Abstract
We used quantitative real-time RT-PCR to not only investigate the mRNA levels of anthrax toxin receptor 1 (ANTXR1) and 2 (ANTXR2) in the murine J774A.1 macrophage cells and different tissues of mice, but also evaluate the effect of anthrax edema toxin and Bacillus anthracis Sterne spores on the expression of mRNA of these receptors. The mRNA transcripts of both receptors was detected in J774A.1 cells and mouse tissues such as the lung, heart, kidney, spleen, stomach, jejunum, brain, skeleton muscle, and skin. The ANTXR2 mRNA level was significantly higher than that of ANTXR1 in J774A.1 cells and all tissues examined. The mRNA expression of both receptors in the lung was the highest among the tissues evaluated. Interestingly, the mRNA expression of both receptors in J774A.1 cells was up-regulated by edema toxin. In addition, ANTXR mRNA expression in the lung was down-regulated after subcutaneous inoculation of B. anthracis Sterne spores as well as after intranasal administration of anthrax toxin-based vaccine BioThrax™. These results suggest that anthrax edema toxin and B. anthracis Sterne spore are involved in the ANTXR mRNA regulation in host cells.
Keywords: Anthrax toxin receptor, Bacillus anthracis, Lung, Macrophage, mRNA, RT-PCR
1. Introduction
Anthrax is a disease resulting from infection by spores of the Gram-positive bacteria Bacillus anthracis. The formation of spores protects B. anthracis and allows it to remain dormant and survive harsh chemical and thermal distress until the local environment becomes more suitable for growth [1]. The disease manifests itself in three ways, resulting from three separate modes of infection. The most common occurrence of anthrax results from cutaneous exposure, where B. anthracis infects through a cut or abrasion on the skin. Secondly, gastrointestinal anthrax occurs through consumption of contaminated food products by gaining entry in the gut. The final and by far most deadly form of anthrax is inhalational or pulmonary anthrax caused by B. anthracis infection through respiratory system [2, 3]. The reason for the extreme severity of inhalational anthrax is unclear. The weaponization of anthrax aims to make use of the pulmonary mode of infection via the mass production of anthrax spores. Clearly the potential of bioterrorism threat involving anthrax underscores the need for investigation into prevention, vaccine development, and research detailing bacterial/host interactions and pathogenesis at the molecular level.
The outer layer of the B. anthracis spore consists of various proteins, polysaccharides, and lipids. Macrophages engulf the spore inadvertently creating an opportunity for germination [1]. The bacterium owes its virulence to the two plasmids pXO2 and pXO1. pXO2 codes for the poly-D-γ-glutamic acid capsule [4, 5]. It has been postulated that the capsule is antiphagocytic and able to facilitate systemic invasion and dissemination within the bloodstream [6]. pXO1 codes for the three anthrax toxin protein components that interact on the surface of mammalian cells: edema factor (EF), a Ca2+- and calmodulin-dependent adenylate cyclase; lethal factor (LF), a Zn2+-metalloprotease; and protective antigen (PA83, 83 kDa). Anthrax toxin assembly begins upon the binding of PA83 to one of two anthrax toxin receptors: anthrax toxin receptor 1 (ANTXR1)/Tumor Endothelial Marker 8 (TEM8), a product of the TEM8 gene originally found to be upregulated in colorectal cancer [3], or anthrax toxin receptor 2 (ANTXR2)/capillary morphogenesis protein 2 (CMG2) [7]. PA83 facilitates the entry of EF and LF into the cell. Upon binding to the toxin receptor, PA83 is cleaved by a cell surface furin into PA63 and subsequently oligomerizes into a heptameric pore that creates binding sites for up to three molecules of EF or LF with nanomolar affinity. The entire receptor-toxin complex is internalized by receptor-mediated endocytosis [7]. Once within the cytosol, EF and LF catalyze reactions that result in toxicity. The combination of LF and PA is called lethal toxin (Letx) which has been shown to cleave members of the mitogen-activated protein kinase kinase (MAPKK) family, including Mek1, Mek2, and MKK isotypes 1–4 and 6–7 [8], leading to host death. Edema toxin (Edtx), a combination of EF and PA, raises the level of cAMP, activates protein kinase A (PKA), disrupts water homeostasis, and inhibits phagocytosis of the bacterium by neutrophils allowing anthrax to evade the immune system [3].
