Abstract
DEAD/H-box NTPases remodel the spliceosome at multiple steps during the pre-mRNA splicing cycle. The RNA-dependent NTPase Prp43 catalyzes dissociation of excised lariat-intron from the spliceosome, but it is unclear how Prp43 couples the energy of ATP hydrolysis to intron release. Here, we report that activation of Prp43’s inherently feeble helicase activity by the splicing factor Ntr1 is required for lariat-intron release. Lethal Prp43 mutants T384A and T384V, which are active for ATP hydrolysis and fail to dissociate lariat-intron from spliceosomes, are refractory to stimulation of RNA unwinding by Ntr1. An N-terminal 120-amino-acid segment of Ntr1 suffices for binding to Prp43 and for stimulating its helicase activity. We identify missense mutations in Prp43 and Ntr1 that disrupt protein–protein interaction and impair Ntr1 enhancement of Prp43 RNA unwinding. Our results demonstrate for the first time that regulating the motor activity of a DEAH-box protein by an accessory factor is critical for mRNA splicing.
Keywords: Pre-mRNA splicing, DEAH-box helicase, Prp43, Ntr1, spliceosome
Nuclear pre-mRNA splicing proceeds via two successive transesterification reactions (Burge et al. 1999). In the first step, the 2′-OH of the branch site adenosine within the intron attacks the phosphodiester at the 5′ exon–intron boundary to form a lariat-intermediate. In the second step, the 3′-OH of the 5′ exon attacks the phosphodiester at the intron–3′ exon junction to form mature mRNA and lariat-intron. Pre-mRNA splicing requires five snRNAs and >80 proteins (Will and Lührmann 2006). The small nuclear RNAs (snRNAs), packaged as snRNPs, assemble together with non-snRNP proteins onto the precursor RNA to form the spliceosome. The snRNPs are involved in identifying the splice junctions and positioning the reactive nucleotides for catalysis (Valadkhan 2005). ATP-dependent remodeling of RNA–RNA and RNA–protein interactions is essential for faithful spliceosome assembly, for the catalytic activation of the splice sites, and for spliceosome disassembly (Staley and Guthrie 1998).
The remodeling events in the spliceosome are catalyzed by DExD/H NTPases that act at discrete stages of the splicing pathway (Staley and Guthrie 1998). Splicing factors Prp2, Prp16, Prp22, and Prp43 are members of the DEAH family of RNA-dependent NTPases (Burgess et al. 1990; Chen and Lin 1990; Company et al. 1991; Arenas and Abelson 1997). They are composed of a central NTPase/helicase domain, characteristic of all DExH proteins, plus an ∼300-amino-acid C-terminal domain unique to the Prp2/Prp16/Prp22/Prp43 clade (Tanner and Linder 2001; Silverman et al. 2003). The N-terminal segments are distinctive in each DEAH-box splicing factor. Understanding how the DEAH proteins carry out their specific functions during the splicing cycle requires knowledge about how they are recruited to the splicing complex, what their biochemical activities are, and whether they are regulated.
Prp43 catalyzes the final step of splicing, the dissociation of lariat-intron and simultaneous disassembly of the spliceosome (Arenas and Abelson 1997; Martin et al. 2002; Tsai et al. 2005). Once released, lariat-intron RNA is cleaved by Dbr1, a branch-specific phosphodiesterase, and the linearized intron is then degraded (Ruskin and Green 1985; Martin et al. 2002). Concomitant with lariat-intron release, the residual spliceosome containing the U2/U6 and U5 snRNPs is separated into individual components (Tsai et al. 2005; Small et al. 2006). Defects in spliceosome disassembly steps are expected to influence subsequent rounds of spliceosome assembly and catalysis, insofar as splicing factors will be sequestered in product complexes and fail to recycle.
The ability to hydrolyze ATP is essential for Prp43’s biological function (Martin et al. 2002; Tanaka and Schwer 2006). Lethal ATPase-defective Prp43 mutants associate in vitro with splicing complexes that contain excised lariat-intron, but they fail to catalyze intron dissociation (Martin et al. 2002). How Prp43 couples the energy of NTP hydrolysis to spliceosome disassembly is unclear. In the case of Prp22, which acts prior to Prp43, it was shown that helicase activity is critical for Prp22-catalyzed release of the mature mRNA from the spliceosome (Schwer and Meszaros 2000). Prp43 exhibits a weak ATP-dependent RNA unwinding activity in vitro. However, this activity does not suffice for its biological function, insofar as Prp43 mutants (T384A and T384V) that retained ATP-dependent unwinding activity were nonetheless lethal and unable to dissociate lariat-intron RNA from spliceosomes in vitro (Tanaka and Schwer 2006). These results suggested that Thr-384 might be critical for interaction of Prp43 with other splicing factors.
At least two essential proteins besides Prp43 are involved in spliceosome disassembly: Ntr1 and Ntr2 (Tsai et al. 2005). Depletion of Ntr1/Ntr2 blocks release of lariat-intron from the spliceosome in vitro. Functional complementation of extracts depleted of Ntr1/Ntr2, or of Prp43, was achieved with an affinity-purified Ntr complex from yeast, hinting that Prp43 was part of the Ntr complex (Tsai et al. 2005). The association of Prp43 with Ntr1/Ntr2 was confirmed by Western blot analysis (Tsai et al. 2005). A physical interaction between Ntr1/Ntr2 and Prp43 was also inferred from two-hybrid and affinity-tagging studies (Gavin et al. 2002; Hazbun et al. 2003; Tsai et al. 2005; Krogan et al. 2006), and genetic data implicate Ntr1 and Prp43 in a turnover pathway for defective spliceosomes (Pandit et al. 2006). These findings suggest that Ntr1 and Prp43 collaborate in the spliceosome.
Here, we address several outstanding questions regarding the physical and functional connection between Prp43 and Ntr1. We report that Prp43 interacts directly with Ntr1 in vitro and that binding to Ntr1 stimulates the helicase activity of Prp43. The Prp43 mutants T384A and T384V that were previously shown to be defective for coupling ATP hydrolysis to intron release are refractory to activation by Ntr1, suggesting that stimulation of Prp43’s weak helicase activity is vital. We identify an N-terminal 120-amino-acid segment of Ntr1 that suffices for Prp43 binding and helicase enhancement and map amino acids in Ntr1 and Prp43 that are components of the Prp43–Ntr1 interface. Although individual changes do not necessarily abolish the proteins’ biological functions, losing more than one constituent of the Prp43–Ntr1 interface is deleterious for cell growth and for lariat-intron release in vitro. We show that the interaction with Ntr1 is important for Prp43’s association with the spliceosome. Together, these results suggest a model whereby Ntr1 aids in the recruitment of Prp43 and then activates Prp43’s helicase activity to disrupt RNA–RNA or RNA–protein contacts that hold the spliceosome together.
Results
Prp43 and Ntr1 proteins form a complex in vitro
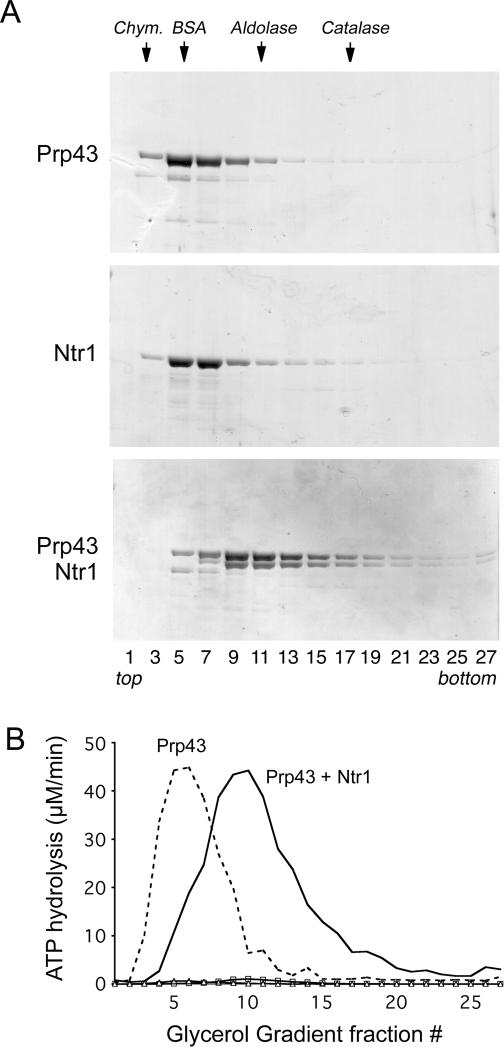
Genome-wide affinity purification and two-hybrid analyses demonstrated that Prp43, Ntr1, and Ntr2 interact in vivo. To determine whether this reflects direct binding, we assessed complex formation in vitro. The purification of recombinant Prp43 protein, as His10-Prp43 and in native tag-free form, was carried out as described (Martin et al. 2002; Tanaka and Schwer 2006). Our attempts to purify the 708-amino-acid Ntr1 protein, fused to an N-terminal His10 or a His10-Smt3 tag, were not successful, insofar as the bacterially expressed Ntr1 polypeptide was mostly insoluble and the small amount present in the soluble fraction was aggregated (data not shown). We found that coexpression of Ntr1 with the 322-amino-acid Ntr2 polypeptide increased the solubility of Ntr1 and prevented aggregation. Thus, His10-Smt3-Ntr1 was produced in bacteria together with untagged Ntr2, and the proteins were isolated from a soluble lysate by nickel-agarose chromatography. The N-terminal His10-Smt3 tag was removed by cleavage with the protease Ulp1, and tag-free native Ntr1 was purified by gel filtration. The predominant species in the preparation was the 83-kDa Ntr1 protein, which sedimented as a single discrete peak in a glycerol gradient (Fig. 1A). Although Ntr2 was not detected in Coomassie blue-stained SDS gels, Western blot analysis of the gradient fractions using Ntr2-specific antibodies revealed that small amounts of the 37-kDa Ntr2 polypeptide had copurified and then cosedimented with Ntr1 (see Supplementary Fig. 1). We estimated by Western blotting that the molar ratio of Ntr1:Ntr2 in the peak fractions was ≥10:1 (data not shown).
Figure 1.
Prp43–Ntr1 interaction. (A) Glycerol gradient sedimentation of Prp43 and Ntr1 proteins. Native Ntr1 (83 kDa) and His10-Prp43 (90 kDa) proteins (200 μg) were sedimented in 15%–30% glycerol gradients, and aliquots (10 μL) of the odd-numbered gradient fractions were analyzed by 8% SDS-PAGE. The Coomassie blue-stained gels are shown. The positions of Prp43 and Ntr1 are indicated at the left. The peak positions of marker proteins chymotrypsinogen (Chym.) (25 kDa), BSA (67 kDa), aldolase (156 kDa), and catalase (232 kDa), sedimented in a parallel gradient, are indicated at the top. (B) ATPase activity. ATP hydrolysis was measured in the absence and presence of poly(A) RNA. ATPase activities (micromolar per minute) are plotted for all fractions from gradients in which Prp43 (dashed line, with RNA cofactor; Δ, without RNA) and Prp43 + Ntr1 (solid line, with RNA; ◽, activity without RNA) were analyzed.
