Abstract
The rod component of the bacterial flagellum polymerizes from the inner membrane across the periplasmic space and stops at a length of 25 nm at the outer membrane. Bushing structures, the P- and L-rings, polymerize around the distal rod and form a pore in the outer membrane. The flagellar hook structure is then added to the distal rod growing outside the cell. Hook polymerization stops after the rod–hook structure reaches ∼80 nm in length. This study describes mutants in the distal rod protein FlgG that fail to terminate rod growth. The mutant FlgG subunits continue to polymerize close to the length of the normal rod–hook structure of 80 nm. These filamentous rod structures have multiple P-rings and fail to form the L-ring pore at the outer membrane. The flagella grow within the periplasm similar to spirochete flagella. This provides a simple method to evolve intracellular flagella as in spirochetes. The mechanism that couples rod growth termination to the ring assembly and outer membrane penetration exemplifies the importance of stopping points in the construction of a complex macromolecular machine that facilitate efficient coupling to the next step in the assembly pathway.
Keywords: Flagellar assembly, spirochete evolution, membrane penetration
The construction of macromolecular complexes poses formidable regulatory hurdles. Many large cellular components assemble by protein polymerization that must achieve a precise final size and subunit composition in order to function properly. An example is the molecular motor that enables bacterial movement (Kojima and Blair 2004). These rotary motors drive the flagella, which are controlled by a signal transduction cascade and allow bacteria to move along chemical gradients (Armitage et al. 2005). The molecular machines that enable this function are assembled from multiple copies of a number of protein types, many with apparent, intrinsic self-assembly properties (Minamino and Namba 2004). Coordination of the flagellar assembly process is aided by genetic mechanisms that manage the logistics of component production (Aldridge and Hughes 2002). Other mechanisms regulate the order in which subunit types are exported into the growing structure (Minamino and Macnab 1999; Minamino et al. 1999; Makishima et al. 2001; Hirano et al. 2003).
Depending on the bacterial species, flagellar filaments are located internal or external to the cell. The spirochete flagella are located in the periplasm, and in addition to the ability to confer motility to the cell, flagella play a cytoskeletal role in maintaining cell shape (Wolgemuth et al. 2006). A fundamental difference between eubacterial flagella and spirochete flagella is the ability of eubacterial flagella to penetrate the outer membrane and elongate outside the cell.
The bacterial flagellum is composed of three main substructures: the basal body (which functions as a transmembrane rotary motor), the hook (which serves as a universal joint permitting articulation between the motor and the filament), and the filament (the propeller) (Berg and Anderson 1973; Macnab 2003). Flagellum assembly initiates with the construction of cytoplasmic membrane-anchored ring structures. The flagellar type III secretion (T3S) system is then assembled in the center of this structure to secrete the axial components. These form a rod structure that transverses the cell envelope followed by the external hook and filament structures. Protein subunits travel through the hollow center of the substructures to the tip of the elongating structure where they assemble into place (Iino 1969; Emerson et al. 1970). In Salmonella, the transition from rod completion to hook elongation is coupled to outer membrane penetration. The flagellar rod is composed of four structural subunits FlgB, FlgC, FlgF, and FlgG with another protein, FliE, thought to act as an adaptor between a planar MS-ring and the axial rod (Homma et al. 1990; Minamino et al. 2000). The FlgB, FlgC, and FlgF proteins form a proximal rod structure that is 10 nm in length. The order of assembly of these subunit types into the proximal rod is not known.
The FlgG protein comprises the distal rod structure that is 15 nm in length (Macnab 2003). Once rod growth terminates, ring structures acting as bushings in the peptidoglycan (P-ring) and lipopolysaccharride (L-ring) form around the distal rod. The L-ring forms a pore in the outer membrane that allows the flagellar structure to continue to grow outside the cell. Outside the cell, hook subunits polymerize to a final hook length of 55 nm, followed by filament polymerization. Hook growth is terminated by the action of a molecular ruler, FliK (Shibata et al. 2007). The length of the hook is directly dependent on FliK length. Insertions or deletions of FliK result in longer and shorter hook lengths, respectively.
The completion of the hook is an important checkpoint in the flagellar assembly pathway (Chilcott and Hughes 2000; Aldridge and Hughes 2002). Upon hook–basal body (HBB) completion, a switch in the specificity of the flagellar secretion system from rod–hook-type substrates to late secretion substrates occurs. In addition, the flagellar-specific transcription factor, σ28, begins transcription of filament and chemosensory genes. FlgM is an anti-σ28 factor that inhibits σ28-dependent transcription prior to HBB completion (Ohnishi et al. 1992). FlgM is also a late secretion substrate. In coordination with HBB completion and the secretion specificity switch, FlgM is secreted from the cell through the completed HBB structure, and σ28 is free to transcribe the late assembly genes now needed (Hughes et al. 1993; Kutsukake 1994). In this way genes whose products polymerize outside the cytoplasm in the final assembly stage after hook completion (in particular, the large external filament) are not transcribed until an HBB structure onto which these late subunits will be added is completed.
Here we report the discovery of a possible mechanism of outer membrane penetration by the growing flagellar structure. The penetration of the outer membrane requires that the rod component of the flagellar motor cease to polymerize at its normal length of 25 nm. We present evidence that the amount of FlgG subunits assembled into a normal rod allows for the formation a single P-ring structure around the distal rod. The formation of a single P-ring around the distal rod at 25 nm in length appears to be essential for L-ring pore formation in the outer membrane. FlgG mutants that allowed continued rod polymerization beyond 25 nm form filamentous rod structures, with multiple P-rings that are unable to assemble L-ring pores in the outer membrane. The inability to penetrate the outer membrane results in the growth of the flagellum in the periplasm akin to spirochete flagella.
Results
Isolation of filamentous rod mutants
The anti-σ28 factor FlgM is secreted through a completed HBB structure to allow σ28-dependent transcription of flagellin subunit genes only when a functional HBB is present within a cell (Karlinsey et al. 2000). In a HBB-defective strain FlgM accumulates and prevents σ28-dependent transcription (Gillen and Hughes 1991). Among mutants selected for reduced intracellular FlgM levels by selection for σ28-dependent transcription in strains deleted for the P- and L-ring structural genes (ΔflgHI) (see Materials and Methods), we obtained loss-of-function mutants in the flk locus encoding a membrane-associated protein of unknown function (Aldridge et al. 2006). Loss of Flk in the ring mutant strains resulted in the export of FlgM into the periplasm where it was degraded (Aldridge et al. 2006). This selection for reduced FlgM anti-σ28 activity in the ring mutants also yielded point mutants in the FlgG rod protein gene (see Materials and Methods), which we have characterized here. From >100 flgG mutants isolated, 18 different alleles were obtained. These resulted in the following amino acid changes in the FlgG protein: P52L, G53R, Δ(A53–Q54–S55–S56), Δ(Q59–T60–T61–L62–P63)∷H, S64P, G65E, G65R, G65V, Δ(Q59–L66), L66P, D117Y, G132R, G133V, G183R, G183W, E189K, N190K, and S197L. The Δ symbol was used to denote a deletion. The Δ[Q59–T60–T61–L62–P63]∷H mutant has a deletion of amino acids 59–63 replaced by histidine residue.