While the interaction of the toxin components has been the subject of intense investigation, less is known regarding the true physiological function (s) of the two anthrax toxin receptors. Discovery of the first anthrax receptor ANTXR1 showed that the first 364 amino acids were identical to TEM8 [9]. Expression of the mouse homolog of ANTXR1 (TEM8) was found to be upregulated in the vasculature of the developing mouse embryo and also shown to be significantly upregulated in human tumor angiogenesis [10, 11]. ANTXR1/TEM8 is expressed in a variety of tissues [12]; however, the precise physiological function of ANTXR1/TEM8 is not known. Shortly after the discovery of ANTXR1, a second anthrax toxin receptor was identified as ANTXR2 (CMG2) [13]. It has the highest degree of homology with TEM8 compared to any protein described to date, including 1) a signal peptide 2) a von Willebrand factor (VWA) type A domain and 3) a type I transmembrane region. The two protein sequences share 40% overall amino acid identity and 60% identity within their VWA/I domains. Different variants of CMG2 are expressed in many tissues [13]. At present, three isoforms of ANTXR1 and four isoforms of ANTXR2 have been described resulting from alternative mRNA splicing [9, 13]. However, information about the correlation of ANTXR expression with anthrax toxins and spore infection is limited.
In this research, we have evaluated the expression levels of ANTXR1 and ANTXR2 mRNA in murine J774A.1 macrophage cells and different tissues of A/J mice in the absence and presence of the anthrax Edtx by using quantitative real time RT-PCR. We also determined the differential mRNA expression of the two receptors resulted from the stimulation by different effectors: the anthrax Edtx, B. anthracis Sterne spores, and an anthrax toxin-based vaccine BioThrax™ (Anthrax Vaccine Adsorbed).
2. Results
2.1. Assessment of mRNA levels of ANTXR1 and ANTXR2 in murine J774A.1 cells
Toward a greater understanding of how macrophages respond to encountering the Edtx, we chose to first investigate the mRNA expression of ANTXR1 and 2 in the J774A.1 cells. Fig. 1A displays the relative abundance of ANTXR2 to ANTXR1 mRNA, with the RT-PCR amplification plot displayed in Fig. 1B. As shown, the expression of ANTXR2 mRNA exceeds that of ANTXR1 mRNA by roughly 90-fold. This result is in qualitative agreement with recent finding by others [14, 15].
Fig. 1.
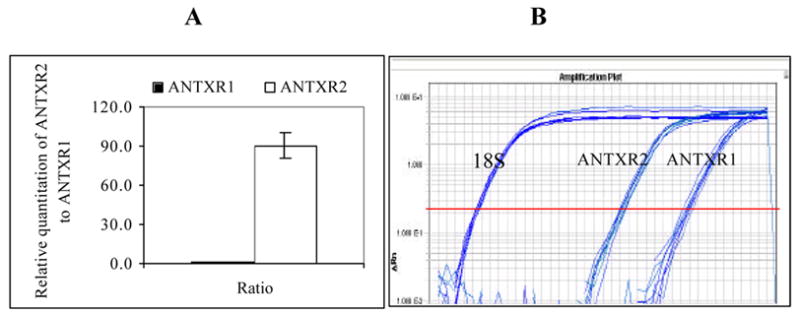
Relative expression level of ANTXR2/ANTXR1 in J774A.1 macrophage cells. Total RNA was isolated from J774A.1 cells and one-step real time RT-PCR was carried out to assess the relative abundance of ANTXR2 mRNA to ANTXR1 mRNA. (A). Ratio of relative quantization of ANTXR2/ANTXR1; (B). Real time RT-PCR amplification plot. (n = 12)
2.2. Effect of edema toxin on ANTXR mRNA expression in J774A.1 cells
To characterize the effect of anthrax Edtx on the gene expression of the toxin receptors, we measured the relative abundance of both ANTXR1 and 2 mRNA when J774A.1 cells were titrated with increasing concentrations of each individual toxin component PA and EF. Fig. 2 depicts the results, with all graphs on the left representing ANTXR1 (Fig. 2A1 and 2B1) and those on the right corresponding to ANTXR2 (Fig. 2A2 and 2B2). Since EF cannot bind ANTXR, it is not surprising that incubation of J774A.1 cells with EF alone had no effect on mRNA expression of either receptor (Fig. 2B1 and 2B2). However, in comparing the effect on expression of mRNA of ANTXR1 and ANTXR2 with varying amounts of PA, a significant change in mRNA expression was found for ANTXR1 but not ANTXR2 when PA concentration in the medium was 1 μg/ml. Since both receptors were present during the experiment, it would appear from this data that the highest amount of PA tested (1 μg/ml) had a stimulating effect on the steady-state mRNA expression of ANTXR1 but not of ANTXR2.