Prp43 by itself sedimented as a discrete monomeric peak (fractions 5–7) coincident with the ATPase activity profile (Fig. 1A,B). When Prp43 was mixed with Ntr1 and the mixture was analyzed by glycerol gradient sedimentation, the proteins cosedimented as a complex in fractions 9–11 (Fig. 1A). The complex sedimented similarly to the marker protein aldolase (156 kDa) that was analyzed in a parallel gradient, indicating that Prp43 and Ntr1 form a heterodimer. A small fraction of the complex also contained Ntr2 that copurified with the Ntr1 protein (see Supplementary Fig. 1). The peak of RNA-dependent ATPase activity coincided with the abundance of the protein complex (Fig. 1B). Complex formation did not influence the ATPase activity of Prp43 insofar as the rates of RNA-dependent ATP hydrolysis by gradient-purified Prp43 and Ntr1–Prp43 complex were virtually identical (Fig. 1B; data not shown). Moreover, Ntr1 did not affect the RNA cofactor requirement and the ATPase activities in the absence of poly(A) were similarly low (≤1 μM/min) in the peak fractions of the Prp43 and Prp43 + Ntr1 gradients (Fig. 1B).
Ntr1 stimulates Prp43’s helicase activity
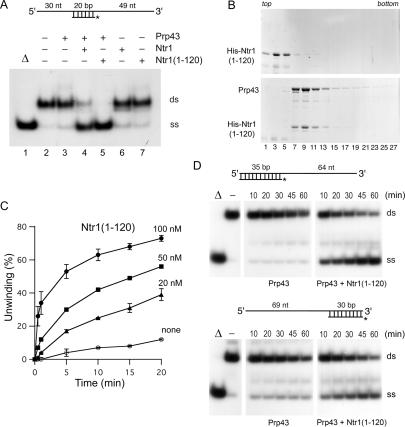
Prp43 disrupts short nucleic acid duplexes with single-stranded RNA (ssRNA) overhangs in an NTP-dependent fashion, albeit only at enzyme concentrations in large excess over the helicase substrate (Tanaka and Schwer 2006). To determine whether Ntr1 influenced the inherently weak helicase activity of Prp43, we measured ATP-dependent disruption of a 20-base-pair (bp) RNA/DNA duplex containing 5′ and 3′ RNA tails (Fig. 2A). When the helicase substrate (2.5 nM) was incubated with Prp43 (100 nM) for 30 min at 37°C, the labeled single-stranded 20-mer, which migrates ahead of the double-stranded substrate in an SDS-containing polyacrylamide gel, constituted 16% of the total labeled nucleic acid (Fig. 2A, lane 3). However, when Ntr1 (100 nM) was added to the reaction mixture containing Prp43, 72% of the helicase substrate was unwound (Fig. 2A, lane 4), indicating that Ntr1 activates the Prp43 helicase. In reactions containing Ntr1 alone, 17% of the labeled nucleic acid migrated as the single-stranded species (Fig. 2A, lane 6), compared with 10% in the reaction without added protein (Fig. 2A, lane 2). In contrast to unwinding by Prp43 and Prp43 + Ntr1, the low level of strand displacement by Ntr1 alone was independent of ATP and incubation time (data not shown), indicating that Ntr1 by itself does not have helicase activity.
Figure 2.
Ntr1 stimulates Prp43 helicase. (A) Unwinding of a 5′/3′-tailed 20-bp RNA/DNA duplex. A schematic drawing of the substrate is shown at the top. The 99-nt top strand is RNA and the 32P-labeled 20-nt bottom strand is DNA. The positions of the duplex substrate (ds) and of the single-strand product of unwinding (ss) are indicated at the right of the panel. The 5′/3′-tailed substrate was incubated with the proteins as specified for 30 min at 37°C. The fraction of the single-stranded 20-mer DNA in the absence of protein (lane 2) constituted 10% of the total 32P-labeled nucleic acid in the reaction (background); Δ (lane 1) marks a reaction that was heated for 3 min at 95°C. (B) Glycerol gradient sedimentation. His10-Smt3-Ntr1(1–120) alone (top panel) and together with Prp43 (bottom panel) was sedimented in 15%–30% glycerol gradients. Aliquots (10 μL) of the odd-numbered gradient fractions were analyzed by 8% SDS-PAGE and the polypeptides were visualized by staining the gels with Coomassie blue. The positions of Prp43 and His10-Smt3-Ntr1(1–120) proteins are indicated at the left. (C) Reaction mixtures containing the 20-bp 5′/3′-tailed substrate (2.5 nM) and 100 nM Prp43 were incubated in the absence (○) or in the presence of 20 nM (▴), 50 nM (◼), and 100 nM (●) of His10-Smt3-Ntr1(1–120) protein. Aliquots were withdrawn after the indicated times and analyzed by native PAGE. The amount of displaced labeled 20-mer was determined as the percentage of total nucleic acid at each time point and adjusted for the background of labeled 20-mer in a reaction carried out without added protein. The values are averages of duplicate measurements; the error bars represent the deviation from the mean. (D) Unwinding of a 3′-tailed 35-bp RNA/DNA duplex (top panel) and a 5′-tailed 30-bp RNA/DNA duplex (bottom panel) as a function of time. Reaction mixtures contained the substrate (2.5 nM) depicted above the panel, 100 nM of the indicated proteins, and 1 mM ATP-Mg2+. Aliquots were withdrawn at the indicated times and analyzed by PAGE. Autoradiographs of the dried gels are shown.
The N-terminal 120-amino-acid segment of Ntr1 suffices to enhance Prp43 helicase activity
The 122-amino-acid N-terminal segment of Ntr1, which contains a glycine-rich G-patch motif, sufficed for interaction with Prp43 in a two-hybrid assay in vivo (Tsai et al. 2005). Here, we produced His10-Smt3-Ntr1(1–120) in bacteria and purified it from a soluble lysate by nickel affinity chromatography. Note that His10-Smt3-Ntr1(1–120) was produced in the absence of Ntr2. The 34-kDa His10-Smt3-Ntr1(1–120) protein sedimented as a discrete peak in a glycerol gradient (Fig. 2B). When mixed with equimolar amounts of Prp43, the sedimentation profile of His10-Smt3-Ntr1(1–120) shifted to a heavier form and peaked with the Prp43 protein in fractions 7–11 (Fig. 2B). Thus, the N-terminal domain of Ntr1 sufficed to interact with Prp43 in vitro.
When we measured ATP-dependent unwinding of a 20-bp 5′/3′-tailed duplex by 100 nM Prp43 in the presence of the N-terminal domain of Ntr1 (100 nM), we found that the single-stranded 20-mer comprised 92% of the total labeled nucleic acid (Fig. 2A, lane 5). Incubation of Ntr1(1–120) alone resulted in a slight increase of single-stranded 20-mer (14% in Fig. 2A, lane 7) compared with protein-free background (10% in Fig. 2A, lane 2), which was independent of ATP (data not shown). We considered that binding of the Ntr1 and Ntr1(1–120) proteins to the substrate might destabilize the 20-bp duplex; however, we have been unable to detect RNA binding by Ntr1 or Ntr1(1–120) using gel-shift and filter-binding assays. Helicase stimulation was dependent on the concentration of Ntr1(1–120) (Fig. 2C). At 100 nM Prp43, increasing concentrations of Ntr1(1–120) from 20–100 nM increased the rate and extent of unwinding. Based on the initial rates of unwinding, Ntr1(1–120) stimulated Prp43 helicase ∼30-fold.
Prp43 by itself does not display strict directionality, but disrupts short duplexes with 5′ or 3′ overhangs (Tanaka and Schwer 2006). To assess whether the Ntr1-stimulated Prp43 helicase exhibited a preference for substrates with 5′ or 3′ overhangs, we measured ATP-dependent unwinding of a 3′-tailed 35-bp and a 5′-tailed 30-bp RNA/DNA duplex (Fig. 2D). Whereas Prp43 alone failed to displace the labeled 35-mer in the 3′-tailed duplex, addition of Ntr1(1–120) enabled Prp43 to unwind nearly all of this substrate in the course of a 60-min reaction (Fig. 2D, top panel). Ntr1(1–120) also enhanced Prp43 unwinding of the 30-bp 5′-tailed duplex (Fig. 2D, bottom panel). Similar stimulation of Prp43 unwinding of 5′- and 3′-tailed substrates was observed with full-length Ntr1 (data not shown).
The lethal Prp43-T384A and Prp43-T384V mutants are unresponsive to stimulation by Ntr1
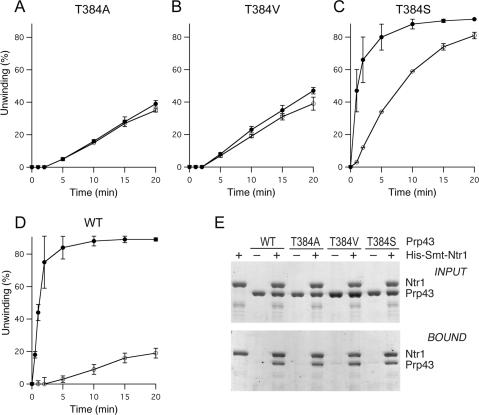
Lethal valine and alanine mutations at residue Thr-384 in motif V of the ATPase/helicase domain did not diminish RNA binding or basal ATP-dependent RNA unwinding activities of Prp43 (Tanaka and Schwer 2006). Here we tested the effect of Ntr1 on ATP-dependent RNA unwinding by the T384A and T384V proteins (Fig. 3). The 5′/3′-tailed 20-bp RNA/DNA duplex (2.5 nM) was incubated with 500 nM T384A or T384V proteins in the absence or presence of 500 nM Ntr1(1–120) (Fig. 3A,B). In parallel, we analyzed the viable T384S mutant and wild-type Prp43 (Fig. 3C,D). Whereas addition of Ntr1(1–120) did not influence the rate and extent of unwinding by T384A and T384V, it stimulated the helicase activity of the T384S protein. From the initial rates, we determined that Ntr1 enhanced unwinding by T384S protein approximately fourfold. The rate enhancement for wild-type Prp43 at 500 nM was ∼30-fold. Although the initial rate of T384S helicase activity in the absence of Ntr1 is higher than that of wild-type Prp43, the T384S mutation does not alleviate the requirement for Ntr1 in vivo or in vitro (data not shown).
Figure 3.
