SUMMARY
A fascinating feature of thyroid hormone (T3) receptors (TR) is that they constitutively bind to promoter regions of T3-response genes, providing dual functions. In the presence of T3, TR activates T3-inducible genes, while unliganded TR represses these same genes. Although this dual function model is well demonstrated at the molecular level, few studies have addressed the presence or the role of unliganded TR-induced repression in physiological settings. Here, we analyze the role of unliganded TR in Xenopus laevis development. The total dependence of amphibian metamorphosis upon T3 provides us a valuable opportunity for studying TR function in vivo. First, we designed a dominant negative form of TR-binding corepressor N-CoR (dnN-CoR) consisting of its receptor interacting domain. We confirmed its dominant negative activity by showing that dnN-CoR competes away the binding of endogenous N-CoR to unliganded TR and relieves unliganded TR-induced gene repression in frog oocytes. Next, we overexpressed dnN-CoR in tadpoles through transgenesis and analyzed its effect on gene expression and development. Quantitative RT-PCR revealed significant derepression of T3-response genes in transgenic animals. In addition, transgenic tadpoles developed faster than wild type siblings, with an acceleration of as much as 7 days out of the 30-day experiment. These data thus provide in vivo evidence for the presence and a role of unliganded TR-induced gene repression in physiological settings and strongly support our earlier model that unliganded TR represses T3-response genes in premetamorphic tadpoles to regulate the progress of development.
Keywords: Xenopus laevis, thyroid hormone receptor, transcriptional regulation, amphibian metamorphosis, derepression
INTRODUCTION
Thyroid hormone receptors (TRs) are believed to mediate the vast majority of the diverse biological effects of thyroid hormone (T3, or 3, 5, 5′-triiodothyronin). TRs belong to the superfamily of nuclear hormone receptors, which also includes steroid hormone receptors and 9-cis retinoic acid receptors (RXR), and most likely function as heterodimers with RXRs (Evans, 1988; Lazar, 1993; Mangelsdorf et al., 1995). One of the unique features of TRs is that they can constitutively, regardless of the ligand availability, bind to T3 response elements (TREs) in the promoter region of T3-response genes. In other words, TRs can adopt either the unliganded or liganded conformation at TREs (Perlman et al., 1982; Sachs and Shi, 2000; Tsai and O'Malley, 1994; Wong and Shi, 1995). Various in vitro studies have led to the dual function model for how TRs regulate target gene transcription. That is, unliganded TRs repress T3-inducible genes, and liganded TRs activate the same genes. The dual effects of TRs are accomplished by recruiting mutually exclusive sets of coregulators to the target promoters (Fondell et al., 1993; Hsia et al., 2001; Puzianowska-Kuznicka et al., 1997; Tsai and O'Malley, 1994; Wong et al., 1995; Zhang and Lazar, 2000). The most studied corepressor complexes, those composed of N-CoR (nuclear corepressor) or SMRT (silencing mediator of retinoid and thyroid hormone receptors), HDAC3 (histone deacetylase 3), TBL1 (transducin beta-like protein 1)/TBLR1 (TBL1-related protein 1), and GPS2 (G-protein pathway suppressor 2), associate with unliganded TRs (Burke and Baniahmad, 2000; Chen and Evans, 1995; Glass and Rosenfeld, 2000; Horlein et al., 1995; Jepsen and Rosenfeld, 2002; Jones et al., 2001; Jones and Shi, 2003; Li et al., 2000; Tomita et al., 2004; Underhill et al., 2000; Wen et al., 2000; Yoon et al., 2003; Zhang et al., 2002; Zhang and Lazar, 2000). The presence of T3 induces a conformational change in TR, promoting the release of corepressor complexes and recruitment of coactivator complexes such as those containing SRCs (steroid receptor coactivators), p300, and pCAF (p300 associated factor) (Ito and Roeder, 2001; McKenna and O'Malley, 2002; Rachez and Freedman, 2001). Despite the wealth of knowledge of the molecular mechanisms of TR action in vitro, it still remains to be determined whether this dual function model works during animal development in vivo.
We have been using frog metamorphosis as a model to examine the mechanism and function of gene regulation by TRs (Buchholz et al., 2006). In tadpoles, T3 production is limited to their metamorphic period, while significant expression of TRα mRNA begins much earlier, around hatching. More importantly, frog metamorphosis is totally dependent on T3, as indicated by the fact that athyroid tadpole cannot go through metamorphosis and exogenous T3 can induce precocious metamorphosis (Dodd and Dodd, 1976; Shi, 1999). Based on the expression profiles of T3 and TR in Xenopus laevis, we have proposed a dual function model for the role of TR in frog development (Buchholz et al., 2006; Sachs et al., 2000). In premetamorphic tadpoles, the unliganded TR recruits corepressors and represses T3-response genes to prevent precocious metamorphosis. During metamorphosis, elevation of endogenous T3 switches unliganded TR to the liganded conformation. The liganded TR, then, recruits coactivators and activates the same T3-response genes to induce various metamorphic changes. We and others have provided several lines of evidence that support this dual function model in vivo. First, TR function is both necessary and sufficient for the causative effects of T3 on metamorphosis (Buchholz et al., 2003; Buchholz et al., 2006; Buchholz et al., 2004; Schreiber et al., 2001). Second, TR binds constitutively to the promoter regions of T3-response genes in premetamorphic tadpoles (Buchholz et al., 2005; Sachs and Shi, 2000). Third, corepressors, N-CoR and SMRT as well as the associated factor TBLR1, are expressed in premetamorphic tadpoles at least by stage 46, the onset of tadpole feeding (Sachs et al., 2002; Tomita et al., 2004) (unpublished observation). More importantly, these corepressors are recruited to TREs in premetamorphic tadpoles and released at the climax of metamorphosis (Sachs et al., 2002; Tomita et al., 2004). Fourth, the coactivator SRC3 is recruited to TREs in T3-treated as well as naturally metamorphosing tadpoles in a gene- and tissue-specific manner (Havis et al., 2003; Paul et al., 2005a; Paul et al., 2005b). Finally, blocking endogenous coactivator recruitment with a dominant negative form of SRC3 through transgenesis inhibits both gene activation by TR and metamorphosis (Paul et al., 2005b). These findings thus indicate that liganded TR-coactivator complexes are recruited to TREs in the metamorphosing tadpoles and these complexes are required for T3-response gene activation as well as metamorphosis. They also demonstrate the recruitment of unliganded TR-corepressor complexes to TREs in premetamorphic tadpoles. Although a recent report suggests that unliganded TR is important for embryonic eye development (Havis et al., 2006), it remains to be determined whether unliganded TR has a general function in postembryonic development to repress T3-response genes in vivo and prevent precocious metamorphosis as the dual function model suggests.
To address this issue, we have generated a dominant negative from of N-CoR (dnN-CoR) consisting of two copies of the receptor interacting domain of Xenopus laevis N-CoR. We show that dnN-CoR can compete away endogenous N-CoR from TR and relieve the repression by unliganded TR in the frog oocyte transcription system. More importantly, we have introduced the dnN-CoR into tadpoles under the control of an inducible promoter through sperm-mediated transgenesis. We demonstrate that induction of the expression of dnN-CoR enhances the expression of various T3-response genes and accelerates development of the transgenic tadpoles as compared with the control wild type siblings. These results support the view that unliganded TRs repress T3-target genes in premetamorphic tadpoles to regulate developmental timing.
RESULTS
dnN-CoRs relieve unliganded TR-induced transcriptional repression in Xenopus laevis oocytes
Our previous studies have shown that N-CoR/SMRT-TBLR1 corepressor complexes are recruited by unliganded TR to target genes in premetamorphic tadpoles (Sachs et al., 2002; Tomita et al., 2004). We thus hypothesized that blocking corepressor recruitment to TR should relieve unliganded TR-induced gene repression in tadpoles and thus affect premetamorphic tadpole development. For this purpose, we designed a dnN-CoR, myc-ID monomer, that is composed of the binding site for TR but lacks binding sites for other corepressors present in the full length Xenopus laevis N-CoR (Fig. 1) (Sachs et al., 2002). Since dimerization of dominant negative constructs of coactivators enhanced the dominant negative activity on TR (B. Paul and Y.-B. Shi, unpublished observation), we reasoned that a myc-ID dimer consisting of two ID monomers connected with a linker peptide (Fig. 1) may be more effective in inhibiting repression by unliganded TR.
