Clontech). For details, see the Supplemental Material.
Protein expression, purification, and NMR spectroscopy
The DNA sequence corresponding to the Drosophila HP1a CSD (residues 132–206) was cloned into the pET-15b vector to produce an N-His6 tagged protein (EMD Biosciences-Novagen). The recombinant protein was overexpressed in BL21(DE3)STAR cells (Invitrogen Corp.), purified using a Ni2+-NTA column, and digested by thrombin to remove the N-His6 tag. The resulting fragment, which contained the additional GSHM residues N-terminal to the HP1a CSD domain, was further purified by size exclusion chromatography and was renumbered to 1–79 in NMR studies. NMR buffer contained 25 mM sodium phosphate, 100 mM KCl, and 10% D2O (pH 7.0). The 13C-/15N-labeled protein sample was prepared by growing bacterial cells in M9 minimal media with 15N-NH4Cl and 13C-glucose as the sole nitrogen and carbon sources. NMR experiments were carried out at 27°C using a Varian INOVA 600-MHz spectrometer. Backbone resonances were collected using a suite of triple-resonance experiments [HNCA/HN(CO)CA, HN(CA)CB/HN(COCA)CB, and HA(CA)NH/HA(CACO)NH] (Clore and Gronenborn 1993; Ferentz and Wagner 2000); these resonances were assigned using PACES (Coggins and Zhou 2003) and were confirmed by manual analysis. NMR data were processed using NMRPIPE (Delaglio et al. 1995) and were analyzed by XEASY (Bartels et al. 1995). NMR titration was performed by adding increasing amounts of the unlabeled PIWI peptide to 15N-labeled HP1a CSD at molar ratios of 0:1, 1:3, 2:3, 1:1, 2:1, and 3:1. A series of 1H–15N HSQC experiments were collected at 27°C on a Varian INOVA 600-MHz spectrometer. The exchange rate of the HP1a CSD/PIWI peptide complex was found to be slow on the NMR time scale, and the binding ratio was estimated to be one PIWI peptide per HP1a CSD dimer.
Drosophila cultures, stocks, and genetic analysis
Mutants Su(var)2-504 and Su(var)2-505 were used for HP1 depletion experiments (Eissenberg et al. 1992). Each of these alleles was balanced over CyO-GFP, and GFP-negative third-instar larvae were selected from heteroallelic crosses for cytological investigation. The piwi2 mutant allele (Lin and Spradling 1997) was likewise balanced over CyO-GFP; GFP-negative third-instar larvae were selected from appropriate crosses for cytological investigation.
Construction of the wild-type and V30A piwi genomic transgenic animals
The piwi V30A mutant transgene is identical to the previously described piwi genomic P-element-based rescue construct, pRC12 (Cox et al. 1998), except that the wild-type V30 codon was replaced with an alanine codon. This was accomplished in the following multistep procedure in which the numbers (in parentheses) refer to nucleotides within the 6484-base-pair (bp) EcoRV–HindIII piwi genomic rescue fragment of pRC12. First, the 1518-bp PmeI(2195)–SwaI(3713) fragment of pRC12 was transferred to a cloning vector (SDF-SP728s) to generate clone SDF164.1. Mutagenesis was accomplished using the Stratagene QuikChange II site-directed mutagenesis kit under conditions recommended by the manufacturer (Stratagene). The following oligonucleotides were used to mutagenize piwi’s V30 codon (GTG) to an alanine codon (GCC), resulting in clone SDF165.1: GV30MTOP, 5′-AATGCTTTCATGCAGCGGGCCAAAGTA TTCAGAGGATCTTC-3′; GV30MBOT, 5′-GAAGATCCTCT GAATACTTTGGCCCGCTGCATGAAAGCATT-3′ (the alanine codon 30 and its reverse complement are indicated in bold). Finally, the internal 1274-bp PflfI(2439)–SwaI(3713) fragment of SDF165.1 was substituted for the wild-type 1274-bp PflfI(2439)–SwaI(3713) fragment of pRC12, resulting in clone SDF178-50. Transgenic animals of pRC12 and SDF178-50 were generated using standard embryo microinjection techniques.