(A–D) Influence of Ntr1 on unwinding by Prp43 mutants. The 5′/3′-tailed 20-bp nucleic acid duplex (2.5 nM), 1 mM ATP-Mg2+, and Prp43 proteins (500 nM) were incubated without Ntr1 (○), or with 500 nM His10-Smt3-Ntr1(1–120) (●). The Prp43 proteins were T384A (A), T384V (B), T384S (C), and wild-type Prp43 (D). Aliquots were withdrawn after the indicated times and analyzed by native PAGE. The amounts of the displaced 20-mer as percentage of total labeled nucleic acid were determined and the values were adjusted for background. Helicase activities (percentage unwinding) are plotted as a function of time. The values are averages from duplicate experiments. (E) Interaction of Prp43 mutant proteins with Ntr1. His10-Smt3-Ntr1 and various Prp43 proteins (10 μg each) were mixed as indicated above the panel prior to adsorption to Ni-agarose. Aliquots of the INPUT mixtures (10%) and the BOUND (20%) material, eluted with 500 mM imidazole, were analyzed by 8% SDS-PAGE. The polypeptides were visualized by staining the gels with Coomassie blue.
We used affinity-chromatography to query whether the T384A and T384V mutations affected Prp43 binding to Ntr1 (Fig. 3E). Wild-type and mutant Prp43 proteins were mixed with His10-Smt3-Ntr1 and the mixtures were then adsorbed to Ni-agarose resin. The resin was collected by centrifugation, the pellets were washed, and the bound proteins eluted and analyzed by SDS-PAGE. The His10-Smt3-Ntr1 protein bound to the resin by virtue of its His10-tag, while the untagged Prp43 proteins did not bind to Ni-agarose in the absence of His10-Smt3-Ntr1. However, when mixed with His10-Smt3-Ntr1, wild-type Prp43 and the T384A, T384V, and T384S mutant polypeptides were all recovered in the bound fractions. These results indicate that the failure of the T384A and T384V helicases to respond to Ntr1 was not due to loss of binding.
The mutational effects on helicase activation correlated well with the mutational effects on Prp43’s function in spliceosome disassembly, insofar as the T384A and T384V mutants, which were unresponsive to Ntr1, were lethal in vivo and did not catalyze release of excised-lariat RNA from the spliceosome in vitro (Tanaka and Schwer 2006). This was in contrast to Prp43-T384S, which was activated by Ntr1, and was functional in vivo and in vitro. Taken together, these findings suggest that activation by Ntr1 is critical for Prp43 function.
Missense mutations in the Ntr1 G-patch diminish binding to Prp43 and helicase activation
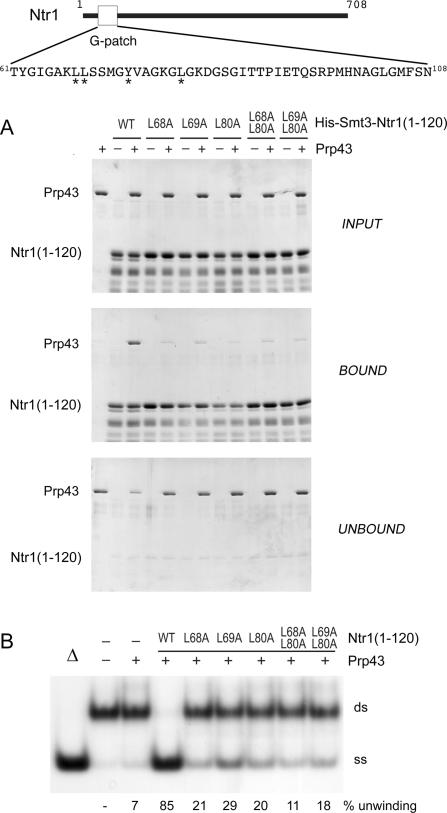
The N-terminal segment of Ntr1 encompassing the G-patch motif sufficed for interaction with Prp43 (Fig. 2B). To assess whether residues that are conserved in the glycine-rich segment of G-patch proteins are important for Ntr1’s interaction with Prp43, we replaced Leu-68, Leu-69, and Leu-80 individually or in pairs by alanine in His10-Smt3-Ntr1(1–120). The mutant polypeptides were partially purified from bacterial lysates and analyzed by affinity-chromatography for binding to native Prp43 (Fig. 4A). When Prp43 was mixed with wild-type His10-Smt3-Ntr1(1–120) prior to adsorption to Ni-agarose, Prp43 was recovered in the bead-bound fraction together with tagged Ntr1(1–120). In contrast, when mixed with the L68A, L69A, L80A, L68A–L80A, and L69A–L80A His10-Smt3-Ntr1(1–120) mutant polypeptides, the bulk of Prp43 was found in the unbound supernatants (Fig. 4A).
Figure 4.
Binding of Ntr1(1–120) to Prp43 is necessary for stimulation. Ntr1 is drawn as a black line, with the rectangle indicating the G-patch motif. The amino acid sequence of the G-patch motif is shown in the expanded view. The stars below the sequence highlight positions that were replaced by alanine. (A) Purified Prp43 (10 μg) and partially purified His10-Smt3-Ntr1(1–120) (30 μg) proteins were mixed as indicated. Aliquots of the input mixtures (INPUT), the proteins eluted from the Ni-NTA agarose (BOUND), and the supernatants after binding (UNBOUND) were analyzed by 8% SDS-PAGE. Polypeptides were visualized by staining with Coomassie blue. The positions of Prp43 and His10-Smt3-Ntr1(1–120) proteins are indicated at the left. (B) Effect of His10-Smt3-Ntr1(1–120) mutant proteins on RNA helicase activity. Reaction mixtures containing 2.5 nM 5′/3′-tailed 20-bp RNA/DNA duplex and 1 mM ATP-Mg2+ were supplemented with proteins (100 nM) as indicated (+), incubated for 30 min at 37°C, and analyzed by native PAGE. (−) indicates that buffer instead of protein was added. The position of duplex substrate (ds) and displaced labeled 20-mer (ss) are indicated at the right. In Δ, the reaction mixture was heated for 3 min at 95°C.
The Ntr1(1–120) mutant proteins that were defective for Prp43 binding were also impaired in their ability to stimulate ATP-dependent Prp43 helicase activity (Fig. 4B). In the presence of wild-type Ntr1(1–120), Prp43 unwound 85% (adjusted for background) of a 20-bp duplex substrate. The L68A, L69A, L80A, L68A–L80A, or L69A–L80A mutant Ntr1(1–120) proteins enabled Prp43 to unwind 21%, 29%, 20%, 11%, or 18% of the substrate, respectively.
Mutational effects on Ntr1 function in vivo
G-patch mutations were introduced in the context of full-length NTR1 and the ability of mutant alleles to complement an ntr1Δ strain was assessed using the plasmid shuffle method. Ntr1 mutants L68A, L69A, Y74A, and L80A grew as well as wild-type cells at 30°C, and they elicited only a mild growth defect at 37°C. The double-alanine mutant L68A–L80A failed to support growth of ntr1Δ cells, and L69A–L80A cells grew poorly and formed only pinpoint colonies after prolonged incubation (data not shown). Thus, although the mutational effects on Ntr1 function in vivo generally paralleled the effects on Prp43 helicase stimulation in vitro, insofar as L68A–L80A, which was lethal, was least effective in enhancing the Prp43 helicase activity, the effects were disproportionately more severe in vitro. Single alanine replacements at conserved residues in the Ntr1 G-patch diminished binding of the purified proteins, but they did not disrupt Ntr1 function in vivo, suggesting that other contacts contribute to Ntr1/Prp43 interaction in the context of the spliceosome.
Genetic interactions between Prp43 and Ntr1
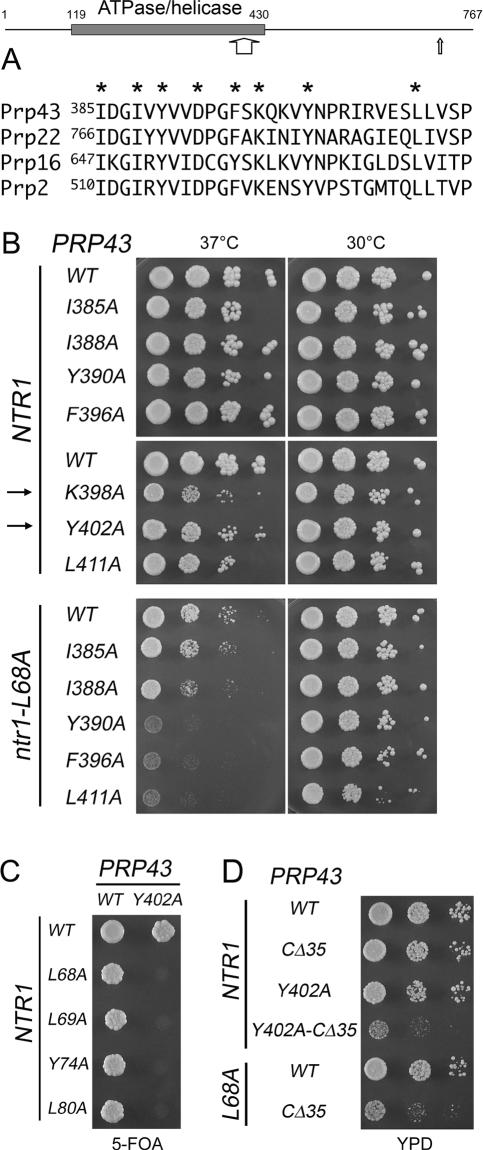
If the L68A–L80A mutant was lethal because it disrupts the Ntr1/Prp43 interaction, then specific mutations in PRP43 might elicit synthetic lethal effects in partially defective, but viable ntr1-L68A cells. Such Prp43 mutations might identify (1) elements of the Prp43–Ntr1 interface, or (2) residues that are important in the absence of Ntr1 binding. We generated a prp43Δ ntr1Δ strain, growth of which is contingent on a URA3 PRP43 NTR1 CEN plasmid. This allowed us to readily screen viable prp43 alleles in combination with ntr1 mutant alleles using the plasmid shuffle method. We primarily chose prp43 alleles that elicited no growth defects or showed only mild phenotypes at 30°C in an NTR1 background (Martin et al. 2002; Tanaka and Schwer 2006) and introduced them together with ntr1-L68A into the prp43Δ ntr1Δ strain (see Supplementary Table 1). Individual transformants were streaked to 5-FOA-containing medium at 30°C to select against cells that retained the URA3 PRP43 NTR1 CEN plasmid. Of 45 prp43 alleles that were tested, 35 showed no growth difference in ntr1-L68A compared with NTR1 cells. Five prp43 alleles slowed the growth of ntr1-L68A cells at 30°C and five prp43 alleles were lethal in conjunction with ntr1-L68A. One of the synthetic lethal mutations mapped within the ATPase/helicase domain (prp43-Y402C). Three other synthetic lethal alleles had Ala-cluster mutations in the C-terminal domain of Prp43 (L439A–Y440A, I648A–R649A–K650A, and W717A–L718A–I719A) and one was a C-terminal truncation of 45 amino acids.
