Abstract
Many neuronal disorders such as lissencephaly, epilepsy, and schizophrenia are caused by the abnormal migration of neurons in the developing brain. The role of the actin cytoskeleton in neuronal migration disorders has in large part remained elusive. Here we show that the F-actin depolymerizing factor n-cofilin controls cell migration and cell cycle progression in the cerebral cortex. Loss of n-cofilin impairs radial migration, resulting in the lack of intermediate cortical layers. Neuronal progenitors in the ventricular zone show increased cell cycle exit and exaggerated neuronal differentiation, leading to the depletion of the neuronal progenitor pool. These results demonstrate that mutations affecting regulators of the actin cytoskeleton contribute to the pathology of cortex development.
Keywords: Cortex development, cofilins, actin cytoskeleton, neuronal migration disorders, cell cycle
Various neuronal diseases such as mental retardation, lissencephaly, epilepsy, and schizophrenia have been linked to neuronal migration defects during development (Ayala et al. 2007). The cerebral cortex is a particularly demanding structure because of the intricate pattern and interplay of cell proliferation and neuronal precursor migration that is required during formation of the cortical layers. While the etiology of the cortical disorder lissencephaly (LIS1) has been studied extensively (Reiner et al. 1993; Feng et al. 2000), the underlying mechanisms of a large number of “neuronal migration disorders” remain elusive.
The actin cytoskeleton is important in the orientation of the cleavage plane (Cowan and Hyman 2004), chromosome congression (Lenart et al. 2005), and cleavage furrow formation (Fishkind and Wang 1993) in many cell types. Furthermore, a role of actin in symmetric–asymmetric cell division, a key event in neuronal cell fate determination, has been proposed (McConnell and Kaznowski 1991; Gotz and Huttner 2005). Considering the importance of actin in growth cone protrusion, neurite extension, and tail retraction, it is surprising that the genetic evidence for a role of actin in “neuronal migration disorders” is rather sparse (Fox et al. 1998; Cappello et al. 2006). Studies in which actin polymerization was completely inhibited by cytochalasin B suggested that control of actin filament length is critical in cortical neuron migration (Rivas and Hatten 1995). In vivo, actin filament length can be regulated by members of the cofilin family of actin filament depolymerizing factors. In neurons and other cell types the cofilins have been shown to regulate migration and chemotaxis (Meberg and Bamburg 2000; Gungabissoon and Bamburg 2003; Ghosh et al. 2004). In humans and the mouse, three highly conserved cofilin genes were identified, and owing to their activity and expression pattern they were named n-cofilin (nonmuscle cofilin), m-cofilin (muscle cofilin), and ADF (actin depolymerizing factor). On the amino acid level the three cofilins are 70%–82% identical and, together with the common genomic organization in three exons, the identical intron localization (Gurniak et al. 2005), and the comparable biochemical properties (Vartiainen et al. 2002), it is justified to consider the three cofilins as members of the same gene family.
Complete deletion of n-cofilin in the mouse leads to impaired neural crest cell migration and a neural tube closure defect (Gurniak et al. 2005). Recent single-nucleotide polymorphism (SNP) studies revealed that the human n-cofilin gene is a genetic modifier for “spina bifida” risk (Zhu et al. 2007), supporting the notion that variations in the human n-cofilin gene can contribute to neuronal disorders.
We generated mouse mutants for ADF and conditional mutants for n-cofilin that allowed us to study the role of actin remodeling in the development of the brain. Here we show for the first time that mutations affecting actin filament formation can contribute to cell cycle and neuronal migration defects in the cerebral cortex that are reminiscent of symptoms in lissencephaly.
Results
N-cofilin and ADF have distinct roles in brain development
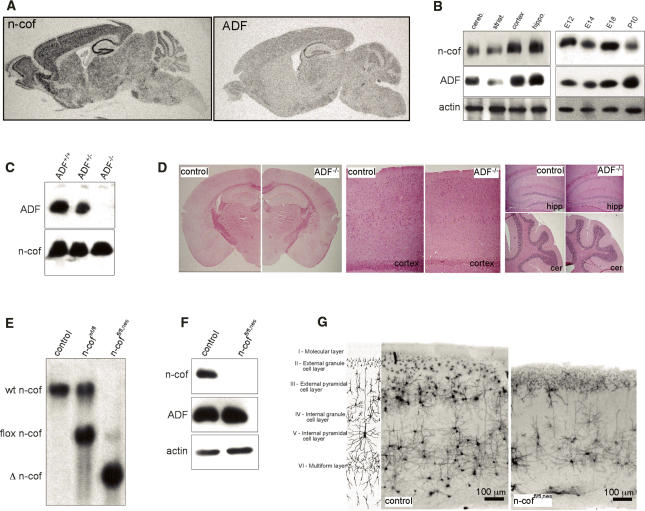
Of the three cofilin genes found in human and mouse, m-cofilin is considered to be the muscle isoform (Ono et al. 1994), while n-cofilin (Matsuzaki et al. 1988) and ADF (Adams et al. 1990) were both shown to be coexpressed in the brain (Meberg et al. 1998). The activity of all three cofilins can be regulated by phosphorylation of Ser3 (Abe et al. 1996). Our expression studies are consistent with the idea that n-cofilin and ADF could have overlapping functions (Hotulainen et al. 2005). In situ hybridization showed comparable pattern of n-cofilin and ADF expression in the adult brain, although the signal was weaker for ADF (Fig. 1A). The expression pattern throughout the cortex, hippocampus, and cerebellum was confirmed on the protein level by Western blot analysis of dissected brain regions (Fig. 1B, left panel). Protein levels in total brain lysates of different stage embryos further showed that n-cofilin and ADF are coexpressed throughout embryonic brain development (Fig. 1B, right panel). However, it should be mentioned that, depending on the developmental stage, we estimated the absolute amount of n-cofilin protein in the brain to be six to 10 times higher than ADF, which is in agreement with previous studies by Minamide et al. (2000).
Figure 1.
Distinct roles of n-cofilin and ADF in cerebral cortex formation. (A) Expression of n-cofilin and ADF shown by radioactive in situ hybridization. The expression patterns of n-cofilin and ADF are comparable, although the ADF signal is weaker. (B, left panel) Protein levels of n-cofilin and ADF in different brain regions (cerebellum, striatum, cortex, hippocampus). (Right panel) Total brain lysates from E12 to postnatal day 10 show coexpression of n-cofilin and ADF in the developing brain. Actin serves as a loading control. (C) ADF is absent from the brain of ADF−/− mice. Western blots of total brain lysates were probed for ADF and n-cofilin. (D) Brain development and morphology is not affected in ADF−/− mice. Nissl staining confirmed normal organization of the cortex, hippocampus (hipp), and cerebellum (cer). (E) Southern blot analysis shows efficient deletion (Δ n-cof) of the conditional n-cofilin allele (flox n-cof, referred to as “fl”) in homozygous conditional mutants carrying the nestin-cre transgene (n-coffl/fl,nes). (F) N-cofilin is absent in the brain of n-coffl/fl,nes mice. Western blot of total brain extracts from postnatal n-coffl/fl,nes mice. ADF levels are not altered. (G) Golgi staining of coronal sections from the cortex of control and n-coffl/fl,nes mice. Cortical layers II–IV and parts of layer V are missing in mutant brains. The cartoon illustrates regular cortical layer organization in the mammalian brain.
Although n-cofilin and ADF are coexpressed, the knockout models that we generated showed distinct roles of both proteins in brain development.
ADF-null mutants were generated by conventional gene targeting (Supplementary Fig. 1A) and homozygous mutants (ADF−/−) were lacking ADF protein in the brain (Fig. 1C). No compensatory up-regulation of n-cofilin was seen in ADF−/− brains. ADF−/− mice were born at the expected ratios and were indistinguishable from control littermates with the exception of a mild hyperplasia of the cornea, similar to what has been observed in a natural ADF mouse mutant (Ikeda et al. 2003). Histological analysis of ADF−/− brains showed no gross alterations of morphology (Fig. 1D), and obvious changes in behavior were not noticeable. From these results we concluded that ADF is dispensable for brain development, or that its role is redundant with n-cofilin. We therefore wanted to address the role of n-cofilin in the brain.