Fig. 2.
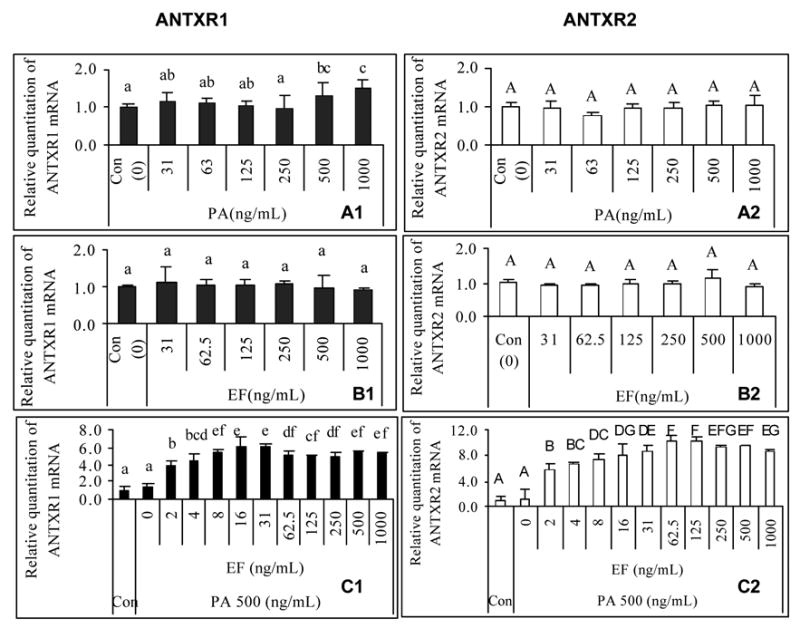
Effect of anthrax edema toxin on expression of ANTXR1 (Panel 1, left) and ANTXR2 (Panel 2, right) mRNA in J774A.1 cells. Total RNA was isolated from J774A.1 cells treated with different concentrations of anthrax toxin components, and then one-step real time RT-PCR was carried out to assess the relative abundance of ANTXR1 and ANTXR2 mRNA as compared to levels of 18S rRNA. One sample from control groups without toxin treatment was designated as a calibrator. Different letters represent significant differences between treatments in the same plot (P < 0.05). (A). PA-treated; (B). EF-treated; (C). constant PA + EF titration; (1). ANTXR1; (2). ANTXR2. (n = 8)
In order to determine what effect the combination of PA and EF (Edtx) would have on the transcription of ANTXR in J774A.1 cells, we observed the effect on mRNA expression by titrating increasing concentrations of EF incubated with a constant amount of PA (500 ng/ml). This resulted in a dramatic increase in relative mRNA expression for both ANTXR1 and ANTXR2 genes (Fig. 2C1 and 2C2). Thus Edtx stimulates the upregulation of ANTXR mRNA expression.
