Abstract
JIP scaffold proteins are implicated in the regulation of protein kinase signal transduction pathways. To test the physiological role of these scaffold proteins, we examined the phenotype of compound mutant mice that lack expression of JIP proteins. These mice were found to exhibit severe defects in N-methyl-D-aspartic acid (NMDA) receptor function, including decreased NMDA-evoked current amplitude, cytoplasmic Ca++, and gene expression. The decreased NMDA receptor activity in JIP-deficient neurons is associated with reduced tyrosine phosphorylation of NR2 subunits of the NMDA receptor. JIP complexes interact with the SH2 domain of cFyn and may therefore promote tyrosine phosphorylation and activity of the NMDA receptor. We conclude that JIP scaffold proteins are critically required for normal NMDA receptor function.
Keywords: JIP, JNK, scaffold protein, signal transduction
The JIP1 scaffold protein is implicated in the assembly and coordination of cJun N-terminal kinase (JNK) signaling modules that include the mitogen-activated protein (MAP) kinase kinase MKK7 and the mixed-lineage kinase group of MAP kinase kinase kinases (Dickens et al. 1997; Whitmarsh et al. 1998). Indeed, studies of JIP1-deficient mice confirm that this scaffold protein is essential for some forms of stress-induced JNK activation (Morrison and Davis 2003). Thus, JIP1 is required for JNK activation and neuronal death in response to ischemia in vitro and stroke in vivo (Whitmarsh et al. 2001; Im et al. 2003) and for JNK-mediated insulin resistance in type II diabetes (Jaeschke et al. 2004).
Although JIP1 is established to be important for JNK regulation, it appears that JIP1 may also have other physiological functions. For example, JIP1 is implicated in both the AKT and SRC signaling pathways (Kim et al. 2003; Nihalani et al. 2007). Furthermore, JIP1 may also act as an adapter protein that links the kinesin-1 motor protein to cargoes (Verhey et al. 2001; Whitmarsh et al. 2001). JIP1 may therefore influence the kinesin-mediated localization of cargo molecules and organelles within the cell. Proposed cargoes are represented by the low-density lipoprotein receptor-related protein LRP8 (Verhey et al. 2001) and organelles, including mitochondria and synaptic vesicles (Horiuchi et al. 2005). Together, these observations suggest that JIP1-deficient mice should have major physiological defects. However, JIP1-deficient mice exhibit only a limited spectrum of defects, including the response to ischemia (Whitmarsh et al. 2001; Im et al. 2003) and metabolic stress (Jaeschke et al. 2004).
The analysis of additional phenotypes of JIP1-deficient mice may be compromised by partial compensation mediated by the scaffold protein JIP2 (Yasuda et al. 1999). JIP1 and JIP2 are structurally related scaffold proteins that share a similar tissue distribution, including high levels of expression in the brain. To test the role of these scaffold proteins, we examined the phenotype of compound mutant mice that lack expression of both JIP1 and JIP2. This analysis led to the discovery that the JIP1/2 group of scaffold proteins is critically required for normal N-methyl-D-aspartic acid (NMDA) receptor function.
Results
Creation of JIP-deficient mice
We disrupted the murine Jip2 gene using homologous recombination in embryonic stem cells (Fig. 1A). Chimeric mice prepared with these cells transmitted the disrupted Jip2 allele through the germline (Fig. 1B,C). The presence of JIP1 in mice that lack expression of JIP2 may result in compensation of phenotypes that might be caused by JIP2 deficiency. Indeed, it was observed that the expression of JIP1 was increased in the brain of Jip2−/− mice compared with wild-type mice (Fig. 1C). We therefore mated JIP1-deficient mice with JIP2-deficient mice to create compound mutant animals (Fig. 2A). Genotype analysis of the double-knockout mice confirmed that the Jip1 and Jip2 genes were disrupted (Fig. 2B), and immunoblot analysis demonstrated that no JIP1 or JIP2 protein was detected (Fig. 2C). As expected (Whitmarsh et al. 2001; Im et al. 2003), JIP deficiency caused defects in JNK activity in primary neurons in vitro (Fig. 2D,E) and the brain in vivo (Fig. 2F).
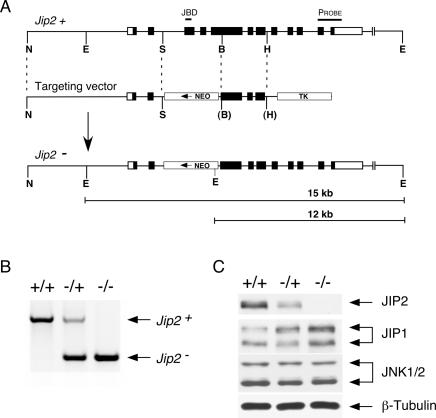
Figure 1.
Targeted disruption of the murine Jip2 gene. (A) The structures of the wild-type Jip2 genomic locus, the targeting vector, and the disrupted Jip2 gene are schematically illustrated. Exons are represented with boxes; noncoding regions (white) and coding regions (black) are indicated. The region that encodes the JNK-binding domain (JBD), the probe used for Southern blot analysis, the NeoR and TK cassettes, and some restriction sites (Not1 [N], EcoR1 [E], Spe1 [S], BamH1 [B], and HindIII [H]) are indicated. (B) Identification of JIP2-deficient mice. Wild-type (+/+), heterozygous (−/+), and homozygous (−/−) knockout mice were identified using PCR analysis of genomic tail DNA. The Jip2−/− mice were slightly smaller than wild-type littermates. The male Jip2−/− mice were found to be infertile. (C) Brain lysates prepared from Jip2+/+, Jip2−/+, and Jip2−/− mice were examined by immunoblot analysis using antibodies to JIP1, JIP2, JNK1/2, and β-Tubulin.
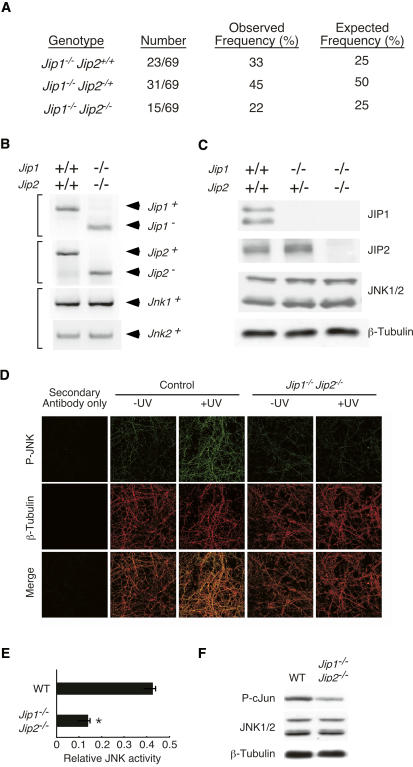
Figure 2.
Creation of Jip1 and Jip2 compound mutant mice. (A) A cohort of 69 animals obtained by mating of Jip1−/− Jip2−/+ mice was examined by genotype analysis. For each genotype group, the number, frequency of mice, and expected frequency based on Mendelian inheritance are presented. The JIP1/2-deficient mice were smaller than littermate control mice, exhibited severe ataxia, and did not survive after weaning. (B) Genotype analysis of wild-type and compound mutant Jip1−/− Jip2−/− mice was performed using PCR primers to examine the Jip1, Jip2, Jnk1, and Jnk2 genes. (C) Protein extracts prepared from the brains of wild-type mice, Jip1−/− Jip2−/+ mice, and Jip1−/− Jip2−/− mice were examined by immunoblot analysis using antibodies to JIP1, JIP2, JNK1/2, and β-Tubulin. (D,E) Primary cortical neurons were exposed to UV radiation (60 J/m2) and incubated for 45 min. The neurons were stained with antibodies to phospho-JNK (green) and β-Tubulin (red). (Bottom panels) Merged images of phospho-JNK and β-Tubulin are shown. (Left panels) Background staining was evaluated in studies of neurons stained without the primary antibody. (D) Representative immunofluorescence images are presented. JNK activation was quantitated by measurement of the ratio of fluorescence intensity of phospho-JNK and β-Tubulin staining (mean ± SD; n = 10). (E) A statistically significant (P < 0.05) reduction of UV-stimulated JNK activation in JIP1/2-deficient neurons compared with wild-type neurons was detected. (F) Protein extracts were prepared from the cerebellum of wild-type mice and Jip1−/− Jip2−/− mice. (E, top panel) JNK activity was measured in an in vitro protein kinase assay using [γ-32P]ATP and c-Jun as substrates. (Bottom panels) Immunoblot analysis confirmed that these lysates contain equal amounts of JNK1/2 and β-Tubulin.