Complete deletion of n-cofilin resulted in an embryonic-lethal phenotype (Gurniak et al. 2005). In order to study n-cofilin in the brain, we therefore generated a conditional knockout of n-cofilin (flox n-cof, or “fl”) using nestin-cre-mediated deletion (Fig. 1E; Supplementary Fig. 1B). Nestin-cre-mediated deletion was shown to occur in all neuronal cell types of the brain starting around embryonic day 10.5 (E10.5) (Tronche et al. 1999). N-cofilin deletion was >95% as judged by Southern blotting and Western blot analysis of postnatal brain extracts (Fig. 1E,F). Unlike the ADF mutants, >90% of homozygous n-cofilin mutants (n-coffl/fl,nes) died within postnatal days 1–3, and few escapees survived up to 30 d with growth retardation, ataxia, and signs of epileptic seizures. The frequency of n-coffl/fl,nes pups obtained at birth suggested no significant loss during embryonic development (data not shown).
N-cofilin is essential for neuronal progenitor migration and layer formation in the cerebral cortex
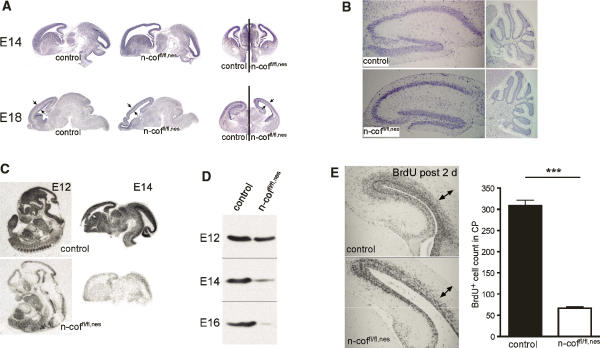
Gross analysis of mutant brains revealed that an overall normal organization at E14 and E18 (Fig. 2A), and in postnatal brain (P20) structures such as the hippocampus and the cerebellum were comparable in mutants and control littermates (Fig. 2B). However, the cerebral cortex had a translucent appearance and enlarged ventricles, suggesting a defect in cortical plate (CP) development. This was confirmed by Golgi staining of coronal sections that revealed that layers II, III, and IV, and large parts of V were missing in n-coffl/fl,nes mice, while the upper molecular layer I and the inner multiform layer VI seemed to be present (Fig. 1G). Such defects in layer formation and enlarged ventricles (Fig. 2A) resemble the pathology described in type 1 lissencephaly (LIS1) patients (Couillard-Despres et al. 2001).
Figure 2.
Radial migration in the cortex is impaired. (A) Brain anatomy in n-coffl/fl,nes embryos at E14 and E18 shows enlarged ventricles and reduced thickness of the CP (see arrows). (B) In postnatal mutant brains (P20) the structure of the hippocampus and cerebellum is comparable with control littermates. (C) Specific deletion of n-cofilin mRNA in the developing brain is shown by in situ hybridization of E12 whole-embryo sections and E14 brain sections. (D) N-cofilin protein levels in total brain lysates of E12, E14, and E16 n-coffl/fl,nes embryos. At E12, n-cofilin levels start decreasing, while at E14–E16, n-cofilin is efficiently deleted. (E) BrdU pulse labeling of E16 embryos and analysis at E18 indicates a cell migration defect in the CP (see arrows). The number of BrdU+ cells translocating into the CP is reduced about sixfold in n-coffl/fl,nes embryos (P < 0.0001).
In all mammals the layering of the cerebral cortex results from a complex sequence and pattern of cell cycle progression in the ventricular zone (VZ), and migration of neuronal precursor cells from there toward the pial surface (Rakic 1982; Nadarajah and Parnavelas 2002). Early in development the first post-mitotic neurons leave the germinal VZ to form the preplate, which is then split by infiltrating CP neurons into an outer marginal zone (MZ) and the inner subplate (SP). The MZ (layer I) and SP (layer VI) are early structures, while the intermediate layers, II–V, are formed between E13 and E19 in an “inside-out” fashion. Neurons born in the deepest layers migrate past the existing layers to form the next outermost layer (Rakic 1978).
The time course of n-cofilin depletion in mutant mice coincides well with the time frame of layer II–V formation and also explains the presence of the earlier-forming layers, I and VI, in n-coffl/fl,nes mice. Reduction of n-cofilin mRNA specifically in the brain was seen at E12 and E14 by in situ hybridization (Fig. 2C). Western blot analysis of E12–E16 brain extracts showed residual n-cofilin at E12 due to the long half-life of the protein, significant reduction of n-cofilin at E14, and almost complete absence at E16 (Fig. 2D). This suggested that a migration defect in neuronal progenitor cells born in the VZ between E13 and E19 could account for the missing intermediate layers.
To support this hypothesis, we performed BrdU pulse labeling of embryos at E16 and analyzed the brains at E18. Thereby, mitotically active cells are marked in the VZ and can then be traced on their trajectory through the CP. In control brains, BrdU-positive neurons moved to the upper cortical layers (Fig. 2E), while in n-coffl/fl,nes embryos only very few BrdU-positive cells were found in the CP, showing that cells are unable to translocate from the VZ to the outer layers.
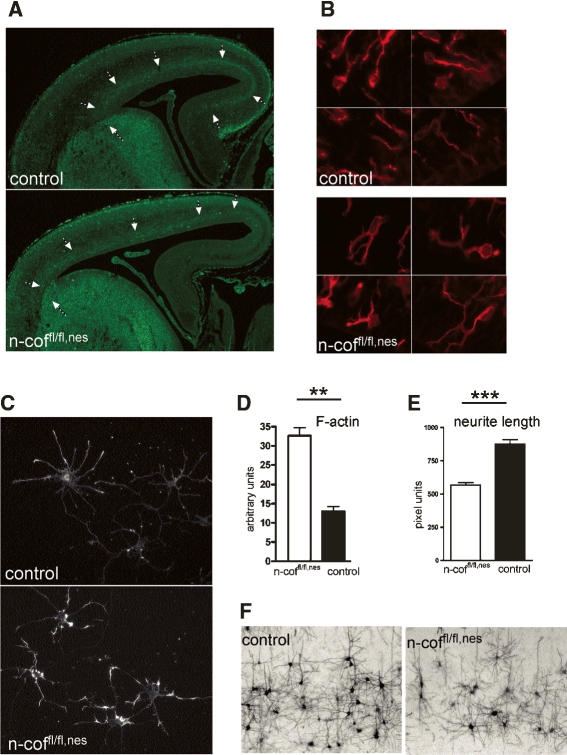
Apart from this radial migration in the cortex, which is considered essential for setting up the layering, a different mode of migration usually referred to as tangential migration also plays an important role in the developing cortex. There is compelling evidence that GABA-producing interneurons reach the cortex in tangentially migrating streams (Anderson et al. 1997). Interneurons originate in the medial and lateral ganglionic eminence and then migrate long distances into the cortex and hippocampus. In the mouse, early tangential migration begins around E11 and peaks around E13.5, a time point when n-cofilin is still detectable in n-coffl/fl,nes brains. Using antibodies for VIAAT (vesicular inhibitory amino acid transporter), we followed the tangential path of GABAerigic neurons in the cortex. Interestingly, tangential migration of neuronal precursors out of the ganglionic eminence into the cortex partially occurred in n-coffl/fl,nes embryos; however, the medial pallium was not reached in mutants (Fig. 3A, arrows). This observation supports the critical role of n-cofilin in cortical neuron migration, but also might explain why the gross morphology of n-coffl/fl,nes brains and structures such as the hippocampus and the cerebellum are intact (see Fig. 2B).
Figure 3.
Tangential migration, cell shape, and actin remodeling in n-coffl/fl,nes neurons. (A) Tangential migration is partially impaired in the cortex of E16 n-cofilin mutants. In control embryos, GABAergic neurons labeled with an anti-VIAAT antibody emerge from the ganglionic eminence and migrate tangentially through the cortex. In n-coffl/fl,nes embryos, tangential migration through the cortex occurs, but GABAergic neurons do not reach the medial pallium (see arrows). (B) Cell shape of differentiating neurons (βIII-tubulin positive) in the VZ of n-coffl/fl,nes embryos is characterized by multiple cell protrusions, while in control embryos most migrating neurons have acquired an elongated bipolar shape. Four different representative fields from the VZ of mutant and control embryos (E16) are shown. (C) Cultured cortical neurons from control and n-coffl/fl,nes embryos were stained with phalloidin for F-actin after 3 d in culture. (D) In mutant cells, F-actin is increased about twofold (P = 0.0012). (E) Reduced average neurite length and outgrowth defects in n-coffl/fl,nes mice (P < 0.0001). (F) In vivo connectivity and outgrowth of pyramidal neurons is impaired, as seen in the cortex of postnatal n-coffl/fl,nes mice. Neurons are visualized by silver staining.