Prp43-Y402C is located between motifs V and VI in a segment that is conserved among DEAH-box splicing factors (Fig. 5A). To determine whether specific amino acid side chains within this region are important for Prp43 function and to ascertain whether the observed synthetic lethal effect is specific for the Tyr-402-Cys change, we introduced alanine in lieu of Tyr-402 and vicinal amino acids Ile-385, Ile-388, Tyr-390, Asp-393, Phe-396, Lys-398, and Leu-411, and then assessed the mutational effects in NTR1 and ntr1-L68A cells (Fig. 5B). Four classes of effects were seen: (1) prp43-D393A was lethal even in an NTR1 background (data not shown). (2) prp43 mutants I385A and I388A grew similar to wild-type PRP43 in ntr1-L68A cells at all temperatures (Fig. 5B; data not shown). (3) Y390A, F396A, and L411A supported growth of NTR1 and ntr1-L68A cells; however, they significantly exacerbated the growth phenotype that was seen in PRP43 ntr1-L68A and PRP43 ntr1-L80A cells at 37°C (Fig. 5B; Supplementary Fig. 2A). (4) prp43 mutants K398A and Y402A, which showed wild-type growth at 30°C in an NTR1 strain (Fig. 5B), were lethal in combination with multiple ntr1 mutant alleles, including L68A (Fig. 5C; data not shown). Thus, the peptide encompassing amino acids 390–411 in Prp43 functionally interacts with Ntr1.
Figure 5.
Genetic interactions between Prp43 and Ntr1. Prp43 is drawn as a horizontal line with the gray rectangle highlighting the ATPase/helicase domain. The numbers refer to the first and last amino acids of Prp43 or of the ATPase/helicase domain. The wide and narrow arrows indicate the positions of the peptide shown in A, and of the C terminus in Prp43(1–722), respectively. (A) Alignment of conserved segments between motifs V and VI in DEAH splicing factors. The stars indicate positions that were replaced by alanine in Prp43. (B) Growth phenotypes of strains containing various PRP43 and NTR1 mutants. prp43Δ ntr1Δ cells harboring the indicated PRP43 and NTR1 alleles were grown in liquid cultures. Cultures were adjusted to A600 of 10−1 and 10-fold serial dilutions (3 μL) were spotted to YPD agar plates. Plates were photographed after 2.5 d at 30°C and 37°C. The arrows at the left indicate prp43 alleles that are lethal in ntr1-L68A. (C) prp43Δ ntr1Δ cells harboring an NTR1 PRP43 URA3 plasmid were transformed with the indicated NTR1 plus PRP43 alleles. His+Trp+ transformants were selected and grown in liquid culture in synthetic medium lacking tryptophan and histidine. Cultures were adjusted to A600 of 0.2 and spotted (5 μL) to 5-FOA-containing agar plates. Photographs of the plates after 4 d of incubation at 30°C are shown. (D) Tenfold serial dilutions of prp43Δ ntr1Δ cells harboring the indicated PRP43 and NTR1 alleles were spotted to YPD medium. Photographs were taken after 2 d of incubation at 30°C.
The finding that changes in either a small segment of the ATPase/helicase domain or the C-terminal domain of Prp43 elicited synthetic lethality with ntr1 mutants suggested that those regions might play redundant roles. To test for functional redundancy between Y402A and the C-terminal 45-amino-acid segment, we introduced the Y402A mutation into the truncated PRP43 gene. We found that prp43-Y402A-CΔ45 failed to support growth of prp43Δ NTR1 cells (data not shown), thereby mimicking the defect elicited by either prp43-Y402A or prp43-CΔ45 in ntr1-L68A cells (Fig. 5C; Supplementary Fig. 2B). Also, the mutational effects of prp43-Y402A-CΔ35, a version in which the C-terminal 35 amino acids were deleted, phenocopied the severe growth defect elicited by prp43-CΔ35 in ntr1-L68A cells (Fig. 5D). These results indicate redundancy between Tyr-402 and the nonessential C-terminal segment of Prp43 for functional interaction with Ntr1.
Tyr-402 in Prp43 is a component of the Ntr1–Prp43 interface
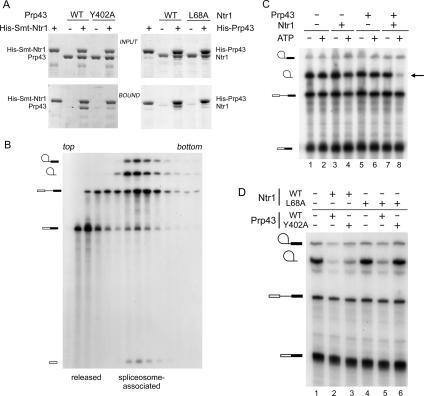
We purified recombinant Prp43-Y402A and measured the protein’s biochemical activities. The ATPase and RNA unwinding activities were similar to wild-type Prp43 (data not shown). In contrast to wild-type Prp43, however, Prp43-Y402A showed reduced binding to Ntr1 in a pull-down assay, and Ntr1 did not stimulate the helicase activity of Prp43-Y402A (Fig. 6A, left panel; data not shown). We propose that a conserved peptide between motifs V and VI in Prp43 contains components of the Ntr1–Prp43-binding interface. The Ntr1 side of the interface involves the G-patch (Fig. 4), and the L68A mutation impaired binding to Prp43 also in the context of full-length Ntr1 (Fig. 6A, right panel).
Figure 6.
(A, left panel) Interaction of Prp43 and Ntr1. His10-Smt3-Ntr1 and wild-type Prp43 or the Y402A proteins (10 μg each) were mixed as indicated above the panel prior to adsorption to Ni-agarose. Aliquots of the INPUT mixtures (10%) and the BOUND (20%) material, eluted with 500 mM imidazole were analyzed by 8% SDS-PAGE. The polypeptides were visualized by staining the gels with Coomassie blue. The right panel shows a similar experiment using His10-Prp43 and tag-free Ntr1 proteins as indicated. (B) Lariat-intron RNA is retained in the Δntr1Δprp43 spliceosomes. Splicing was carried out in Δntr1Δprp43 extract for 20 min at 28°C. The reaction mixture (100 μL) was analyzed by sedimentation in a 15%–40% glycerol gradient. Fractions were collected from the top of the gradient and aliquots of odd-numbered fractions (3–29) were analyzed by denaturing PAGE. An autoradiogram of the gel is shown. (C) Reconstitution of intron release activity. 32P-labeled actin pre-mRNA was incubated in Δntr1Δprp43 extract in the presence of ATP (2 mM) for 20 min at 28°C. Then ATP was depleted from the reaction mixture by endogenous hexokinase upon addition of glucose (2.5 mM) during a 10-min incubation. Aliquots were then supplemented with proteins (±ATP) as indicated at the top of the panel. The reactions were halted after 10 min and analyzed by denaturing PAGE. An autoradiogram of the dried gel is shown. The symbols at the left indicate the positions of the following RNA species (from top to bottom): lariat-intermediate, lariat-intron, pre-mRNA, and mRNA. (D) Lariat-intron release activity of Ntr1 and Prp43 mutant proteins. 32P-labeled actin pre-mRNA was reacted in Δntr1Δprp43m extract (prepared from cell that express Prp43-Y402A-TAP and Ntr1TAP) in the presence of ATP (2 mM) for 20 min at 28°C. Aliquots of the mixture were then supplemented with proteins as indicated at the top of the panel. The reactions were halted after 10 min and analyzed by denaturing PAGE. An autoradiogram of the dried gel is shown.
Reconstitution of lariat-intron release activity
Prior studies showed that depletion of Ntr1/Ntr2 or Prp43 from yeast extracts resulted in accumulation of excised lariat-intron RNA (Tsai et al. 2005). The release defect was alleviated when depleted extracts were supplemented with Ntr complex affinity-purified from yeast extracts. Here, we sought to reconstitute activity with purified recombinant components. Splicing extract was prepared from a yeast strain in which both Ntr1 and Prp43 were expressed as fusion proteins carrying C-terminal TAP tags (Rigaut et al. 1999). This allowed for simultaneous removal of Ntr1-TAP and Prp43-TAP by binding to IgG sepharose. 32P-labeled ACT1 pre-mRNA was incubated in Δntr1Δprp43 extract in the presence of ATP-Mg2+ for 20 min and the mixture was then sedimented in a 15%–40% glycerol gradient to determine whether the splicing products were released or retained in spliceosomes. Aliquots of odd-numbered gradient fractions were analyzed by denaturing PAGE (Fig. 6B). The bulk of the spliced mRNA sedimented close to the top of the gradient signifying that it was released. In contrast, excised lariat-intron RNA was spliceosome associated, insofar as it sedimented together with the products of the first transesterification reaction, the 5′ exon and lariat-intermediate (fractions 15–19). Thus, simultaneous depletion of Ntr1 and Prp43 blocked spliceosome disassembly after mRNA was released, similar to depletion of Ntr1/Ntr2 (Tsai et al. 2005), or inhibition by ATPase-defective Prp43 proteins (Martin et al. 2002).
To assess the requirements for restoring lariat-intron release activity to Δntr1Δprp43 extracts, ACT1 pre-mRNA was reacted for 20 min and then ATP was depleted by endogenous hexokinase upon addition of glucose. Aliquots of the mixture were supplemented with recombinant Prp43 and Ntr1 proteins and incubated further in the presence or absence of added ATP-Mg2+ (Fig. 6C). The levels of excised lariat-intron RNA served as a gauge for activity, insofar as spliceosome-associated RNA is protected from debranching and degradation, while released lariat-intron is rapidly degraded in extracts (Martin et al. 2002). Functional complementation of Δntr1Δprp43 extract, indicated by low levels of lariat-intron RNA, was achieved by supplementing Prp43 and Ntr1 proteins plus ATP (Fig. 6C, lane 8). Addition of Prp43 protein alone (+ATP or −ATP) did not lead to a decrease in the level of lariat-intron RNA (Fig. 6C, lanes 5,6), showing that the activity was dependent on Ntr1. Ntr1(1–120) failed to complement the reaction (data not shown). Addition of Ntr1 alone (+ATP) did not suffice for efficient intron release (Fig. 6C, lane 4), although the lariat-intron levels (normalized to mRNA) were reproducibly 20%–30% lower than those in reactions without added protein (Fig. 6C, lane 4; data not shown), indicating that Prp43 might not have been completely depleted. Using Western blot analysis with α-Prp43 antibodies, we found that the Δntr1Δprp43 extract contained residual Prp43-TAP protein (10%–15% compared with the amount seen in undepleted extract, with a detection limit of ∼5%) (data not shown). In summary, our results show that efficient lariat-intron release in Δntr1Δprp43 extracts depends on Prp43 and Ntr1 proteins and on ATP. The ATP requirement was satisfied by all common NTPs and dNTPs, but not the nonhydrolyzable ATP analog AMP-PCP (data not shown). ATPase-deficient Prp43 proteins, or the T384A and T384V mutant proteins, were ineffectual for lariat-intron release (data not shown). Although purified recombinant Ntr2 protein could not substitute for Ntr1 (data not shown), we cannot rule out the possibility that Ntr2 was required in addition to Ntr1. If Ntr2 was codepleted by association with Ntr1, then the small amounts of Ntr2 that are present in our preparation of recombinant full-length Ntr1, might suffice for functional complementation.