For PEV assays involving the piwi wild-type and V30A transgenes, stable fly lines were created with each transgene in a genetic background including the piwi2 mutation balanced over CyO and variegating white reporters embedded either in the pericentric chromatin (118E-10) or the banded portion of the fourth chromosome (39C-12). Six sets of 10 flies of each genotype (five male/five female) were selected at random for pigment assays performed as detailed in previous publications (Rabinow et al. 1991). Digital images of variegating eye phenotypes were selected to represent median eye pigmentation for each genotypic population.
Embryo preparation, polytene chromosome preparation, and RNase treatment
Embryos were collected and prepared for immunostaining using standard methods (Sullivan et al. 2000). Chromosomes were prepared as described previously (Stephens et al. 2004). RNase treatment of chromosomes was carried out in situ in the salivary glands. Briefly, dissected glands were incubated at room temperature with the appropriate RNase in Cohen Buffer (Alfageme et al. 1976) for 10 min. Ribonuclease digest concentrations were DNase-free RNase A (10 μg/mL; Promega Corp.), recombinant RNase H (0.05 U/μL; Ambion, Inc.), and recombinant RNase III (0.05 U/μL; New England Biolabs). All reactions were quenched by the addition of yeast tRNA (100 μg/mL), and the glands were transferred to formaldehyde fixative for 10 min and squashed.
Immunostaining of polytene chromosomes and embryos
Rabbit sera against PIWI C-terminal peptide (C)SIPQNALEKKFYYL were prepared and affinity-purified as described previously (Caudy et al. 2002) and was used at 1:200 on polytene chromosomes and embryos. Rabbit sera against the N-terminal portion of the protein (four tested) were not effective in this assay. The monoclonal serum C1A9 against HP1 (James et al. 1989) was used throughout this investigation at 1:20 to detect HP1a in polytene chromosomes and at 1:100 in embryos. Polyclonal antibodies against other HP1 isoforms were raised in guinea pigs with peptide antigens (HP1b antigen, CPESIRSKRKSFLEDDTEEQK; HP1c antigen, CRHIAMRMKGVPEELRLAASR) representing unique portions of each protein. Rabbit anti-H3dimethylK9 (catalog no. 07-441, Upstate Biotechnology) was used at 1:25. Indirect immunofluorescent staining of polytene chromosomes from third instar larvae of D. melanogaster was performed as described (Stephens et al. 2004). Secondary antibodies (Invitrogen-Molecular Probes) were labeled with Alexa Fluor 488 or 594. Competition of anti-PIWI antibodies with peptide antigens was accomplished by incubation of diluted antiserum with 50 μg/mL peptide antigen for 30 min at room temperature prior to staining. Controls were prepared with antisera incubated with an equivalent quantity of a peptide unrelated to the specific antigen.
ChIP and quantitative PCR
Purification of nuclei from adult myc-piwi flies was performed following standard procedures. Briefly, adult whole flies were ground in liquid nitrogen, and the powder was suspended in buffer A+ and homogenized sequentially in a ground-glass homogenizer (Pyrex) and a Wheaton Dounce homogenizer. The homogenate was filtered through two layers of Miracloth. The resulting filtrate was layered on a 2-mL buffer AS cushion and centrifuged at 2200g for 10 min at 4°C. The nuclear pellet was resuspended in buffer A+ by Dounce homogenization with a loose pestle and centrifuged as previously. The resulting pellet was resuspended in buffer A by Dounce homogenization with a tight pestle and centrifuged as previously. Buffer A: 60 mM KCl, 15 mM NaCl, 1 mM EDTA, 0.1 mM EGTA, 20 mM HEPES (pH 7.5), 15 mM spermine, 0.15 mM spermidine, 1 mM DTT, 1× Complete Mini, EDTA-free proteinase inhibitor cocktail; buffer AS: buffer A with 0.3 M sucrose; buffer A+: buffer A with 0.5% NP40.