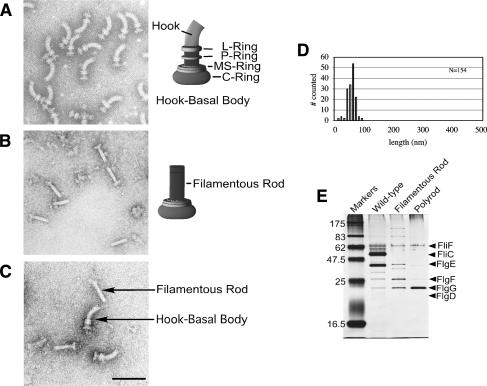
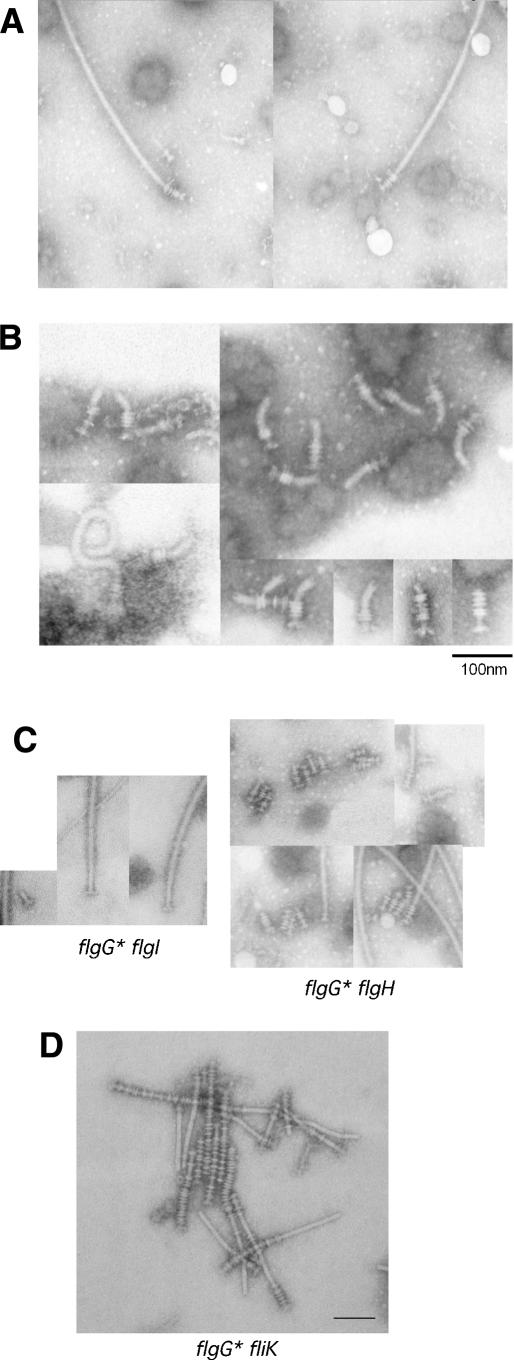
The FlgG protein has never previously been implicated in gene regulation. Because FlgG is a rod structural gene, the mutants were examined by electron microscopy to determine if the flgG regulatory mutants (flgG*) affected HBB formation. Flagellar basal structures were examined from a wild-type strain (Fig. 1A) and from a flgG* mutant strain lacking P- and L-rings (Fig. 1B,C) that was isolated from the genetic selection for reduced intracellular FlgM levels (above). The flgG* mutants resulted in extended rods, termed filamentous rods, that grow to a length approaching that of a normal rod–hook structure (Fig. 1C,D). Flagellar basal structures isolated from the flgG* mutants contained excess FlgG protein relative to the FliF MS-ring protein in the filamentous rod mutant (Fig. 1E). This indicated that FlgG polymerization was extended by the flgG* regulatory mutations.
Figure 1.
The flgG regulatory mutants result in a filamentous rod phenotype. Structures isolated from flgG* regulatory mutants. For visualization by electron microscopy, cells and isolated flagellar structures were stained with 1% PTA (pH 7 or pH 5) and observed with a JEOL 1200Ex electron microscope at 80 kV. Electron micrographs of isolated HBB structures from wild-type cells (SJW1103) (A) and from the ring-defective (ΔflgHI) flgG regulatory (flgG*) double mutants (TH5931) (B), and a mixture of HBB and filamentous rod structures from strains SJW1103 and TH5931 (C). (D) The rod length measurements of isolated flagellar–basal structures from a ring mutant strain with a FlgG*-rod (TH5931). (E) SDS-PAGE analysis of protein from flagellar structures isolated from wild-type cells (SJW1103) including filaments, filamentous rod structures from strains lacking filaments (TH5931, FlgG*-rod), and polyrod structures from strains lacking filaments (TH9709, ΔfliK FlgG*-rod). The concentration of FliF in each fraction was first determined and the amount of extract loaded was based on equal amounts of FliF.
FlgM is exported into the periplasm in flgG* mutants
The flgG* mutants were isolated in a selection for reduced FlgM activity. One possibility was that FlgM was secreted into the periplasm in flgG* mutants as was shown for the flk-null mutants (Aldridge et al. 2006). FlgM export into the periplasm was tested using the secretion of a FlgM–β-lactamase (FlgM–Bla) hybrid protein into the periplasm as a reporter. When β-lactamase, lacking its secretion signal, is fused to the C terminus of FlgM and the FlgM–Bla hybrid is exported through the flagellum into the periplasm, it confers ampicillin resistance (ApR) to the cell (Aldridge et al. 2006). Unlike native FlgM, the FlgM–Bla chimera is stable in the periplasm (Aldridge et al. 2006). Resistance to Ap was tested in strains expressing the FlgM–Bla fusion in wild-type (HBB+), ring mutant and flgG* ring double-mutant strains. In cases where FlgM–Bla was secreted completely from the cell (HBB+ strain), or not secreted from the cytoplasm at all (ring mutant with a wild-type rod structure), the cells were ApS, indicating the FlgM–Bla was not in the periplasm (Table 1). However, the presence of the flgG* mutation (filamentous rod) conferred ApR to the ring mutant cells indicating export of FlgM–Bla into the periplasm (Table 1). Therefore, it seems likely that the reduced intracellular FlgM level was a consequence of export of FlgM in ring-defective strains where it entered the periplasm and was degraded.
Table 1.
Secretion of FlgM–Bla into the periplasm of filamentous rod mutants
The secretion of the FlgM–Bla fusion into the periplasm confers ApR in cells with the flgG filamentous rod mutation (flgG*).
aAmpicillin resistance (R) or sensitivity (S) was measured as growth on MacConkey medium supplemented with sodium ampicillin (30 μg/mL). FlgM–Bla secretion assays were performed as described by Aldridge et al. (2006).