The JIP1/2-deficient mice were severely ataxic. Altered cerebellar function is a frequent cause of ataxia (Grusser-Cornehls and Baurle 2001). We therefore examined the cerebellum of wild-type and JIP1/2-deficient mice. This analysis demonstrated that the cerebellar morphology of JIP1/2-deficient mice exhibited the expected foliation and localization of Purkinje cells. However, defects in Purkinje cell dendritic arborization were found in the JIP1/2-deficient mice (Supplementary Fig. S1A,B). In contrast, no defects in the morphology of cortical or hippocampal neurons were detected (Supplementary Fig. S1B).
The Purkinje cell defect in dendritic arborization observed in JIP1/2-deficient mice could be cell autonomous; alternatively, this defect may be a consequence of decreased neurotrophin signaling or decreased synaptic activity (Sotelo 2004). Purkinje cells are an important source of mitogen (sonic hedgehog) that is required for the proliferation of cerebellar granule neurons (CGN) (Sotelo 2004). We therefore anticipated that JIP1/2-deficient mice may exhibit defects in CGN proliferation. However, examination of CGN in the external granular layer demonstrated no defects in proliferation (Supplementary Fig. S1A). Nevertheless, defects in CGN, which make mutiple synaptic connections to Purkinje cells, could contribute to the altered Purkinje cell dendritic arborization observed in JIP1/2-deficient mice.
Effect of JIP deficiency on kinesin cargo localization
We examined primary CGN isolated from wild-type and JIP1/2-deficient mice. No differences in morphology between wild-type and mutant CGN were detected (Figs. 3, 4; Supplementary Fig. S2). The JIP1 and JIP2 proteins can bind the light chain of kinesin-1, and this interaction results in microtubule-directed transport of JIP proteins to growth cones in cultured neurons (Verhey et al. 2001; Whitmarsh et al. 2001). It is therefore possible that JIP1/2 may function as adapter proteins that recruit cargo proteins to kinesin-1. Potential cargoes include the low-density lipoprotein receptor-related protein LRP8 (Verhey et al. 2001). However, no defect in the localization of LRP8 to neuronal growth cones was detected (Fig. 3A). LRP8 is known to localize to the apical surface of epithelial cells in the epididymus, a tissue that expresses JIP2 but not JIP1 (Stockinger et al. 2000). We therefore examined LRP8 localization in the epididymus of wild-type and JIP2-deficient mice. No defect in the apical localization of LRP8 in the epididymal epithelial cells of JIP-deficient mice was detected (Fig. 3B). These data suggest that JIP1/2 may play either no role, or a redundant role, as adapter proteins in kinesin-mediated subcellular localization of LRP8.
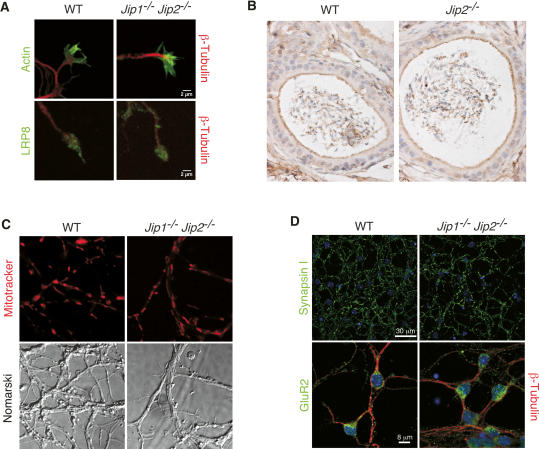
Figure 3.
Effect of JIP deficiency on cargo localization. (A) Immunofluorescence microscopy of primary neurons prepared from wild-type (WT) and Jip1−/− Jip2−/− mice demonstrated a similar localization of LRP8 in growth cones. Representative images are presented of the growth cones of primary cortical neurons stained with phalloidin (filamentous actin, green) and an antibody to β-Tubulin (red) (top panels) or with antibodies to LRP8 (green) and β-Tubulin (red) (bottom panels). (B) Sections of the epididymis were examined by staining with an antibody to LRP8 (brown). No difference in LRP8 localization between wild-type and Jip2−/− mice was detected. (C) The mitochondria of wild-type and Jip1−/− Jip2−/− CGN were labeled with a dye (Mitotracker), and a confocal microscope was used to acquire fluorescence (top panels) and Nomarski (bottom panels) images. No difference in the localization of mitochondria between wild-type and JIP1/2-deficient CGN was detected. (D) Wild-type and Jip1−/− Jip2−/− CGN were stained with an antibody to synapsin I (top panels) or an antibody to GluR2 plus β-Tubulin (bottom panels). Nuclear DNA was stained with DAPI (blue).
Figure 4.
Defective NMDA responses in Jip1/2 compound mutant mice. (A,B) Phase contrast images of primary cultures of wild-type and Jip1−/− Jip2−/− CGN are illustrated in A. (B) The CGN were labeled with calcein and imaged by fluorescence microscopy. (C,D) CGN labeled with the calcium indicator dye Fluo-4 AM were treated with NMDA (50 μM), and cytoplasmic calcium was examined by fluorescence microscopy. (C) The images presented represent cells prior to NMDA treatment (control) and after treatment with NMDA. (D) The intracellular Ca++ concentration is presented as the mean ± SD (n = 10), and statistically significant differences (P < 0.05) are indicated with an asterisk. (E) The effect of treatment of wild-type CGN with the selective NMDA receptor antagonist APV (12.5 μM) was examined. The maximum NMDA-induced Ca++ response was examined using the indicator dye Fluo-4 AM (mean ± SD; n = 10). Statistically significant differences (P < 0.01) are indicated with an asterisk. (F) The effect of KCl-mediated depolarization on the Ca++ response of wild-type and JIP1/2-deficient CGN was examined. The CGN were treated with KCl (70 mM final concentration), and the maximum Ca++ response was examined using the indicator dye Fluo-4 AM (mean ± SD; n = 10). No statistically significant differences (P < 0.05) were detected. (G,H) NMDA-regulated gene expression was examined in primary cultures of CGN prepared from wild-type and compound mutant Jip1−/− Jip2−/− mice. The neurons were treated with NMDA (100 μM) for the indicated times, and RNA was isolated. The expression of c-Fos and BDNF mRNA was examined by quantitative PCR (TaqMan assay) and is presented as the mean ± SD (n = 3). Statistically significant differences are indicated with asterisks (P < 0.01).
The JIP1/2 scaffold proteins have also been implicated as adapter molecules that mediate kinesin-mediated localization of organelles, including mitochondria and synaptic vesicles (Horiuchi et al. 2005). However, no defects in the subcellular distribution of mitochondria or the synaptic vesicle marker Synapsin I were observed in JIP1/2-deficient CGN (Fig. 3C,D). JIP1 and JIP2 therefore do not play a nonredundant role in determining the subcellular localization of mitochondria or synaptic vesicles. Control studies demonstrated that JIP1/2-deficiency also did not affect the subcellular localization of the glutamate receptor GluR2 (Fig. 3D).