Synergy between Prp43-Y402A and Ntr1-L68A in vitro
If the observed genetic interactions between Prp43 and Ntr1 were important for the proteins’ functions during spliceosome disassembly, then Prp43-Y402A and Ntr1-L68A proteins should not promote lariat-intron release in Δntr1Δprp43 extract. Because depletion of Prp43 was incomplete, we prepared extract from a yeast strain expressing Ntr1-TAP and Prp43-Y402A-TAP. Thus, any residual Prp43 protein in the Δntr1Δprp43m extract was the Y402A mutant. Labeled ACT1 pre-mRNA was incubated in Δntr1Δprp43m extract in the presence of ATP for 20 min. Aliquots of the mixture were then supplemented with various combinations of proteins and further incubated for 10 min (Fig. 6D). Addition of wild-type Ntr1 and Prp43 proteins resulted in low levels of lariat-intron RNA, indicating that the intron was efficiently released (Fig. 6D, lane 2). Lariat-intron was also released, albeit less efficiently, when the splicing mixtures were supplemented with wild type Ntr1 and Prp43-Y402A (Fig. 6D, lane 3), or with Ntr1-L68A and wild type Prp43 (Fig. 6D, lane 5). In contrast, Ntr1-L68A failed to trigger lariat-intron release alone or together with Prp43-Y402A (Fig. 6D, lanes 4,6). We conclude that the Prp43–Ntr1 interaction is important for the final step of splicing and that in the context of the spliceosome, losing more than one constituents of the Prp43–Ntr1 interface is necessary to disrupt the proteins’ functional interaction.
Interaction with Ntr1 is important for Prp43’s association with the spliceosome
The allele-specific synthetic lethal effects described above point to the functional requirement for (1) a small segment (amino acids 390–411) within the ATPase/helicase domain of Prp43, (2) residues within an ∼300-amino-acid C-terminal region conserved between Prp43 and the other DEAH-box splicing factors, and (3) the final 45-amino-acid segment that is unique to Prp43, when interaction with Ntr1 is impaired. Whereas residues in the ATPase/helicase domain directly contribute to Ntr1 binding (Fig. 6A), truncating Prp43 by 45 or 55 amino acids did not abolish binding with Ntr1(1–120) in vitro (see Supplementary Fig. 3), suggesting that Prp43’s C-terminal region might be involved in other contacts within the spliceosome or contribute otherwise to Prp43 activity.
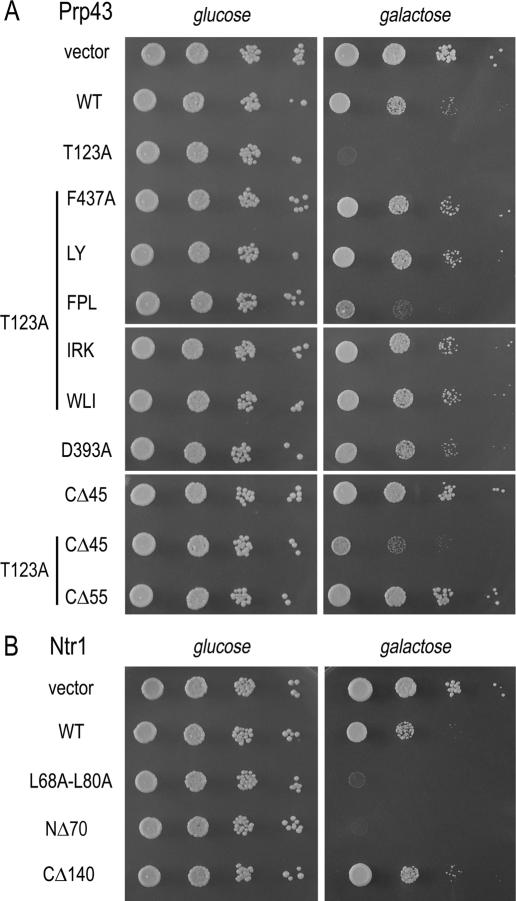
We sought to assess whether Prp43’s spliceosome binding was affected by mutations that depended on wild-type Ntr1 function. Prior studies showed that catalytically inactive Prp43 mutants, including the ATPase-defective T123A, retain the ability to bind to spliceosomes and thus lead to dominant-negative growth inhibition when overexpressed in wild-type cells (Martin et al. 2002; Tanaka and Schwer 2006). We introduced prp43 mutations into GAL1-prp43-T123A and transformed the plasmids into wild-type cells (Fig. 7A). All strains grew equally well on glucose-containing medium. Galactose-induced expression of T123A prevented growth of wild-type cells (Fig. 7A). The F437A, L439A–Y440A, I648A–R649A–K650A, or W717A–L718A–I719A mutations, relieved the dominant-negative effect of T123A. The triplealanine mutations F520A–P521A–L522A and T544A–I545A–V546A, which showed no genetic interaction with NTR1 (see Supplementary Table 1), failed to alleviate T123A’s dominant-negative effects (Fig. 7A; data not shown). The galactose-induced Prp43 protein levels, gauged by Western blot analyses, were similar in strains containing wild-type or the various Prp43 mutants (data not shown), arguing against the formal possibility that loss of growth inhibition was due to nonspecific effects, such as unfolding and degradation of the T123A protein in the presence of C-terminal mutations. Rather, we suggest that the residues in the C-terminal domain of Prp43 that, when replaced by alanine, alleviate the dominant-negative effect of T123A and are lethal in ntr1-L68A cells, play a role in Prp43’s spliceosome binding.
Figure 7.
Dominant-negative effects in vivo. Wild-type strains harboring an empty vector or plasmids (TRP1 CEN) with the indicated prp43 alleles (A), and ntr1 mutants (B) under the transcriptional control of the GAL1 promoter were grown in liquid raffinose-containing medium. The cultures were adjusted to A600 of 0.1 and 10-fold serial dilutions were spotted to synthetic agar medium lacking tryptophan and containing glucose or galactose. The plates were photographed after incubation for 2.5 d (glucose) or 4 d (galactose) at 30°C.
In contrast to nonfunctional prp43 alleles that disrupt the ATPase/helicase functions of Prp43, overexpression of the lethal D393A allele did not prevent cell growth, indicating that D393A did not effectively compete with wild-type Prp43 (Fig. 7). Because Asp-393 is located at the putative Prp43/Ntr1 interface (Fig. 5), this finding suggests that direct binding to Ntr1 is important for Prp43’s association with the spliceosome.
Mutations of the 717WLI919 tripeptide are situated within a 10-amino-acid segment (713VRPEWLIEIA722) that defines the C-terminal margin of essentiality (Martin et al. 2002). Overexpression of the lethal prp43(1–712) allele did not prevent growth of wild-type cells (data not shown) and truncating the T123A mutant by 55 amino acids (to terminate at amino acid 712) relieved its dominant-negative effect (Fig. 7A). Control experiments showed that deleting the C-terminal 55-amino-acid segment did not impact on expression levels or nuclear localization of Prp43(1–712) (data not shown). Although the C-terminal 55-amino-acid segment of Prp43 appears to contribute to spliceosome binding, it is unlikely to do so through direct interaction with Ntr1, because the Prp43(1–712) protein remained competent to bind Ntr1 (Supplementary Fig. 3). Taken together, these findings suggest that Prp43 associates with the spliceosome via direct binding to Ntr1 and also via other contacts that are mediated by residues in the C-terminal domain, including the 717WLI919 tripeptide.
The C-terminal 45-amino-acid segment unique to Prp43 is essential only when interaction with Ntr1 is impaired. Dominant-negative growth inhibition by T123A was not alleviated when the final 45 amino acids were removed (Fig. 7A), suggesting that the peptide is not involved in spliceosome binding, but rather affects Prp43 activity. It is noteworthy that the slight growth defect observed upon overexpression of wild-type Prp43 was ameliorated by truncating the C-terminal 45 amino acids in Prp43 (Fig. 7A).
Dominant-negative effects of Ntr1 mutants
We used similar approaches to assess whether Ntr1 might depend on its interaction with Prp43 for spliceosome binding. Wild-type NTR1 and the lethal alleles L68A–L80A, ntr1(71–708), and ntr1(1–568) were placed under the transcriptional control of the GAL1 promoter and introduced into wild-type cells (Fig. 7B). Overexpression of wild-type Ntr1 and a nonfunctional C-terminal truncation mutant elicited a slight growth defect (Fig. 7B, cf. vector and wild type). However, galactose-induced overexpression of the L68A–L80A mutant, which failed to interact with Prp43 in vitro, and of NΔ70, a mutant that lacks part of its N-terminal G-patch motif, completely blocked growth of wild-type cells. The finding that the Ntr1 G-patch mutants block the function of wild-type Ntr1 is consistent with the idea that Ntr1 can bind to the spliceosome irrespective of Prp43.
Discussion
Prp43 binds to ssRNA with high affinity and, propelled by NTP hydrolysis, the enzyme can unwind short nucleic acid duplexes in vitro (Tanaka and Schwer 2006). Here we report that Prp43’s feeble helicase activity is enhanced by binding to Ntr1. This study demonstrates that the motor activity of a DEAH-box splicing factor is regulated by an accessory protein and, more importantly, it shows that the stimulation is crucial for mRNA splicing. We propose that Ntr1 aids in the recruitment of Prp43 to the splicing complex and that it then activates the Prp43 helicase to disrupt RNA–RNA or RNA–protein contacts in the spliceosome to dissociate lariat-intron RNA.
With the exception of vaccinia virus NPH-II, a processive DExH-box helicase (Shuman 1992; Jankowsky et al. 2000), the RNA unwinding activities of other characterized DExD/H box enzymes are either weak or undetectable in vitro, prompting speculations about the requirement for specific cofactors or substrates (Silverman et al. 2003). Accessory factors that enhance RNA helicase activity have been described for the translation initiation factor eIF4A, the exon-junction complex component eIF4AIII, and hepatitis C virus NS3 (Howe et al. 1999; Rogers et al. 2001; Noble and Song 2007). For example, the DEAD-box helicase eIF4A is stimulated by eIF4B and by the scaffold protein eIF4G within the eIF4F complex. Enhancement of RNA unwinding is, at least in part, accomplished by increasing the RNA binding and ATPase activities of eIF4A. The eIF4B protein directly provides an RNA-binding function, and structural data suggest that eIF4G functions to hold the two helicase domains of the eIF4A protein in a conformation that is favorable for RNA and ATP binding (Rogers et al. 2001; Oberer et al. 2005). The RNA unwinding activity of NS3 is stimulated by NS4A, which enhances the loading of NS3 onto RNA (Howe et al. 1999; Pang et al. 2002).