Filamentous rod length is controlled by FliK
The finding that FlgM–Bla was secreted into the periplasm in the flgG* mutants strains (Table 1) suggested that the flagellar secretion substrate specificity had switched from rod–hook-type substrates to late secretion substrates, which includes FlgM. Normally, the secretion specificity switch occurs when hook length has reached 55 ± 6 nm (Hirano et al. 1994). In addition, the length distribution of filamentous rods in Figure 1D shows a relatively sharp peak that is similar to the sharp peak in hook length distribution. This suggests that some length control mechanism is preventing further growth of the filamentous rod structures. The secretion specificity switch is controlled by the FlhB component of the flagellar T3S system (Ferris and Minamino 2006). In order to switch from hook–rod secretion to late secretion, FlhB must undergo an autocleavage event and interact with the FliK protein (Ferris et al. 2005). Null alleles of fliK result in hooks of uncontrolled lengths or polyhooks (Patterson-Delafield et al. 1973). FliK was shown to measure hook length in the flagellar system by a molecular ruler mechanism (Shibata et al. 2007). Insertions and deletions in FliK resulted in longer and shorter hooks. It has been proposed that during the process of secretion, FliK makes a pause through an interaction of its secreted N terminus with the hook-capping protein FlgD (Moriya et al. 2006). According to this model, during the pause in secretion, the C terminus of FliK is in close proximity to the FlhB component of the flagellar secretion apparatus then the secretion specificity switch would presumably occur. Normally this occurs at a hook length of 55 nm (Hirano et al. 1994). Secretion of FliK is not necessary for the secretion specificity switch to occur since overexpression of FliK deleted for its secretion signal would catalyze the switch (Hirano et al. 2005). However, under this condition of FliK overexpression, hook length control was abolished. This implies that the process of secretion and the length of FliK increase the local concentration of the FliK C terminus in the vicinity of FlhB at the optimal length of the hook structure to switch secretion specificity from hook subunits to late secretion substrates.
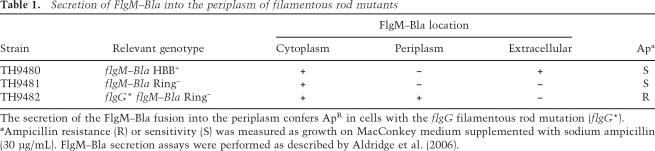
We decided to examine the effect of fliK-null alleles on rod length control in the filamentous rod mutants. If FliK was responsible for the apparent controlled length in the filamentous rods (Fig. 1D), then loss of FliK might result in rods of uncontrolled lengths. This was what we observed. Strain TH9709 [ΔfliK∷tetRA ΔflgHI flgG*5667(G65V)] was constructed by deleting the fliK gene and replacing it with a tetracycline resistance cassette in a filamentous rod mutant strain (flgG*5667) deleted for the P- and L-ring structural genes, flgH and flgI. The resulting rod structures, termed polyrods, are much longer than the original filamentous rod structures isolated from the isogenic fliK+ strain, occasionally reaching up to 1 micron in length (Fig. 2A). The polyrods exhibited a substantial increase in FlgG protein levels relative to the other basal body proteins, such as FliF, indicating the polyrods were composed primarily of FlgG subunits (Fig. 1E). About 25% of the polyrod structures have attached polyhook structures in the FlgG-G65V mutant background (Fig. 2B). Unexpectedly, the polyrods with attached hooks exhibit a peak in measured lengths. The significance of this result is not clear at this time.
Figure 2.
Electron micrographs of structures isolated from a filamentous rod-producing strain lacking the hook length control protein FliK {TH9709 [ΔfliK flgG*(G65V) ΔflgHI]}. (A) Isolated polyrod structures. (B) Isolated polyrod structures with attached polyhooks. The rod length measured includes the distal rod only; 10 nm must be added to each measurement for total rod length (proximal [10 nm] + distal).
Modeling the FlgG structure based on homology with FlgE
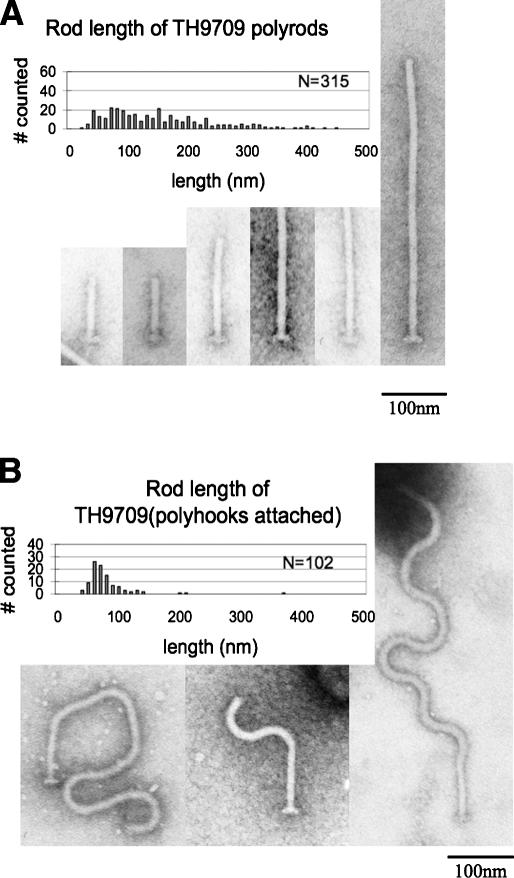
Filamentous rod structures resulted from single amino acid substitutions in FlgG. This suggested a simple mechanism leading to the cessation of FlgG-rod growth and provided an important clue to the design of the flagellar structure. The FlgG amino acid sequence was shown to have a high degree of identity with the flagellar hook protein (FlgE), which is assembled just after FlgG-rod completion (Homma et al. 1990). Predicted secondary structure analysis shown in Figure 3A suggests that these proteins have a high degree of structural identity as well. This allowed the modeling of the filamentous rod mutations on a three-dimensional FlgG structure. There are two significant differences between the FlgG-rod and FlgE-hook sequences. First, the FlgG-rod has an insertion of 18 amino acids (residues 46–65 of FlgG) not present in FlgE, where the majority of the filamentous rod mutations occurred (amino acids 52–66). Second, FlgE-hook has two insertions of 16 amino acids and a stretch of 146 amino acids in the middle of the protein that is not present in FlgG (Fig. 3A). The structure of FlgE has been determined (Samatey et al. 2004). We modeled the FlgG-rod sequence onto the FlgE-hook structure that had previously been solved (Fig. 3B; Samatey et al. 2004). Unfortunately, the first 70 amino acids of FlgE-hook were not structured, which corresponds to the first 90 amino acids in FlgG, and where a number of filamentous rod mutations were located (amino acids 52–66 of FlgG). However, two filamentous rod mutant sites that include the G183R/G183W and S197L mutations, reside close to each other at the very bottom of the predicted FlgG structural model, and two other filamentous rod mutant sites that include the D117Y, G132R, and G133V mutations, are located close to each other in the middle of the structure (Fig. 3B). This allowed us to propose mechanisms for FlgG stop-polymerization. The 52- to 66-amino-acid region of one FlgG subunit could interact with the bottom region of a second FlgG subunit stacked on top of it at residues G183 and S197 to stop FlgG polymerization. The isolation of mutants at positions D117 and G132 would indicate an effect of these residues on this interaction.
Figure 3.