JIP-deficient neurons exhibit defects in NMDA receptor signaling
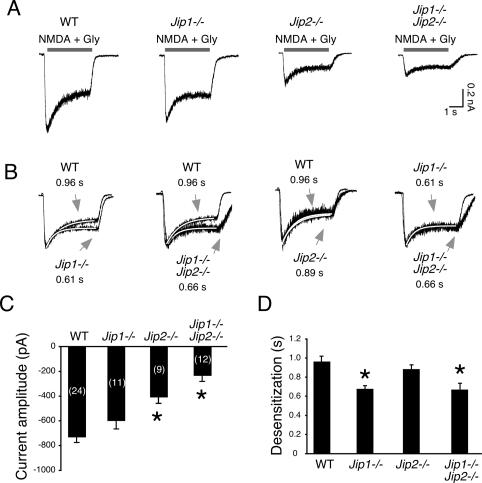
To test the function of JIP1/2-deficient CGN, we examined the response of these neurons to the glutamate receptor agonist NMDA. Treatment with NMDA caused an increase in the cytoplasmic concentration of Ca++ in wild-type neurons, but not in JIP1/2-deficient neurons (Fig. 4C,D). We confirmed that this Ca++ response was initiated by the NMDA receptor by demonstrating that the selective NMDA receptor antagonist D(-)-2-amino-5-phosphonopentanoic acid (APV) strongly suppressed the NMDA-induced Ca++ response (Fig. 4E). The reduced NMDA-induced Ca++ response of JIP1/2-deficient neurons (Fig. 4C,D) may reflect a defect in NMDA receptor signaling or may be caused by a more generalized defect in Ca++ homeostasis. To distinguish between these possibilities, we examined the effect of depolarization caused by addition of KCl to the culture medium. These studies indicated that KCl-induced depolarization caused similar increases in cytoplasmic Ca++ in both wild-type and JIP1/2-deficient neurons (Fig. 4F). These data indicate that JIP1/2-deficient neurons may have defects in NMDA receptor signaling. Indeed, the mutant neurons exhibited severely reduced NMDA-induced gene expression. Thus, NMDA increased the expression of brain-derived neurotrophic factor (BDNF) and c-Fos mRNA in wild-type neurons, but this response was suppressed in JIP1/2-deficient neurons (Fig. 4G,H). These data demonstrate that JIP1/2-deficient neurons may exhibit a major defect in signal transduction mediated by NMDA receptors. To confirm this conclusion, we performed electrophysiological analysis of NMDA receptor function in wild-type and JIP-deficient neurons. NMDA-induced whole-cell currents in JIP1/2-deficient CGN exhibited markedly decreased current amplitude and an increased rate of current decay (desensitization) compared with wild-type neurons (Fig. 5). Analysis of neurons isolated from mice lacking expression of single JIP isoforms demonstrated that JIP1 and JIP2 have partially redundant functions, but JIP2 primarily governs the magnitude of the initial current amplitude, and JIP1 primarily delays the subsequent decay of the NMDA-induced current (Fig. 5).
Figure 5.
Electrophysiological analysis of CGNs from JIP-deficient mice. (A) NMDA-induced whole-cell currents in primary CGN isolated from wild-type, Jip1−/−, Jip2−/−, and compound mutant Jip1−/− Jip2−/− mice are illustrated. (B) The amplitude of the NMDA-induced whole-cell current was normalized for each genotype and superimposed to compare the rate of current decay (desensitization). The rate was calculated by fitting the data to a single exponential and the time constant (in seconds) is presented. (C,D) The NMDA current amplitude (C) and the rate of desensitization (D) were quantitated (mean ± SD; n = 9–24). The current amplitude and desensitization were significantly reduced in JIP-deficient neurons compared with wild-type neurons (P < 0.0005, indicated with asterisks).
The decreased whole-cell NMDA receptor current observed in JIP1/2-deficient neurons could be the result of a defect in NMDA receptor expression. However, immunoblot analysis of the NR1, NR2A, and NR2B subunits of the NMDA receptor indicated that the JIP1/2-deficient cerebellum does not express lower levels of NMDA receptor subunits than wild-type mice (Fig. 6A). These data were confirmed by measurement of NMDA receptor subunit mRNA expression in primary cultures of CGN (Fig. 6B). Reduced NMDA receptor expression therefore does not account for the decreased NMDA receptor function observed in JIP1/2-deficient mice. However, it is possible that the cell surface location of NMDA receptors may be decreased in JIP1/2-deficient neurons compared with wild-type neurons. To test this hypothesis, we examined the cell surface expression of the obligate NMDA receptor subunit NR1 by immunofluorescence analysis. Studies of nonpermeabilized cells demonstrated that both wild-type and JIP1/2-deficient neurons expressed a similar amount of cell surface NR1 (Fig. 6C,D). The cell surface expression of Reelin was also similar in wild-type and JIP1/2-deficient neurons (Fig. 6D). Control studies using permeabilized cells indicated that the distribution of the total population of NR1 was similar in wild-type and JIP1/2-deficient neurons (Fig. 6D). However, the amount of NR1 detected by immunoblot analysis (Fig. 6A) and immunofluorescence microscopy of permeabilized neurons (Fig. 6D) of JIP1/2-deficient neurons was slightly larger than that detected in wild-type neurons. Since the cell surface expression of NR1 was not affected by JIP1/2 deficiency (Fig. 6C), these data indicate that the mutant neurons express a higher amount of intracellular NR1. Together, these data indicate that the reduced NMDA receptor function observed in JIP1/2-deficient neurons (Fig. 5) was not caused by decreased expression of NMDA receptors.
Figure 6.
NMDA receptor expression in JIP-deficient mice. (A,B) The expression of the NR1, NR2A, and NR2B subunits of the NMDA receptor in the cortex and cerebellum of wild-type (WT) and compound mutant Jip1−/− Jip2−/− (KO) mice (6 and 10 d old) was examined by immunoblot analysis, shown in A. (B) NMDA receptor subunit mRNA expression in CGN was measured by quantitative RT–PCR. The data presented represent the mean ± SD (n = 5). No statistically significant differences between the wild-type and mutant CGN were detected. The expression of NR2C, which is expressed in the cerebellum later in development (Sotelo 2004), was not detected by immunoblot analysis. β-Tubulin expression was examined as a loading control. (C,D) Wild-type and compound mutant Jip1−/− Jip2−/− CGNs were fixed and processed for immunofluorescence analysis under nonpermeabilizing conditions (D, top and bottom rows) or permeabilizing conditions (D, middle row). The localization of NR1, β-Tubulin, and Reelin was examined by confocal fluorescence microscopy. Staining with β-Tubulin was employed as a control to confirm whether or not the neurons were permeabilized. (C) The cell surface staining of NR1 was quantitated by measurement of puncta per square micrometer (mean ± SD; n = 10). No statistically significant difference between wild-type and mutant neurons was detected (P < 0.05).
JIP-deficient neurons exhibit defects in NMDA receptor tyrosine phosphorylation
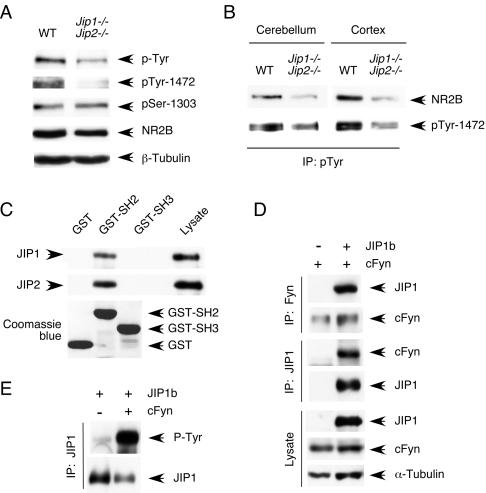
What mechanism might account for the requirement of JIP scaffold proteins for the normal function of the NMDA receptor? It is established that the NMDA receptor is subject to regulatory phosphorylation of NR1 and NR2 subunits on Ser residues by several protein kinases (including PKA, CAMK II, and CK2) (Llansola et al. 2005) and also Tyr phosphorylation of NR2 subunits mediated by members of the SRC family (Salter and Kalia 2004). The reduced NMDA receptor function in JIP1/2-deficient neurons might therefore be mediated by altered regulatory phosphorylation of the NMDA receptor. To test this hypothesis, we examined NR2B phosphorylation in wild-type and JIP1/2-deficient neurons. Immunoblot analysis demonstrated that JIP deficiency caused markedly decreased Tyr phosphorylation of NR2B (Fig. 7A,B), an NMDA receptor substrate that is established to be phosphorylated by the cFyn tyrosine kinase in vivo (Nakazawa et al. 2001). In contrast, no difference in NR2B phosphorylation on Ser1303 (Omkumar et al. 1996) was detected (Fig. 7A). Loss of tyrosine phosphorylation of NR2B is established to cause reduced NMDA receptor signaling (Salter and Kalia 2004) and may therefore contribute to the decreased NMDA receptor function observed in JIP1/2-deficient neurons (Fig. 5).