Figure 1. Schematic representation of dominant negative constructs of Xenopus laevis N-CoR.

Xenopus laevis N-CoR is a corepressor protein composed of 2498 amino acids. N-CoR can interact with nuclear hormone receptors including TR via the receptor interacting domain (ID) near the C-terminus. The repressor interacting domains (RDs) located in the N-terminal part are required for the recruitment of other corepressors such as TBL1/TBLR1 and HDAC3. The dominant negative forms, dnN-CoRs, used in this study are shown below. The myc-ID monomer comprises the ID (amino acids 1988-2349) fused to an N-terminal peptide containing myc tag and nuclear localizing sequences (NLS). The other dnN-CoR, myc-ID dimer, consists of two direct repeats of the ID separated by a linker sequence.
To examine the effect of the dnN-CoRs on TR-dependent transcription, we utilized the reconstituted frog oocyte system (Wong and Shi, 1995). In vitro transcribed mRNAs encoding FLAG-TRα and RXR were microinjected into the cytoplasm of the oocytes with or without mRNAs encoding myc-tagged dnN-CoRs. Two hours later, a reporter construct that contains the T3-dependent Xenopus TRβ promoter driving the expression of firefly luciferase was microinjected into the nuclei of the same oocytes together with a control plasmid containing the tk promoter driving the expression of Renilla luciferase. After overnight incubation in the presence or absence of T3, oocytes were lysed and assayed for luciferase activities. The ratio of firefly luciferase activity to Renilla luciferase activity was determined as a measure of the transcription level from the reporter. As shown in Fig. 2A, in the absence of T3, TRα/RXR overexpression reduced the reporter gene transcription (unliganded TR-induced repression). In the presence of T3, TRα/RXR overexpression increased the reporter gene transcription (liganded TR-induced activation). The dnN-CoRs relieved the gene repression by unliganded TR in a dose-dependent manner without affecting the expression of the reporter in the absence of TR (Fig. 2 and data not shown). This derepression was more dramatic with myc-ID dimer than the monomer (Fig. 2A, left panel). On the other hand, the dnN-CoRs had no significant effect on liganded TR-induced reporter gene activation (Fig. 2A, right panel), as expected. Given the greater effect of the dimer, we used myc-ID dimer for the rest of the study.
Figure 2. The dnN-CoRs derepress a T3-repsonse promoter by binding to TR and blocking endogenous N-CoR binding in the reconstituted frog oocyte system.
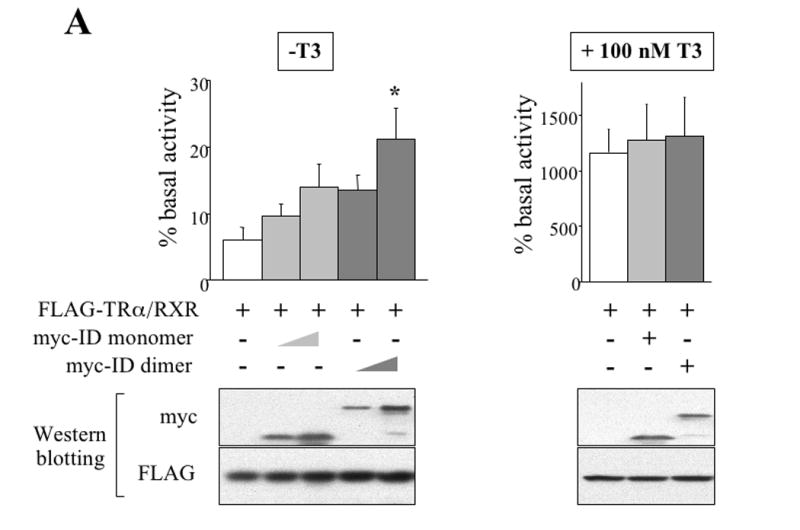

(A) The dimeric dnN-CoR relieves reporter gene repression by unliganded TR more effectively than the monomeric dnN-CoR. The mRNAs for FLAG-TRα/RXR and myc-tagged dnN-CoRs (myc-ID dimer or monomer) were injected into the cytoplasm of the frog oocytes. The firefly luciferase reporter vector together with the control Renilla luciferase plasmid was then injected into the nucleus. After overnight incubation with or without T3, the oocytes were lysed and assayed for luciferase activities. The ratio of firefly luciferase activity to Renilla luciferase activity was determined as a measure of the reporter gene transcription level. The result from each group was expressed as a percentage of the basal transcription level that was obtained from the oocytes without TRα/RXR mRNA injection. The experiment was repeated 8 times and the combined results are presented here. The same oocyte samples used in luciferase assay were subjected to Western blotting with anti-myc and anti-FLAG antibodies. Representative results were shown in the lower panels, confirming the expression of dnN-CoRs and FLAG-TRα. Note that both dnN-CoRs relieved the unliganded TR-induced repression, but the derepression was more potent with the myc-ID dimer (left graph, * p<0.05). Neither dnN-CoR affected the liganded-TR induced reporter gene activation (right graph).
(B) myc-ID dimer interacts with unliganded TRα in frog oocytes. The mRNAs for FLAG-TRα/RXR and myc-ID dimer were injected into the cytoplasm of oocytes as indicated. After overnight incubation with or without 100 nM T3, the oocytes were lysed and subjected to immunoprecipitation (IP) with anti-FLAG antibody against TR. Pre-IP lysates and IP samples were immunoblotted with anti-FLAG and anti-myc antibodies. Note that myc-ID dimer was co-immunoprecipitated with FLAG-TRα in the sample without T3 treatment (lane 3) but not in the T3-treated sample (lane 4).
(C) myc-ID dimer competes with the endogenous N-CoR for binding to unliganded TR. The mRNAs for FLAG-TRα/RXR and myc-ID dimer were injected into the cytoplasm of oocytes as indicated. After overnight incubation with or without 100 nM T3, the oocyte lysates were subjected to IP with anti-FLAG antibody. Pre-IP lysates and IP samples were immunoblotted with anti-FLAG, anti-N-CoR, and anti-myc antibodies. Similar to myc-ID dimer, endogenous N-CoR was co-immunoprecipitated with FLAG-TRα only in the sample without T3 treatment (compare lane 2 and 3 in the second panel). The myc-ID dimer overexpression reduced the amount of endogenous N-CoR co-immunoprecipitated with FLAG-TRα (compare lane 2 and 4 in the second panel). The experiments in Fig. 2 were repeated at least 3 times with similar results.
To analyze the molecular mechanism underlying the observed derepression by myc-ID dimer, co-immunoprecipitation assay was conducted. The mRNAs encoding FLAG-TRα/RXR and/or myc-ID dimer were injected into the cytoplasm of the oocytes. After overnight incubation in the absence or the presence of T3, oocytes were lysed and the extracts were incubated with anti-FLAG antibody to pull down the protein complex that contains FLAG-TRα. As intended, the interaction of myc-ID dimer with TRα was detected in the oocytes that had been incubated in the absence of T3 but not in the presence of T3 (Fig. 2B). Consistent with our previous study (Tomita et al., 2004), the recruitment of endogenous N-CoR to TRα was detected in the absence of T3 but not in the presence of T3 (Fig. 2C). Co-expression of myc-ID dimer reduced the amount of endogenous N-CoR bound to the unliganded TR (Fig. 2C), demonstrating that myc-ID dimer functions as a dominant negative form of N-CoR.
Induction of myc-ID dimer expression by heat shock in transgenic tadpoles
To investigate the effect of myc-ID dimer overexpression on frog development, the transgenesis vector, pH-myc-ID dimer-CG, was introduced into tadpoles. In this double-promoter construct, the expression of myc-ID dimer is under the control of heat shock-inducible promoter and GFP expression is driven by the eye specific γ-crystallin promoter (Fig 3A). This allows us to rear both wild type and transgenic animals together in the same tank to avoid possible variations in treatment conditions, because transgenic animals can be easily identified at the end of the treatment by their green eyes under a fluorescent microscope (Fig. 3B).
Figure 3. Transgenic analysis of the effects of overexpressing myc-ID dimer on animal development.