Modeling FlgG and filamentous rod mutants on the three-dimensional structure of FlgE (hook). (A) Alignment of FlgG-rod and FlgE-hook sequences. The FlgG-rod protein is 260 amino acids in length. Of these, 38% correspond to identical residues (in red) in the corresponding FlgE sequences throughout the FlgG protein, while the predicted structural conservation (blue H for α-helix for FlgE and light blue for FlgG) is nearly identical (Homma et al. 1990). The exceptions are a large insertion in FlgE-hook relative to FlgG that defines a complete and separate domain from the core of the FlgE protein (Samatey et al. 2004). In addition, FlgG contains an 18-amino-acid insertion after the corresponding residue 43 of FlgE. This region includes the majority of the FlgG changes that are defective in the stop-polymerization mechanism (shaded in green). (B) An FlgG structural model based on FlgE. A three-dimensional structure of FlgG based on the crystal structure of FlgE (Samatey et al. 2004). The FlgE crystal structure is missing the N- and C-terminal regions corresponding to residues 1–91 from the N terminus of FlgG and residues 223–260 from the C terminus, and thus these regions could not be included in the model. We suggest that the region including the majority of FlgG* mutations (residues 52–66, drawn in a green oval) are located near the top of the FlgG subunit where it interacts with residues G183 and S197, located at the bottom of the structure during FlgG polymerization. A three-dimensional model of FlgG was constructed using the comparative modeling module of the Protinfo server (http://protinfo.compbio.washington.edu), which has been shown to work well in the CASP protein structure prediction experiments (Hung and Samudrala 2003; Hung et al. 2005). The structure of FlgE was used as a template for the modeling the FlgG sequence excluding the N- and C-terminal insertions (Samatey et al. 2004). Images were produced using the Molscript (Kraulis 1991) and Raster3D software (Merritt and Murphy 1994).
Rod length control is required for normal P- and L-ring formation
The filamentous rod mutants were originally isolated in a strain deleted for the P- and L-ring structural genes (ΔflgHI). Strains were constructed that carried the flgG* mutant alleles and functional ring structural genes (flgH+I+). These strains were found to be defective in motility. Upon flagella purification, intact flagellar structures obtained from these strains had filamentous rods fused to filaments (Fig. 4A). Figure 4B shows isolated rod–hook–filament structures obtained after depolymerization of the associated filaments by low pH treatment. Unexpectedly, the rings associated with the basal structures varied from two to six per structure, and appeared to depend on the length of the individual filamentous rods (Fig. 4B). Depending on the flgG* allele, the structures had either shorter hooks or no apparent hook structures at all.
Figure 4.
Flagellar structures from a flgG* mutant strain. (A) Intact filament structures were isolated from a strain (TH9614) that carried a single flgG* allele (flgG*5664 [G53C]) and examined by electron microscopy. The two smaller structures visible in the left micrograph are virulence-associated type III needle structures that copurify with flagellar filaments. (B) Intact filaments were isolated from a strain (TH9616) that carried a single flgG* allele (flgG*5671 [P52L]), the filaments were depolymerized by acid treatment, and the final flagellar basal structures were examined by electron microscope. (C) The flgG*5671 (P52L) allele was introduced into strains with single null alleles in flgI (P-ring defect, strain TH10281) or flgH (L-ring defect, strain TH10280). (Right panel) The flgG*5671 mutant rod in the flgH (L-ring defect) mutant strain (TH10280) has many rings attached, presumably P-rings. (Left panel) The flgG*5671 mutant rod in the flgI (P-ring defect) mutant strain (TH102801) has no rings. (D) A flgG* mutant strain that is deleted for fliK and expresses functional P- and L-rings, TH12354 (flgG*5667 [G65V] ΔfliK6137::tetRA), produces polyrod structures with many attached rings.
It was not known what controlled P- and L-ring formation. These results suggest that some aspect of ring formation is coupled to rod growth termination. To determine if the polyrings were composed of P- or L-rings or a mixture of both structures, a flgG* allele was introduced into separate flgI (P-ring) and flgH (L-ring) mutant strains. The flgG* flgH double mutant still produced filamentous rod–filament structures with polyrings, while the flgG* flgI double mutant produced filamentous rod–filament structures without rings (Fig. 4C). This demonstrated that the polyring structures were multiple P-rings surrounding the filamentous rod structures. These results also suggest that the FlgG stop-polymerization mechanism is coupled to the formation of a single P-ring structure in wild-type flagella. We also tested the effect of a fliK-null allele in a flgG*(G65V) strain with functional P- and L-ring structural genes (flgH+ flgI+). As expected, filamentous rods with many attached P-rings were observed (Fig. 4D).
Finally, 16 FlgG* filamentous rod alleles (P52L, G53C, G53R, Δ54–57, Q59H-Δ60–63, G65V, G65V, G65R, Δ65–66, L66P, D117Y, G132R, G183R, G183W, E189K, and S197L) were examined in strains with functional P- and L-ring structural genes (flgH+ flgI+). Three of these alleles (G53R, Δ54–57, and G183R) did not possess ring structures. These are apparently defective in both rod stop-polymerization and P-ring formation. It is possible that these residues are important in the polymerization of P-ring structures on normal rods. This remains to be determined.
Rod length control is required for outer membrane penetration of the flagellum
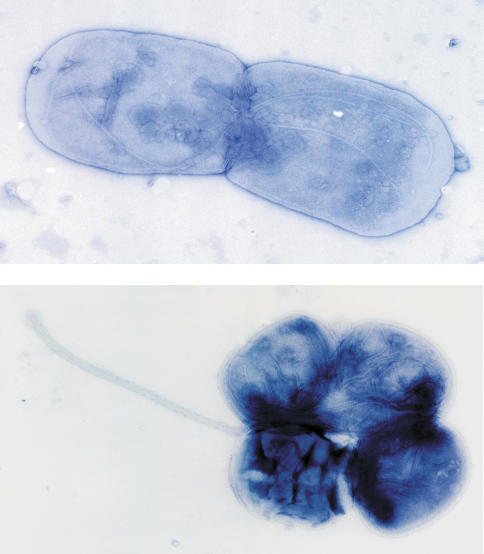
Although intact rod–filament structures could be obtained from strains with only the flgG* alleles, no visible flagellar filament structures were detected growing from the surfaces of these cells. However, upon closer examination after osmotic shock treatment, flagella could be visualized in the flgG* strains, but instead of growing outside the cell body, the flagella were observed growing between the inner and outer cell membranes (Fig. 5). Some isolated filaments were observed protruding from cells, but these were always sheathed in membrane (Fig. 5). We conclude that the filamentous rod mutant alleles produced flagella unable to penetrate the cell’s outer membrane. These results suggest that the evolution of spirochete flagella, which grow in the periplasmic space (Charon and Goldstein 2002), can occur from extracellular flagella by limited mutational events. Here, we show conversion of extracellular flagella of Salmonella to periplasmic flagella can arise by single amino acid changes in the FlgG rod subunits.
Figure 5.
Growth of flagella in the periplasm in flgG* mutant strains. Strains that carry only a flgG* allele TH9613 [flgG*5662 (G65E)] (top) and TH10080 [flgG*5670 (G183R)] (bottom) were subject to lysozyme treatment in order to visualize flagellar structures growing in the periplasmic space by electron microscopy. (Bottom) Occasionally, membrane-enveloped flagellar filaments structures were observed.