Figure 7.
JIP deficiency causes defects in NMDA receptor tyrosine phosphorylation. (A,B) The amount of NR2B tyrosine phosphorylation was examined in cerebellar extracts prepared from wild-type and compound mutant Jip1−/− Jip2−/− mice (6 d old) by immunoblot analysis of cell lysates (A) or phospho-Tyr immunoprecipitates (B) with antibodies to phospho-Tyr, phospho-Tyr-1472 NR2B, phospho-Ser1303 NR2B, and total NR2B. (C) The SH2 domain of cFyn binds to JIP proteins. Immobilized GST, GST-cFyn-SH2, and GST-cFyn-SH3 were incubated with cell lysates. Bound JIP1 and JIP2 were detected by immunoblot analysis. (D,E) Coimmunoprecitation analysis of JIP1b with cFyn. Recombinant JIP1b and cFyn were expressed in COS cells. The cell lysate and JIP1b or cFyn immunoprecipitates were examined by immunoblot analysis.
Regulatory tyrosine phosphorylation of NR2A and NR2B is largely mediated by the cFyn tyrosine kinase (Tezuka et al. 1999; Nakazawa et al. 2001). Previous studies have demonstrated that JIP proteins may act as scaffolds for components of the JNK, AKT, and SRC signaling pathways (Whitmarsh et al. 1998; Kim et al. 2003; Nihalani et al. 2007). In addition, it is established that JIP proteins are localized at post-synaptic densities in neurons (Pellet et al. 2000). Indeed, immunofluorescence analysis of CGN demonstrated the peripheral localization of JIP proteins (Supplementary Fig. S3). Thus, JIP proteins, like other kinase scaffolds (Smith et al. 2006), can form multifunctional complexes. We therefore tested whether JIP proteins might also interact with cFyn. Indeed, JIP1 was tyrosine-phosphorylated and cFyn coimmunoprecipitated with JIP1 (Fig. 7D,E). Deletion analysis indicated that the SH2 domain of cFyn mediates the interaction with JIP1 and JIP2 (Fig. 7C). Together, these data suggest that JIP proteins may function to recruit the cFyn tyrosine kinase to NMDA receptor complexes.
Discussion
The results of this study demonstrate that JIP scaffold proteins are important for the normal activity of NMDA receptors. It is established that JIP proteins are localized at post-synaptic densities in neurons (Pellet et al. 2000), but the mechanism that accounts for the functional interaction of JIP proteins with NMDA receptors is unclear. One possibility is that the PTB domain of JIP1 and JIP2 may contribute to this regulatory process. Indeed, several ligands for this PTB domain have been described, including the low-density lipoprotein receptor-related protein LRP8 (Stockinger et al. 2000), the Rac exchange factor Tiam1 (Buchsbaum et al. 2002), and the Ras exchange factor Ras-Grf (Buchsbaum et al. 2002). Interestingly, all three of these proteins (LRP8, Tiam1, and Ras-Grf) are known to bind NMDA receptors (Krapivinsky et al. 2003; Beffert et al. 2005; Tolias et al. 2005). These proteins may therefore recruit JIP/cFyn complexes to the NMDA receptor to regulate NR2 subunit tyrosine phosphorylation.
The interaction of LRP8 with JIP1/2 is particularly intriguing because it is established that the site of interaction of the JIP1/2 PTB domain with the cytoplasmic domain of LRP8 is encoded by exon 19 (Stockinger et al. 2000). This is an alternative spliced exon that is selectively included in Lrp8 mRNA in response to neuronal activity (Beffert et al. 2005). Thus, only exon 19-positive LRP8 can bind to the JIP1/2 PTB domain (Stockinger et al. 2000). Gene targeting studies have demonstrated that ablation of Lrp8 exon 19 results in mice that exhibit defects in NMDA receptor signaling associated with markedly decreased NR2 subunit tyrosine phosphorylation (Beffert et al. 2005). Exon 19-positive LRP8 may therefore regulate NMDA receptor activity. Interestingly, the defect in NMDA receptor signaling and NR2 subunit phosphorylation caused by loss of Lrp8 exon 19 (Beffert et al. 2005) is similar to that caused by compound mutation of JIP1/2 (Fig. 5). Together, these data suggest that the physical interaction between the JIP1/2 PTB domain and the segment of the LRP8 cytoplasmic tail encoded by Lrp8 exon 19 is functionally significant. Indeed, it is possible that JIP/cFyn complexes may mediate the effects of LRP8 on NMDA receptors. Engagement of cell surface LRP8 by its ligand Reelin may trigger this regulatory pathway to control NMDA receptor function (Beffert et al. 2005).
The postnatal role of LRP8 to regulate NMDA receptor activity requires Lrp8 exon 19 (Beffert et al. 2005). However, LRP8 also plays an important developmental role in determining the positioning of neurons in the brain (Trommsdorff et al. 1999). Mice lacking the ligand Reelin (reeler mice) or mice with targeted ablation of the Lrp8 gene exhibit marked defects in neuronal positioning (Trommsdorff et al. 1999). However, neither targeted ablation of the alternatively spliced Lrp8 exon 19 (Beffert et al. 2005) nor compound deficiency of JIP1/2 (Supplementary Fig. S1A) causes a reeler-like neuronal positioning defect during development. Thus, the JIP1/2 PTB domain interaction with the cytoplasmic tail of LRP8 does not play a role in the early developmental function of LRP8 to regulate neuronal positioning.
A critical role for cFyn in NMDA receptor regulation is established by the finding that cFyn−/− mice exhibit severely reduced tyrosine phosphorylation of NR2A (Tezuka et al. 1999) and NR2B (Nakazawa et al. 2001) and display major defects in long-term potentiation and spatial learning (Grant et al. 1992). Nevertheless, other members of the SRC family of tyrosine kinases (including Lck, Lyn, Src, and Yes) have also been implicated in the regulation of NMDA receptor function (Salter and Kalia 2004). cFyn may therefore represent only one member of a larger group of SRC family kinases that are recruited by JIP scaffold proteins. This function of JIP proteins may be coordinated with the actions of other scaffold proteins that can bind SRC family kinases and are present within post-synaptic densities (e.g., PSD-95 or RACK1) (Tezuka et al. 1999; Yaka et al. 2002). Since Jip1−/− and Jip2−/− neurons exhibit different defects in NMDA receptor function (Fig. 5), it is possible that these JIP scaffold proteins may cooperate by influencing different steps in the regulatory process. It is also possible that the structurally unrelated scaffold proteins JIP3 and JIP4 may also contribute to NMDA receptor regulation.
In conclusion, we demonstrate that JIP1/2 scaffold proteins represent novel components of the NMDA receptor regulatory network required for normal NMDA-mediated signal transduction.