Here we find that binding to Ntr1 enables Prp43 to unwind a relatively stable 3′-tailed 35-bp duplex, raising the possibility that Ntr1 affects processivity of the Prp43 helicase. Ntr1 does not alter Prp43’s requirement for a ssRNA overhang to afford duplex unwinding, and stimulation is not at the level of ATP hydrolysis, insofar as Ntr1, which has no catalytic activity on its own, does not influence Prp43’s ATPase activity in the presence or absence of an RNA cofactor. Although Prp43 binds to the helicase substrate with nanomolar affinity in the absence of ATP, we have not detected a higher-order complex in native gels when Ntr1 or Ntr1(1–120) proteins were included in the binding reactions. Further studies will be necessary to determine how Ntr1 stimulates Prp43’s helicase activity, but at first glance, the mechanism is likely to be distinct from the mechanisms employed by other RNA helicases and their partners. The stimulatory effect of Ntr1 appears to be specific to Prp43, insofar as Ntr1 does not enhance the helicase activity of Prp22, a DEAH-box helicase that functions during the mRNA release step (our unpublished data). There is no evidence to date that Prp22 helicase depends on an accessory factor, and mutational studies show that there is an excellent correlation between Prp22’s ability to unwind nucleic acid duplexes and to elicit release of mRNA from the spliceosome (Schwer and Meszaros 2000; Campodonico and Schwer 2002; Schneider et al. 2004).
Two lines of evidence support the conclusion that the functional interaction with Ntr1 is critical for Prp43’s role during lariat-intron release. First, mutations in Prp43 that uncouple ATP hydrolysis from Ntr1-stimulated helicase activity, cause lethality and ablate lariat-intron release in vitro. Such mutations do not affect binding of Prp43 to Ntr1 or to the spliceosome. Second, mutations in Prp43 and Ntr1 that impair protein–protein interaction, and hence stimulated helicase activity, are deleterious to intron release and cell growth. Within the Ntr1 protein, constituents of the Ntr1/Prp43 interface include amino acids that are conserved in G-patch proteins (Fig. 4). Glycine-rich motifs are present in many proteins implicated in RNA processing events, including splicing (Aravind and Koonin 1999; Jurica and Moore 2003; Behzadnia et al. 2007).
The G-patch splicing factor Spp2 interacts with Prp2, a DEAH-box NTPase required for the first transesterification reaction of pre-mRNA splicing (Roy et al. 1995). Within the G-patch motif of Spp2, Leu-109 (corresponding to Leu-68 in Ntr1) was shown to be important for interaction with Prp2 in a two-hybrid assay (Silverman et al. 2004). On the Prp2 side, a short 11-amino-acid peptide (845DCLVIPKEEWL855) within the C-terminal domain is critical for interaction with Spp2. Because the same 11-amino-acid segment is also important for Prp2’s ability to bind to the spliceosome (Edwalds-Gilbert et al. 2004) it was proposed that Prp2 associates with the spliceosome by binding to Spp2. Thus, the interaction with Spp2 likely contributes to the functional specificity of Prp2 during the first transesterification step (Silverman et al. 2004). It is not reported whether binding to Spp2 activates the enzymatic activities of Prp2, an NTPase, for which helicase activity has not been detected (Kim et al. 1992).
Analogous to Prp2/Spp2, direct binding to Ntr1 might be important for the recruitment of Prp43 to the spliceosome. Consistent with this idea, Boon et al. (2006) reported that an ATPase-deficient Prp43 mutant protein fails to bind to spliceosomes produced in extract that was substantially depleted of Ntr1. We show here that Prp43’s association with the spliceosome relies on direct interaction with Ntr1 and also on other contacts (Fig. 7A). Allele-specific synthetic lethal effects reveal that residues in the C-terminal domain of Prp43 are necessary for spliceosome binding and Prp43 activity when Ntr1/Prp43 binding is compromised by mutations in the putative binding interface. For example, the 717WLI719 tripeptide within the 713VRPEWLIEIA722 segment contributes to spliceosome binding, whereas the C-terminal 45-amino-acid segment unique to Prp43 does not play a role in spliceosome binding, but otherwise affects Prp43 activity.
In vitro depletion/reconstitution assays show that Prp43 and Ntr1 are needed to release excised lariat-intron RNA from the spliceosome in an NTP-dependent fashion (Fig. 6A; Tsai et al. 2005), arguing against a functional requirement prior to this step. Consistent with their principal roles occurring after both catalytic steps of splicing are complete and mRNA is released, yeast Prp43 and Ntr1 proteins are, by immunoprecipitation and affinity-purification analyses, primarily found associated with complexes that contain excised lariat-intron RNAs and the U2/U6 and U5 snRNAs (James et al. 2002; Martin et al. 2002; Tsai et al. 2005; Boon et al. 2006; Pandit et al. 2006; Small et al. 2006). Affinity-purification studies revealed that Prp43 is also a component of preribosomal complexes and conditional inactivation of prp43 mutants demonstrated a role for Prp43 in pre-rRNA processing, and hence, ribosome biogenesis (Lebaron et al. 2005; Combs et al. 2006; Leeds et al. 2006). If Prp43 functions similarly during pre-rRNA processing as it does during splicing, then its role might depend on stimulation by Gno1, a G-patch protein that, like Prp43, is a component of multiple preribosomal complexes (Lebaron et al. 2005).
Although Ntr1 is mainly associated with splicing complexes that contain excised lariat-intron RNAs, Ntr1 has been detected in spliceosomes that were stalled prior to the first or second transesterification step in vitro, possibly indicating an earlier role (Tsai et al. 2005; Pandit et al. 2006). In addition to releasing lariat-intron RNA as part of the normal splicing cycle, Ntr1 and Prp43 have been implicated in a turnover pathway for defective spliceosomes (Pandit et al. 2006). This proposal comes from the finding that several ntr1 alleles (also called spp382) and prp43 mutants with reduced activity suppress the temperature-sensitive growth phenotype of prp38-1, a mutant that impairs spliceosome activation by slowing the release of U1 and U4 snRNA (Xie et al. 1998). It is thought that reducing the activity of the disassembly factors Ntr1/Prp43 allows more time for compromised splicing complexes to proceed and thereby alleviate the growth defect in prp38-1 cells. Our finding that residues in the G-patch of Ntr1 are critical for direct interaction with Prp43 provides a plausible explanation for the observation that ntr1 alleles (spp383-2 and spp382-3) with G-patch mutations fail to suppress temperature sensitivity of prp38-1 cells (Pandit et al. 2006) and argues that Ntr1 and Prp43 cooperate similarly in both disassembly processes.
We propose that Ntr1, which can associate with spliceosomes independent of Prp43 (Fig. 7B; Tsai et al. 2005; Pandit et al. 2006), aids in the recruitment of Prp43 and then stimulates the helicase activity of Prp43 to effect spliceosome disassembly. DExD/H box NTPases can remodel RNP assemblies by disrupting RNA helices and displacing proteins from RNA (Jankowsky and Bowers 2006). The biochemical characteristics of Prp43 are consistent with a scenario in which the enzyme binds to ssRNA and translocates, energized by NTP hydrolysis, to disrupt RNA/RNA or RNA/protein contacts in the spliceosome. The challenge now is to identify the molecular target for Ntr1-stimulated helicase activity.
Materials and methods
Targeted mutagenesis and analysis of Prp43 and Ntr1 mutants in vivo
Mutations were introduced into the PRP43 and NTR1 genes using the two-stage PCR overlap extension method. The mutated PRP43 and NTR1 DNA fragments were inserted into p358-Prp43 (TRP1 CEN) (Martin et al. 2002) and p413-Ntr1 (HIS3 CEN) (see the Supplemental Material for a description of plasmids and strains), respectively, in lieu of the wild-type gene. The plasmids carrying the PRP43 mutant alleles were transformed into the prp43Δ strain that carries the PRP43 on a CEN URA3 plasmid. Trp+ transformants were selected and streaked to 5-FOA medium to select cells that had lost the PRP43 URA3 plasmid. The plasmids carrying the NTR1 mutant alleles were transformed into the ntr1Δ strain containing NTR1 on a CEN URA3 plasmid. His+ transformants were selected and streaked to 5-FOA medium to select cells that had lost the NTR1 URA3 plasmid. The ability of the mutant alleles to support growth on 5-FOA was assessed at 18°C, 30°C, and 37°C.
Expression and purification of Ntr1 proteins
A DNA fragment spanning the Ntr1 ORF was inserted into pET28-His10Smt3 (a gift from Dr. Christopher Lima, Sloan-Kettering Institute, New York). For coexpression of His10-Smt3-Ntr1 and untagged Ntr2, plasmid pET16-Ntr2/His10-Smt3-Ntr1 (see the Supplemental Material) was transformed into Escherichia coli BL21-Codon Plus (DE3) (Novagen). Cultures were inoculated from single colonies and maintained in logarithmic growth at 37°C in LB medium containing ampicillin (100 μg/mL). [For expression of Ntr1-L68A, pET28-His10Smt3-Ntr1-L68A was transformed together with tag-free pET16-Ntr2 into BL21-Codon Plus (DE3) cells. Cultures were grown in LB medium containing ampicillin (100 μg/mL) and kanamycin (50 μg/mL).] When the A600 of a 1-L culture reached 0.6–0.8, the culture was chilled for 30 min on ice, IPTG was added to a final concentration of 0.4 mM, and the culture were incubated for 16 h at 17°C with constant shaking. Cells were harvested by centrifugation and stored at −20°C.
The procedures for the purification of Ntr1 were carried out at 4°C and are described in detail in the Supplemental Material.
Purification of Ntr1(1–120) proteins
DNA fragments spanning the coding sequence for Ntr1 (1–120) and the mutants L68A, L69A, L80A, L68A–L80A, and L69A–L80A were inserted into pET28-His10Smt3 for expression in E. coli BL21-Codon Plus (DE3) cells. His10-Smt3-Ntr1(1–120) and the mutant variants were purified from soluble lysates by Ni-NTA chromatography as described in the Supplemental Material. Ntr1(1–120)-containing eluates were dialysis against 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 10% glycerol. The final yields of His10-Smt3-Ntr1(1–120) proteins were between 10 and 16 mg from 200-mL cultures.