Discussion
Insights for rod and hook subunit polymerization by the eubacterial flagellum
The flagellum is a helical structure containing 11 protofilaments (Macnab 2003). The 11 subunits are added per two turns of the helix. The proximal rod is estimated to contain six subunits each of FlgB, FlgC, and FlgF subunits, which is consistent with the polymerization of one turn each. The distal rod is estimated to contain 26 subunits, which is consistent with four turns or two stacks of FlgG subunits within the 11 protofilaments. There is likely to be interactions between FlgB, FlgC, and FlgF that allow polymerization of one turn each and no more. The stacking mechanism for the FlgG subunits that make up the distal rod must be different to allow polymerization to continue to four turns of the helix. It must also be different from the hook, which polymerizes to ∼120 subunits. The characterization of the filamentous FlgG mutants reported here suggests that the FlgG and hook subunits have polymerization capabilities that are more closely related to each other than those of the proximal rod subunits because single amino acid substitution mutations in FlgG result in a polymerization phenotype that is identical to that of the hook. It seems probable that FlgG and hook evolved from a duplication of an ancestral gene and that FlgG gained the ability to stop polymerization after one FlgG subunit was added on top of another. This would be the case for two stacks of FlgG in the 11 protofilaments of the flagellum.
Remarkably, the FlgG filamentous rod mutants (FlgG*) grew to a final length close to that of a normal rod–hook structure (Fig. 1C,D). This suggested the existence of a length control mechanism that prevented filamentous rod polymerization beyond the length of a normal rod–hook structure. Our data show that the final length of the filamentous rods is determined by the hook–length control protein, FliK. It is known that the loss of FliK results in continuous hook growth (polyhook). Similarly, in the absence of FliK, the FlgG* mutants exhibited continuous rod growth (polyrod). FliK is supposed to be a molecular ruler for the hook (Shibata et al. 2007). Our results demonstrate that the flgG* mutations bring rod length under FliK control. Furthermore, these results suggest that FliK is not specific for hook length control, but a molecular ruler for the first 70–80 nm of the flagellar structure, whether that structure is rod–hook or extended rod.
Outer membrane penetration is coupled to P–L-ring assembly on normal rod
The isolation of Salmonella mutants that could grow periplasmic flagella was surprising. The fact that these mutants resulted from single amino acid substitutions suggests that evolution of periplasmic flagella from extracellular flagella might be a relatively straightforward process. However, we do not know that periplasmic flagella did evolve from extracellular flagella. A major distinction between spirochete and eubacterial flagella is the ability of the eubacterial flagellum to penetrate the outer membrane. The flgG* mutants lost both the ability to terminate rod polymerization and the ability to couple rod growth termination to outer membrane penetration of the growing structure. Our data supports a model that rod growth must terminate at a length that facilitates a perpendicular positioning of the rod tip to the outermembrane. Continued rod polymerization beyond 25 nm might place the rod tip in a position that is more parallel to the outer membrane. A perpendicular positioning might be necessary for the L-ring pore to form properly around the distal rod and within the outer membrane for continued flagellar growth outside the cell. We propose a three-step process for outer membrane penetration. First, the distal rod component FlgG ceases to polymerize after a second FlgG subunit has polymerized on top of a first. This results in a rod length of 25 nm that places the rod in a position that is perpendicular to the outer membrane. Second, the P-ring polymerizes around a stack of two FlgG subunits. Also, if P-ring formation is slow relative to rod polymerization, a complete cessation of rod growth would allow for all the time needed for the P-ring to polymerize around the distal rod. Finally, the L-ring polymerizes within the outer membrane on top of the assembled P-ring to form a pore for the flagellum.
It was unexpected to find that the flagella of Salmonella flgG* mutants could grow between the inner and outer membranes similar to what is seen in spirochete flagella. Whether or not a similar FlgG-dependent mechanism also operates to confine flagella to the periplasmic space in spirochetes remains to be determined. Many spirochete flagella lack P- and L-rings altogether (Charon and Goldstein 2002). A BLAST search revealed that at least two species of spirochetes, Borrelia burgdorferi and Borrelia garnii, possess a P-ring structural gene (flgI), but no flgH gene to encode an L-ring. However, a different spirochete genus, Leptospira, do carry both P- and L-ring structural genes. Thus, the presence of an L-ring structural gene in a Gram-negative bacterium does not correlate with growth of the flagella outside the cell. It would be interesting to determine if the periplasmic flagella of Leptospira possess L-rings. Perhaps there are conditions that allow Leptospira to grow extracellular flagella and only under these conditions are L-rings produced.
The formation of P-rings on the flagellar rod
We propose that P-rings form around stacks of FlgG subunits in the distal rod. However, the extra P-rings we observed on the filamentous rods or the longer polyrods were not evenly spaced or absent altogether (Fig. 4). We suspect that in the flgG* mutants P-ring subunits are limited relative to FlgG subunits. The P-rings subunits are secreted into the periplasm by a different secretion system (Sec) than flagellum-secreted FlgG subunits and they must compete for secretion with all other Sec-dependent substrates. In the filamentous rod strains (flgG*), there is at least fivefold more FlgG subunits assembled into filamentous rod structures than in wild-type rod structures and even more in the polyrod strains (flgG* fliK−). Thus, we presume that there is a large excess of assembled FlgG subunits to available P-ring subunits in the filamentous rod and polyrod strains, and this could account for the limited number and uneven spacing of P-rings on the filamentous rods and polyrods.
Model for transition from rod to hook polymerization coupled to ring assembly and outer membrane penetration
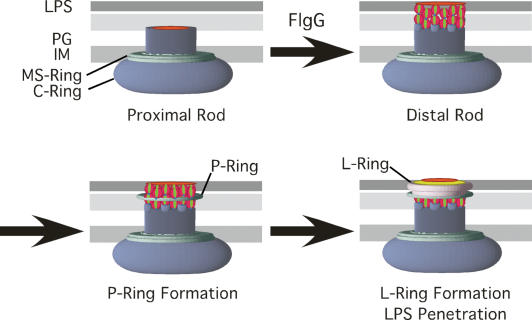
The results presented here provide the foundation for a model, presented in Figure 6, for a structural checkpoint in flagellum assembly that couples rod completion, P- and L-ring assembly, outer membrane penetration, and initiation of flagellar growth outside the cell. The number of FlgG subunits at the distal end of the rod has been estimated to be ∼26 subunits (Jones et al. 1990; Makishima et al. 2001). Two stacks of FlgG subunits in the 11-protofilament flagellar structure would add up to 22 subunits, which is in reasonable agreement with this estimate. We envision at least three mechanisms that would allow FlgG polymerization to stop to terminate rod growth. All mechanisms would include interactions between residues at the bottom (Gly-183/Ser-197) and top (the 52- to 66-amino-acid region) of FlgG subunits. One mechanism predicts that the stacking of one FlgG residue onto another results in a conformational change of the second FlgG protein that prevents further interactions with additional FlgG subunits. The second mechanism predicts that a slight rotation might occur perpendicular to the central core axis when one FlgG residue stacks on the other, resulting from the interactions between residues Gly-183/Ser-197 and residues 52–66 and transmitted through the intervening region (including residues D117, G132, and G133). The degree of rotation may be tolerated in one stack, but no further. The stacking models are supported by the fact that flgG* mutations reside at the region of the folded subunit that corresponds to the outside of the final polymerized rod structure from the top to the bottom of the FlgG folded subunit modeled to the homologous FlgE structure (Fig. 3B).
Figure 6.