Materials and methods
Mice
A 129/SvJ strain Jip2 genomic clone was isolated from a λFixII phage library (Stratagene) by hybridization analysis using a probe derived from a Jip2 cDNA. A targeting vector designed to disrupt the Jip2 gene (Fig. 1A) was constructed using standard techniques. Embryonic stem (ES) cells (strain 129SvJ) were electroporated with this vector and selected with 200 μg/mL G418 and 2 μM gangcyclovir. Jip2−/+ ES cell clones identified by Southern blot analysis were injected into C57BL/6J blastocysts to create chimeric mice that were used to obtain germline transmission of the disrupted Jip2 allele. The Jip1 (Whitmarsh et al. 2001) and Jip2 knockout mice were backcrossed to the C57BL/6J strain (Jackson Laboratories). Homozygous and compound mutant mice were obtained by breeding. Immunoblot analysis indicated that the Jip1 and Jip2 gene disruption caused loss of expression of full-length JIP1/2 proteins and did not result in the detection of JIP protein fragments (Supplementary Fig. S4). The mice were housed in a facility accredited by the American Association for Laboratory Animal Care (AALAC). The Institutional Animal Care and Use Committee (IACUC) of the University of Massachusetts approved all studies using animals.
Genotyping
Genomic DNA was examined using a PCR assay to detect the wild-type and disrupted Jip1 alleles (Whitmarsh et al. 2001). Wild-type and disrupted Jip2 alleles were detected using two different assays. First, Southern blot analysis of EcoR1 restricted genomic DNA was performed by probing with a 300-base-pair (bp) fragment of the Jip2 gene that was isolated by PCR using the primers 5′-CTTCATCACCAAACACCCGCT-3′ and 5′-GTCC TTGCTCAAGCCACAGT-3′. The wild-type and disrupted Jip2 alleles were detected as 15-kb and 12-kb bands, respectively. Second, a PCR assay was employed using the primers 5′-GTC CCACAAGCTCCGATCCA-3′, 5′-CCTGCATCCTCAGAGT CCGA-3′, and 5′-GTAGAAGGTGGCGCGAAGGG-3′. The wild-type and disrupted Jip2 alleles were amplified to yield 626-bp and 288-bp DNA fragments, respectively.
Morphology
Histology was performed using 7-μm sections prepared from tissue fixed in 10% formalin for 24 h, dehydrated, and embedded in paraffin. Immunohistochemistry was performed by staining tissue sections with the following antibodies: LRP8 (provided by J. Nimpf, Medical University of Vienna, Vienna, Austria), calbindin D (Sigma), GFAP (Chemicon), and BrdU (BioGenex). Immunocomplexes were detected using a biotinylated secondary antibody (Biogenex), streptavidin-conjugated horseradish peroxidase (Biogenex), and the substrate 3,3′-diaminobenzidene (Vector Laboratories), followed by brief counterstaining with Mayer’s hematoxylin (Sigma). Rapid Golgi staining was performed with a kit (FD NeuroTechnologies) using the manufacturer’s recommendations.
Primary neurons
Embryonic day 16.5 mouse embryos were used to isolate primary cortical neurons. Briefly, cortices were placed in ice-cold Hank’s buffered saline solution containing 20 mM HEPES (pH 7.3; HBSS) and minced with a sterile razor blade. Trypsin (Invitrogen) and DNase I (Sigma) were added to a final concentration of 0.25% and 1 mg/mL, respectively, and the tissue was incubated at 37°C (20 min). The trypsin solution was removed, and the minced tissue was triturated in neurobasal media containing DNase I (1 mg/mL; Sigma) and soybean trypsin inhibitor (0.5 mg/mL; Sigma) to obtain a single-cell suspension. Cortical neurons were seeded in poly-D-lysine/laminin-coated chamber slides (Becton Dickenson) in neurobasal medium containing B27 supplements, 1% glutamine, 1% penicillin/streptomycin, and 10% fetal calf serum (Invitrogen). The following day, the neurons were washed and refed with serum-free neurobasal medium containing B27, glutamine, and antibiotics (Invitrogen). The cortical neurons were cultured for 7 d in vitro prior to analysis.
Cerebella from postnatal day 6 mouse pups were used to isolate primary CGN. Briefly, cerebella were placed in ice-cold HBSS and minced with a sterile razor blade. Trypsin and DNase I were added to a final concentration of 1% and 1 mg/mL, respectively, and the tissue was incubated at 37°C (3 min). The trypsin solution was removed, and the minced tissue was triturated in 1.0 mL of neurobasal media containing DNase I (1 mg/mL) and soybean trypsin inhibitor (0.5 mg/mL) to obtain a single-cell suspension. Dissociated neurons were centrifuged (180g, 10 min, 4°C) through a cushion of 4% bovine serum albumin (BSA) in HBSS. The cells were resuspended in neurobasal medium containing B27 supplements, 1% glutamine, 1% penicillin/streptomycin, 25 mM glucose, and 25 mM KCl, and seeded in poly-D-lysine/laminin-coated chamber slides (Becton Dickenson) or dishes (MatTek). The CGN were cultured for 7 d in vitro prior to measurement of Ca++ responses and for 12 d in vitro for other studies.
Biochemical assays
Immunoblot analysis was performed using antibodies to cFyn (Biolegends), JIP1 and JIP2 (Yasuda et al. 1999), the C terminus of JIP1/2 (Santa Cruz Biotechnology), the C-terminal SH3 domain of JIP1/2 (Zymed), JNK1/2 (Pharmingen), phospho-JNK (Cell Signaling), phosphotyrosine 4G10 (Upstate Biotechnology), Flag tag (Sigma), α-Tubulin (Sigma), β-Tubulin (Covance), NR1 (BD Pharmingen), NR2A and NR2B (Chemicon), phospho-Ser1303-NR2B (Upstate Biotechnology), phospho-Tyr1472-NR2B (Abcam), and T7 tag (Novagen). JNK activity was measured in an in vitro kinase assay using c-Jun as the substrate (Whitmarsh and Davis 2001) and was quantitated using a PhosphorImager (Molecular Dynamics). Pull-down assays were performed using bacterially expressed GST, GST-cFyn-SH2, or GST-cFyn-SH3 (provided by A. Veillette, Clinical Research Institute of Montreal, Montreal, Quebec, Canada) immobilized on GSH-Sepharose using methods described previously (Whitmarsh et al. 2001). Quantitative PCR analysis of mRNA expression was performed using TaqMan reagents and an Applied Biosystems real-time PCR machine using the manufacturer’s recommendations. Duplex PCR of the target mRNA and GAPDH mRNA in a single well was performed, and the data are presented as the normalized ratio of [target mRNA]/[GAPDH mRNA].
Transfection assays
COS7 cells (American Type Culture Collection) were cultured in DMEM supplemented with 5% fetal bovine serum, transiently transfected with expression vectors for cFyn (provided by A. Veillette), JIP1 (Whitmarsh et al. 1998), or JIP2 (Yasuda et al. 1999), and were harvested at 48 h post-transfection.
Immunofluorescence analysis
CGNs were fixed by incubation with 4% paraformaldehyde at 22°C (20 min). The cells were permeabilized with 0.1% Triton X-100 or methanol (−20°C) for 5 min and incubated in blocking buffer for 1 h (1% BSA, 2% normal goat serum in PBS). For analysis of surface expression of NR1 and Reelin, cells were fixed under nonpermeabilizing conditions by incubating with 4% paraformaldehyde and 4% sucrose for 8 min at 22°C followed by incubation (1 h) in blocking buffer. Neurons were then incubated in blocking buffer containing primary antibodies GluR2 (Chemicon), LRP8 (provided by J. Nimpf), JIP1 (Santa Cruz Biotechnology), phospho-JNK (Promega), β-Tubulin (Covance), NR1 (Chemicon), Synapsin-1 (Novus), and Reelin (Chemicon). The primary antibodies were detected by incubation with anti-mouse or anti-rabbit Ig conjugated to Alexa Fluor 488 or 633 (Molecular Probes). Filamentous actin (phalloidin), mitochondria (Mitotracker), cytoplasm (calcein), and DNA (DAPI) were detected by staining (Molecular Probes). Fluorescence was visualized using a Leica TCS SP2 confocal microscope equipped with a 405-nm diode laser. Potentially overlapping emissions were collected separately using sequential scanning. Fluorescence was quantitated using ImageJ (W.S. Rasband, 1997–2007; http://rsbinfonihgov/ij) and Image Pro Plus 5.0 (Media Cybernetics) software.