(A) Schematic representation of the construct used for transgenesis. The heat shock-inducible promoter drives the expression of the myc-ID dimer transgene. The construct also harbors GFP driven by γ-crystallin promoter as a marker to identify transgenic animals.
(B) A transgenic (Tg) and wild type (WT) Xenopus laevis tadpole at stage 46. The presence of GFP in the eye (arrows) indicates the presence of the transgene, myc-ID dimer, in the Tg tadpole. The arrowheads indicate the auto-fluorescence, likely due to the yolk remaining in the tadpoles.
(C) Experimental scheme using myc-ID dimer transgenic tadpoles. WT and Tg tadpoles at stage 46 (about 10 days old, shortly after feeding begins at stage 45) were heat shocked for 1 h at 33-34 °C on days 1 through 30. Some of the tadpoles were also treated with 1 nM of T3 for the first 5 days. The developmental stages of the tadpoles were examined every 5 days. Note that the experiment ended after 30 days, when the tadpoles reached stage 55 and plasma thyroid hormones (T3, 3, 5, 3′-triiodo-L-thyronine, and T4, 3, 5, 3′, 5′-tetraiodo-L-thyronine) become detectable, which would lead to the dissociation of both dnN-CoR and endogenous corepressors from TR.
Previously, we reported that the metamorphic process is blocked in the transgenic tadpoles overexpressing a dominant negative form of SRC3 (dnSRC3) that can compete away endogenous coactivator binding to liganded TR, indicating that the coactivator recruitment is required for the frog metamorphosis (Paul et al., 2005b). Interestingly, in such transgenic animals, there was little upregulation of T3-response gene when treated with T3, even though corepressor release occurred in the T3-treated transgenic animals. These results indicate that corepressor release alone is not sufficient to induce high levels of gene expression and metamorphosis. Therefore, we speculated that the possible effect of myc-ID dimer may not be very dramatic and a large number of animals would be needed to detect any phenotypic changes. Since it was difficult to obtain sufficient number of transgenic animals directly from the transgenesis procedure (F0 generation), we produced tadpoles of the F1 generation by crossing F0 transgenic frogs. Equal numbers of wild type and transgenic offspring from the same mating were reared in the same container. We chose to induce the expression of myc-ID dimer at the onset of feeding stage (stage 46), when TRα expression is high and the tadpoles are competent to respond to exogenous T3 (Shi et al., 1994; Tata, 1966; Yaoita and Brown, 1990). On the other hand, because endogenous T3 accumulates by stage 55 (Leloup et al., 1981) (Fig. 3C) and TR would thus take the liganded conformation that would not be affected by myc-ID dimer, we ended our experiments at this stage (about 30 days after stage 46).
To verify the induction of myc-ID dimer expression in transgenic animals, we subjected wild type and transgenic tadpoles at stage 46 to daily heat shock treatment together. The wild type and transgenic animals were separated after indicated number of days based on the presence of green eyes in the transgenic animals under a fluorescent microscope. Total RNA was then isolated from the whole animals and subjected to RT-PCR. The very low level of basal expression of myc-ID dimer present in transgenic animals before heat shock was increased after heat shock treatment of 1 or more days, while no expression was seen in wild type siblings (Fig. 4A). The expression of myc-ID dimer protein was also confirmed in transgenic animals but not in the wild type siblings after heat shock treatment (Fig. 4B).
Figure 4. Heat shock induces the expression of the myc-ID dimer transgene.

(A) The expression of the myc-ID dimer mRNA is induced in the transgenic tadpoles after heat shock. Transgenic tadpoles were identified by the GFP in the eyes at the end of the treatments. Total RNAs were isolated from individual wild type (WT) and transgenic (Tg) animals before (0) and after 1, 3, 5, 10 days of heat shock with or without T3 treatment. The cDNAs reverse-transcribed from the total RNAs were subjected to PCR using primers specific for the myc-ID dimer or ribosomal protein L8 (rpL8) (Shi and V. C.-T. Liang, 1994) as an internal control. Shown here is representative of three independent experiments with similar results. Each band corresponds to one WT or Tg animal heat-shocked for indicated number of days.
(B) Immunoprecipitation (IP) analysis confirms the expression of myc-ID dimer protein in the transgenic animals after heat shock treatment. The protein lysates from WT and Tg animals after 30 days of heat shock were immunoprecipitated with anti-myc antibody. IP samples were immunoblotted with anti-myc antibody (upper panel). Pre-IP samples were blotted with β-actin antibody (lower panel) as a loading control.
Transgenic expression of myc-ID dimer leads to the upregulation of T3-response genes
To study the effects of myc-ID dimer on gene expression and tadpole development, we first determined whether under our experimental conditions, tadpoles as early as stage 46 were able to properly respond to T3. Thus, we treated wild type tadpoles at stage 46 with or without 1 nM of T3 treatment. Total RNA was isolated from the whole animals and RT-qPCR was carried out to analyze the expression of several known T3-response genes. The expression of TRβ, stromelysin-3 (ST3), and T3-responsive basic leucine zipper transcription factor (TH/bZip), all of which are known direct T3-response genes (Ishizuya-Oka et al., 1997; Patterton et al., 1995; Ranjan et al., 1994), were significantly higher in T3-treated tadpoles than in non-treated counterparts after 1 day of T3 treatment and remained higher after 3 and 5 days (Fig. 5A). The upregulation of Xenopus hedgehog (xhh), which is also a direct but organ-specific T3-response gene (Stolow and Shi, 1995), became significant after 5 days of T3 treatment (Fig. 5A). The expression of HoxA1, which is known to be a retinoic acid (RA)-response gene (Kolm and Sive, 1995), was not affected by T3 treatment (Fig. 5A). As the developmental stages of premetamorphic tadpoles during this period are largely determined by the size and morphology of the hind limbs, we examined hind limb of T3-treated and control animals. As shown in Fig. 5B, after 5 days of T3 treatment, limb development was noticeably accelerated compared to animals without T3 treatment, although much less dramatic than that observed when higher levels of T3 are used. These results thus confirmed that functional TR is present by stage 46 and release of repression by unliganded TR during T3 treatment may contribute to gene activation and acceleration of limb development.
Figure 5. A low level of T3 (1nM) can activate T3-inducible genes and metamorphosis in early premetamorphic tadpoles.

(A) T3 activates various T3-response genes in early premetamorphic tadpoles. Wild type tadpoles at stage 46 were reared in the water with or without 1 nM T3. Total RNA was isolated from individual animals after 0, 1, 3, and 5 days. Each group had four individual animals. The cDNAs reverse-transcribed from the total RNAs were subjected to quantitative PCR. T3-response genes TRβ, stromelysin-3 (ST3), T3-responsive basic leucine zipper transcription factor (TH/bZip), and sonic hedgehog (xhh), were examined. A retinoic acid-response gene (HoxA1) was analyzed as a negative control. The expression level of each gene was normalized to that of rpL8 and expressed in arbitrary units. Note that expression of T3-response genes was significantly upregulated by T3 treatment (* p<0.05), whereas HoxA1 was unaffected. The xhh gene is an organ-specific T3-response gene and thus the upregulation was not very dramatic when analyzed in whole animals (Stolow and Shi, 1995). Similar results were obtained from another experiment with an independent batch of animals.
(B) T3 accelerates hind limb development in premetamorphic tadpoles. A wild type tadpole reared in (1 nM) T3-contaning water for 5 days had larger hind limbs (arrowhead) compared to a control tadpole without T3 treatment.
Next, to examine possible effect of myc-ID dimer on gene expression in premetamorphic tadpoles, total RNA was isolated from wild type and transgenic tadpoles after indicated heat shock treatment and RT-qPCR for known T3-response genes was performed. The expression levels of TRβ, ST3, TH/bZip, and xhh in transgenic animals tended to be higher than those in wild type counterparts at most of the time points (Fig. 6). Despite the small sample size (4 tadpoles/group), statistically significant upregulation (p<0.05) was observed for ST3 and xhh and similar results were obtained from another independent experiment (not shown). On the other hand, the RA-response gene, HoxA1, showed no significant upregulation in transgenic animals as compared with wild type siblings throughout the treatment (Fig. 6). While the effect of the dnN-CoR was expectedly much less than T3, which induces both derepression and activation (Fig. 5), these results support that unliganded TR participates in gene repression in premetamorphic tadpoles.