Unwinding assays
The 3′- and 5′-tailed RNA–DNA substrates were prepared as described (Tanaka and Schwer 2005). The 99-nucleotide (nt) RNA strand (5′-GGGCGAAUUGGGCCCUCUAGAUGCAUG CUCGAGCGGCCGCCAGUGUGAUGGAUAUCUGCAGAA UUCGCCCUUAAACCAUAAUCAUACCAGUUUUGGCAG-3′) was synthesized by in vitro transcription, and it was annealed at a 1:3 molar ratio of RNA:DNA to the 5′ 32P-labeled DNA oligonucleotides 5′-CATCACACTGGCGGCCGCTC-3′ to generate a 3′/5′-tailed 20-bp duplex, 5′-CGCTCGAGCATG CATCTAGAGGGCCCAATTCGCCC-3′ to yield a 3′-tailed 35-bp duplex, and 5′-CTGCCAAAACTGGTATGATTATGGT TTAAG-3′ to yield a 5′-tailed 30-bp duplex. The relative stability of the 35-bp RNA/DNA duplex, calculated using nearest neighbor parameters (Sugimoto et al. 1995), was −57.8 kcal/mol. The relative stability of the 30-bp duplex with 5′ overhang was −34.6 kcal/mol, similar to that of the generic 20-bp 5′/3′-tailed duplex (−33.6 kcal/mol). The annealed substrates were gel-purified by native 6% PAGE. Reaction mixtures (10 μL) containing 40 mM Tris-HCl (pH 7.0), 2 mM DTT, 1 mM ATP-Mg2+, 2.5 nM RNA/DNA substrate, and indicated amount of Prp43 and Ntr1 or Ntr1(1–120) were incubated at 37°C. Reactions were halted by transfer to ice and addition of 5-μL of loading buffer (100 mM Tris-HCl at pH 7.4, 5 mM EDTA, 0.5% SDS, 50% glycerol, 0.1% [w/v] bromophenol blue, xylene cyanol dyes). Samples were analyzed by electrophoresis through an 8% polyacrylamide gel in 40 mM Tris-borate, 0.5 mM EDTA, and 0.1% SDS. The 32P-labeled substrate and products were visualized by autoradiography of the dried gel and quantified using a PhosphorImager. The relative value of 32P-label in the single-standed species was determined as a percentage of total nucleic acid in each lane. The loss of the overall signal over extended periods of incubation at 37°C, due to an unidentified activity present in the protein preparation despite several purification steps, similarly affected the single-stranded and double-stranded species.
Glycerol gradient sedimentation
Protein preparations were applied to 5.2 mL of 15%–30% glycerol gradients containing 150 mM NaCl, 50 mM Tris/HCl (pH 7.4), 2 mM DTT, 1 mM EDTA, and 0.1% Triton X-100. The gradients were centrifuged at 47,000 rpm for 20 h at 4°C in a Sorvall AH650 rotor. Aliquots (195 μL) were collected from the tops of the gradients and analyzed by SDS-PAGE.
In vitro splicing assays
Yeast whole-cell extracts were prepared by grinding in liquid nitrogen, and splicing reactions using 32P-labeled actin pre-mRNA were carried out as described (Ansari and Schwer 1995). Depleted extracts Δntr1Δprp43 and Δntr1Δprp43m were prepared from BJ-NTR1-TAP/PRP43-TAP and HW-prp43-Y402A-TAP/NTR1-TAP whole-cell extracts, respectively, as follows: Four-hundred microliters of extract were applied to an IgG Sepharose column (200-μL packed volume) that had been equilibrated with PBS (150 mM NaCl, 10 mM NaPO4 at pH 7.2) containing 10% glycerol. The tagged proteins bound to IgG through the Protein A peptide in the TAP tag, the flow-through fraction (by gravity) constituted depleted extracts used for in vitro splicing assays.
Affinity chromatography to measure Prp43–Ntr1 binding in vitro
Recombinant proteins were mixed in binding buffer (150 mM NaCl, 50 mM imidazole, 50 mM Tris-HCl at pH 7.4, 10% glycerol) in a final volume of 100 μL. Aliquots (10 μL) were withdrawn to serve as inputs. Ni-NTA agarose (100 μL of a 50% slurry) was equilibrated in binding buffer and added to the proteins, and the mixtures were gently mixed for 1 h at 4°C. The resin was collected by centrifugation and the supernatant (unbound) was removed. The resins were washed three times with 100 μL of binding buffer. Bound proteins were eluted with 100 μL of binding buffer containing 500 mM imidazole. The samples were analyzed by 8% SDS-PAGE followed by staining with Coomassie brilliant blue.
ATPase assay
Reaction mixtures (100 μL) contained 40 mM Tris-HCl (pH 8.0), 2 mM DTT, 1 mM ATP-Mg2+, and 10 μL of each glycerol gradient fraction. To measure RNA-dependent ATPase activities, 0.1 mM poly(A) (given as the concentration of AMP) was included and the reaction mixtures were incubated for 2–5 min at 22°C. The reaction mixtures without RNA were incubated for 40 min. Reactions were quenched by adding 1 mL of Biomol Green reagent (Biomol Research Laboratories), and the absorbance (A620) was measured after 20 min at 22°C. The amounts of free phosphate were calculated by interpolation of the A620 values to a phosphate standard curve.
Acknowledgments
This work was supported by the National Institutes of Health grant GM50288.
Footnotes
Supplemental material is available at http:/www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1580507
References
- Ansari A., Schwer B., Schwer B. SLU7 and a novel activity, SSF1, act during the PRP16-dependent step of yeast pre-mRNA splicing. EMBO J. 1995;14:4001–4009. doi: 10.1002/j.1460-2075.1995.tb00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L., Koonin E.V., Koonin E.V. G-patch: A new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Sci. 1999;24:342–344. doi: 10.1016/s0968-0004(99)01437-1. [DOI] [PubMed] [Google Scholar]
- Arenas J.E., Abelson J.N., Abelson J.N. Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl. Acad. Sci. 1997;94:11798–11802. doi: 10.1073/pnas.94.22.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadnia N., Golas M.M., Hartmuth K., Sander B., Kastner B., Deckert J., Dube P., Will C.L., Urlaub H., Stark H., Golas M.M., Hartmuth K., Sander B., Kastner B., Deckert J., Dube P., Will C.L., Urlaub H., Stark H., Hartmuth K., Sander B., Kastner B., Deckert J., Dube P., Will C.L., Urlaub H., Stark H., Sander B., Kastner B., Deckert J., Dube P., Will C.L., Urlaub H., Stark H., Kastner B., Deckert J., Dube P., Will C.L., Urlaub H., Stark H., Deckert J., Dube P., Will C.L., Urlaub H., Stark H., Dube P., Will C.L., Urlaub H., Stark H., Will C.L., Urlaub H., Stark H., Urlaub H., Stark H., Stark H., et al. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 2007;26:1737–1748. doi: 10.1038/sj.emboj.7601631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon K.L., Auchynnikava T., Edwalds-Gilbert G., Barrass J.D., Droop A.P., Dez C., Beggs J.D., Auchynnikava T., Edwalds-Gilbert G., Barrass J.D., Droop A.P., Dez C., Beggs J.D., Edwalds-Gilbert G., Barrass J.D., Droop A.P., Dez C., Beggs J.D., Barrass J.D., Droop A.P., Dez C., Beggs J.D., Droop A.P., Dez C., Beggs J.D., Dez C., Beggs J.D., Beggs J.D. Yeast ntr1/spp382 mediates prp43 function in postspliceosomes. Mol. Cell. Biol. 2006;26:6016–6023. doi: 10.1128/MCB.02347-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C.B., Tuschl T.H., Sharp P.A., Tuschl T.H., Sharp P.A., Sharp P.A. Splicing of precursors to mRNAs by the spliceosomes. In: Gesteland R.F., et al., editors. RNA world II. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1999. pp. 525–560. [Google Scholar]
- Burgess S., Couto J.R., Guthrie C., Couto J.R., Guthrie C., Guthrie C. A putative ATP binding protein influences the fidelity of branchpoint recognition in yeast splicing. Cell. 1990;60:705–717. doi: 10.1016/0092-8674(90)90086-t. [DOI] [PubMed] [Google Scholar]
- Campodonico E., Schwer B., Schwer B. ATP-dependent remodeling of the spliceosome: Intragenic suppressors of release-defective mutants of Saccharomyces cerevisiae Prp22. Genetics. 2002;160:407–415. doi: 10.1093/genetics/160.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.H., Lin R.J., Lin R.J. The yeast PRP2 protein, a putative RNA-dependent ATPase, shares extensive sequence homology with two other pre-mRNA splicing factors. Nucleic Acids Res. 1990;18:6447. doi: 10.1093/nar/18.21.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs D.J., Nagel R.J., Ares M., Stevens S.W., Nagel R.J., Ares M., Stevens S.W., Ares M., Stevens S.W., Stevens S.W. Prp43p is a DEAH-box spliceosome disassembly factor essential for ribosome biogenesis. Mol. Cell. Biol. 2006;26:523–534. doi: 10.1128/MCB.26.2.523-534.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Company M., Arenas J., Abelson J., Arenas J., Abelson J., Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- Edwalds-Gilbert G., Kim D.H., Silverman E., Lin R.J., Kim D.H., Silverman E., Lin R.J., Silverman E., Lin R.J., Lin R.J. Definition of a spliceosome interaction domain in yeast Prp2 ATPase. RNA. 2004;10:210–220. doi: 10.1261/rna.5151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A.C., Bosche M., Krause R., Grandi P., Marziuch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Bosche M., Krause R., Grandi P., Marziuch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Krause R., Grandi P., Marziuch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Grandi P., Marziuch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Marziuch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Rick J.M., Michon A.M., Cruciat C.M., Michon A.M., Cruciat C.M., Cruciat C.M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Hazbun T.R., Malmstrom L., Anderson S., Graczyk G.J., Fox B., Riffle M., Sardin B.A., Aranda J.D., McDonald W.H., Chiu C.H., Malmstrom L., Anderson S., Graczyk G.J., Fox B., Riffle M., Sardin B.A., Aranda J.D., McDonald W.H., Chiu C.H., Anderson S., Graczyk G.J., Fox B., Riffle M., Sardin B.A., Aranda J.D., McDonald W.H., Chiu C.H., Graczyk G.J., Fox B., Riffle M., Sardin B.A., Aranda J.D., McDonald W.H., Chiu C.H., Fox B., Riffle M., Sardin B.A., Aranda J.D., McDonald W.H., Chiu C.H., Riffle M., Sardin B.A., Aranda J.D., McDonald W.H., Chiu C.H., Sardin B.A., Aranda J.D., McDonald W.H., Chiu C.H., Aranda J.D., McDonald W.H., Chiu C.H., McDonald W.H., Chiu C.H., Chiu C.H., et al. Assigning function to yeast proteins by integration of technologies. Mol. Cell. 2003;12:1353–1365. doi: 10.1016/s1097-2765(03)00476-3. [DOI] [PubMed] [Google Scholar]