One gram of purified nuclei was suspended in 0.5 mL of ChIP buffer (50 mM HEPES at pH 7.5, 60 mM KCl, 15 mM NaCl, 2 mM MgCl2, 1 mM EDTA, 0.1% deoxycholate.Na2, 1% Triton X-100, 1 mM DTT, 1× Complete Mini, EDTA-free proteinase inhibitor cocktail) and cross-linked with 1% formaldehyde for 15 min at room temperature with continuous rotation. The cross-linking reaction was quenched by the addition of glycine to a final concentration of 0.125 M for 5 min. Cross-linked nuclei were fragmented by sonication on ice (Branson Digital Sonifier 450, output 30%, 15 sec per pulse, 30 rounds), and insoluble material was removed by centrifugation at 12,000g for 10 min at 4°C. Nuclear lysate was incubated overnight at 4°C with anti-MYC monoclonal antibody (1:50). Fifty microliters of salmon sperm DNA/protein G agarose beads (Upstate Biotechnology) were added, and samples were incubated for an additional 2 h at 4°C. Beads were collected and washed once with low-salt RIPA buffer (50 mM HEPES at pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS), once with high-salt RIPA buffer (50 mM HEPES at pH 7.5, 500 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS), once with LiCl washing buffer (50 mM HEPES at pH 7.5, 0.25 M LiCl, 1 mM EDTA, 1% NP-40, 1% Deoxychloate.Na2), and once with TE buffer. Cross-links were reversed overnight at 65°C, followed by proteinase K digestion. DNA was purified from the beads by phenol/chloroform extraction and ethanol precipitation.
Quantitative PCR was conducted on a Roche LightCycler 2.0 system with the Roche SYBR Green Master mix using the following primer sets: F element (forward, 5′-ATCACGGTA GAAGAGCCGCA-3′; reverse, 5′-TGTGAAGACGGATTTT CAGCTC-3′), 1360 element (forward, 5′-GGAGCTCTGCG TATAGCCAACTT-3′; reverse, 5′-ACCTAAACCGCCGAGT CCTG-3′); RpL32 (forward, 5′-CGATCTCGCCGCAGTAAAC 3′; reverse, 5′-CTTCATCCGCCACCAGTCG-3′). The quantities of F element and 1360 DNA precipitated by anti-MYC antibody were normalized against those of RpL32. These ratios are further normalized against those from ChIP input DNA, which gives the relative enrichments of PIWI at F element and 1360 sequences.
Immunoblot analysis
Western blot analysis was performed by standard methods. The rabbit antibody against PIWI was used at 1:2000. Chemiluminescent detection of HRP-conjugated goat secondary antibodies was performed by standard methods. Peptide competition was performed using the same ratio of antiserum:antigen described for polytene chromosome-staining experiments above. Results are shown in Supplementary Figure 1.
Immunoprecipitation
Nuclear sonicate from 6- to 18-h Oregon R embryos was diluted to 1 g of embryo equivalent per milliliter in IP lysis buffer (50 mM Tris at pH 7.5, 150 mM NaCl, 1.0 mM EDTA, 0.1% Triton X-100, 10% glycerol, 1.0 mM DTT, 1× protease inhibitor cocktail). Lysate from 0.25 g of embryo was incubated with 5 μL of the appropriate antibody for 4 h at 4°C with rotation. Antibodies were either rabbit anti-PIWI-C or rabbit anti-HP1a (WA184). Fifty microliters of protein A Sepharose (50% slurry in IP lysis buffer) were added to this mixture and incubated for 1 h at 4°C with rotation, after which immunoprecipitates were collected and washed four times with 500-μL volumes of wash buffer (50 mM Tris at pH 7.5, 150 mM NaCl, 1.0 mM EDTA, 1% NP40, 0.1% SDS, 1.0 mM DTT, 1× protease inhibitor cocktail). Immunoprecipitates were denatured and solubilized by boiling in 1× Laemmli sample buffer, fractionated, blotted, and probed with either rabbit anti-PIWI-C (1:2000) or mouse anti-HP1a (C1A9, 1:1000).