Figure 6. Expression of myc-ID dimer derepresses T3-response genes in premetamorphic tadpoles.

Wild type (WT) and myc-ID dimer transgenic (Tg) tadpoles at stage 46 were heat shocked for 1 hour per day. Total RNA was isolated from individual animals before (day 0) and after 1 and 5 days of heat shock treatment. Each group had four individuals. The cDNAs reverse-transcribed from the total RNAs were subjected to qPCR analysis as in Figure 5. White bars represent expression levels in WT animals and black bars represent those in Tg tadpoles. Note that expression of T3-response genes (TRβ, ST3, TH/bZip, and xhh) in Tg animals tended to be higher than WT counterparts at most of time points (*p<0.05), whereas such tendency was not seen in HoxA1 (slightly lower at one time point), a retinoic acid-response gene. Similar results were obtained from another experiment with an independent batch of animals.
Transgenic expression of myc-ID dimer accelerates premetamorphic tadpole development
To study the effects of dnN-CoR on development, both wild type and transgenic tadpoles from the same mating were heat shocked every day starting from stage 46 and reared together. The developmental stages of the tadpoles (10 tadpoles for each group) were determined every 5 days for up to 30 days, until stage 55 when endogenous T3 levels become detectable (Fig. 3C). Although no significant stage difference was observed between wild type and transgenic animals for the first 15 days, the stages of the transgenic animals were significantly more advanced after 20 to 30 days, the end of the experiment (Fig. 7A and Table 2). As shown in Fig. 7B, hind limbs of a representative transgenic animal were larger and morphologically more advanced than those of a wild type sibling (it is worth pointing out that the animal stage is largely determined by the size and morphology of the hind limbs during this premetamorphic period). Compared to the wild type siblings, the transgenic animals accelerated their development by about 1 stage (stage 55.3 vs. 54.4) (Table 2) or 6 days out of the total development time of about 22 days needed to go from stage 46 to 54 for the wild type animals under normal conditions (Nieuwkoop and Faber, 1956), which would extrapolate to about 7 days out of the 30 days under our experimental conditions. This is a very significant acceleration in development considering that it only takes about 2 months from fertilization to the end of metamorphosis with about one month involved in metamorphic transition. These results thus support the view that unliganded TR represses gene expression to prevent precocious metamorphosis.
Figure 7. Transgenic expression of myc-ID dimer accelerates tadpole development.

(A) Wild type (WT) and myc-ID dimer transgenic (Tg) sibling tadpoles were heat shocked every day starting from stage 46. Developmental stages of 10 WT and 10 Tg tadpoles were examined every 5 days. The average developmental stages of the animals at different time points were plotted (also see Table 2). Note that stages of Tg animals were significantly more advanced after 20 days of heat shock (* p<0.05). At the end of the experiments, the transgenic animals accelerated their development by about 1 stage compared to the wild type siblings. Similar acceleration was observed in another experiment with an independent batch of animals.
(B) Heat shocked Tg tadpoles develop to more advanced stages. Representative WT and Tg tadpoles are shown after 25 days of heat shock treatment. The right panel is a magnified image of the boxed area in the left panel, showing that limb buds of a Tg animal were larger and more advanced (around early stage 53) than those of a WT sibling (around early stage 52) (arrowheads).
Table 2.
Developmental acceleration by the myc-ID dimer transgene
| Days of heat shock | Stage (mean±SE) | Acceleration in Development | ||
|---|---|---|---|---|
| WT | Tg | Stage | Days | |
| 0 | 46.0±0 | 46.0±0 | 0 | 0 |
| 5 | 47.8±0.4 | 47.8±0.4 | 0 | 0 |
| 10 | 49.8±0.4 | 50.0±0 | 0.2 | 0.6 |
| 15 | 50.1±0.3 | 50.5±0.5 | 0.4 | 1.1 |
| 20 | 51.8±0.6 | 52.5±0.5 | 0.7 | 2.9 |
| 25 | 53.4±0.96 | 54.2±0.4 | 0.8 | 2.9 |
| 30 | 54.4±0.96 | 55.3±0.64 | 0.9 | 6.8 |
The developmental stages of the animals at different time points were tabulated with the standard errors (SE). The acceleration in development of transgenic animals (Tg) relative to wild type ones (WT) is the difference in the average stage numbers of the two groups. The acceleration in days was calculated based on the differences in the developmental stages and the developmental time according to [Nieuwkoop, 1956 #78] after normalizing against the actual time that the wild type animals took to reach the average stage (as shown) from stage 46 (e.g., it took 30 days under our experimental condition to go from stage 46 to 54.4 but 24 days according to [Nieuwkoop, 1956 #78]. Thus, the difference of 5.4 days between WT animals at stage 54.4 and Tg animals at 55.3 based on [Nieuwkoop, 1956 #78] would be equivalent to 6.8 days under our experimental conditions). Note that by the end of the experiment, the transgenic animals accelerated their development by about 0.9 stage compared to the wild type siblings or about 7 days (the time needed for stage 54.4 wild type tadpoles to reach stage 55.3) out of the 30-day experiment under our conditions. The conclusion was supported by another experiment with an independent batch of animals.
DISCUSSION
T3 has important roles in vertebrate development. These developmental effects are believed to be mediated by TRs. Although TRs have been demonstrated to function in the unliganded state in vitro, it has been difficult to obtain in vivo evidence to support a physiological role of unliganded TR during development. Here, by using the total dependence of frog metamorphosis as a model, we provide the first in vivo evidence for a role of unliganded TR in postembryonic development at the organismal level.
A developmental role of unliganded TR has also been implicated from a number of studies in mouse. First, the circulating level of T3 in the fetus is very low and dramatically increases after birth (Hadj-Sahraoui et al., 2000; Morreale de Escobar et al., 1994). In contrast, considerable amount of TR is expressed in fetal tissues (Nagasawa et al., 1997), suggesting that most of TR is likely to be present in the unliganded conformation during the fetal period. Second, Pax8−/− knockout mice, which lack thyroid follicular cells and exhibit severe congenital hypothyroidism, die in the early postnatal weeks due to multiple defects in intestine, bone, and brain (Mansouri et al., 1998). On the other hand, complete TR knockout mice (TRα0/0β−/−) are viable and have milder phenotype than Pax8−/− mice (Flamant et al., 2002; Flamant and Samarut, 2003). This discrepancy suggests some deleterious influence of unliganded TR on murine development, which is also supported by the fact that the lethal phenotype of Pax8−/− mice can be rescued by concomitant inactivation of TRα (Flamant et al., 2002). Finally, Mai et al. reported that heart rates and the expression of some cardiac T3-response genes of TRα knockout (TRα0/0) fetal mice in TRα0/0 mothers are higher than those of wild type (TRα+/+) fetuses in wild type (TRα+/+) mothers, suggesting repressing effects of unliganded TRα on cardiac function in embryonic life under physiological conditions (Mai et al., 2004). In this report, however, the authors were not able to compare TRα0/0 fetuses with TRα+/+ littermates from mothers of the same genetic background (i.e., both from TRα0/+ mothers) because of technical difficulties. Intra-uterine lives of fetuses are dependent on their mother and fetal conditions are affected by those of the mothers. Therefore, it might be difficult to completely rule out the possibility that the difference between TRα0/0 and TRα+/+ fetuses is derived from their distinct intra-uterine environments.