2.3. ANTXR mRNA expression levels in mice
The picture of overall expression of ANTXR1 and ANTXR2 mRNA in different tissues is incomplete, so our next goal was to describe the distribution of mRNA transcripts for these genes in the mouse. After performing RT-PCR analysis, Fig. 3 shows that both receptors’ mRNA is expressed in all mouse tissues that were examined. Although these results are in conflict with earlier results in which expression of ANTXR1 (TEM8) was restricted to tumor endothelium and developing vasculature [10], we are in general agreement with a more recent examination of mouse tissues showing widespread expression of ANTXR1 mRNA [12]. Fig. 3A shows clearly that the level of expression of ANTXR1 mRNA in the lung far exceeds that in any other tissue. With respect to the ANTXR2, the level of mRNA expression is also highest in the lung among all tissues analyzed (Fig. 3B). As there were mRNA of ANTXR2 detected in all tissues examined here, data in Fig. 3B also qualitatively agrees with previous findings [13]. However, the overall expression of ANTXR2 mRNA in mouse tissues is much more evenly distributed than that of ANTXR1 mRNA.
Fig. 3.
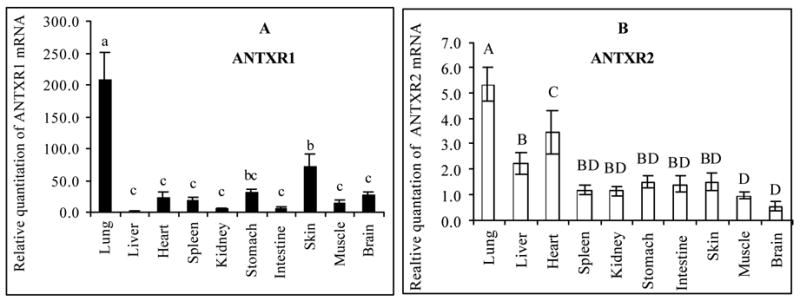
Differential expression of ANTXR1 (Panel A) and ANTXR2 (Panel B) mRNA in mouse tissues. Total RNA was isolated from A/J mouse tissues and one-step real-time RT-PCR was carried out to assess relative abundance of ANTXR1 and ANTXR2 mRNA as compared to 18S rRNA. A mouse liver total RNA sample purchased from Ambion was designated as a calibrator. Different letters represent significant differences between treatments in the same plot (P < 0.05). (n = 8)
To illustrate the difference in relative mRNA expression levels between ANTXR2 and ANTXR1, we determined the expression ratio of both receptors in our study. Fig. 4 shows that the mRNA level of ANTXR2 always exceeded that of ANTXR1 in all tissues examined. The most obvious difference in the expression ratio of these two receptors is seen for the liver, which is ~65-fold higher. This is the only tissue examined in our study that approaches the drastic difference in expression seen in the macrophage J774A.1 cell line (Fig. 1). The ratio of ANTXR2 and ANTXR1 mRNA is approximately the same (4:1) in the lung and skin, the two major sites for B. anthracis entry to cause inhalational and cutaneous anthrax, respectively. ANTXR2 mRNA expression in the intestine, another site for pathogen entry to cause digestive anthrax, is nearly 20-fold higher than that of ANTXR1 mRNA.
Fig. 4.
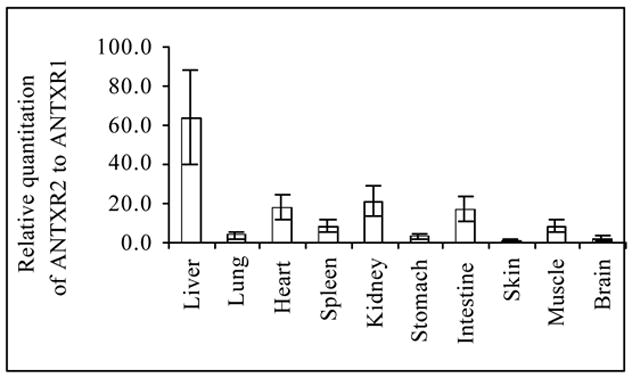
The mRNA expression level of ANTXR2 is always higher than that of ANTXR1 in all mouse tissues examined. Relative expression levels using data obtained from the experiments in Fig. 3 are presented to compare the ratio of mRNA expression of both receptors in mouse tissues (n = 8).