A model of a checkpoint in flagellum assembly. A FlgG stop-polymerization signal couples rod completion to P- and L-ring assembly and outer membrane penetration of the growing flagellum. Two stacks of FlgG subunits polymerize onto the proximal rod to complete the rod structure. A P-ring forms around two layers of FlgG subunits. This is followed by L-ring formation on the P-ring and within the outer membrane, which effectively opens a hole in the outer membrane to allow polymerization of the flagellum outside the cell. (IM) Inner membrane; (PG) peptidoglycan; (LPS) lipopolysaccharide.
Another mechanism is based on the similarity between wild-type, stacked FlgG structures and filamentous rod structures to the two forms of tobacco mosaic virus (TMV) coat protein. Purified TMV coat protein exists in cylindrical two-layer stacks of 17-fold rotational symmetry presumably corresponding to 17 subunits per stack while RNA-associated TMV coat protein is filamentous (Díaz-Avalos and Caspar 1998). Thus, TMV coat protein can transition from a two-stack cylindrical form to a filamentous helical form depending on the presence of the RNA genome. It is possible that stacking FlgG subunits during rod construction go through a similar transition from cylindrical stacking to a filamentous stacking upon the initiation of hook subunit polymerization on the FlgG stacks. The cylindrical form would constitute the stop-polymerization mechanism, but then the addition of hook subunits could change the conformation to a helical form allowing a continuous filamentous flagellar structure that transition from rod to hook to filament subunits.
Requirements for efficient organelle assembly
The results of this study suggest that the efficient assembly of the bacterial flagellum occurs through a fundamental mechanism in the assembly of organelle structures: the requirement for discrete, independent stop–start construction signals so that substructures are created in a programmed order. Each of the rod components incorporates a stop-polymerization mechanism signal, which results in a single turn of each of the FlgB, FlgC, and FlgF subunits within the axial rod and two complete layers (four turns) of the FlgG subunits. The P-ring forms around the assembled FlgG subunits. By having a mechanism to stop rod polymerization, the P-ring subunits have time to assemble around the completed rod, followed by L-ring formation. The discovery of the stop-polymerization for FlgG subunits in the flagellar rod resulted from a genetic selection for mutants that affect regulation of flagellar gene expression. This selection was done in a strain deleted for the P- and L-ring structural genes (ΔflgHI) and missing both flagellin genes. Further investigation of the flgG* alleles in an otherwise wild-type flagellar strain led to the discovery that the transition from rod completion to hook growth initiation outside the cell is coupled to the formation of the P- and L-ring structures on the completed rod and in membrane penetration of the flagellum followed by continued growth outside the cell. The flgG* mutants in the otherwise wild-type flagellar strain background exhibited flagella that grew in the periplasm and membranous flagella were observed to occasionally extend from these cells as had been reported in spirochetes (Charon et al. 1992). We would like to suggest that in building complex structures, such as the bacterial flagellum, it is critical to have distinct, coupled start–stop mechanisms to assemble substructures in a programmed order. The discovery reported here may provide a new paradigm for all organelle assembly processes.
Materials and methods
Bacterial strains, media, and standard genetic manipulations
Bacterial strains used in this study originated from Salmonella enterica serovar typhimurium strain LT2. Media, growth conditions, transductional methods, and motility assays were described (Aldridge et al. 2006). The generalized transducing phage of S. typhimurium P22 HT105/1 int-201 was used in all transductional crosses (Davis et al. 1980).
Genetic selection for filamentous rod mutants in ring mutant strains
Strain TH4987 (ΔflgHI958 motA5461∷MudJ flhAts) carries a transcriptional fusion of the lac operon (MudJ) under expression of the σ28-dependent motA promoter. It also carries an in-frame deletion of the P- and L-ring structural genes (ΔflgHI) and a temperature-sensitive mutation in an integral membrane component of the flagellar T3S apparatus (flhAts). This strain is Lac− because in the absence of the P- and L-rings (ΔflgHI), the anti-σ28-factor FlgM is not secreted from the cell, and σ28-dependent transcription of the motA promoter (and thus the motA-lac operon reporter construct motA∷MudJ) does not occur. At 30°C the strain is FlhA+ resulting in a functional flagellar secretion apparatus, the flagellar rod structure forms and hook elongation initiates, but the structure fails to extend beyond the outer membrane due to the missing P- and L-ring (ΔflgHI) components. In strains defective only in P- and L-ring formation, loss of either FlgM or Flk would result in σ28-dependent transcription from the motA promoter (Lac+) (Karlinsey et al. 1997). At 42°C the strain is FlhA−. The flagellar secretion apparatus is not functional without FlhA. Only loss of FlgM allows σ28-dependent transcription in strains defective in FlhA (Karlinsey et al. 1997). To isolate Flk-bypass mutants, strain TH4987 was plated on minimal lactose (E-Lac) medium and incubated at 30°C for 48 h. The 30°C Lac+ colonies were screened for those that were Lac− at 42°C by replica printing. The Lacts alleles allow σ28-dependent motA transcription at 30°C (FlgHI-negative FlhA+), but not at 42°C (FlgHI-negative FlhA-negative). The Lacts mutants were screened for linkage to the flk locus by bacteriophage P22-mediated transduction to the flk region of the chromosome as described (Karlinsey et al. 1997). Those not linked to the flk region were kept as Flk-bypass mutants (flkBP). Insertions of transposon Tn10dTc linked to the flkBP alleles followed by DNA sequence analysis was performed as described for the isolation of Tn10dTc linked to the serT locus (Chevance et al. 2006). DNA sequence analysis located the linked Tn10dTc insertions to the flg region of the chromosome. Fine mapping and DNA sequence analysis of the flkBP alleles revealed them to be mutated in the distal rod structural gene flgG.
Electron microscopy
For visualization by electron microscopy, cells and isolated flagellar structures were stained with 1% phosphotungstic acid (PTA) (pH 7 or pH 5) and observed with a JEOL 1200Ex electron microscope at 80 kV.
FlgG structure modeling
A three-dimensional model of the flgG was constructed using the comparative modeling module of the Protinfo server (http://protinfo.compbio.washington.edu), which has been shown to work well in the CASP protein structure prediction experiments (Hung and Samudrala 2003; Hung et al. 2005). The structure of FlgE was used as a template for the modeling the flgG sequence excluding the N- and C-terminal insertions (Samatey et al. 2004). Images were produced using the Molscript (Kraulis 1991) and Raster3D software (Merritt and Murphy 1994).
Acknowledgments
We thank Howard Berg, Ariel Blocker, Tohru Minamino, Kevin Plaxco, and John Roth for critically reading the manuscript prior to submission, and David Blair and Sandy Parkinson for useful discussions. Finally, we acknowledge that Takashi Fujii in Keiichi Namba’s laboratory first observed the formation of polyrings on filamentous rods, and that Kazuhiro Kutsukake first observed filamentous rod structures from Flk (RflH)-bypass mutants (personal communications to K.T.H.). This work was supported by Public Health Service grant GM56141 from the National Institutes of Health (to K.T.H.) and from CREST (to S.-I.A.).