Cytoplasmic calcium
Neurons cultured in poly-D-lysine/laminin-coated glass-bottom dishes (MatTec) were labeled by incubation (15 min) in neurobasal medium supplemented with 1 μM Ca++ indicator dye Fluo-4 AM (Molecular Probes) at 37°C. The cells were washed and incubated (15 min) in neurobasal medium prior to analysis. The cells were imaged using a Leica TCS SP2 scanning confocal microscope (lex = 488 nm, lem = 500–560 nm) at 37°C with maximum continuous scan rate (512 × 512 pixels, 1.5 sec per frame). After a 10-frame baseline scan, the cells were treated with 50 μM NMDA plus 200 μM glycine or depolarized by addition of KCl (70 mM final concentration) during continuous imaging. Each experiment was terminated by addition of 10 μM 4-Bromo-A23187 (Calbiochem) to determine the maximum fluorescence intensity and then 10 mM EGTA to determine the minimum fluorescence intensity. The free intracellular calcium concentration was calculated using the equation [Ca2+]free = Kd[F − Fmin]/[Fmax − F], where Kd Fluo-4 is 345 nM (Molecular Probes), and Fmin and Fmax are the fluorescence intensities after treatment with 4-Bromo-A23187 and EGTA, respectively (Grynkiewicz et al. 1985).
Electrophysiology
We used standard whole-cell recording (Hamill et al. 1981). Patch electrodes, pulled from borosilicate capillary glass on a horizontal Brown-Flaming puller (Sutter Instruments), were fire-polished to a final resistance of 3–5 MΩ, and filled with 90 mM Cs2SO4, 10 mM CsCl, 11 mM EGTA, 10 mM HEPES, and 0.5 mM CaCl2 (pH 7.35 with CsOH; osmolarity, 290–295 mOsM). Recordings in voltage-clamp mode (−60 mV) were acquired at a sampling rate of 20 kHz and filtered at 6 kHz with an EPC-10 double amplifier (HEKA Electronik). Series resistance, between 10 and 15 MΩ, was 70% compensated. Potentials were not corrected for liquid junction potentials, estimated to be +2 mV.
Recording dishes were superfused with Mg++ and PO4-free Hank’s buffered saline (120 mM NaCl, 10 mM KCl, 2.2 mM CaCl2, 5 mM Glucose, 10 mM HEPES at pH 7.35). Drugs were applied locally by gravity at a rate of 1.5 mL/sec, using a rapid three-barrel capillary superfusion device (Warner Instrument) with the pipette tips (700 μm diameter) placed ∼200 μm from the recorded cell. The pipette assembly was attached to a motor, allowing fast lateral motions controlled by the acquisition software (PatchMaster 2.01; HEKA Electronik). A drug onset time of 20 msec for the application system was determined by measuring the change in tip potential of the recording pipette filled with intracellular solution as the perfusion was switched from a normal to a 1:2 dilution of the extracellular recording solution.
After recording a stable current, CGN were exposed four consecutive times (4 sec) to 500 μM NMDA (Sigma) plus 200 μM glycine (Sigma), with a stimulation interval of 30 sec to avoid possible current inactivation. We included 1 μM strychnine (Sigma) in control and NMDA/glycine solutions to block the possible activation of taurine receptors by glycine (Martin and Siggins 2002). NMDA currents from wild-type and knockout neurons were recorded on the same day. To ensure that differences in NMDA current amplitude between groups were not caused by neurons with a larger membrane surface area, we measured the membrane capacitance of each neuron as part of our routine acquisition protocol. Peak NMDA current amplitudes and desensitization rates averaged from four consecutive traces were analyzed with PatchMaster 2.05 (HEKA Electronik). The NMDA current was fitted with a single exponential using ∼40 iterations.
Statistical comparisons of the individual data sets employed standard ANOVA and Tukey honest significant difference tests for independent samples with unequal Ns as found in the basic statistics module of Statistica (version 7.1, Statsoft).
Acknowledgments
We thank Johannes Nimpf and Andre Veillette for providing essential reagents; Li-Huei Tsai for discussions; Stephen Jones for assistance with blastocyst injections; Dan Kilpatrick and George Gagnon for advice concerning primary neuron cultures; Linda Evangelista for ES cell culture; Tammy Barrett, Vicky Benoit, Beth Doran, Jian-Hua Liu, and Judith Reilly for expert technical assistance; and Kathy Gemme for administrative assistance. These studies were supported by a grant from the National Institutes of Health. Core facilities used by these studies were supported by the NIDDK Diabetes and Endocrinology Research Center (DK52530) at the University of Massachusetts. N.K. was the recipient of a fellowship from the Charles A. King Trust, Bank of America, Co-Trustee. R.A.F. and R.J.D. are Investigators of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1563107
References
- Beffert U., Weeber E.J., Durudas A., Qiu S., Masiulis I., Sweatt J.D., Li W.P., Adelmann G., Frotscher M., Hammer R.E., Weeber E.J., Durudas A., Qiu S., Masiulis I., Sweatt J.D., Li W.P., Adelmann G., Frotscher M., Hammer R.E., Durudas A., Qiu S., Masiulis I., Sweatt J.D., Li W.P., Adelmann G., Frotscher M., Hammer R.E., Qiu S., Masiulis I., Sweatt J.D., Li W.P., Adelmann G., Frotscher M., Hammer R.E., Masiulis I., Sweatt J.D., Li W.P., Adelmann G., Frotscher M., Hammer R.E., Sweatt J.D., Li W.P., Adelmann G., Frotscher M., Hammer R.E., Li W.P., Adelmann G., Frotscher M., Hammer R.E., Adelmann G., Frotscher M., Hammer R.E., Frotscher M., Hammer R.E., Hammer R.E., et al. Modulation of synaptic plasticity and memory by Reelin involves differential splicing of the lipoprotein receptor Apoer2. Neuron. 2005;47:567–579. doi: 10.1016/j.neuron.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Buchsbaum R.J., Connolly B.A., Feig L.A., Connolly B.A., Feig L.A., Feig L.A. Interaction of Rac exchange factors Tiam1 and Ras-GRF1 with a scaffold for the p38 mitogen-activated protein kinase cascade. Mol. Cell. Biol. 2002;22:4073–4085. doi: 10.1128/MCB.22.12.4073-4085.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickens M., Rogers J.S., Cavanagh J., Raitano A., Xia Z., Halpern J.R., Greenberg M.E., Sawyers C.L., Davis R.J., Rogers J.S., Cavanagh J., Raitano A., Xia Z., Halpern J.R., Greenberg M.E., Sawyers C.L., Davis R.J., Cavanagh J., Raitano A., Xia Z., Halpern J.R., Greenberg M.E., Sawyers C.L., Davis R.J., Raitano A., Xia Z., Halpern J.R., Greenberg M.E., Sawyers C.L., Davis R.J., Xia Z., Halpern J.R., Greenberg M.E., Sawyers C.L., Davis R.J., Halpern J.R., Greenberg M.E., Sawyers C.L., Davis R.J., Greenberg M.E., Sawyers C.L., Davis R.J., Sawyers C.L., Davis R.J., Davis R.J. A cytoplasmic inhibitor of the JNK signal transduction pathway. Science. 1997;277:693–696. doi: 10.1126/science.277.5326.693. [DOI] [PubMed] [Google Scholar]