The free-living tadpoles and the requirement of T3 for metamorphosis make frogs a valuable model to analyze the possible role of TR in vertebrate development. In contrast to mice where maternal T3 may reach the fetus, TRs are unliganded throughout premetamorphosis in tadpoles that live independently of their mother (although some T3 may exist during embryogenesis (Dubois et al., 2006), unliganded TR is also required for embryogenesis of the eyes prior to the formation of a free-living tadpole (Havis et al., 2006)). Making use of the fact that N-CoR binds to TR in the absence of T3 to facilitate gene repression by unliganded TR, we have generated two dominant negative forms of N-CoR, myc-ID monomer and myc-ID dimer. Using the reconstituted frog oocyte system, where we previously demonstrated that unliganded TR recruits N-CoR /SMRT-TBLR1 complexes to mediate target gene repression (Tomita et al., 2004), we have shown that both forms of dnN-CoR could relieve unliganded TR-induced gene repression. As we intended, the derepression was more potent with myc-ID dimer than with the monomer. Furthermore, our immunoprecipitation results suggest that the derepression is due to the competition between dnN-CoR and endogenous N-CoR for binding to unliganded TR. Interestingly, even the dimer could only partially derepress the transcriptional repression by unliganded TR. One possible explanation could be that affinity of dnN-CoRs for unliganded TR is lower than that of endogenous N-CoR in the context of a full functional corepressor complex, resulting in incomplete competition and derepression. Alternatively, unliganded TR may be able to repress gene expression even in the absence of N-CoR/SMRT or related corepressor complexes, such as by binding directly to basal transcription proteins like TFIIB (Baniahmad et al., 1993). Regardless, given the preponderant role of TR in metamorphosis and the ability of relatively small changes in gene expression to significantly affect development, we examined whether this level of derepression could affect frog development.
Comparison of myc-ID dimer-overexpressing tadpoles and wild type siblings revealed that the expression of several early, direct T3-response genes, TRβ, ST3, TH/bZip, and xhh, is indeed higher in transgenic animals than in wild type siblings. Although the number of tadpoles used in each treatment group is limited (4 tadpoles/group), the differences are statistically significant at some of the time points and similar results were obtained in another independent set of experiments (data not shown). Similar to the results from the frog oocyte system, the increase in gene expression caused by myc-ID dimer in the transgenic animals is much less than that caused by T3 treatment of wild type animals. In addition, myc-ID dimer had little effect on the expression of these T3-response genes in T3-treated animals, i.e., T3 treatment leads to similar levels of expression of these genes in the transgenic and wild type animals (data not shown), consistent with the fact that myc-ID dimer does not bind to TR in the presence of T3. These findings strongly suggest that unliganded TR repressed T3-response genes during development, although the extent of repression appears to be relatively small and may vary among different genes. Interestingly, while myc-ID dimer led to an increase in expression of T3 response genes, it failed to do so on a RA-response gene, HoxA1. Our previous study has shown that unliganded RA receptor also recruits N-CoR to the target promoters to mediate gene repression in the frog oocyte (Tomita et al., 2003). The lack of change in HoxA1 gene expression in transgenic animals is consistent with the fact that bioactive retinoids are present in developing Xenopus laevis embryos (Blumberg et al., 1996) and supports the specificity of myc-ID dimer action.
It may not be surprising that the observed changes in gene expression and development caused by the dnN-CoR transgene are relatively small and vary among individual animals. Compared to in vitro and tissue culture cell studies, where one can routinely use large amounts of materials of identical background and properties, in vivo studies in animals are limited by the ability to use only a small number of animals and by the developmental asynchrony among individual animals. In this regard, it is of interest to note that in mouse embryos, the TRα knockout-induced increases in the expression of several T3-response genes in the heart, where TRα is the dominant TR form, were also relatively small and with large variation among different animals (Mai et al., 2004). Nonetheless, TRα knockout causes a small but significant increase in heart rate of the mouse embryos (Mai et al., 2004). Likewise, the relatively small changes in the expression of T3 response genes caused by the dnN-CoR transgene appear to have a significant effect on tadpole development. The transgenic expression of myc-ID dimer accelerated development within 20 days of its expression. During the developmental period between stage 46 and 55 when it is possible to study TR repression under physiological conditions, the most important developmental landmark is the growth and differentiation of the hind limbs. By the end of the 30-day experiment, transgenic animals accelerated their development by as much as 7 days. Such a dramatic change is consistent with the fact that tadpoles as early as stage 46 can be induced to accelerate development with even a low level of T3 (Fig. 5), supporting a critical role of unliganded TR in coordinating premetamorphic animal development and preventing precocious metamorphosis, which would be deleterious to animal health and survival in the wild.
While the dnN-CoR is likely to interact with other nuclear hormone receptors or transcription factors that bind to the same region of N-CoR as TRs and affect their function, we believe that the observed developmental effect is specific to TR for several reasons. 1) Once a tadpole is formed upon the completion of embryogenesis, only TR plays such a preponderant role in regulating the developmental transition into a frog. 2) The roles of other receptors or transcription factors in metamorphosis are either unknown or secondary to TR action and no other hormones or chemicals can affect metamorphosis in the absence of T3 and without affecting T3 pathways. 3) The presence of ligands for other nuclear receptors, such as RA, also prevents the dnN-CoR from affecting the expression of their target genes as we have shown for the HoxA1 gene. All these make frog development a unique model that allows us to correlate the developmental phenotype to the action of T3-TR system.
In summary, by introducing dnN-CoR into premetamorphic tadpoles, we have provided direct in vivo evidence for the presence as well as a role of unliganded TR-induced gene repression in a physiological setting at the organismal level. The dnN-CoR functions by competing away the binding of endogenous N-CoR to unliganded TR and derepressing T3-response genes in premetamorphic tadpoles. Most importantly, the expression of the dnN-CoR accelerates premetamorphic tadpole development, mimicking low dose T3 treatments. Our data thus provide one of the strongest pieces of in vivo evidence for the concept that unliganded TR represses T3-response genes to prevent precocious metamorphosis.
EXPERIMENTAL PROCEDURES
Cloning and constructs
Two dominant negative forms of N-CoR (dnN-CoRs), myc-ID monomer and dimer, were generated by PCR amplification of the sequences encompassing the receptor interaction domain (ID) of Xenopus laevis N-CoR (amino acids 1988 to 2357). The myc-ID monomer was generated to encode a fusion protein with an N-terminal myc epitope tag followed by the simian virus 40 (SV40) nuclear localization signal (NLS), using a forward primer containing the sequences encoding these fusion peptides in frame with the ID sequence of Xenopus laevis N-CoR. To facilitate cloning, the restriction site for EcoRI or XhoI was added to the 5′ end of forward primer or reverse primer, respectively. The primers used were forward primer 5′CATCATGAATTCACCGGTATGGAACAAAAACTCATCTCAGAAGAGGATCTGCCAAAGAAGAAGCGTAAGGTAGCGCACACAAAGTCACGCTAT3′ (the myc tag is underlined and the NLS is shown in boldface) and reverse primer 5′ATGATGCTCGAGTTATGATTGGTTCCCTTGTGGTACTCC3′. For expression and detection in Xenopus laevis oocytes, the PCR product was cloned into the EcoRI and XhoI sites of pSP64RI (S. Sato, National Institute of Diabetes and Digestive and Kidney Diseases), which contains the 5′ and 3′-untranslated regions of the Xenopus laevis β-globin gene flanking the multiple cloning sites.
The myc-ID dimer contained two copies of the ID sequence connected by a linker sequence. For this purpose, the ID sequence was amplified using a forward primer 5′CATCATCTCGAGGGAGGAGGAGGATCTGGAGGAGGAGGATCTGCGCACACAAAGT CACGCTAT3′ (the linker sequence is underlined) and the same reverse primer used for generating the ID monomer. The PCR product was then cloned into the XhoI site of pSP64RI-myc-ID monomer.
For transgenesis, myc-ID dimer was subcloned into the vector pCGHG (Fu et al., 2002) under the control of the heat shock-inducible promoter, replacing the original green fluorescent protein (GFP) fragment at this location, to produce the double-promoter construct pH-myc-ID dimer-CG, which also has the gene for GFP driven by the eye lens-specific γ-crystallin promoter.
Animals
Adult Xenopus laevis animals were purchased from Nasco (Fort Atkinson, WI) for oocyte assays and transgenesis as described below. Developmental stages were determined according to Nieuwkoop and Faber (Nieuwkoop and Faber, 1956).