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1571607
References
- Aldridge P., Hughes K.T., Hughes K.T. Regulation of flagellar assembly. Curr. Opin. Microbiol. 2002;5:160–165. doi: 10.1016/s1369-5274(02)00302-8. [DOI] [PubMed] [Google Scholar]
- Aldridge P., Karlinsey J.E., Becker E., Chevance F.F., Hughes K.T., Karlinsey J.E., Becker E., Chevance F.F., Hughes K.T., Becker E., Chevance F.F., Hughes K.T., Chevance F.F., Hughes K.T., Hughes K.T. Flk prevents premature secretion of the anti-σ factor FlgM into the periplasm. Mol. Microbiol. 2006;60:630–643. doi: 10.1111/j.1365-2958.2006.05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage J.P., Holland I.B., Jenal U., Kenny B., Holland I.B., Jenal U., Kenny B., Jenal U., Kenny B., Kenny B. ‘Neural networks’ in bacteria: Making connections. J. Bacteriol. 2005;187:26–36. doi: 10.1128/JB.187.1.26-36.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg H.C., Anderson R.A., Anderson R.A. Bacteria swim by rotating their flagellar filaments. Nature. 1973;245:380–382. doi: 10.1038/245380a0. [DOI] [PubMed] [Google Scholar]
- Charon N.W., Goldstein S.F., Goldstein S.F. Genetics of motility and chemotaxis of a fascinating group of bacteria: The spirochetes. Annu. Rev. Genet. 2002;36:47–73. doi: 10.1146/annurev.genet.36.041602.134359. [DOI] [PubMed] [Google Scholar]
- Charon N.W., Goldstein S.F., Block S.M., Curci K., Ruby J.D., Kreiling J.A., Limberger R.J., Goldstein S.F., Block S.M., Curci K., Ruby J.D., Kreiling J.A., Limberger R.J., Block S.M., Curci K., Ruby J.D., Kreiling J.A., Limberger R.J., Curci K., Ruby J.D., Kreiling J.A., Limberger R.J., Ruby J.D., Kreiling J.A., Limberger R.J., Kreiling J.A., Limberger R.J., Limberger R.J. Morphology and dynamics of protruding spirochete periplasmic flagella. J. Bacteriol. 1992;174:832–840. doi: 10.1128/jb.174.3.832-840.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevance F.F., Karlinsey J.E., Wozniak C.E., Hughes K.T., Karlinsey J.E., Wozniak C.E., Hughes K.T., Wozniak C.E., Hughes K.T., Hughes K.T. A little gene with big effects: A serT mutant is defective in flgM gene translation. J. Bacteriol. 2006;188:297–304. doi: 10.1128/JB.188.1.297-304.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilcott G.S., Hughes K.T., Hughes K.T. The coupling of flagellar gene expression to flagellar assembly in Salmonella typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 2000;64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R.W., Botstein D., Roth J.R., Botstein D., Roth J.R., Roth J.R. Advanced bacterial genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1980. [Google Scholar]
- Diaz-Avalos R., Caspar D.L., Caspar D.L. Structure of the stacked disk aggregate of tobacco mosaic virus protein. Biophys. J. 1998;74:595–603. doi: 10.1016/S0006-3495(98)77818-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson S.U., Tokuyasu K., Simon M.I., Tokuyasu K., Simon M.I., Simon M.I. Bacterial flagella: Polarity of elongation. Science. 1970;169:190–192. doi: 10.1126/science.169.3941.190. [DOI] [PubMed] [Google Scholar]
- Ferris H.U., Minamino T., Minamino T. Flipping the switch: Bringing order to flagellar assembly. Trends Microbiol. 2006;14:519–526. doi: 10.1016/j.tim.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Ferris H.U., Furukawa Y., Minamino T., Kroetz M.B., Kihara M., Namba K., Macnab R.M., Furukawa Y., Minamino T., Kroetz M.B., Kihara M., Namba K., Macnab R.M., Minamino T., Kroetz M.B., Kihara M., Namba K., Macnab R.M., Kroetz M.B., Kihara M., Namba K., Macnab R.M., Kihara M., Namba K., Macnab R.M., Namba K., Macnab R.M., Macnab R.M. FlhB regulates ordered export of flagellar components via autocleavage mechanism. J. Biol. Chem. 2005;280:41236–41242. doi: 10.1074/jbc.M509438200. [DOI] [PubMed] [Google Scholar]
- Gillen K.L., Hughes K.T., Hughes K.T. Negative regulatory loci coupling flagellin synthesis to flagellar assembly in Salmonella typhimurium. J. Bacteriol. 1991;173:2301–2310. doi: 10.1128/jb.173.7.2301-2310.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Yamaguchi S., Oosawa K., Aizawa S.-I., Yamaguchi S., Oosawa K., Aizawa S.-I., Oosawa K., Aizawa S.-I., Aizawa S.-I. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J. Bacteriol. 1994;176:5439–5449. doi: 10.1128/jb.176.17.5439-5449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Minamino T., Namba K., Macnab R.M., Minamino T., Namba K., Macnab R.M., Namba K., Macnab R.M., Macnab R.M. Substrate specificity classes and the recognition signal for Salmonella type III flagellar export. J. Bacteriol. 2003;185:2485–2492. doi: 10.1128/JB.185.8.2485-2492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Shibata S., Ohnishi K., Tani T., Aizawa S., Shibata S., Ohnishi K., Tani T., Aizawa S., Ohnishi K., Tani T., Aizawa S., Tani T., Aizawa S., Aizawa S. N-terminal signal region of FliK is dispensable for length control of the flagellar hook. Mol. Microbiol. 2005;56:346–360. doi: 10.1111/j.1365-2958.2005.04615.x. [DOI] [PubMed] [Google Scholar]
- Homma M., Kutsukake K., Hasebe M., Iino T., Macnab R.M., Kutsukake K., Hasebe M., Iino T., Macnab R.M., Hasebe M., Iino T., Macnab R.M., Iino T., Macnab R.M., Macnab R.M. FlgB, FlgC, FlgF and FlgG. A family of structurally related proteins in the flagellar basal body of Salmonella typhimurium. J. Mol. Biol. 1990;211:465–477. doi: 10.1016/0022-2836(90)90365-S. [DOI] [PubMed] [Google Scholar]
- Hughes K.T., Gillen K.L., Semon M.J., Karlinsey J.E., Gillen K.L., Semon M.J., Karlinsey J.E., Semon M.J., Karlinsey J.E., Karlinsey J.E. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- Hung L.H., Samudrala R., Samudrala R. PROTINFO: Secondary and tertiary protein structure prediction. Nucleic Acids Res. 2003;31:3296–3299. doi: 10.1093/nar/gkg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung L.H., Ngan S.C., Liu T., Samudrala R., Ngan S.C., Liu T., Samudrala R., Liu T., Samudrala R., Samudrala R. PROTINFO: New algorithms for enhanced protein structure predictions. Nucleic Acids Res. 2005;33 (Web server issue):77–80. doi: 10.1093/nar/gki403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino T. Genetics and chemistry of bacterial flagella. Bacteriol. Rev. 1969;33:454–475. doi: 10.1128/br.33.4.454-475.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C.J., Macnab R.M., Okino H., Aizawa S., Macnab R.M., Okino H., Aizawa S., Okino H., Aizawa S., Aizawa S. Stoichiometric analysis of the flagellar hook–(basal-body) complex of Salmonella typhimurium. J. Mol. Biol. 1990;212:377–387. doi: 10.1016/0022-2836(90)90132-6. [DOI] [PubMed] [Google Scholar]