RT–PCR
RT–PCR was performed to examine piwi expression in different developmental stages and tissues. Total RNA was extracted from embryo, third instar larvae, pupae, ovary, testis, adult female carcass, and adult male carcass. The primer used in the RT reaction was 5′-CAAACTGCTGGCACTCGT-3′, and primers for the subsequent PCR reaction were sense primer (5′-TAC TTCCCGAGGTAGTGGTGATGG-3′) and antisense primer (5′-CCACGGCTTTCGCTCTGTCG-3′).
Acknowledgments
We gratefully acknowledge Lori Wallrath and Carrie Hines (University of Iowa) for HP1a point mutant clones and unpublished HP1a data; Roger Brent (The Molecular Sciences Institute) for Yeast Two-Hybrid materials; and Jorg Grosshans (Max Planck Institute) for Drosophila Yeast Two-Hybrid libraries. Thanks also to Gina Volpi for excellent technical assistance. This work is supported by NIH grant HD33760 to H.L., by NIH grant GM073190 to S.C.R.E., and by a Whitehead Foundation grant to P.Z.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1564307
References
- Alfageme C.R., Rudkin G.T., Cohen L.H., Rudkin G.T., Cohen L.H., Cohen L.H. Locations of chromosomal proteins in polytene chromosomes. Proc. Natl. Acad. Sci. 1976;73:2038–2042. doi: 10.1073/pnas.73.6.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels C., Xia T., Billeter M., Güntert P., Wüthrich K., Xia T., Billeter M., Güntert P., Wüthrich K., Billeter M., Güntert P., Wüthrich K., Güntert P., Wüthrich K., Wüthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biol. NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- Brasher S.V., Smith B.O., Fogh R.H., Nietlispach D., Thiru A., Nielsen P.R., Broadhurst R.W., Ball L.J., Murzina N.V., Laue E.D., Smith B.O., Fogh R.H., Nietlispach D., Thiru A., Nielsen P.R., Broadhurst R.W., Ball L.J., Murzina N.V., Laue E.D., Fogh R.H., Nietlispach D., Thiru A., Nielsen P.R., Broadhurst R.W., Ball L.J., Murzina N.V., Laue E.D., Nietlispach D., Thiru A., Nielsen P.R., Broadhurst R.W., Ball L.J., Murzina N.V., Laue E.D., Thiru A., Nielsen P.R., Broadhurst R.W., Ball L.J., Murzina N.V., Laue E.D., Nielsen P.R., Broadhurst R.W., Ball L.J., Murzina N.V., Laue E.D., Broadhurst R.W., Ball L.J., Murzina N.V., Laue E.D., Ball L.J., Murzina N.V., Laue E.D., Murzina N.V., Laue E.D., Laue E.D. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 2000;19:1587–1597. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J., Dus M., Kellis M., Sachidanandam R., Hannon G.J., Kellis M., Sachidanandam R., Hannon G.J., Sachidanandam R., Hannon G.J., Hannon G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Caudy A.A., Myers M., Hannon G.J., Hammond S.M., Myers M., Hannon G.J., Hammond S.M., Hannon G.J., Hammond S.M., Hammond S.M. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes & Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore G.M., Gronenborn A.M., Gronenborn A.M. NMR of proteins. CRC Press; Boca Raton, FL: 1993. [Google Scholar]
- Coggins B.E., Zhou P., Zhou P. PACES: Protein sequential assignment by computer-assisted exhaustive search. J. Biomol. NMR. 2003;26:93–111. doi: 10.1023/a:1023589029301. [DOI] [PubMed] [Google Scholar]