In n-cofilin-null neurons, actin filament turnover and neurite outgrowth are impaired
Radial migration and layer formation are affected in n-coffl/fl,nes embryos, and our results suggest that impaired remodeling of the actin cytoskeleton and cell shape contributes to these defects. We noticed that the morphology of differentiating neurons (βIII-tubulin-positive cells) in the VZ of n-coffl/fl,nes embryos was different (for an overview, see Fig. 5D, below), and a detailed analysis showed that the majority of cells in mutants had multiple cell protrusions, while in control embryos the migrating neurons in the VZ were characterized by an elongated bipolar shape (Fig. 3B). This suggests that neurons lacking n-cofilin are impaired in the transition from a multipolar to a bipolar migrating cell or that cell shape is disturbed per se. Further studies on cultured cortical neurons confirmed the autonomous role of n-cofilin in controlling shape and motility. F-actin heavily accumulated in mutant neurons (Fig. 3C), and quantitative measurements revealed a more than twofold increase in filamentous actin (Fig. 3D). Assuming that in the brain under normal conditions ∼40% of cellular actin is F-actin (Pilo Boyl et al. 2007), this finding suggests that in mutant neurons most cellular G-actin is converted to F-actin. Motile responses such as neurite outgrowth are thus impaired in cultured n-coffl/fl,nes neurons (Fig. 3E; Supplementary Fig. 2A), and also in situ the length of neuronal processes and connectivity of pyramidal cells was diminished (Fig. 3F). These data show that n-cofilin is essential in regulating remodeling of the actin cytoskeleton in neurons, consistent with previous reports showing the importance of n-cofilin in neural crest cell migration (Gurniak et al. 2005), chemotaxis (Ghosh et al. 2004), and neurite outgrowth (Meberg and Bamburg 2000).
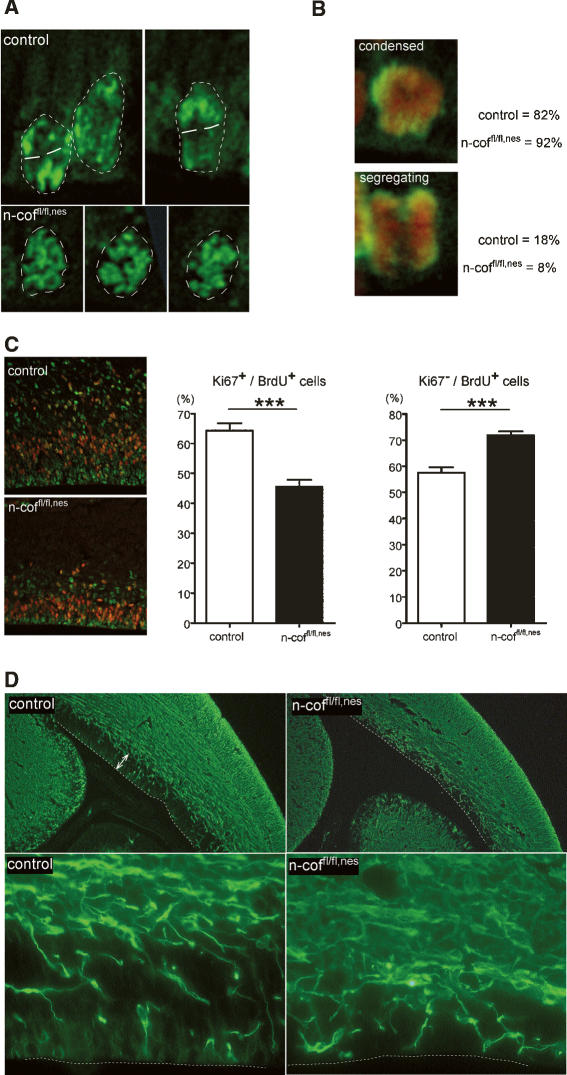
Figure 5.
Chromosome segregation is impaired and cell cycle exit is increased in the VZ of n-coffl/fl,nes embryos. (A) pH3 staining of dividing cells visualizes chromosome organization. Nuclei are marked by a dashed line. Control nuclei showed polarized chromosome figures with a defined cleavage plane (white lines), while in n-coffl/fl,nes nuclei chromosomes showed a mostly random organization. (B) Mitotic figures were classified according to the images shown, and “condensed” chromatin versus “segregating” figures were scored (pH3 in green, DNA in red). Eighteen percent of control cells, but only 8% of mutant cells, showed segregating chromosomes. (C) Cell cycle profiles of progenitor cells in the VZ. After pulse labeling with BrdU at E16, brain sections were double-stained for Ki67 (red) and BrdU (green). The numbers of Ki67+/BrdU+ and Ki67−/BrdU+ cells are presented as percentages of BrdU+ cells (see histograms). In mutants, fewer cells enter the cell cycle, as indicated by the smaller Ki67+/BrdU+ population (P < 0.0001), while more cells exit the cell cycle as shown by the increased Ki67−/BrdU+ population (P = 0.004). (D) Neuronal differentiation is increased in the VZ of n-coffl/fl,nes mice. E16 brains were stained using a neuronal differentiation marker (βIII-tubulin). The VZ in control embryos (white arrows) contains few differentiating neurons, while in n-coffl/fl,nes brains the VZ is depleted of progenitors and is instead populated with differentiated βIII-tubulin-positive neurons. For better orientation, the ventricle border is marked by a white dashed line.
Interestingly, the activity of the n-cofilin homolog ADF is neither sufficient nor essential to drive actin filament turnover in neurons. This is a surprising and unexpected result, since n-cofilin and ADF show ∼83% homology (73% identity) on the amino acid level and both share G-actin binding and F-actin depolymerizing activities. Furthermore, ADF was shown to be about four times more potent in depolymerizing F-actin (Yeoh et al. 2002), which should allow compensation despite the lower levels of ADF protein in neurons. Also, deletion of both ADF alleles and one additional n-cofilin allele in the same mouse (ADF−/−, n-cofwt/fl,nes), thereby reducing levels to one single n-cofilin allele, yielded viable and phenotypically normal mice, arguing against a simple effect of cofilin levels as the reason for the different mutant phenotypes. Our results show specificity of both proteins in regulating neuronal migration and brain development, and we propose two likely explanations for this specificity. One would be that n-cofilin and ADF function in different, as-yet-unidentified pathways, and the second would be that the regulation of n-cofilin and ADF activity—for example, by phosphorylation—is more complex than predicted.
Cell cycle progression and interkinetic nuclear migration are regulated by n-cofilin
Other mouse models with defects in the cerebral cortex have shown that, apart from cell migration, cell cycle progression is another critical determinant of cortex development (McConnell and Kaznowski 1991; Feng and Walsh 2004). In neurons the balance of symmetric versus asymmetric cell division is of utmost importance for controlling cell fate (Gotz and Huttner 2005). Owing to the function of the actin cytoskeleton in cell polarization, it is conceivable that alterations of actin dynamics by deleting n-cofilin will have an impact on cell cycle progression. Cell cycle-associated changes in cofilin activity, by the cofilin–phosphatase slingshot (Kaji et al. 2003) and chronophin (Gohla et al. 2005), further suggest a link to cell cycle control.