- Grant S.G., O’Dell T.J., Karl K.A., Stein P.L., Soriano P., Kandel E.R., O’Dell T.J., Karl K.A., Stein P.L., Soriano P., Kandel E.R., Karl K.A., Stein P.L., Soriano P., Kandel E.R., Stein P.L., Soriano P., Kandel E.R., Soriano P., Kandel E.R., Kandel E.R. Impaired long-term potentiation, spatial learning, and hippocampal development in fyn mutant mice. Science. 1992;258:1903–1910. doi: 10.1126/science.1361685. [DOI] [PubMed] [Google Scholar]
- Grusser-Cornehls U., Baurle J., Baurle J. Mutant mice as a model for cerebellar ataxia. Prog. Neurobiol. 2001;63:489–540. doi: 10.1016/s0301-0082(00)00024-1. [DOI] [PubMed] [Google Scholar]
- Grynkiewicz G., Poenie M., Tsien R.Y., Poenie M., Tsien R.Y., Tsien R.Y. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- Hamill O.P., Marty A., Neher E., Sakmann B., Sigworth F.J., Marty A., Neher E., Sakmann B., Sigworth F.J., Neher E., Sakmann B., Sigworth F.J., Sakmann B., Sigworth F.J., Sigworth F.J. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Arch. 1981;391:85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- Horiuchi D., Barkus R.V., Pilling A.D., Gassman A., Saxton W.M., Barkus R.V., Pilling A.D., Gassman A., Saxton W.M., Pilling A.D., Gassman A., Saxton W.M., Gassman A., Saxton W.M., Saxton W.M. APLIP1, a kinesin binding JIP-1/JNK scaffold protein, influences the axonal transport of both vesicles and mitochondria in Drosophila. Curr. Biol. 2005;15:2137–2141. doi: 10.1016/j.cub.2005.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im J.Y., Lee K.W., Kim M.H., Lee S.H., Ha H.Y., Cho I.H., Kim D., Yu M.S., Kim J.B., Lee J.K., Lee K.W., Kim M.H., Lee S.H., Ha H.Y., Cho I.H., Kim D., Yu M.S., Kim J.B., Lee J.K., Kim M.H., Lee S.H., Ha H.Y., Cho I.H., Kim D., Yu M.S., Kim J.B., Lee J.K., Lee S.H., Ha H.Y., Cho I.H., Kim D., Yu M.S., Kim J.B., Lee J.K., Ha H.Y., Cho I.H., Kim D., Yu M.S., Kim J.B., Lee J.K., Cho I.H., Kim D., Yu M.S., Kim J.B., Lee J.K., Kim D., Yu M.S., Kim J.B., Lee J.K., Yu M.S., Kim J.B., Lee J.K., Kim J.B., Lee J.K., Lee J.K., et al. Repression of phospho-JNK and infarct volume in ischemic brain of JIP1-deficient mice. J. Neurosci. Res. 2003;74:326–332. doi: 10.1002/jnr.10761. [DOI] [PubMed] [Google Scholar]
- Jaeschke A., Czech M.P., Davis R.J., Czech M.P., Davis R.J., Davis R.J. An essential role of the JIP1 scaffold protein for JNK activation in adipose tissue. Genes & Dev. 2004;18:1976–1980. doi: 10.1101/gad.1216504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A.H., Sasaki T., Chao M.V., Sasaki T., Chao M.V., Chao M.V. JNK-interacting protein 1 promotes Akt1 activation. J. Biol. Chem. 2003;278:29830–29836. doi: 10.1074/jbc.M305349200. [DOI] [PubMed] [Google Scholar]
- Krapivinsky G., Krapivinsky L., Manasian Y., Ivanov A., Tyzio R., Pellegrino C., Ben-Ari Y., Clapham D.E., Medina I., Krapivinsky L., Manasian Y., Ivanov A., Tyzio R., Pellegrino C., Ben-Ari Y., Clapham D.E., Medina I., Manasian Y., Ivanov A., Tyzio R., Pellegrino C., Ben-Ari Y., Clapham D.E., Medina I., Ivanov A., Tyzio R., Pellegrino C., Ben-Ari Y., Clapham D.E., Medina I., Tyzio R., Pellegrino C., Ben-Ari Y., Clapham D.E., Medina I., Pellegrino C., Ben-Ari Y., Clapham D.E., Medina I., Ben-Ari Y., Clapham D.E., Medina I., Clapham D.E., Medina I., Medina I. The NMDA receptor is coupled to the ERK pathway by a direct interaction between NR2B and RasGRF1. Neuron. 2003;40:775–784. doi: 10.1016/s0896-6273(03)00645-7. [DOI] [PubMed] [Google Scholar]
- Llansola M., Sanchez-Perez A., Cauli O., Felipo V., Sanchez-Perez A., Cauli O., Felipo V., Cauli O., Felipo V., Felipo V. Modulation of NMDA receptors in the cerebellum. 1. Properties of the NMDA receptor that modulate its function. Cerebellum. 2005;4:154–161. doi: 10.1080/14734220510007996. [DOI] [PubMed] [Google Scholar]
- Martin G., Siggins G.R., Siggins G.R. Electrophysiological evidence for expression of glycine receptors in freshly isolated neurons from nucleus accumbens. J. Pharmacol. Exp. Ther. 2002;302:1135–1145. doi: 10.1124/jpet.102.033399. [DOI] [PubMed] [Google Scholar]
- Morrison D.K., Davis R.J., Davis R.J. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu. Rev. Cell Dev. Biol. 2003;19:91–118. doi: 10.1146/annurev.cellbio.19.111401.091942. [DOI] [PubMed] [Google Scholar]
- Nakazawa T., Komai S., Tezuka T., Hisatsune C., Umemori H., Semba K., Mishina M., Manabe T., Yamamoto T., Komai S., Tezuka T., Hisatsune C., Umemori H., Semba K., Mishina M., Manabe T., Yamamoto T., Tezuka T., Hisatsune C., Umemori H., Semba K., Mishina M., Manabe T., Yamamoto T., Hisatsune C., Umemori H., Semba K., Mishina M., Manabe T., Yamamoto T., Umemori H., Semba K., Mishina M., Manabe T., Yamamoto T., Semba K., Mishina M., Manabe T., Yamamoto T., Mishina M., Manabe T., Yamamoto T., Manabe T., Yamamoto T., Yamamoto T. Characterization of Fyn-mediated tyrosine phosphorylation sites on GluR ε 2 (NR2B) subunit of the N-methyl-D-aspartate receptor. J. Biol. Chem. 2001;276:693–699. doi: 10.1074/jbc.M008085200. [DOI] [PubMed] [Google Scholar]
- Nihalani D., Wong H., Verma R., Holzman L.B., Wong H., Verma R., Holzman L.B., Verma R., Holzman L.B., Holzman L.B. Src family kinases directly regulate JIP1 module dynamics and activation. Mol. Cell. Biol. 2007;27:2431–2441. doi: 10.1128/MCB.01479-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omkumar R.V., Kiely M.J., Rosenstein A.J., Min K.T., Kennedy M.B., Kiely M.J., Rosenstein A.J., Min K.T., Kennedy M.B., Rosenstein A.J., Min K.T., Kennedy M.B., Min K.T., Kennedy M.B., Kennedy M.B. Identification of a phosphorylation site for calcium/calmodulindependent protein kinase II in the NR2B subunit of the N-methyl-D-aspartate receptor. J. Biol. Chem. 1996;271:31670–31678. doi: 10.1074/jbc.271.49.31670. [DOI] [PubMed] [Google Scholar]