- Cox D.N., Choa A., Baker J., Chang L., Qiaom D., Lin H., Choa A., Baker J., Chang L., Qiaom D., Lin H., Baker J., Chang L., Qiaom D., Lin H., Chang L., Qiaom D., Lin H., Qiaom D., Lin H., Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes & Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D.N., Chao A., Lin H., Chao A., Lin H., Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- Cryderman D.E., Grade S.K., Li Y., Fanti L., Pimpinelli S., Wallrath L.L., Grade S.K., Li Y., Fanti L., Pimpinelli S., Wallrath L.L., Li Y., Fanti L., Pimpinelli S., Wallrath L.L., Fanti L., Pimpinelli S., Wallrath L.L., Pimpinelli S., Wallrath L.L., Wallrath L.L. Role of Drosophila HP1 in euchromatic gene expression. Dev. Dyn. 2005;232:767–774. doi: 10.1002/dvdy.20310. [DOI] [PubMed] [Google Scholar]
- De Lucia F., Ni J.-Q., Vaillant C., Sun F.L., Ni J.-Q., Vaillant C., Sun F.L., Vaillant C., Sun F.L., Sun F.L. HP1 modulates the transcription of cell-cycle regulators in Drosophila melanogaster. Nucleic Acids Res. 2005;33:2852–2858. doi: 10.1093/nar/gki584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A., Vuister G.W., Zhu G., Pfeifer J., Bax A., Zhu G., Pfeifer J., Bax A., Pfeifer J., Bax A., Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Eissenberg J.C., Morris G.D., Reuter G., Hartnett T., Morris G.D., Reuter G., Hartnett T., Reuter G., Hartnett T., Hartnett T. The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics. 1992;131:345–352. doi: 10.1093/genetics/131.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanti L., Dorer D.R., Berloco M., Henikoff S., Pimpinelli S., Dorer D.R., Berloco M., Henikoff S., Pimpinelli S., Berloco M., Henikoff S., Pimpinelli S., Henikoff S., Pimpinelli S., Pimpinelli S. Heterochromatin protein 1 binds transgene arrays. Chromosoma. 1998;107:286–292. doi: 10.1007/s004120050310. [DOI] [PubMed] [Google Scholar]
- Fanti L., Berloco M., Piacentini L., Pimpinelli S., Berloco M., Piacentini L., Pimpinelli S., Piacentini L., Pimpinelli S., Pimpinelli S. Chromosomal distribution of heterochromatin protein 1 (HP1) in Drosophila: A cytological map of euchromatic HP1 binding sites. Genetica. 2003;117:135–147. doi: 10.1023/a:1022971407290. [DOI] [PubMed] [Google Scholar]
- Ferentz A.E., Wagner G., Wagner G. NMR spectroscopy: A multifaceted approach to macromolecular structure. Q. Rev. Biophys. 2000;33:29–65. doi: 10.1017/s0033583500003589. [DOI] [PubMed] [Google Scholar]
- Grewal S.I., Jia S., Jia S. Heterochromatin revisited. Nat. Rev. Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Gunawardane L.S., Saito K., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C., Saito K., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C., Kawamura Y., Nagami T., Siomi H., Siomi M.C., Nagami T., Siomi H., Siomi M.C., Siomi H., Siomi M.C., Siomi M.C. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Hall T.M. Structure and function of argonaute proteins. Structure. 2005;13:1403–1408. doi: 10.1016/j.str.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Haynes K., Caudy A., Collins L., Elgin S.C., Caudy A., Collins L., Elgin S.C., Collins L., Elgin S.C., Elgin S.C. Element 1360 and RNAi components contribute to HP1-dependent silencing of a pericentric reporter. Curr. Biol. 2006;16:2222–2227. doi: 10.1016/j.cub.2006.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines K.A. “Mechanisms of gene silencing by Heterochromatin Protein 1.” Ph.D. thesis, University of Iowa, Iowa City, IA. 2006. [Google Scholar]