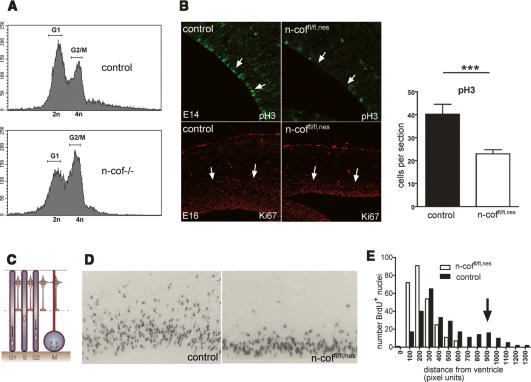
Measurements of DNA content in embryonic cells lacking n-cofilin (Gurniak et al. 2005) provided direct evidence for the importance of n-cofilin for cell cycle progression. The increased number of 4n cells suggested a role of n-cofilin for G2/M-phase transition (Fig. 4A). Encouraged by this result from total knockout embryos, we next tested the hypothesis that, in n-coffl/fl,nes brains, alterations in cell cycle progression could contribute to the defects seen in cerebral cortex formation. Indeed, a block or delay in cell proliferation was confirmed in the VZ of E16 n-coffl/fl,nes embryos by the decrease in Ki67 and phospho-histoneH3 (pH3)-positive cells (Fig. 4B). Ki67 is a nuclear factor that is expressed in proliferating cells, and histoneH3 phosphorylation is coupled to chromosome condensation during mitosis.
Figure 4.
Cell cycle block and impaired interkinetic nuclear migration in the VZ of n-coffl/fl,nes embryos. (A) DNA content of total embryonic cells harvested from n-cof−/− embryos (Gurniak et al. 2005) suggests that n-cofilin is required for G2/M transition, as indicated by the increase in cells with a 4n DNA content. (B) Cell proliferation is impaired in the cortex of n-coffl/fl,nes embryos, as shown by the decreased number of pH3-positive and Ki67-positive cells in the VZ of E14 and E16 embryos, respectively. Quantitation of pH3-positive cells in the VZ of E14 embryos is depicted in the right panel (P = 0.0094). (C) Cartoon of interkinetic nuclear migration (adapted by permission from Macmillan Publishers Ltd.: Nature Reviews Molecular Cell Biology, Gotz and Huttner 2005, © 2005) to indicate the correlation with cell cycle. (D) Short-term pulse labeling with BrdU (60 min) in the VZ allows us to follow interkinetic nuclear movement. Note that in n-coffl/fl,nes embryos vertical translocation of nuclei is impaired. (E) Quantitation of nuclear translocation in the VZ of control embryos shows a second peak of S-phase nuclei distal from the ventricles (see arrow), while in n-coffl/fl,nes embryos this peak is absent.
One peculiar characteristic of dividing cells in the VZ is the interkinetic nuclear migration, a vertical movement of the nucleus along the cell axis in which the position of the nucleus was shown to correlate with the G1, S, and G2 phases (Fig. 4C; Takahashi et al. 1999). Little is known about the mechanisms that underlie nuclear migration, and the requirement for Nde1 (Feng and Walsh 2004) and Lis1 (Tsai et al. 2005) argues for a role of microtubules, while data from cdc42 mutant mice also implicated the actin cytoskeleton (Cappello et al. 2006).
To clarify whether actin filament turnover is important, we analyzed nuclear migration in n-coffl/fl,nes embryos using a short-pulse-labeling protocol with BrdU and subsequent scoring of nuclear position in the VZ (Fig. 4D). In control embryos, two populations of BrdU+ nuclei could be identified—one distributed close to the ventricle corresponding to cells in G1/G2, and a second one of vertically migrated nuclei representing S-phase cells (Fig. 4E). Vertically migrated nuclei were absent in n-coffl/fl,nes embryos, suggesting an important role of n-cofilin in interkinetic nuclear migration. However, the presence of BrdU-positive cells in the VZ indicate that, in n-coffl/fl,nes embryos, DNA synthesis can occur independently from nuclear migration, which is in agreement with previous observations made in cytochalasin B-treated cells, showing that interkinetic nuclear migration can be uncoupled from DNA synthesis (Messier and Auclair 1974). Interkinetic nuclear oscillations have been shown to be required for radial glia cell division (Tsai et al. 2005), which is in agreement with our findings of a reduced mitotic index in n-coffl/fl,nes embryos.
Increased cell cycle exit and alteration of cell fate in the cortex of n-cofilin mutant mice
Later events in G2/M phase, possibly chromosome segregation and cytokinesis, must account for the decreased mitotic index in the cortex of n-coffl/fl,nes embryos. Chromosome condensation and segregation in the VZ can be studied using pH3 as a marker for condensed chromatin. N-coffl/fl,nes embryos preferentially showed nuclei with condensed chromatin, while in control embryos a significant number of cells with polarized, segregating chromosomes were seen (Fig. 5A). A quantitative analysis of mitotic figures supported this initial observation. In control embryos, 18% of pH3-positive cells in the VZ showed “segregating chromosomes,” while in n-coffl/fl,nest embryos only 8% were counted (Fig. 5B). Conversely, the number of cells with “condensed-unpolarized chromatin” was increased in n-coffl/fl,nes embryos. This result suggests that n-coffl/fl,nes cells might have problems setting up the cleavage plane, which is critical for the decision on symmetric versus asymmetric cell division.
Two further observations point to an imbalance of symmetric–asymmetric cell division in n-coffl/fl,nes embryos. First, in the VZ of mutant embryos fewer cells enter the cell cycle as judged by the concomitant incorporation of BrdU and expression of Ki67 (Ki67+/BrdU+) (Fig. 5C), while more cells exit the cell cycle (Ki67−/BrdU+). As a consequence, proliferative symmetric divisions decrease and differentiation occurs at the expense of the available progenitor pool.
Such a depletion in the progenitor compartment is supported by the pattern of the neuronal differentiation marker βIII-tubulin in the VZ of n-coffl/fl,nes embryos. While in control embryos the VZ shows few differentiating βIII-tubulin-positive cells, most of the proliferative zone in n-coffl/fl,nes embryos has been replaced by βIII-tubulin-positive neurons (Fig. 5D).
We conclude from these findings that the imbalance of cell homeostasis in conjunction with the impaired ability of differentiated neurons to migrate away from the VZ leads to the lack of cortical layers in n-coffl/fl,nes embryos and the expansion of the cortical area. This is illustrated by the reduction in volume and an increase in the surface of the cortex (Fig. 2A; Supplementary Fig. 2B). Unlike primates, mice do not have a gyrencephalic cortex that allows the accommodation of an increase in area, and the surface expansion occurring in n-coffl/fl,nes embryos leads to a mid-line crossing and stacking of the hemispheres (Supplementary Fig. 2C). It is noteworthy that cortical expansion in mammals has been attributed to changes in the ratio of symmetric versus asymmetric cell division in the VZ (Kriegstein et al. 2006).
Discussion
Here we describe a striking analogy in the defects and alterations caused by deletion of n-cofilin and LIS1—the gene mutated in lissencephaly. Radial migration, neural progenitor division, interkinetik nuclear oscillations, progression from multipolar to migratory bipolar neurons, as well as neurite outgrowth are impaired in n-coffl/fl,nes embryos, similar to experiments in which LIS1 expression was ablated (Tsai et al. 2005). This suggests that both proteins function through pathways that ultimately control the same cellular processes. There are a number of interesting implications from these congruent findings. Regulation of F-actin levels by n-cofilin is crucial for neuronal functions in cortex formation, and ADF or m-cofilin are apparently not sufficient to compensate. But how do alterations of F-actin organization in n-coffl/fl,nes embryos ultimately lead to the observed defects? One possibility is that actin-based motor molecules like myosin IIB are inhibited by the increased F-actin levels, and myosin II activity has been shown to be important for establishing cell migration (Vicente-Manzanares et al. 2007) as well as nucleokinesis (Schaar and McConnell 2005). Alternatively, n-cofilin might be part of the signaling pathways inducing cell polarity such as the small GTPases (Govek et al. 2005) and the PAR complexes. Neuronal polarity has been shown to be critical for glia-guided migration and brain development (Solecki et al. 2006). An intimate association of actin polymerization, cdc42, and PAR complexes was suggested by several reports (Zmuda and Rivas 2000; Schwamborn and Puschel 2004), and deletion of n-cofilin might affect this delicate signaling network. For example, Par3 has been shown to inhibit LIM kinase, thereby decreasing cofilin phosphorylation and activating the cofilin depolymerizing function (Chen and Macara 2006), while rac1 and cdc42 can have the opposite effect by increasing cofilin phosphorylation (Sumi et al. 1999). Our finding of increased multipolar neurons in the VZ of n-coffl/fl,nes embryos (see Fig. 3B) is in agreement with a role of n-cofilin in cell polarity.