2.4. Effects of B. anthracis Sterne spores and anthrax toxin-based vaccine on ANTXR mRNA expression
After establishing basal levels of ANTXR mRNA expression in mouse tissues and observing that ANTXR2 mRNA is the more abundant ANTXR in all tissues examined, we proceeded to investigate the regulation of these receptors’ mRNA expression. Toward illuminating the response of each receptor under B. anthracis infection and an in vivo setting, we evaluated how the mRNA expression of ANTXR1 and 2 changed when mice were subcutaneously challenged with 100 × LD50 of B. anthracis Sterne spores. A/J mice are susceptible to the Sterne strain, a nonencapsulated attenuated strain lacking the poly-D-γglutamic acid capsule [16, 17]. Infection with the live spores results in anthrax symptoms in 3–4 days and death between 5–10 days. This experiment attempts to monitor changes in ANTXR mRNA expression en route to a fatal exposure to anthrax spore infection.
Due to the large difference in relative change of expression for ANTXR1 mRNA in this experiment, the results for the lung are presented separately in Fig. 5A, while results for all other tissues are shown in Fig. 5B. The expression of ANTXR1 mRNA in the lung drops nearly 5-fold upon subcutaneous challenge with spores (Fig. 5A). When the other tissues were examined for ANTXR1 mRNA expression levels after administration with the Sterne strain, transcripts of ANTXR1 mRNA either dropped nearly 2–4 fold (the brain, heart, kidney, muscle, skin, and stomach), or they remained relatively unchanged (the intestine, liver, and spleen) (Fig. 5B).
Fig. 5.
Subcutaneous injection of B. anthracis Sterne spores to mice significantly affects relative expression of ANTXR1 and ANTXR2 mRNA. After A/J mice were administrated subcutaneously with 100 × LD50 B. anthracis Sterne spores, total RNA was isolated from mice tissues at 0, 24 and 48 h to assess relative abundance of ANTXR1 and ANTXR2 mRNA to 18S rRNA by one-step quantitative real time RT-PCR. A mouse liver total RNA sample purchased from Ambion was designated as a calibrator. (A). Changes in relative expression of ANTXR1 mRNA in mice lungs; (B). Changes in relative expression of ANTXR1 mRNA in all remaining mice tissues; (C). Changes in relative expression of ANTXR2 mRNA in all mice tissues examined. * P < 0.05, ** P < 0.01 compared with the controls (untreated mice) (n = 8).
Next, we examined the effect of subcutaneous injection of the Sterne strain on expression of ANTXR2 mRNA. Fig. 5C shows that across all of the tissues examined, the only significant change was seen in the lung (2.2- fold decrease). Taken together, there appear to be different responses in mRNA expression of each receptor after a subcutaneous injection of the Sterne strain spores. The mRNA expression of both receptors in the lungs are sensitive to spore infection, but in the rest of the mouse, ANTXR1 mRNA levels change considerably while those of ANTXR2 mRNA do not. This data suggests that in an infection with live spores, ANTXR1 is more sensitive to downregulation than ANTXR2. Fig. 5 represents an interesting outcome after subcutaneous injection of Sterne spores: ANTXR mRNA expression in the lungs are most affected despite no direct delivery of the spores to that location. This may imply that anthrax toxins traversed to the lung through the bloodstream or through macrophages engulfed the spores.