- Hiragami K., Festenstein R., Festenstein R. Heterochromatin protein 1: A pervasive controlling influence. Cell. Mol. Life Sci. 2005;62:2711–2726. doi: 10.1007/s00018-005-5287-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S.A., Taverna S.D., Zhang Y., Briggs S.D., Li J., Eissenberg J.C., Allis C.D., Khorasanizadeh S., Taverna S.D., Zhang Y., Briggs S.D., Li J., Eissenberg J.C., Allis C.D., Khorasanizadeh S., Zhang Y., Briggs S.D., Li J., Eissenberg J.C., Allis C.D., Khorasanizadeh S., Briggs S.D., Li J., Eissenberg J.C., Allis C.D., Khorasanizadeh S., Li J., Eissenberg J.C., Allis C.D., Khorasanizadeh S., Eissenberg J.C., Allis C.D., Khorasanizadeh S., Allis C.D., Khorasanizadeh S., Khorasanizadeh S. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James T.C., Eissenberg J.C., Craig C., Dietrich V., Hobson A., Elgin S.C., Eissenberg J.C., Craig C., Dietrich V., Hobson A., Elgin S.C., Craig C., Dietrich V., Hobson A., Elgin S.C., Dietrich V., Hobson A., Elgin S.C., Hobson A., Elgin S.C., Elgin S.C. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur. J. Cell Biol. 1989;50:170–180. [PubMed] [Google Scholar]
- Lechner M.S., Schultz D.C., Negorev D., Maul G.G., Rauscher F.J., Schultz D.C., Negorev D., Maul G.G., Rauscher F.J., Negorev D., Maul G.G., Rauscher F.J., Maul G.G., Rauscher F.J., Rauscher F.J. The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem. Biophys. Res. Commun. 2005;331:929–937. doi: 10.1016/j.bbrc.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Lee C. LOOK: A software system for integrated macromolecular sequence—Structure analysis and modeling. Molecular Applications Group, Palo Alto, CA. 1993. [Google Scholar]
- Lei E.P., Corces V.G., Corces V.G. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat. Genet. 2006;38:936–941. doi: 10.1038/ng1850. [DOI] [PubMed] [Google Scholar]
- Lin H. piRNAs in the germ line. Science. 2007;316:397. doi: 10.1126/science.1137543. [DOI] [PubMed] [Google Scholar]
- Lin H., Spradling A.C., Spradling A.C. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Lu B.Y., Ma J., Eissenberg J.C., Ma J., Eissenberg J.C., Eissenberg J.C. Developmental regulation of heterochromatin-mediated gene silencing in Drosophila. Development. 1998;125:2223–2234. doi: 10.1242/dev.125.12.2223. [DOI] [PubMed] [Google Scholar]
- Maison C., Bailly D., Peters A.H., Quivy J.P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Bailly D., Peters A.H., Quivy J.P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Peters A.H., Quivy J.P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Quivy J.P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Taddei A., Lachner M., Jenuwein T., Almouzni G., Lachner M., Jenuwein T., Almouzni G., Jenuwein T., Almouzni G., Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- Matzke M.A., Birchler J.A., Birchler J.A. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- Megosh H., Cox D.N., Campbell C., Lin H., Cox D.N., Campbell C., Lin H., Campbell C., Lin H., Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr. Biol. 2006;16:1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- Noma K., Sugiyama T., Cam H., Verdel A., Zofall M., Jia S., Moazed D., Grewal S.I., Sugiyama T., Cam H., Verdel A., Zofall M., Jia S., Moazed D., Grewal S.I., Cam H., Verdel A., Zofall M., Jia S., Moazed D., Grewal S.I., Verdel A., Zofall M., Jia S., Moazed D., Grewal S.I., Zofall M., Jia S., Moazed D., Grewal S.I., Jia S., Moazed D., Grewal S.I., Moazed D., Grewal S.I., Grewal S.I. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat. Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M., Bhadra U., Birchler J.A., Bhadra U., Birchler J.A., Birchler J.A. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol. Cell. 2002;9:315–327. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M., Leibovitch B.A., Gandhi S.G., Rao M., Bhadra U., Birchler J.A., Elgin S.C., Leibovitch B.A., Gandhi S.G., Rao M., Bhadra U., Birchler J.A., Elgin S.C., Gandhi S.G., Rao M., Bhadra U., Birchler J.A., Elgin S.C., Rao M., Bhadra U., Birchler J.A., Elgin S.C., Bhadra U., Birchler J.A., Elgin S.C., Birchler J.A., Elgin S.C., Elgin S.C. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- Peng J.C., Karpen G.H., Karpen G.H. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat. Cell Biol. 2007;9:25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piacentini L., Fanti L., Berloco M., Perrini B., Pimpinelli S., Fanti L., Berloco M., Perrini B., Pimpinelli S., Berloco M., Perrini B., Pimpinelli S., Perrini B., Pimpinelli S., Pimpinelli S. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J. Cell Biol. 2003;161:707–714. doi: 10.1083/jcb.200303012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinow L., Nguyen-Huynh A., Birchler J.A., Nguyen-Huynh A., Birchler J.A., Birchler J.A. A trans-acting regulatory gene that inversely affects the expression of the white, brown and scarlet loci in Drosophila. Genetics. 1991;129:463–480. doi: 10.1093/genetics/129.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Nishida K.M., Mori T., Kawamura Y., Miyoshi K., Nagami T., Siomi H., Siomi M.C., Nishida K.M., Mori T., Kawamura Y., Miyoshi K., Nagami T., Siomi H., Siomi M.C., Mori T., Kawamura Y., Miyoshi K., Nagami T., Siomi H., Siomi M.C., Kawamura Y., Miyoshi K., Nagami T., Siomi H., Siomi M.C., Miyoshi K., Nagami T., Siomi H., Siomi M.C., Nagami T., Siomi H., Siomi M.C., Siomi H., Siomi M.C., Siomi M.C. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes & Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarot E., Payen-Groschene G., Bucheton A., Pelisson A., Payen-Groschene G., Bucheton A., Pelisson A., Bucheton A., Pelisson A., Pelisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–1321. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smothers J.F., Henikoff S., Henikoff S. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr. Biol. 2000;10:27–30. doi: 10.1016/s0960-9822(99)00260-2. [DOI] [PubMed] [Google Scholar]
- Smothers J.F., Henikoff S., Henikoff S. The hinge and chromo shadow domain impart distinct targeting of HP1-like proteins. Mol. Cell. Biol. 2001;21:2555–2569. doi: 10.1128/MCB.21.7.2555-2569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens G.E., Craig C.A., Li Y., Wallrath L.L., Elgin S.C., Craig C.A., Li Y., Wallrath L.L., Elgin S.C., Li Y., Wallrath L.L., Elgin S.C., Wallrath L.L., Elgin S.C., Elgin S.C. Immunofluorescent staining of polytene chromosomes: Exploiting genetic tools. Methods Enzymol. 2004;376:372–393. doi: 10.1016/S0076-6879(03)76025-X. [DOI] [PubMed] [Google Scholar]
- Sullivan W., Ashburner M., Hawley R.S., Ashburner M., Hawley R.S., Hawley R.S. Drosophila protocols. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. [Google Scholar]