Another exciting, as-yet-unexplored possibility is a cross-talk of n-cofilin with the microtubule network and dynein in perhaps jointly regulating centrosome orientation. Such a link might also provide a rational for the specific defects seen in cell cycle progression upon deletion of n-cofilin. An association of cofilin with centrosome movement and cytokinesis has been suggested by studies in Drosophila (Gunsalus et al. 1995) and Xenopus (Abe et al. 1996), and a direct link of cofilin to cell cycle signaling is conceivable (Gohla et al. 2005). Our conditional n-cofilin mutant mice provide a powerful tool to further address this aspect of mammalian cell division.
Our n-coffl/fl,nes mutant mice reproduce symptoms that have been associated with human neuronal disorders, and they are the first example to provide evidence that proteins affecting actin filament dynamics are valid candidates in “neuronal migration disorders” such as mental retardation, epilepsy, and schizophrenia.
To date, only a handful of target genes are known to cause human congenital disorders linked to neuronal migration defects (Ayala et al. 2007). Although in humans no null mutations in the n-cofilin gene have been identified yet, mutations in a regulator of cofilin activity—the LIM-kinase1—have been discussed as contributing factors in Williams syndrome, a milder form of mental retardation (Frangiskakis et al. 1996; Gray et al. 2006). Furthermore, recent SNP studies revealed that the human n-cofilin gene is a genetic modifier for “spina bifida” risk (Zhu et al. 2007), suggesting that variations in the human n-cofilin gene contribute to neuronal disorders.
Materials and methods
Generation of n-cofilin mutant mice
Brain-specific deletion of n-cofilin was achieved by crossing the conditional n-coffl allele (Supplementary Fig. 1B; Gurniak et al. 2005) with a transgenic cre-expressing line, driven by the nesting promoter (Tronche et al. 1999). Animal care was conducted according to the institutional guidelines that are in compliance with international laws and policies (European Community Council Directive 86 609, Official Journal L358, December 18, 1986).
Generation of ADF mutant mice
The β-gal cassette was inserted as a 4.2-kbp BamHI/SmaI fragment into the BsmI site of exon 2 of the ADF gene (Supplementary Fig. 1A). As a selection marker, a floxed neo cassette was inserted into the NcoI site located immediately downstream from the β-gal cassette. The neo cassette was removed by crossing with a cre-deleter mouse line (Schwenk et al. 1995). Insertion of the β-gal cassette inactivated the ADF gene.
Neuronal culture
Cortical neurons were prepared from E17 embryos as described previously (Dotti and Banker 1987) and maintained according to the procedure recommended for Neurobasal-A medium (Life Technologies). For morphological studies, neurons were plated on laminin-treated glass coverslips.
For F-actin labeling, cells were fixed for 10 min with 4% paraformaldehyde/PBS, permeabilized with 0.1% Triton X-100/PBS, and stained for 30 min with Alexa-conjugated phalloidin (Molecular Probes). Quantitative F-actin measurements were performed using the MetaMorph software and the Neurite Outgrowth module (Universal Imaging).
Biochemistry
Embryonic and postnatal brains were dissected and homogenized in 50 mM NaCl, 20 mM Tris-HCl (pH 7.5), and 1% Triton X-100 using a tight-fitting Potter-Elvehjem homogenizer. Ten micrograms of protein extract were separated by SDS-PAGE and proteins were transferred onto ImmobilionP. Membranes were probed with antibodies against actin (mab224-236, J. Faix), n-cofilin (KG40, W. Witke), and ADF (GV-13, Sigma), followed by secondary antibodies conjugated to horseradish peroxidase (Pierce). Signals were detected using the ECL chemiluminescence reaction (Amersham Pharmacia).
Histology and immunohistochemistry
Embryos and brains at different embryonic states were dissected, fixed overnight in 4% paraformaldehyde, and embedded in paraffin. For morphological evaluation, sections were stained with hematoxilin-eosin.
In situ hybridization using 35S-CTP-labeled probes on 8-μm paraffin sections was performed according to procedures described previously (Gurniak et al. 2005). Briefly, sections were dewaxed, rehydrated, digested with proteinase K, and hybridized with 35S-CTP-labeled probe at 62°C. Post-hybridization washes in 20% formamide and 0.5× SSC were done at 60°C. Full-length cDNA was used to generate probes for both n-cofilin and ADF.
For Golgi-Cox impregnation of brain tissue, mice were anesthetized and perfused intracardially with 0.9% saline. Brains were dissected and impregnated by using a standard Golgi-Cox solution (1% potassium dichromate, 1% mercuric chloride, 0.8% potassium chromate) according to the method described by Glaser and Van der Loos (1981). The brains immersed in the Golgi-Cox solution were stored for 6 d at room temperature, immersed in a sucrose solution (30%) for 2 d, and then sectioned coronally (200 μm) using a Vibratome. Sections were mounted on gelatinized slides, stained according to the Gibb and Kolb method (Gibb and Kolb 1998), and covered with Permount.
Immunohistochemistry using the neuronal marker βIII-tubulin was performed on paraffin sections and antibody staining was carried out according to the manufacturer’s instructions (Dako). The antibodiy against VIAAT to label GABAergic neurons in early mouse development (Dumoulin et al. 1999) was kindly provided by B. Gasnier.
Cell cycle analysis
For BrdU labeling, pregnant females received a single injection of 100 μg of BrdU per gram of body weight. At the respective time points, the brains were dissected from the embryos, fixed, and processed for paraffin embedding. BrdU-labeled cells were detected on 8-μm sections using the Roche BrdU detection kit II.
For neuronal migration studies, embryos were pulsed at E16 and harvested 2 d later. To quantify the number of BrdU-positive cells migrating out from the proliferative zone into the CP, we counted the BrdU-positive cells in the CP. For interkinetic nuclear migration studies, E14 embryos were pulsed with BrdU for 60 min. The radial distance of BrdU-positive nuclei from the VZ was measured using the MetaMorph Image analysis software (Universal Imaging). At least 250 cells were counted for both genotypes on consecutive sections over a total tissue thickness of 100 μm. All experiments were repeated independently three times. Graphs were designed using Graph Pad (Prism Software).
For cell cycle exit studies, BrdU-labeled embryos were harvested at E16, 1 h after injection. Fixed brains were embedded, sectioned, and double-labeled for Ki67 (which is a nuclear transcription factor expressed in proliferating cells from S phase to M phase) (Dako), and BrdU (Molecular Probes). The ratios of Ki67+/BrdU+ and Ki67−/BrdU+ cells in the VZ were determined as described previously (Lien et al. 2006).
For chromosome condensation and segregation experiments, sections were stained for pH3 (Upstate Biotechnology) and the DNA dye propidium iodide. Images were acquired on a Leica confocal microscope and mitotic figures were categorized into “segregating chromosomes,” and “condensed chromatin.” At least 150 mitotic figures were analyzed per genotype.
The DNA content of total embryonic cells prepared from trypsinized E10.5 n-cofilin-null embryos (Gurniak et al. 2005) was determined by FACS analysis using propidium iodide as DNA dye.