Luciferase assays using frog oocytes
pSP64-FLAG-TRα (Toμιτα ετ αλ., 2004), pSP64-RXR (Wong and Shi, 1995), pSP64RI-myc-ID monomer and dimer were used to synthesize the corresponding mRNAs with a SP6 in vitro transcription kit (mMESSAGE mMACHINE; Ambion). The mRNAs for FLAG-TRα/RXR (5.75 ng/oocyte each) and/or mRNA for myc-RID monomer or dimer (5.75-23 ng/oocyte) were injected into the cytoplasm of 12 Xenopus laevis stage VI oocytes. The reporter plasmid DNA (0.33 ng/oocyte), which contained the T3-dependent Xenopus TRβ promoter driving the expression of the firefly luciferase (Amano et al., 2002), was injected into the oocyte nucleus, together with a control construct that contains the herpes simplex virus tk promoter driving the expression of Renilla luciferase (0.03 ng/oocyte). Following overnight incubation at 18°C in the absence or presence of 100 nM T3, oocytes were prepared for luciferase assay with the Dual-Luciferase Reporter Assay system (Promega) according to the manufacturer's recommendations. To verify protein expression from the injected mRNAs, the same oocyte lysates were subjected to Western blotting using anti-FLAG antibody (Sigma) for detection of FLAG-TRα and anti-myc antibody (Invitrogen) for detection of myc-ID monomer or dimer. These experiments were repeated 8 times and inter-group comparisons were done with the one-way analysis of variance (ANOVA) followed by Scheffe's F-test.
Co-immunoprecipitation using frog oocytes
The above-prepared mRNAs for FLAG-TRα/RXR (23 ng/oocyte each) and/or mRNA for myc-ID dimer (23 ng/oocyte) were injected into the cytoplasm of 20 Xenopus laevis stage VI oocytes. After overnight incubation at 18°C with or without 100nM of T3, the oocytes were Lysed In Ip Buffer (20 Mm HEPES, pH 7.5, 5 Mm KCl, 1.5 Mm MgCl2, 1 Mm EGTA, 10 Mm glycerophosphate, 50 mM NaCl, 0.1% NP-40, 1 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, and protease inhibitor mixture (Roche Applied Science)). After centrifugation at 14,000 rpm for 10 min at 4°C, a part of the supernatant was kept as the input sample and the rest was used for immunoprecipitation with Ezview Red ANTI-FLAG M2 Affinity Gel (Sigma). Each lysate was incubated with the gel for 4 hours and washed three times in the same IP buffer. The immunoprecipitates were boiled in sodium dodecyl sulfate (SDS) loading buffer, separated on an SDS-polyacrylamide gel, and immunoblotted with anti-FLAG, anti-myc, or anti-NCoR antibody. The anti-NCoR antibody was raised against Xenopus N-CoR N terminus fragment (amino acids 155–264) (Sachs et al., 2002) and does not cross-react with the dnN-CoRs (data not shown).
Transgenesis and animal treatment
The double promoter construct, pH-myc-ID dimer-CG, was introduced into Xenopus laevis through the restriction enzyme-mediated integration method (Kroll and Amaya, 1996). Transgenic animals were identified by GFP expression in the eyes due to the presence of the second promoter, the γ-crystallin promoter, driving the expression of GFP in the lens.
To obtain a sufficient number of transgenic animals for the following analyses, the transgenic animals were allowed to develop to reproductive ages and mated with each other. The same numbers of the wild type and transgenic offspring from the same mating were reared in the same container throughout the following experiments to ensure identical conditions of treatment for wild type and transgenic animals. These tadpoles were fed daily, and water was changed every other day. The tadpoles were photographed under bright field and fluorescence with a green fluorescence filter, and the image were merged to show GFP expression in transgenic animals.
To study the effect of the transgene, myc-ID dimer, on frog development, premetamorphic tadpoles at stage 46 (about 10 days old) were heat shocked for 1 h at 33-34 °C on days 1 through 30. During the first 5 days, some of the tadpoles were also treated with 1 nM of T3. The developmental stages of 10 tadpoles from each group were examined every 5 days. Inter-group comparisons of the developmental stages were done with the Mann-Whitney test.
Protein extraction from tadpoles and detection of myc-ID dimer
To verify the induction of myc-ID dimer protein in transgenic animals, wild type and transgenic tadpoles after 30 days of heat shock treatment were homogenized on ice in lysis buffer (20 mM HEPES, pH 7.5, 5 mM KCl, 1.5 mM MgCl2, 1 mM EGTA, 10 mM glycerophosphate, 150 mM NaCl, 1% NP-40, 1 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, and protease inhibitor mixture). After centrifugation at 14,000 rpm for 10 min at 4°C, a part of the supernatant was kept as the input sample and the rest was used for immunoprecipitation with Ezview Red Anti-c-Myc Affinity Gel (Sigma). Each lysate was incubated with the beads for 4 h and washed three times in the same lysis buffer. The immunoprecipitates were boiled in SDS loading buffer, separated on an SDS-polyacrylamide gel, and immunoblotted with anti-myc antibody to detect protein expression of myc-ID dimer. Input samples were blotted with anti-β actin antibody (Abcam Inc., Cambridge, MA) as a loading control.
RNA isolation from tadpoles and reverse transcription
Total RNA was extracted individually from 4 wild type and 4 transgenic tadpoles before and after 1, 3, 5, and 10 days of heat shock with or without T3 treatment using Trizol reagent (Invitrogen) as recommended by the manufacturer. To generate first-strand cDNA, 2 μg of the total RNA was reverse-transcribed with random primers using the High-Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA) at 25 °C for 10 min and then at 37 °C for 2 h. The first-strand cDNA was diluted with distilled water to a final volume of 100 μl for use in polymerase chain reaction (PCR) and quantitative PCR (qPCR) as described below.
PCR
To verify the induction of myc-ID dimer mRNA in transgenic animals, PCR was performed in 10 μl of Taq Buffer, containing 1 μl of the above prepared cDNA solution, 25mM of MgCl2, 2 mM dNTPs, and 0.5 U of rTaq DNA polymerase (Promega) with 0.5 μM each of forward and reverse primers for the myc-ID dimer or ribosomal protein L8 (rpL8) (Shi and Liang, 1994) as a control. The sequences of the primers used were 5′ATGGAACAAAAACTCATCTC 3′ (the myc tag sequence) and 5′GGAGGAGGAGGATCTGGAGG 3′ (the linker sequence) for myc-ID dimer and 5′CGTGGTGCTCCTCTTGCCAAG3′ and 5′GACGACCAGTACGACGAGCAG3′ for rpL8. The conditions for amplification were: 5 minutes at 94 °C, 30 cycles of incubation for 30 seconds at 94 °C, annealing for 30 seconds at 55 °C and incubation for 1 minute at 72 °C, followed by a final extension for 5 minutes at 72 °C. PCR products were resolved by electrophoresis on 1 % agarose gels and viewed by ethidium bromide staining.
qPCR
To compare the expression levels of various T3-response genes in wild type and transgenic animals with or without 1 nM of T3 treatment, qPCR was carried out on an ABI 7000 system (Applied Biosystems) using forward and reverse specific primers and TaqMan MGB probes per manufacturer's instruction. The cDNAs were synthesized from RNA isolated from individual wild type and transgenic tadpoles as described above. The genes examined were TRβ, T3-responsive basic leucine zipper transcription factor (TH/bZIP), sonic hedgehog (xhh), stromelysin-3 (ST3), and HoxA1. Expression level of each gene was normalized to that of the control gene rpL8. The primers used are summarized in Table 1. Twelve fivefold serial dilutions from cDNA prepared from the wild type tadpoles at stage 50 were used to generate the standard curve. Results from the experimental samples were within the range of the standard curve. Comparison between untreated and T3-treated animals or between wild type and transgenic animals were done with the unpaired t-test.
Table 1.