- Sun F.L., Haynes K., Simpson C.L., Lee S.D., Collins L., Wuller J., Eissenberg J.C., Elgin S.C., Haynes K., Simpson C.L., Lee S.D., Collins L., Wuller J., Eissenberg J.C., Elgin S.C., Simpson C.L., Lee S.D., Collins L., Wuller J., Eissenberg J.C., Elgin S.C., Lee S.D., Collins L., Wuller J., Eissenberg J.C., Elgin S.C., Collins L., Wuller J., Eissenberg J.C., Elgin S.C., Wuller J., Eissenberg J.C., Elgin S.C., Eissenberg J.C., Elgin S.C., Elgin S.C. cis-Acting determinants of heterochromatin formation on Drosophila melanogaster chromosome four. Mol. Cell. Biol. 2004;24:8210–8220. doi: 10.1128/MCB.24.18.8210-8220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiru A., Nietlispach D., Mott H.R., Okuwaki M., Lyon D., Nielsen P.R., Hirshberg M., Verreault A., Murzina N.V., Laue E.D., Nietlispach D., Mott H.R., Okuwaki M., Lyon D., Nielsen P.R., Hirshberg M., Verreault A., Murzina N.V., Laue E.D., Mott H.R., Okuwaki M., Lyon D., Nielsen P.R., Hirshberg M., Verreault A., Murzina N.V., Laue E.D., Okuwaki M., Lyon D., Nielsen P.R., Hirshberg M., Verreault A., Murzina N.V., Laue E.D., Lyon D., Nielsen P.R., Hirshberg M., Verreault A., Murzina N.V., Laue E.D., Nielsen P.R., Hirshberg M., Verreault A., Murzina N.V., Laue E.D., Hirshberg M., Verreault A., Murzina N.V., Laue E.D., Verreault A., Murzina N.V., Laue E.D., Murzina N.V., Laue E.D., Laue E.D. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 2004;23:489–499. doi: 10.1038/sj.emboj.7600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y., Zamore P.D., Zamore P.D. Perspective: Machines for RNAi. Genes & Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Vagin V.V., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P.D., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P.D., Li C., Seitz H., Gvozdev V., Zamore P.D., Seitz H., Gvozdev V., Zamore P.D., Gvozdev V., Zamore P.D., Zamore P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez M.A., Liu J., Hannon G.J., Parker R., Liu J., Hannon G.J., Parker R., Hannon G.J., Parker R., Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes & Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Vastenhouw N.L., Brunschwig K., Okihara K.L., Müller F., Tijsterman M., Plasterk R.H., Brunschwig K., Okihara K.L., Müller F., Tijsterman M., Plasterk R.H., Okihara K.L., Müller F., Tijsterman M., Plasterk R.H., Müller F., Tijsterman M., Plasterk R.H., Tijsterman M., Plasterk R.H., Plasterk R.H. Gene expression: Long-term gene silencing by RNAi. Nature. 2006;442:882. doi: 10.1038/442882a. [DOI] [PubMed] [Google Scholar]
- Vaucheret H. Post-transcriptional small RNA pathways in plants: Mechanisms and regulation. Genes & Dev. 2007;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- Verdel A., Moazed D., Moazed D. RNAi-directed assembly of heterochromatin in fission yeast. FEBS Lett. 2005;579:5872–5878. doi: 10.1016/j.febslet.2005.08.083. [DOI] [PubMed] [Google Scholar]
- Volpe T.A., Kidner C., Hall I.M., Teng G., Grewal S.I., Martienssen R.A., Kidner C., Hall I.M., Teng G., Grewal S.I., Martienssen R.A., Hall I.M., Teng G., Grewal S.I., Martienssen R.A., Teng G., Grewal S.I., Martienssen R.A., Grewal S.I., Martienssen R.A., Martienssen R.A. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wallrath L.L., Elgin S.C., Elgin S.C. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes & Dev. 1995;9:1263–1277. doi: 10.1101/gad.9.10.1263. [DOI] [PubMed] [Google Scholar]