Acknowledgments
We thank Drs. C. Gross, P. Pilo-Boyl, and M. Rust for critical reading of the manuscript; Dr. T. Iwata for helpful discussion; J. Kelly-Barrett for ES cell injections; Dr. F. Jönsson for genotyping; and B. Gasnier for the VIAAT antibody. G.C.B. was supported by a FEBS post-doctoral fellowship.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.434307
References
- Abe H., Obinata T., Minamide L.S., Bamburg J.R., Obinata T., Minamide L.S., Bamburg J.R., Minamide L.S., Bamburg J.R., Bamburg J.R. Xenopus laevis actin-depolymerizing factor/cofilin: A phosphorylation-regulated protein essential for development. J. Cell Biol. 1996;132:871–885. doi: 10.1083/jcb.132.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams M.E., Minamide L.S., Duester G., Bamburg J.R., Minamide L.S., Duester G., Bamburg J.R., Duester G., Bamburg J.R., Bamburg J.R. Nucleotide sequence and expression of a cDNA encoding chick brain actin depolymerizing factor. Biochemistry. 1990;29:7414–7420. doi: 10.1021/bi00484a009. [DOI] [PubMed] [Google Scholar]
- Anderson S.A., Eisenstat D.D., Shi L., Rubenstein J.L., Eisenstat D.D., Shi L., Rubenstein J.L., Shi L., Rubenstein J.L., Rubenstein J.L. Interneuron migration from basal forebrain to neocortex: Dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- Ayala R., Shu T., Tsai L.H., Shu T., Tsai L.H., Tsai L.H. Trekking across the brain: The journey of neuronal migration. Cell. 2007;128:29–43. doi: 10.1016/j.cell.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Cappello S., Attardo A., Wu X., Iwasato T., Itohara S., Wilsch-Brauninger M., Eilken H.M., Rieger M.A., Schroeder T.T., Huttner W.B., Attardo A., Wu X., Iwasato T., Itohara S., Wilsch-Brauninger M., Eilken H.M., Rieger M.A., Schroeder T.T., Huttner W.B., Wu X., Iwasato T., Itohara S., Wilsch-Brauninger M., Eilken H.M., Rieger M.A., Schroeder T.T., Huttner W.B., Iwasato T., Itohara S., Wilsch-Brauninger M., Eilken H.M., Rieger M.A., Schroeder T.T., Huttner W.B., Itohara S., Wilsch-Brauninger M., Eilken H.M., Rieger M.A., Schroeder T.T., Huttner W.B., Wilsch-Brauninger M., Eilken H.M., Rieger M.A., Schroeder T.T., Huttner W.B., Eilken H.M., Rieger M.A., Schroeder T.T., Huttner W.B., Rieger M.A., Schroeder T.T., Huttner W.B., Schroeder T.T., Huttner W.B., Huttner W.B., et al. The Rho-GTPase cdc42 regulates neural progenitor fate at the apical surface. Nat. Neurosci. 2006;9:1099–1107. doi: 10.1038/nn1744. [DOI] [PubMed] [Google Scholar]
- Chen X., Macara I.G., Macara I.G. Par-3 mediates the inhibition of LIM kinase 2 to regulate cofilin phosphorylation and tight junction assembly. J. Cell Biol. 2006;172:671–678. doi: 10.1083/jcb.200510061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couillard-Despres S., Winkler J., Uyanik G., Aigner L., Winkler J., Uyanik G., Aigner L., Uyanik G., Aigner L., Aigner L. Molecular mechanisms of neuronal migration disorders, quo vadis? Curr. Mol. Med. 2001;1:677–688. doi: 10.2174/1566524013363195. [DOI] [PubMed] [Google Scholar]
- Cowan C.R., Hyman A.A., Hyman A.A. Asymmetric cell division in C. elegans: Cortical polarity and spindle positioning. Annu. Rev. Cell Dev. Biol. 2004;20:427–453. doi: 10.1146/annurev.cellbio.19.111301.113823. [DOI] [PubMed] [Google Scholar]
- Dotti C.G., Banker G.A., Banker G.A. Experimentally induced alteration in the polarity of developing neurons. Nature. 1987;330:254–256. doi: 10.1038/330254a0. [DOI] [PubMed] [Google Scholar]
- Dumoulin A., Rostaing P., Bedet C., Levi S., Isambert M.F., Henry J.P., Triller A., Gasnier B., Rostaing P., Bedet C., Levi S., Isambert M.F., Henry J.P., Triller A., Gasnier B., Bedet C., Levi S., Isambert M.F., Henry J.P., Triller A., Gasnier B., Levi S., Isambert M.F., Henry J.P., Triller A., Gasnier B., Isambert M.F., Henry J.P., Triller A., Gasnier B., Henry J.P., Triller A., Gasnier B., Triller A., Gasnier B., Gasnier B. Presence of the vesicular inhibitory amino acid transporter in GABAergic and glycinergic synaptic terminal boutons. J. Cell Sci. 1999;112:811–823. doi: 10.1242/jcs.112.6.811. [DOI] [PubMed] [Google Scholar]
- Feng Y., Walsh C.A., Walsh C.A. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004;44:279–293. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Feng Y., Olson E.C., Stukenberg P.T., Flanagan L.A., Kirschner M.W., Walsh C.A., Olson E.C., Stukenberg P.T., Flanagan L.A., Kirschner M.W., Walsh C.A., Stukenberg P.T., Flanagan L.A., Kirschner M.W., Walsh C.A., Flanagan L.A., Kirschner M.W., Walsh C.A., Kirschner M.W., Walsh C.A., Walsh C.A. LIS1 regulates CNS lamination by interacting with mNudE, a central component of the centrosome. Neuron. 2000;28:665–679. doi: 10.1016/s0896-6273(00)00145-8. [DOI] [PubMed] [Google Scholar]
- Fishkind D.J., Wang Y.L., Wang Y.L. Orientation and three-dimensional organization of actin filaments in dividing cultured cells. J. Cell Biol. 1993;123:837–848. doi: 10.1083/jcb.123.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J.W., Lamperti E.D., Eksioglu Y.Z., Hong S.E., Feng Y., Graham D.A., Scheffer I.E., Dobyns W.B., Hirsch B.A., Radtke R.A., Lamperti E.D., Eksioglu Y.Z., Hong S.E., Feng Y., Graham D.A., Scheffer I.E., Dobyns W.B., Hirsch B.A., Radtke R.A., Eksioglu Y.Z., Hong S.E., Feng Y., Graham D.A., Scheffer I.E., Dobyns W.B., Hirsch B.A., Radtke R.A., Hong S.E., Feng Y., Graham D.A., Scheffer I.E., Dobyns W.B., Hirsch B.A., Radtke R.A., Feng Y., Graham D.A., Scheffer I.E., Dobyns W.B., Hirsch B.A., Radtke R.A., Graham D.A., Scheffer I.E., Dobyns W.B., Hirsch B.A., Radtke R.A., Scheffer I.E., Dobyns W.B., Hirsch B.A., Radtke R.A., Dobyns W.B., Hirsch B.A., Radtke R.A., Hirsch B.A., Radtke R.A., Radtke R.A., et al. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998;21:1315–1325. doi: 10.1016/s0896-6273(00)80651-0. [DOI] [PubMed] [Google Scholar]
- Frangiskakis J.M., Ewart A.K., Morris C.A., Mervis C.B., Bertrand J., Robinson B.F., Klein B.P., Ensing G.J., Everett L.A., Green E.D., Ewart A.K., Morris C.A., Mervis C.B., Bertrand J., Robinson B.F., Klein B.P., Ensing G.J., Everett L.A., Green E.D., Morris C.A., Mervis C.B., Bertrand J., Robinson B.F., Klein B.P., Ensing G.J., Everett L.A., Green E.D., Mervis C.B., Bertrand J., Robinson B.F., Klein B.P., Ensing G.J., Everett L.A., Green E.D., Bertrand J., Robinson B.F., Klein B.P., Ensing G.J., Everett L.A., Green E.D., Robinson B.F., Klein B.P., Ensing G.J., Everett L.A., Green E.D., Klein B.P., Ensing G.J., Everett L.A., Green E.D., Ensing G.J., Everett L.A., Green E.D., Everett L.A., Green E.D., Green E.D., et al. LIM-kinase1 hemizygosity implicated in impaired visuospatial constructive cognition. Cell. 1996;86:59–69. doi: 10.1016/s0092-8674(00)80077-x. [DOI] [PubMed] [Google Scholar]
- Ghosh M., Song X., Mouneimne G., Sidani M., Lawrence D.S., Condeelis J.S., Song X., Mouneimne G., Sidani M., Lawrence D.S., Condeelis J.S., Mouneimne G., Sidani M., Lawrence D.S., Condeelis J.S., Sidani M., Lawrence D.S., Condeelis J.S., Lawrence D.S., Condeelis J.S., Condeelis J.S. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 2004;304:743–746. doi: 10.1126/science.1094561. [DOI] [PubMed] [Google Scholar]