- Howe A.Y., Chase R., Taremi S.S., Risano C., Beyer B., Malcolm B., Lau J.Y., Chase R., Taremi S.S., Risano C., Beyer B., Malcolm B., Lau J.Y., Taremi S.S., Risano C., Beyer B., Malcolm B., Lau J.Y., Risano C., Beyer B., Malcolm B., Lau J.Y., Beyer B., Malcolm B., Lau J.Y., Malcolm B., Lau J.Y., Lau J.Y. A novel recombinant single-chain hepatitis C virus NS3–NS4A protein with improved helicase activity. Protein Sci. 1999;8:1332–1341. doi: 10.1110/ps.8.6.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James S.A., Turner W., Schwer B., Turner W., Schwer B., Schwer B. How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA. 2002;8:1068–1077. doi: 10.1017/s1355838202022033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E., Bowers H., Bowers H. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res. 2006;34:4181–4188. doi: 10.1093/nar/gkl410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E., Gross C.H., Shuman S., Pyle A.M., Gross C.H., Shuman S., Pyle A.M., Shuman S., Pyle A.M., Pyle A.M. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- Jurica M.S., Moore M.J., Moore M.J. Pre-mRNA splicing: Awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Smith J., Claude A., Lin R.J., Smith J., Claude A., Lin R.J., Claude A., Lin R.J., Lin R.J. The purified yeast pre-mRNA splicing factor PRP2 is an RNA-dependent NTPase. EMBO J. 1992;11:2319–2326. doi: 10.1002/j.1460-2075.1992.tb05291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.J., Cagney G., Yu H., Zhong G., Guo X., Iqnatchenko A., Li J., Pu S., Datta N., Tikuisis A.B., Cagney G., Yu H., Zhong G., Guo X., Iqnatchenko A., Li J., Pu S., Datta N., Tikuisis A.B., Yu H., Zhong G., Guo X., Iqnatchenko A., Li J., Pu S., Datta N., Tikuisis A.B., Zhong G., Guo X., Iqnatchenko A., Li J., Pu S., Datta N., Tikuisis A.B., Guo X., Iqnatchenko A., Li J., Pu S., Datta N., Tikuisis A.B., Iqnatchenko A., Li J., Pu S., Datta N., Tikuisis A.B., Li J., Pu S., Datta N., Tikuisis A.B., Pu S., Datta N., Tikuisis A.B., Datta N., Tikuisis A.B., Tikuisis A.B., et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Lebaron S., Froment C., Fromont-Racine M., Rain J.C., Monsarrat B., Caizergues-Ferrer M., Henry Y., Froment C., Fromont-Racine M., Rain J.C., Monsarrat B., Caizergues-Ferrer M., Henry Y., Fromont-Racine M., Rain J.C., Monsarrat B., Caizergues-Ferrer M., Henry Y., Rain J.C., Monsarrat B., Caizergues-Ferrer M., Henry Y., Monsarrat B., Caizergues-Ferrer M., Henry Y., Caizergues-Ferrer M., Henry Y., Henry Y. The splicing ATPase prp43p is a component of multiple preribosomal particles. Mol. Cell Biol. 2005;25:9269–9282. doi: 10.1128/MCB.25.21.9269-9282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds N.B., Small E.C., Hiley S.L., Hughes T.R., Staley J.P., Small E.C., Hiley S.L., Hughes T.R., Staley J.P., Hiley S.L., Hughes T.R., Staley J.P., Hughes T.R., Staley J.P., Staley J.P. The splicing factor Prp43p, a DEAH box ATPase, functions in ribosome biogenesis. Mol. Cell Biol. 2006;26:513–522. doi: 10.1128/MCB.26.2.513-522.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A., Schneider S., Schwer B., Schneider S., Schwer B., Schwer B. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 2002;277:17743–17750. doi: 10.1074/jbc.M200762200. [DOI] [PubMed] [Google Scholar]
- Noble C.G., Song H., Song H. MLN51 stimulates the RNA-helicase activity of eIF4AIII. PLoS ONE. 2007;3:e303. doi: 10.1371/journal.pone.0000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberer M., Marintchev A., Wagner G., Marintchev A., Wagner G., Wagner G. Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes & Dev. 2005;19:2212–2223. doi: 10.1101/gad.1335305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit S., Lynn B., Rymond B.C., Lynn B., Rymond B.C., Rymond B.C. Inhibition of a spliceosome turnover pathway suppresses splicing defects. Proc. Natl. Acad. Sci. 2006;103:13700–13705. doi: 10.1073/pnas.0603188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang P.S., Jankowsky E., Planet P.J., Pyle A.M., Jankowsky E., Planet P.J., Pyle A.M., Planet P.J., Pyle A.M., Pyle A.M. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J. 2002;21:1168–1176. doi: 10.1093/emboj/21.5.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B., Rutz B., Wilm M., Mann M., Séraphin B., Wilm M., Mann M., Séraphin B., Mann M., Séraphin B., Séraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Rogers G.W., Richter N.J., Lima W.F., Merrick W.C., Richter N.J., Lima W.F., Merrick W.C., Lima W.F., Merrick W.C., Merrick W.C. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 2001;276:30914–30922. doi: 10.1074/jbc.M100157200. [DOI] [PubMed] [Google Scholar]
- Roy J., Kim K., Maddock J.R., Anthony J.G., Woolford J.L., Kim K., Maddock J.R., Anthony J.G., Woolford J.L., Maddock J.R., Anthony J.G., Woolford J.L., Anthony J.G., Woolford J.L., Woolford J.L. The final stages of spliceosome maturation require Spp2p that can interact with the DEAH box protein Prp2p and promote step 1 of splicing. RNA. 1995;1:375–390. [PMC free article] [PubMed] [Google Scholar]
- Ruskin B., Green M., Green M. An RNA processing activity that debranches RNA lariats. Science. 1985;229:135–140. doi: 10.1126/science.2990042. [DOI] [PubMed] [Google Scholar]
- Schneider S., Campodonico E., Schwer B., Campodonico E., Schwer B., Schwer B. Motifs IV and V in the DEAH box splicing factor Prp22 are important for RNA unwinding, and helicase-defective Prp22 mutants are suppressed by Prp8. J. Biol. Chem. 2004;279:8617–8626. doi: 10.1074/jbc.M312715200. [DOI] [PubMed] [Google Scholar]
- Schwer B., Meszaros T., Meszaros T. RNA helicase dynamics in pre-mRNA splicing. EMBO J. 2000;19:6582–6591. doi: 10.1093/emboj/19.23.6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S. Vaccinia virus RNA helicase: An essential enzyme related to the DE-H family of RNA-dependent NTPases. Proc. Natl. Acad. Sci. 1992;89:10935–10939. doi: 10.1073/pnas.89.22.10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman E., Edwalds-Gilbert G., Lin R.J., Edwalds-Gilbert G., Lin R.J., Lin R.J. DExD/H-box proteins and their partners: Helping RNA helicases unwind. Gene. 2003;312:1–16. doi: 10.1016/s0378-1119(03)00626-7. [DOI] [PubMed] [Google Scholar]
- Silverman E.J., Maeda A., Wei J., Smith P., Beggs J.D., Lin R.J., Maeda A., Wei J., Smith P., Beggs J.D., Lin R.J., Wei J., Smith P., Beggs J.D., Lin R.J., Smith P., Beggs J.D., Lin R.J., Beggs J.D., Lin R.J., Lin R.J. Interaction between a G-patch protein and a spliceosomal DEXD/H-box ATPase that is critical for splicing. Mol. Cell. Biol. 2004;24:10101–10110. doi: 10.1128/MCB.24.23.10101-10110.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small E.C., Leggett S.R., Winans A.A., Staley J.P., Leggett S.R., Winans A.A., Staley J.P., Winans A.A., Staley J.P., Staley J.P. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol. Cell. 2006;23:389–399. doi: 10.1016/j.molcel.2006.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley J.P., Guthrie C., Guthrie C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Sugimoto N., Nakano S., Katoh M., Matsumura A., Nakamuta H., Ohmichi T., Yoneyama M., Sasaki M., Nakano S., Katoh M., Matsumura A., Nakamuta H., Ohmichi T., Yoneyama M., Sasaki M., Katoh M., Matsumura A., Nakamuta H., Ohmichi T., Yoneyama M., Sasaki M., Matsumura A., Nakamuta H., Ohmichi T., Yoneyama M., Sasaki M., Nakamuta H., Ohmichi T., Yoneyama M., Sasaki M., Ohmichi T., Yoneyama M., Sasaki M., Yoneyama M., Sasaki M., Sasaki M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- Tanaka N., Schwer B., Schwer B. Characterization of the NTPase, RNA-binding, and RNA helicase activities of the DEAH-box splicing factor Prp22. Biochemistry. 2005;44:9795–9803. doi: 10.1021/bi050407m. [DOI] [PubMed] [Google Scholar]
- Tanaka N., Schwer B., Schwer B. Mutations in PRP43 that uncouple RNA-dependent NTPase activity and pre-mRNA splicing function. Biochemistry. 2006;45:6510–6521. doi: 10.1021/bi052656g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner N.K., Linder P., Linder P. DExD/H box RNA helicases: From generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- Tsai R.T., Fu R.H., Yeh F.L., Tseng C.K., Lin Y.C., Huang Y.H., Cheng S.C., Fu R.H., Yeh F.L., Tseng C.K., Lin Y.C., Huang Y.H., Cheng S.C., Yeh F.L., Tseng C.K., Lin Y.C., Huang Y.H., Cheng S.C., Tseng C.K., Lin Y.C., Huang Y.H., Cheng S.C., Lin Y.C., Huang Y.H., Cheng S.C., Huang Y.H., Cheng S.C., Cheng S.C. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes & Dev. 2005;19:2991–3003. doi: 10.1101/gad.1377405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadkhan S. snRNAs as the catalysts of pre-mRNA splicing. Curr. Opin. Chem. Biol. 2005;9:603–608. doi: 10.1016/j.cbpa.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Will C.L., Lührmann R., Lührmann R. Spliceosome structure and function. In: Gesteland R.F., Cech T.R., Atkins J.F., Cech T.R., Atkins J.F., Atkins J.F., editors. The RNA world. Cold Spring Harbor Press; Cold Spring Harbor, NY: 2006. pp. 369–400. [Google Scholar]
- Xie J., Beickman K., Otte E., Rymond B.C., Beickman K., Otte E., Rymond B.C., Otte E., Rymond B.C., Rymond B.C. Progression through the spliceosome cycle requires Prp38p function for U4/U6 snRNA dissociation. EMBO J. 1998;17:2938–2946. doi: 10.1093/emboj/17.10.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]