- Karlinsey J.E., Pease A.J., Winkler M.E., Bailey J.L., Hughes K.T., Pease A.J., Winkler M.E., Bailey J.L., Hughes K.T., Winkler M.E., Bailey J.L., Hughes K.T., Bailey J.L., Hughes K.T., Hughes K.T. The flk gene of Salmonella typhimurium couples flagellar P- and L-ring assembly to flagellar morphogenesis. J. Bacteriol. 1997;179:2389–2400. doi: 10.1128/jb.179.7.2389-2400.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlinsey J.E., Shugo Tanaka S., Bettenworth V., Yamaguchi S., Boos W., Aizawa S.I., Hughes K.T., Shugo Tanaka S., Bettenworth V., Yamaguchi S., Boos W., Aizawa S.I., Hughes K.T., Bettenworth V., Yamaguchi S., Boos W., Aizawa S.I., Hughes K.T., Yamaguchi S., Boos W., Aizawa S.I., Hughes K.T., Boos W., Aizawa S.I., Hughes K.T., Aizawa S.I., Hughes K.T., Hughes K.T. Completion of the hook–basal body of the Salmonella typhimurium flagellum is coupled to FlgM secretion and fliC transcription. Mol. Microbiol. 2000;37:1220–1231. doi: 10.1046/j.1365-2958.2000.02081.x. [DOI] [PubMed] [Google Scholar]
- Kojima S., Blair D., Blair D. The bacterial flagellar motor: Structure and function of a complex molecular machine. Int. Rev. Cytol. 2004;233:93–134. doi: 10.1016/S0074-7696(04)33003-2. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. Molscript: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- Kutsukake K. Excretion of the anti-σ factor through a flagellar substructure couples flagellar gene expression with flagellar assembly in Salmonella typhimurium. Mol. Gen. Genet. 1994;243:605–612. doi: 10.1007/BF00279569. [DOI] [PubMed] [Google Scholar]
- Macnab R.M. How bacteria assemble flagella. Annu. Rev. Microbiol. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- Makishima S., Komoriya K., Yamaguchi S., Aizawa S.I., Komoriya K., Yamaguchi S., Aizawa S.I., Yamaguchi S., Aizawa S.I., Aizawa S.I. Length of the flagellar hook and the capacity of the type III export apparatus. Science. 2001;291:2411–2413. doi: 10.1126/science.1058366. [DOI] [PubMed] [Google Scholar]
- Merritt E.A., Murphy M.E., Murphy M.E. Raster3D version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- Minamino T., Macnab R.M., Macnab R.M. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 1999;181:1388–1394. doi: 10.1128/jb.181.5.1388-1394.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T., Namba K., Namba K. Self-assembly and type III protein export of the bacterial flagellum. J. Mol. Microbiol. Biotechnol. 2004;7:5–17. doi: 10.1159/000077865. [DOI] [PubMed] [Google Scholar]
- Minamino T., Doi H., Kutsukake K., Doi H., Kutsukake K., Kutsukake K. Substrate specificity switching of the flagellum-specific export apparatus during flagellar morphogenesis in Salmonella typhimurium. Biosci. Biotechnol. Biochem. 1999;63:1301–1303. doi: 10.1271/bbb.63.1301. [DOI] [PubMed] [Google Scholar]
- Minamino T., Yamaguchi S., Macnab R.M., Yamaguchi S., Macnab R.M., Macnab R.M. Interaction between FliE and FlgB, a proximal rod component of the flagellar basal body of Salmonella. J. Bacteriol. 2000;182:3029–3036. doi: 10.1128/jb.182.11.3029-3036.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya N., Minamino T., Hughes K.T., Macnab R.M., Namba K., Minamino T., Hughes K.T., Macnab R.M., Namba K., Hughes K.T., Macnab R.M., Namba K., Macnab R.M., Namba K., Namba K. The type III flagellar export specificity switch is dependent on FliK ruler and a molecular clock. J. Mol. Biol. 2006;359:466–477. doi: 10.1016/j.jmb.2006.03.025. [DOI] [PubMed] [Google Scholar]
- Ohnishi K., Kutsukake K., Suzuki H., Iino T., Kutsukake K., Suzuki H., Iino T., Suzuki H., Iino T., Iino T. A novel transcriptional regulatory mechanism in the flagellar regulon of Salmonella typhimurium: An anti σ factor inhibits the activity of the flagellum-specific σ factor, σF. Mol. Microbiol. 1992;6:3149–3157. doi: 10.1111/j.1365-2958.1992.tb01771.x. [DOI] [PubMed] [Google Scholar]
- Patterson-Delafield J., Martinez R.J., Stocker B.A., Yamaguchi S., Martinez R.J., Stocker B.A., Yamaguchi S., Stocker B.A., Yamaguchi S., Yamaguchi S. A new fla gene in Salmonella typhimurium—flaR—and its mutant phenotype—superhooks. Arch. Mikrobiol. 1973;90:107–120. doi: 10.1007/BF00414513. [DOI] [PubMed] [Google Scholar]
- Samatey F.A., Matsunami H., Imada K., Nagashima S., Shaikh T.R., Thomas D.R., Chen J.Z., Derosier D.J., Kitao A., Namba K., Matsunami H., Imada K., Nagashima S., Shaikh T.R., Thomas D.R., Chen J.Z., Derosier D.J., Kitao A., Namba K., Imada K., Nagashima S., Shaikh T.R., Thomas D.R., Chen J.Z., Derosier D.J., Kitao A., Namba K., Nagashima S., Shaikh T.R., Thomas D.R., Chen J.Z., Derosier D.J., Kitao A., Namba K., Shaikh T.R., Thomas D.R., Chen J.Z., Derosier D.J., Kitao A., Namba K., Thomas D.R., Chen J.Z., Derosier D.J., Kitao A., Namba K., Chen J.Z., Derosier D.J., Kitao A., Namba K., Derosier D.J., Kitao A., Namba K., Kitao A., Namba K., Namba K. Structure of the bacterial flagellar hook and implication for the molecular universal joint mechanism. Nature. 2004;431:1062–1068. doi: 10.1038/nature02997. [DOI] [PubMed] [Google Scholar]
- Shibata S., Takahashi N., Chevance F.F.V., Karlinsey J.E., Hughes K.T., Aizawa S.-I., Takahashi N., Chevance F.F.V., Karlinsey J.E., Hughes K.T., Aizawa S.-I., Chevance F.F.V., Karlinsey J.E., Hughes K.T., Aizawa S.-I., Karlinsey J.E., Hughes K.T., Aizawa S.-I., Hughes K.T., Aizawa S.-I., Aizawa S.-I. FliK regulates flagellar hook length as an internal ruler. Mol. Microbiol. 2007;64:1405–1415. doi: 10.1111/j.1365-2958.2007.05750.x. [DOI] [PubMed] [Google Scholar]
- Wolgemuth C.W., Charon N.W., Goldstein S.F., Goldstein R.E., Charon N.W., Goldstein S.F., Goldstein R.E., Goldstein S.F., Goldstein R.E., Goldstein R.E. The flagellar cytoskeleton of the spirochetes. J. Mol. Microbiol. Biotechnol. 2006;11:221–227. doi: 10.1159/000094056. [DOI] [PubMed] [Google Scholar]