- Gibb R., Kolb B., Kolb B. A method for vibratome sectioning of Golgi-Cox stained whole rat brain. J. Neurosci. Methods. 1998;79:1–4. doi: 10.1016/s0165-0270(97)00163-5. [DOI] [PubMed] [Google Scholar]
- Glaser E.M., Van der Loos H., Van der Loos H. Analysis of thick brain sections by obverse-reverse computer microscopy: Application of a new, high clarity Golgi-Nissl stain. J. Neurosci. Methods. 1981;4:117–125. doi: 10.1016/0165-0270(81)90045-5. [DOI] [PubMed] [Google Scholar]
- Gohla A., Birkenfeld J., Bokoch G.M., Birkenfeld J., Bokoch G.M., Bokoch G.M. Chronophin, a novel HAD-type serine protein phosphatase, regulates cofilin-dependent actin dynamics. Nat. Cell Biol. 2005;7:21–29. doi: 10.1038/ncb1201. [DOI] [PubMed] [Google Scholar]
- Gotz M., Huttner W.B., Huttner W.B. The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Govek E.E., Newey S.E., Van Aelst L., Newey S.E., Van Aelst L., Van Aelst L. The role of the Rho GTPases in neuronal development. Genes & Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- Gray V., Karmiloff-Smith A., Funnell E., Tassabehji M., Karmiloff-Smith A., Funnell E., Tassabehji M., Funnell E., Tassabehji M., Tassabehji M. In-depth analysis of spatial cognition in Williams syndrome: A critical assessment of the role of the LIMK1 gene. Neuropsychologia. 2006;44:679–685. doi: 10.1016/j.neuropsychologia.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Gungabissoon R.A., Bamburg J.R., Bamburg J.R. Regulation of growth cone actin dynamics by ADF/cofilin. J. Histochem. Cytochem. 2003;51:411–420. doi: 10.1177/002215540305100402. [DOI] [PubMed] [Google Scholar]
- Gunsalus K.C., Bonaccorsi S., Williams E., Verni F., Gatti M., Goldberg M.L., Bonaccorsi S., Williams E., Verni F., Gatti M., Goldberg M.L., Williams E., Verni F., Gatti M., Goldberg M.L., Verni F., Gatti M., Goldberg M.L., Gatti M., Goldberg M.L., Goldberg M.L. Mutations in twinstar, a Drosophila gene encoding a cofilin/ADF homologue, result in defects in centrosome migration and cytokinesis. J. Cell Biol. 1995;131:1243–1259. doi: 10.1083/jcb.131.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurniak C.B., Perlas E., Witke W., Perlas E., Witke W., Witke W. The actin depolymerizing factor n-cofilin is essential for neural tube morphogenesis and neural crest cell migration. Dev. Biol. 2005;278:231–241. doi: 10.1016/j.ydbio.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Hotulainen P., Paunola E., Vartiainen M.K., Lappalainen P., Paunola E., Vartiainen M.K., Lappalainen P., Vartiainen M.K., Lappalainen P., Lappalainen P. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol. Biol. Cell. 2005;16:649–664. doi: 10.1091/mbc.E04-07-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S., Cunningham L.A., Boggess D., Hawes N., Hobson C.D., Sundberg J.P., Naggert J.K., Smith R.S., Nishina P.M., Cunningham L.A., Boggess D., Hawes N., Hobson C.D., Sundberg J.P., Naggert J.K., Smith R.S., Nishina P.M., Boggess D., Hawes N., Hobson C.D., Sundberg J.P., Naggert J.K., Smith R.S., Nishina P.M., Hawes N., Hobson C.D., Sundberg J.P., Naggert J.K., Smith R.S., Nishina P.M., Hobson C.D., Sundberg J.P., Naggert J.K., Smith R.S., Nishina P.M., Sundberg J.P., Naggert J.K., Smith R.S., Nishina P.M., Naggert J.K., Smith R.S., Nishina P.M., Smith R.S., Nishina P.M., Nishina P.M. Aberrant actin cytoskeleton leads to accelerated proliferation of corneal epithelial cells in mice deficient for destrin (actin depolymerizing factor) Hum. Mol. Genet. 2003;12:1029–1037. doi: 10.1093/hmg/ddg112. [DOI] [PubMed] [Google Scholar]
- Kaji N., Ohashi K., Shuin M., Niwa R., Uemura T., Mizuno K., Ohashi K., Shuin M., Niwa R., Uemura T., Mizuno K., Shuin M., Niwa R., Uemura T., Mizuno K., Niwa R., Uemura T., Mizuno K., Uemura T., Mizuno K., Mizuno K. Cell cycle-associated changes in Slingshot phosphatase activity and roles in cytokinesis in animal cells. J. Biol. Chem. 2003;278:33450–33455. doi: 10.1074/jbc.M305802200. [DOI] [PubMed] [Google Scholar]
- Kriegstein A., Noctor S., Martinez-Cerdeno V., Noctor S., Martinez-Cerdeno V., Martinez-Cerdeno V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat. Rev. Neurosci. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- Lenart P., Bacher C.P., Daigle N., Hand A.R., Eils R., Terasaki M., Ellenberg J., Bacher C.P., Daigle N., Hand A.R., Eils R., Terasaki M., Ellenberg J., Daigle N., Hand A.R., Eils R., Terasaki M., Ellenberg J., Hand A.R., Eils R., Terasaki M., Ellenberg J., Eils R., Terasaki M., Ellenberg J., Terasaki M., Ellenberg J., Ellenberg J. A contractile nuclear actin network drives chromosome congression in oocytes. Nature. 2005;436:812–818. doi: 10.1038/nature03810. [DOI] [PubMed] [Google Scholar]
- Lien W.H., Klezovitch O., Fernandez T.E., Delrow J., Vasioukhin V., Klezovitch O., Fernandez T.E., Delrow J., Vasioukhin V., Fernandez T.E., Delrow J., Vasioukhin V., Delrow J., Vasioukhin V., Vasioukhin V. αE-catenin controls cerebral cortical size by regulating the hedgehog signaling pathway. Science. 2006;311:1609–1612. doi: 10.1126/science.1121449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki F., Matsumoto S., Yahara I., Yonezawa N., Nishida E., Sakai H., Matsumoto S., Yahara I., Yonezawa N., Nishida E., Sakai H., Yahara I., Yonezawa N., Nishida E., Sakai H., Yonezawa N., Nishida E., Sakai H., Nishida E., Sakai H., Sakai H. Cloning and characterization of porcine brain cofilin cDNA. Cofilin contains the nuclear transport signal sequence. J. Biol. Chem. 1988;263:11564–11568. [PubMed] [Google Scholar]
- McConnell S.K., Kaznowski C.E., Kaznowski C.E. Cell cycle dependence of laminar determination in developing neocortex. Science. 1991;254:282–285. doi: 10.1126/science.254.5029.282. [DOI] [PubMed] [Google Scholar]
- Meberg P.J., Bamburg J.R., Bamburg J.R. Increase in neurite outgrowth mediated by overexpression of actin depolymerizing factor. J. Neurosci. 2000;20:2459–2469. doi: 10.1523/JNEUROSCI.20-07-02459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meberg P.J., Ono S., Minamide L.S., Takahashi M., Bamburg J.R., Ono S., Minamide L.S., Takahashi M., Bamburg J.R., Minamide L.S., Takahashi M., Bamburg J.R., Takahashi M., Bamburg J.R., Bamburg J.R. Actin depolymerizing factor and cofilin phosphorylation dynamics: Response to signals that regulate neurite extension. Cell Motil. Cytoskeleton. 1998;39:172–190. doi: 10.1002/(SICI)1097-0169(1998)39:2<172::AID-CM8>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Messier P.E., Auclair C., Auclair C. Effect of cytochalasin B on interkinetic nuclear migration in the chick embryo. Dev. Biol. 1974;36:218–223. doi: 10.1016/0012-1606(74)90206-1. [DOI] [PubMed] [Google Scholar]
- Minamide L.S., Striegl A.M., Boyle J.A., Meberg P.J., Bamburg J.R., Striegl A.M., Boyle J.A., Meberg P.J., Bamburg J.R., Boyle J.A., Meberg P.J., Bamburg J.R., Meberg P.J., Bamburg J.R., Bamburg J.R. Neurodegenerative stimuli induce persistent ADF/cofilin-actin rods that disrupt distal neurite function. Nat. Cell Biol. 2000;2:628–636. doi: 10.1038/35023579. [DOI] [PubMed] [Google Scholar]