Primers for qPCR
| Probe | Forward primer | Reverse primer | TaqMan MGB probe |
|---|---|---|---|
| TRβ | CAAGAGTTGTTGATTTTGCCAAAAAGC | ACATGATCTCCATACAACAGCCTTT | CTGCCATGTGAAGACC |
| TH/bZip | GGACAAGTGAGAAGCAAGAACAAG | GGGTTGGGCAAGGAACAAG | TTCTGCTGCCCATTCAG |
| xhh | CGAGTCCAAAGCTCATATTCACTGT | AGCACCCGCCAGACTTG | CCACTGAGTTCTCTGCTTTG |
| ST3 | CACTTGTAGCCATTGTATCACACTCA | GCCATGATCTTTCTGAGGCTTTTC | ATGCATTCTCACAAGCTGT |
| Hox A1 | TGAAAGTGAAGAGAAATCCACCCAAA | GCTGGCCGGCATAACCATATT | CCCCGGCCTTACCTG |
| rpL8 | AGAAGGTCATCTCATCTGCAAACAG | CAATACGACCACCTCCAGCAA | CAACCCCAACAATAGCT |
Acknowledgments
This research was supported by the Intramural Research Program of the National Institute of Child Health and Human Development, NIH. Y. Sato was supported in part by JSPS (NIH) fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amano T, Leu K, Yoshizato K, Shi YB. Thyroid hormone regulation of a transcriptional coactivator in Xenopus laevis: implication for a role in postembryonic tissue remodeling. Dev Dyn. 2002;223:526–35. doi: 10.1002/dvdy.10075. [DOI] [PubMed] [Google Scholar]
- Baniahmad A, Ha I, Reinberg D, Tsai S, Tsai MJ, O'Malley BW. Interaction of human thyroid hormone receptor beta with transcription factor TFIIB may mediate target gene derepression and activation by thyroid hormone. Proc Natl Acad Sci U S A. 1993;90:8832–6. doi: 10.1073/pnas.90.19.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg B, Bolado JJ, Derguini F, Craig AG, Moreno TA, Chakravarti D, Heyman RA, Buck J, Evans RM. Novel retinoic acid receptor ligands in Xenopus embryos. P NATL ACAD SCI USA. 1996;93:4873–4878. doi: 10.1073/pnas.93.10.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz DR, Hsia SC, Fu L, Shi YB. A dominant-negative thyroid hormone receptor blocks amphibian metamorphosis by retaining corepressors at target genes. Mol Cell Biol. 2003;23:6750–8. doi: 10.1128/MCB.23.19.6750-6758.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz DR, Paul BD, Fu L, Shi YB. Molecular and developmental analyses of thyroid hormone receptor function in Xenopus laevis, the African clawed frog. Gen Comp Endocrinol. 2006;145:1–19. doi: 10.1016/j.ygcen.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Buchholz DR, Paul BD, Shi YB. Gene-specific changes in promoter occupancy by thyroid hormone receptor during frog metamorphosis. Implications for developmental gene regulation. J Biol Chem. 2005;280:41222–8. doi: 10.1074/jbc.M509593200. [DOI] [PubMed] [Google Scholar]
- Buchholz DR, Tomita A, Fu L, Paul BD, Shi YB. Transgenic analysis reveals that thyroid hormone receptor is sufficient to mediate the thyroid hormone signal in frog metamorphosis. Mol Cell Biol. 2004;24:9026–37. doi: 10.1128/MCB.24.20.9026-9037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke LJ, Baniahmad A. Co-repressors 2000. Faseb J. 2000;14:1876–88. doi: 10.1096/fj.99-0943rev. [DOI] [PubMed] [Google Scholar]
- Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–7. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- Dodd MHI, Dodd JM. The biology of metamorphosis. In: Lofts B, editor. Physiology of the amphibia. Academic Press; New York: 1976. pp. 467–599. [Google Scholar]
- Dubois GM, ebillot A, Kuiper GGJM, Verhoelst CHJ, Darras VM, Visser TJ, Demeneix BA. Deiodinase Activity Is Present in Xenopus laevis during Early Embryogenesis. Endocrinology. 2006;147:4941–4949. doi: 10.1210/en.2006-0609. [DOI] [PubMed] [Google Scholar]
- Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–95. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamant F, Poguet AL, Plateroti M, Chassande O, Gauthier K, Streichenberger N, Mansouri A, Samarut J. Congenital hypothyroid Pax8(−/−) mutant mice can be rescued by inactivating the TRalpha gene. Mol Endocrinol. 2002;16:24–32. doi: 10.1210/mend.16.1.0766. [DOI] [PubMed] [Google Scholar]
- Flamant F, Samarut J. Thyroid hormone receptors: lessons from knockout and knock-in mutant mice. Trends Endocrinol Metab. 2003;14:85–90. doi: 10.1016/s1043-2760(02)00043-7. [DOI] [PubMed] [Google Scholar]
- Fondell JD, Roy AL, Roeder RG. Unliganded thyroid hormone receptor inhibits formation of a functional preinitiation complex: implications for active repression. Genes Dev. 1993;7:1400–10. doi: 10.1101/gad.7.7b.1400. [DOI] [PubMed] [Google Scholar]
- Fu L, Buchholz D, Shi YB. Novel double promoter approach for identification of transgenic animals: A tool for in vivo analysis of gene function and development of gene-based therapies. Mol Reprod Dev. 2002;62:470–6. doi: 10.1002/mrd.10137. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–41. [PubMed] [Google Scholar]
- Hadj-Sahraoui N, Seugnet I, Ghorbel MT, Demeneix B. Hypothyroidism prolongs mitotic activity in the post-natal mouse brain. Neurosci Lett. 2000;280:79–82. doi: 10.1016/s0304-3940(00)00768-0. [DOI] [PubMed] [Google Scholar]
- Havis E, Le Mevel S, Dubois GM, Shi DL, Scanlan TS, Demeneix BA, Sachs LM. Unliganded thyroid hormone receptor is essential for Xenopus laevis eye development. EMBO J. 2006;25:4943–4951. doi: 10.1038/sj.emboj.7601356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havis E, Sachs LM, Demeneix BA. Metamorphic T3-response genes have specific co-regulator requirements. EMBO. 2003 doi: 10.1038/sj.embor.embor908. Reports 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Hsia SC, Wang H, Shi YB. Involvement of chromatin and histone acetylation in the regulation of HIV-LTR by thyroid hormone receptor. Cell Res. 2001;11:8–16. doi: 10.1038/sj.cr.7290061. [DOI] [PubMed] [Google Scholar]
- Ishizuya-Oka A, Ueda S, Shi YB. Temporal and spatial regulation of a putative transcriptional repressor implicates it as playing a role in thyroid hormone-dependent organ transformation. Dev Genet. 1997;20:329–37. doi: 10.1002/(SICI)1520-6408(1997)20:4<329::AID-DVG4>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Ito M, Roeder RG. The TRAP/SMCC/Mediator complex and thyroid hormone receptor function. Trends Endocrinol Metab. 2001;12:127–134. doi: 10.1016/s1043-2760(00)00355-6. [DOI] [PubMed] [Google Scholar]
- Jepsen K, Rosenfeld MG. Biological roles and mechanistic actions of co-repressor complexes. J Cell Sci. 2002;115:689–98. doi: 10.1242/jcs.115.4.689. [DOI] [PubMed] [Google Scholar]
- Jones PL, Sachs LM, Rouse N, Wade PA, Shi YB. Multiple N-CoR complexes contain distinct histone deacetylases. J Biol Chem. 2001;276:8807–11. doi: 10.1074/jbc.C000879200. [DOI] [PubMed] [Google Scholar]
- Jones PL, Shi YB. N-CoR-HDAC corepressor complexes: roles in transcriptional regulation by nuclear hormone receptors. Curr Top Microbiol Immunol. 2003;274:237–68. doi: 10.1007/978-3-642-55747-7_9. [DOI] [PubMed] [Google Scholar]
- Kolm PJ, Sive HL. Regulation of the Xenopus labial homeodomain genes, HoxA1 and HoxD1: activation by retinoids and peptide growth factors. Dev Biol. 1995;167:34–49. doi: 10.1006/dbio.1995.1005. [DOI] [PubMed] [Google Scholar]
- Kroll KL, Amaya E. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development. 1996;122:3173–83. doi: 10.1242/dev.122.10.3173. [DOI] [PubMed] [Google Scholar]
- Lazar MA. Thyroid hormone receptors: multiple forms, multiple possibilities. Endocr Rev. 1993;14:184–93. doi: 10.1210/edrv-14-2-184. [DOI] [PubMed] [Google Scholar]
- Leloup I, Buscaglia M, de Luze A. [Modulation of T4 to T3 conversion. Comparative aspects (author's transl)] Ann Endocrinol (Paris) 1981;42:454–60. [PubMed] [Google Scholar]