- Pellet J.B., Haefliger J.A., Staple J.K., Widmann C., Welker E., Hirling H., Bonny C., Nicod P., Catsicas S., Waeber G., Haefliger J.A., Staple J.K., Widmann C., Welker E., Hirling H., Bonny C., Nicod P., Catsicas S., Waeber G., Staple J.K., Widmann C., Welker E., Hirling H., Bonny C., Nicod P., Catsicas S., Waeber G., Widmann C., Welker E., Hirling H., Bonny C., Nicod P., Catsicas S., Waeber G., Welker E., Hirling H., Bonny C., Nicod P., Catsicas S., Waeber G., Hirling H., Bonny C., Nicod P., Catsicas S., Waeber G., Bonny C., Nicod P., Catsicas S., Waeber G., Nicod P., Catsicas S., Waeber G., Catsicas S., Waeber G., Waeber G., et al. Spatial, temporal and subcellular localization of islet-brain 1 (IB1), a homologue of JIP-1, in mouse brain. Eur. J. Neurosci. 2000;12:621–632. doi: 10.1046/j.1460-9568.2000.00945.x. [DOI] [PubMed] [Google Scholar]
- Salter M.W., Kalia L.V., Kalia L.V. Src kinases: A hub for NMDA receptor regulation. Nat. Rev. Neurosci. 2004;5:317–328. doi: 10.1038/nrn1368. [DOI] [PubMed] [Google Scholar]
- Smith F.D., Langeberg L.K., Scott J.D., Langeberg L.K., Scott J.D., Scott J.D. The where’s and when’s of kinase anchoring. Trends Biochem. Sci. 2006;31:316–323. doi: 10.1016/j.tibs.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Sotelo C. Cellular and genetic regulation of the development of the cerebellar system. Prog. Neurobiol. 2004;72:295–339. doi: 10.1016/j.pneurobio.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Stockinger W., Brandes C., Fasching D., Hermann M., Gotthardt M., Herz J., Schneider W.J., Nimpf J., Brandes C., Fasching D., Hermann M., Gotthardt M., Herz J., Schneider W.J., Nimpf J., Fasching D., Hermann M., Gotthardt M., Herz J., Schneider W.J., Nimpf J., Hermann M., Gotthardt M., Herz J., Schneider W.J., Nimpf J., Gotthardt M., Herz J., Schneider W.J., Nimpf J., Herz J., Schneider W.J., Nimpf J., Schneider W.J., Nimpf J., Nimpf J. The reelin receptor ApoER2 recruits JNK-interacting proteins-1 and -2. J. Biol. Chem. 2000;275:25625–25632. doi: 10.1074/jbc.M004119200. [DOI] [PubMed] [Google Scholar]
- Tezuka T., Umemori H., Akiyama T., Nakanishi S., Yamamoto T., Umemori H., Akiyama T., Nakanishi S., Yamamoto T., Akiyama T., Nakanishi S., Yamamoto T., Nakanishi S., Yamamoto T., Yamamoto T. PSD-95 promotes Fyn-mediated tyrosine phosphorylation of the N-methyl-D-aspartate receptor subunit NR2A. Proc. Natl. Acad. Sci. 1999;96:435–440. doi: 10.1073/pnas.96.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolias K.F., Bikoff J.B., Burette A., Paradis S., Harrar D., Tavazoie S., Weinberg R.J., Greenberg M.E., Bikoff J.B., Burette A., Paradis S., Harrar D., Tavazoie S., Weinberg R.J., Greenberg M.E., Burette A., Paradis S., Harrar D., Tavazoie S., Weinberg R.J., Greenberg M.E., Paradis S., Harrar D., Tavazoie S., Weinberg R.J., Greenberg M.E., Harrar D., Tavazoie S., Weinberg R.J., Greenberg M.E., Tavazoie S., Weinberg R.J., Greenberg M.E., Weinberg R.J., Greenberg M.E., Greenberg M.E. The Rac1-GEF Tiam1 couples the NMDA receptor to the activity-dependent development of dendritic arbors and spines. Neuron. 2005;45:525–538. doi: 10.1016/j.neuron.2005.01.024. [DOI] [PubMed] [Google Scholar]
- Trommsdorff M., Gotthardt M., Hiesberger T., Shelton J., Stockinger W., Nimpf J., Hammer R.E., Richardson J.A., Herz J., Gotthardt M., Hiesberger T., Shelton J., Stockinger W., Nimpf J., Hammer R.E., Richardson J.A., Herz J., Hiesberger T., Shelton J., Stockinger W., Nimpf J., Hammer R.E., Richardson J.A., Herz J., Shelton J., Stockinger W., Nimpf J., Hammer R.E., Richardson J.A., Herz J., Stockinger W., Nimpf J., Hammer R.E., Richardson J.A., Herz J., Nimpf J., Hammer R.E., Richardson J.A., Herz J., Hammer R.E., Richardson J.A., Herz J., Richardson J.A., Herz J., Herz J. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999;97:689–701. doi: 10.1016/s0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- Verhey K.J., Meyer D., Deehan R., Blenis J., Schnapp B.J., Rapoport T.A., Margolis B., Meyer D., Deehan R., Blenis J., Schnapp B.J., Rapoport T.A., Margolis B., Deehan R., Blenis J., Schnapp B.J., Rapoport T.A., Margolis B., Blenis J., Schnapp B.J., Rapoport T.A., Margolis B., Schnapp B.J., Rapoport T.A., Margolis B., Rapoport T.A., Margolis B., Margolis B. Cargo of kinesin identified as JIP scaffolding proteins and associated signaling molecules. J. Cell Biol. 2001;152:959–970. doi: 10.1083/jcb.152.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmarsh A.J., Davis R.J., Davis R.J. Analyzing JNK and p38 mitogen-activated protein kinase activity. Methods Enzymol. 2001;332:319–336. doi: 10.1016/s0076-6879(01)32212-7. [DOI] [PubMed] [Google Scholar]
- Whitmarsh A.J., Cavanagh J., Tournier C., Yasuda J., Davis R.J., Cavanagh J., Tournier C., Yasuda J., Davis R.J., Tournier C., Yasuda J., Davis R.J., Yasuda J., Davis R.J., Davis R.J. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science. 1998;281:1671–1674. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- Whitmarsh A.J., Kuan C.Y., Kennedy N.J., Kelkar N., Haydar T.F., Mordes J.P., Appel M., Rossini A.A., Jones S.N., Flavell R.A., Kuan C.Y., Kennedy N.J., Kelkar N., Haydar T.F., Mordes J.P., Appel M., Rossini A.A., Jones S.N., Flavell R.A., Kennedy N.J., Kelkar N., Haydar T.F., Mordes J.P., Appel M., Rossini A.A., Jones S.N., Flavell R.A., Kelkar N., Haydar T.F., Mordes J.P., Appel M., Rossini A.A., Jones S.N., Flavell R.A., Haydar T.F., Mordes J.P., Appel M., Rossini A.A., Jones S.N., Flavell R.A., Mordes J.P., Appel M., Rossini A.A., Jones S.N., Flavell R.A., Appel M., Rossini A.A., Jones S.N., Flavell R.A., Rossini A.A., Jones S.N., Flavell R.A., Jones S.N., Flavell R.A., Flavell R.A., et al. Requirement of the JIP1 scaffold protein for stress-induced JNK activation. Genes & Dev. 2001;15:2421–2432. doi: 10.1101/gad.922801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaka R., Thornton C., Vagts A.J., Phamluong K., Bonci A., Ron D., Thornton C., Vagts A.J., Phamluong K., Bonci A., Ron D., Vagts A.J., Phamluong K., Bonci A., Ron D., Phamluong K., Bonci A., Ron D., Bonci A., Ron D., Ron D. NMDA receptor function is regulated by the inhibitory scaffolding protein, RACK1. Proc. Natl. Acad. Sci. 2002;99:5710–5715. doi: 10.1073/pnas.062046299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda J., Whitmarsh A.J., Cavanagh J., Sharma M., Davis R.J., Whitmarsh A.J., Cavanagh J., Sharma M., Davis R.J., Cavanagh J., Sharma M., Davis R.J., Sharma M., Davis R.J., Davis R.J. The JIP group of mitogen-activated protein kinase scaffold proteins. Mol. Cell. Biol. 1999;19:7245–7254. doi: 10.1128/mcb.19.10.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]