- Nadarajah B., Parnavelas J.G., Parnavelas J.G. Modes of neuronal migration in the developing cerebral cortex. Nat. Rev. Neurosci. 2002;3:423–432. doi: 10.1038/nrn845. [DOI] [PubMed] [Google Scholar]
- Ono S., Minami N., Abe H., Obinata T., Minami N., Abe H., Obinata T., Abe H., Obinata T., Obinata T. Characterization of a novel cofilin isoform that is predominantly expressed in mammalian skeletal muscle. J. Biol. Chem. 1994;269:15280–15286. [PubMed] [Google Scholar]
- Pilo Boyl P., Di Nardo A., Mulle C., Sassoe-Pognetto M., Panzanelli P., Mele A., Kneussel M., Costantini V., Perlas E., Massimi M., Di Nardo A., Mulle C., Sassoe-Pognetto M., Panzanelli P., Mele A., Kneussel M., Costantini V., Perlas E., Massimi M., Mulle C., Sassoe-Pognetto M., Panzanelli P., Mele A., Kneussel M., Costantini V., Perlas E., Massimi M., Sassoe-Pognetto M., Panzanelli P., Mele A., Kneussel M., Costantini V., Perlas E., Massimi M., Panzanelli P., Mele A., Kneussel M., Costantini V., Perlas E., Massimi M., Mele A., Kneussel M., Costantini V., Perlas E., Massimi M., Kneussel M., Costantini V., Perlas E., Massimi M., Costantini V., Perlas E., Massimi M., Perlas E., Massimi M., Massimi M., et al. Profilin2 contributes to synaptic vesicle exocytosis, neuronal excitability, and novelty-seeking behavior. EMBO J. 2007;26:2991–3002. doi: 10.1038/sj.emboj.7601737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Neuronal migration and contact guidance in the primate telencephalon. Postgrad. Med. J. 1978;54 (Suppl. 1):25–40. [PubMed] [Google Scholar]
- Rakic P. Early developmental events: Cell lineages, acquisition of neuronal positions, and areal and laminar development. Neurosci. Res. Program Bull. 1982;20:439–451. [PubMed] [Google Scholar]
- Reiner O., Carrozzo R., Shen Y., Wehnert M., Faustinella F., Dobyns W.B., Caskey C.T., Ledbetter D.H., Carrozzo R., Shen Y., Wehnert M., Faustinella F., Dobyns W.B., Caskey C.T., Ledbetter D.H., Shen Y., Wehnert M., Faustinella F., Dobyns W.B., Caskey C.T., Ledbetter D.H., Wehnert M., Faustinella F., Dobyns W.B., Caskey C.T., Ledbetter D.H., Faustinella F., Dobyns W.B., Caskey C.T., Ledbetter D.H., Dobyns W.B., Caskey C.T., Ledbetter D.H., Caskey C.T., Ledbetter D.H., Ledbetter D.H. Isolation of a Miller-Dieker lissencephaly gene containing G protein β-subunit-like repeats. Nature. 1993;364:717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- Rivas R.J., Hatten M.E., Hatten M.E. Motility and cytoskeletal organization of migrating cerebellar granule neurons. J. Neurosci. 1995;15:981–989. doi: 10.1523/JNEUROSCI.15-02-00981.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar B.T., McConnell S.K., McConnell S.K. Cytoskeletal coordination during neuronal migration. Proc. Natl. Acad. Sci. 2005;102:13652–13657. doi: 10.1073/pnas.0506008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwamborn J.C., Puschel A.W., Puschel A.W. The sequential activity of the GTPases Rap1B and Cdc42 determines neuronal polarity. Nat. Neurosci. 2004;7:923–929. doi: 10.1038/nn1295. [DOI] [PubMed] [Google Scholar]
- Schwenk F., Baron U., Rajewsky K., Baron U., Rajewsky K., Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solecki D.J., Govek E.E., Tomoda T., Hatten M.E., Govek E.E., Tomoda T., Hatten M.E., Tomoda T., Hatten M.E., Hatten M.E. Neuronal polarity in CNS development. Genes & Dev. 2006;20:2639–2647. doi: 10.1101/gad.1462506. [DOI] [PubMed] [Google Scholar]
- Sumi T., Matsumoto K., Takai Y., Nakamura T., Matsumoto K., Takai Y., Nakamura T., Takai Y., Nakamura T., Nakamura T. Cofilin phosphorylation and actin cytoskeletal dynamics regulated by rho- and Cdc42-activated LIM-kinase 2. J. Cell Biol. 1999;147:1519–1532. doi: 10.1083/jcb.147.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Bhide P.G., Goto T., Miyama S., Caviness V.S., Bhide P.G., Goto T., Miyama S., Caviness V.S., Goto T., Miyama S., Caviness V.S., Miyama S., Caviness V.S., Caviness V.S. Proliferative behavior of the murine cerebral wall in tissue culture: Cell cycle kinetics and checkpoints. Exp. Neurol. 1999;156:407–417. doi: 10.1006/exnr.1999.7023. [DOI] [PubMed] [Google Scholar]
- Tronche F., Kellendonk C., Kretz O., Gass P., Anlag K., Orban P.C., Bock R., Klein R., Schutz G., Kellendonk C., Kretz O., Gass P., Anlag K., Orban P.C., Bock R., Klein R., Schutz G., Kretz O., Gass P., Anlag K., Orban P.C., Bock R., Klein R., Schutz G., Gass P., Anlag K., Orban P.C., Bock R., Klein R., Schutz G., Anlag K., Orban P.C., Bock R., Klein R., Schutz G., Orban P.C., Bock R., Klein R., Schutz G., Bock R., Klein R., Schutz G., Klein R., Schutz G., Schutz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- Tsai J.W., Chen Y., Kriegstein A.R., Vallee R.B., Chen Y., Kriegstein A.R., Vallee R.B., Kriegstein A.R., Vallee R.B., Vallee R.B. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J. Cell Biol. 2005;170:935–945. doi: 10.1083/jcb.200505166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen M.K., Mustonen T., Mattila P.K., Ojala P.J., Thesleff I., Partanen J., Lappalainen P., Mustonen T., Mattila P.K., Ojala P.J., Thesleff I., Partanen J., Lappalainen P., Mattila P.K., Ojala P.J., Thesleff I., Partanen J., Lappalainen P., Ojala P.J., Thesleff I., Partanen J., Lappalainen P., Thesleff I., Partanen J., Lappalainen P., Partanen J., Lappalainen P., Lappalainen P. The three mouse actin-depolymerizing factor/cofilins evolved to fulfill cell-type-specific requirements for actin dynamics. Mol. Biol. Cell. 2002;13:183–194. doi: 10.1091/mbc.01-07-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Zareno J., Whitmore L., Choi C.K., Horwitz A.F., Zareno J., Whitmore L., Choi C.K., Horwitz A.F., Whitmore L., Choi C.K., Horwitz A.F., Choi C.K., Horwitz A.F., Horwitz A.F. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J. Cell Biol. 2007;176:573–580. doi: 10.1083/jcb.200612043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh S., Pope B., Mannherz H.G., Weeds A., Pope B., Mannherz H.G., Weeds A., Mannherz H.G., Weeds A., Weeds A. Determining the differences in actin binding by human ADF and cofilin. J. Mol. Biol. 2002;315:911–925. doi: 10.1006/jmbi.2001.5280. [DOI] [PubMed] [Google Scholar]
- Zhu H., Ebot Enow J.O., Ma C., Shaw G.M., Lammer E.J., Finnell R.H., Ebot Enow J.O., Ma C., Shaw G.M., Lammer E.J., Finnell R.H., Ma C., Shaw G.M., Lammer E.J., Finnell R.H., Shaw G.M., Lammer E.J., Finnell R.H., Lammer E.J., Finnell R.H., Finnell R.H. Association between CFL1 gene polymorphisms and spina bifida risk in a California population. BMC Med. Genet. 2007;8:12. doi: 10.1186/1471-2350-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmuda J.F., Rivas R.J., Rivas R.J. Actin filament disruption blocks cerebellar granule neurons at the unipolar stage of differentiation in vitro. J. Neurobiol. 2000;43:313–328. doi: 10.1002/1097-4695(20000615)43:4<313::aid-neu1>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]