- Li J, Wang J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. Embo J. 2000;19:4342–50. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai W, Janier MF, Allioli N, Quignodon L, Chuzel T, Flamant F, Samarut J. Thyroid hormone receptor alpha is a molecular switch of cardiac function between fetal and postnatal life. Proc Natl Acad Sci U S A. 2004;101:10332–7. doi: 10.1073/pnas.0401843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–9. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri A, Chowdhury K, Gruss P. Follicular cells of the thyroid gland require Pax8 gene function. Nat Genet. 1998;19:87–90. doi: 10.1038/ng0598-87. [DOI] [PubMed] [Google Scholar]
- McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–74. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- Morreale de Escobar G, Calvo R, Escobar del Rey F, Obregon MJ. Thyroid hormones in tissues from fetal and adult rats. Endocrinology. 1994;134:2410–5. doi: 10.1210/endo.134.6.8194467. [DOI] [PubMed] [Google Scholar]
- Nagasawa T, Suzuki S, Takeda T, DeGroot LJ. Thyroid hormone receptor beta 1 expression in developing mouse limbs and face. Endocrinology. 1997;138:1276–81. doi: 10.1210/endo.138.3.5022. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J, editors. Normal table of Xenopus laevis. 1. North Holland Publishing; Amsterdam, The Netherlands: 1956. [Google Scholar]
- Patterton D, Hayes WP, Shi YB. Transcriptional activation of the matrix metalloproteinase gene stromelysin-3 coincides with thyroid hormone-induced cell death during frog metamorphosis. Dev Biol. 1995;167:252–62. doi: 10.1006/dbio.1995.1021. [DOI] [PubMed] [Google Scholar]
- Paul BD, Buchholz DR, Fu L, Shi YB. Tissue- and gene-specific recruitment of steroid receptor coactivator-3 by thyroid hormone receptor during development. J Biol Chem. 2005a;280:27165–27172. doi: 10.1074/jbc.M503999200. [DOI] [PubMed] [Google Scholar]
- Paul BD, Fu L, Buchholz DR, Shi YB. Coactivator recruitment is essential for liganded thyroid hormone receptor to initiate amphibian metamorphosis. Mol Cell Biol. 2005b;25:5712–24. doi: 10.1128/MCB.25.13.5712-5724.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman AJ, Stanley F, Samuels HH. Thyroid hormone nuclear receptor. Evidence for multimeric organization in chromatin. J Biol Chem. 1982;257:930–8. [PubMed] [Google Scholar]
- Puzianowska-Kuznicka M, Damjanovski S, Shi YB. Both thyroid hormone and 9-cis retinoic acid receptors are required to efficiently mediate the effects of thyroid hormone on embryonic development and specific gene regulation in Xenopus laevis. Mol Cell Biol. 1997;17:4738–49. doi: 10.1128/mcb.17.8.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachez C, Freedman LP. Mediator complexes and transcription. Curr Opin Cell Biol. 2001;13:274–80. doi: 10.1016/s0955-0674(00)00209-x. [DOI] [PubMed] [Google Scholar]
- Ranjan M, Wong J, Shi YB. Transcriptional repression of Xenopus TR beta gene is mediated by a thyroid hormone response element located near the start site. J Biol Chem. 1994;269:24699–705. [PubMed] [Google Scholar]
- Sachs LM, Damjanovski S, Jones PL, Li Q, Amano T, Ueda S, Shi YB, Ishizuya-Oka A. Dual functions of thyroid hormone receptors during Xenopus development. Comp Biochem Physiol B Biochem Mol Biol. 2000;126:199–211. doi: 10.1016/s0305-0491(00)00198-x. [DOI] [PubMed] [Google Scholar]
- Sachs LM, Jones PL, Havis E, Rouse N, Demeneix BA, Shi YB. Nuclear receptor corepressor recruitment by unliganded thyroid hormone receptor in gene repression during Xenopus laevis development. Mol Cell Biol. 2002;22:8527–38. doi: 10.1128/MCB.22.24.8527-8538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs LM, Shi YB. Targeted chromatin binding and histone acetylation in vivo by thyroid hormone receptor during amphibian development. Proc Natl Acad Sci U S A. 2000;97:13138–43. doi: 10.1073/pnas.260141297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber AM, Das B, Huang H, Marsh-Armstrong N, Brown DD. Diverse developmental programs of Xenopus laevis metamorphosis are inhibited by a dominant negative thyroid hormone receptor. Proc Natl Acad Sci U S A. 2001;98:10739–44. doi: 10.1073/pnas.191361698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YB. Amphibian Metamorphosis: From morphology to molecular biology. John Wiley & Sons, Inc.; New York: 1999. p. 288. [Google Scholar]
- Shi YB, Liang VCT. Cloning and characterization of the ribosomal protein L8 gene from Xenopus laevis. Biochimica et Biophysica Acta. 1994;1217:227–228. doi: 10.1016/0167-4781(94)90042-6. [DOI] [PubMed] [Google Scholar]
- Shi YB, Liang VCT, Parkison C, Cheng SY. Tissue-dependent developmental expression of a cytosolic thyroid hormone protein gene in Xenopus: its role in the regulation of amphibian metamorphosis. FEBS Letters. 1994;355:61–64. doi: 10.1016/0014-5793(94)01173-7. [DOI] [PubMed] [Google Scholar]
- Shi YB, VCT Liang VCT. Cloning and characterization of the ribosomal protein L8 gene from Xenopus laevis. Biochimica et Biophysica Acta. 1994;1217:227–228. doi: 10.1016/0167-4781(94)90042-6. [DOI] [PubMed] [Google Scholar]
- Stolow MA, Shi YB. Xenopus sonic hedgehog as a potential morphogen during embryogenesis and thyroid hormone-dependent metamorphosis. Nucleic Acids Res. 1995;23:2555–62. doi: 10.1093/nar/23.13.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tata JR. Requirement for RNA and protein synthesis for induced regression of the tadpole tail in organ culture. Develop Biol. 1966;13:77–94. doi: 10.1016/0012-1606(66)90050-9. [DOI] [PubMed] [Google Scholar]
- Tomita A, Buchholz DR, Obata K, Shi YB. Fusion protein of retinoic acid receptor alpha with promyelocytic leukemia protein or promyelocytic leukemia zinc finger protein recruits N-CoR-TBLR1 corepressor complex to repress transcription in vivo. J Biol Chem. 2003;278:30788–95. doi: 10.1074/jbc.M303309200. [DOI] [PubMed] [Google Scholar]
- Tomita A, Buchholz DR, Shi YB. Recruitment of N-CoR/SMRT-TBLR1 corepressor complex by unliganded thyroid hormone receptor for gene repression during frog development. Mol Cell Biol. 2004;24:3337–46. doi: 10.1128/MCB.24.8.3337-3346.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MJ, O'Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–86. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- Underhill C, Qutob MS, Yee SP, Torchia J. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J Biol Chem. 2000;275:40463–70. doi: 10.1074/jbc.M007864200. [DOI] [PubMed] [Google Scholar]
- Wen YD, Perissi V, Staszewski LM, Yang WM, Krones A, Glass CK, Rosenfeld MG, Seto E. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc Natl Acad Sci U S A. 2000;97:7202–7. doi: 10.1073/pnas.97.13.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J, Shi YB. Coordinated regulation of and transcriptional activation by Xenopus thyroid hormone and retinoid X receptors. J Biol Chem. 1995;270:18479–83. doi: 10.1074/jbc.270.31.18479. [DOI] [PubMed] [Google Scholar]
- Wong J, Shi YB, Wolffe AP. A role for nucleosome assembly in both silencing and activation of the Xenopus TR beta A gene by the thyroid hormone receptor. Genes Dev. 1995;9:2696–711. doi: 10.1101/gad.9.21.2696. [DOI] [PubMed] [Google Scholar]
- Yaoita Y, Brown DD. A correlation of thyroid hormone receptor gene expression with amphibian metamorphosis. Genes Dev. 1990;4:1917–24. doi: 10.1101/gad.4.11.1917. [DOI] [PubMed] [Google Scholar]
- Yoon HG, Chan DW, Huang ZQ, Li J, Fondell JD, Qin J, Wong J. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. Embo J. 2003;22:1336–46. doi: 10.1093/emboj/cdg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell. 2002;9:611–23. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- Zhang J, Lazar MA. The mechanism of action of thyroid hormones. Annu Rev Physiol. 2000;62:439–66. doi: 10.1146/annurev.physiol.62.1.439. [DOI] [PubMed] [Google Scholar]