BioThrax™ (Anthrax Vaccine Adsorbed) is the only currently licensed human vaccine against anthrax in the USA [18]. PA is the major component of the vaccine and antibodies against PA have been shown to protect guinea pigs against lethal challenge with anthrax toxin [19]. In addition, the vaccine contains undefined quantities of LF, EF, and other somatic components of B. anthracis [20, 21]. To expand our investigation on regulation of ANTXR mRNA expression, we intranasally administered BioThrax™ into A/J mice. Fig. 6 shows that expression of both receptors mRNA dropped approximately 35% 48 hours after only a single dose of 30 μl BioThrax™. Interestingly, regulation of ANTXR2 mRNA expression occurred at the 24-hour time point whereas regulation from ANTXR1 mRNA was only detected at the 48-hour time point. ANTXR2 mRNA expression appears to be more sensitive than ANTXR1 does in a direct intranasal administration of the effecter that represents an antigen presentation designed to elicit immunity via an anthrax toxin-based vaccine.
Fig. 6.
Effect on relative expression of ANTXR mRNA in the lung after intranasal administration of BioThrax™ vaccine to mice. Mice were inoculated with 30 μl of BioThrax™ vaccine by an intranasal route. Total RNA was isolated from lungs at 24 and 48 h after treatment to assess relative abundance of ANTXR1 and ANTXR2 mRNA as compared to 18S rRNA by one-step quantitative real-time RT-PCR. One of the samples from the control groups (treated with vehicle buffer) at same time point was designated as a calibrator. * P < 0.05, ** P < 0.01, compared with the controls (n = 4).
3. Discussion
In order for an invading pathogen to cause infection in a host, it must find a way to circumvent innate and adaptive immune mechanisms. The challenge for B. anthracis is to have an ability to bypass the obstacles of host entry: the skin, the mucosal lining of the respiratory tract, and the intestine. The macrophage is an important component in the life cycle of B. anthracis spore germination. The phagocytic nature of the macrophage is designed to engulf foreign bacteria that have bypassed the aforementioned lines of defense, and yet it appears that the macrophage acts as an enabler of a successful infection of B. anthracis spores by creating an environment that facilitates their conversion to a germinating state [6, 14]. Previous research has shown that B. anthracis spore germinates in the macrophage allowing the bacteria to multiply and spread into the bloodstream, attaining levels of 109 B. anthraci bacteria per milliliter of blood [22]. At the epicenter of this life cycle are the three anthrax toxin components, EF, LF, and PA, and the two anthrax receptors, ANTXR1 and ANTXR2. Recent work has demonstrated that macrophage susceptibility to spore challenge is dependent on ANTXR expression [14]. Our report herein allows for an expansion of our understanding of anthrax pathogenesis from the vantage point of ANTXR expression by providing new information about bacterial/host interaction as well as protein/receptor function and regulation.
Others have shown that PA binds to ANTXR2 (CMG2) with an extremely high affinity of KD = 170 pM or 780 pM in the presence of Mg2+ or Ca2+ ions, respectively [23], and that the binding affinity of ANTXR1 (TEM8) for PA is nearly 1000-fold lower than that of ANTXR2 (CMG2) [24]. In qualitative agreement with another recent study [14, 15], our work shows that ANTXR2 is by far the predominate receptor in the J774A.1 macrophage cells and mouse tissues. Taken together, these observations suggest that the ANTXR2 receptor is a significant means of entry for anthrax toxins.
It was no surprise that individually titrating the ANTXR with EF or PA had no discernable effect on receptor expression in macrophage cells (Fig. 2A and 2B). However, results with combination of these two toxin components are illuminating when viewed in light of regulation of the ANTXR. Most revealing is the fact that Edtx can dramatically upregulate mRNA expresion of both receptors (Fig. 2C). Recent research by Maldonado-Arocho et al. also demonstrated that Edtx induces ANTXR expression in monocyte-derived cells [25]. Since it is known that EF is an adenylate cyclase which elevates intracellular levels of cAMP, our data could be interpreted to suggest that increased cAMP level in cells stimulate increased transcription of both ANTXR1 and ANTXR2 genes. By inducing the production of more receptors, Edtx may promote increased entry of both EF and LF into the cell.
Edtx not only impairs phagocyte function [22], but also causes cytotoxicity and tissue necrosis [26, 27]. The N-terminal fragment of LF (LFn) has been shown to enter cells without PA resulting in a hypothesis that LF needs PA binding not for entry, but to function as a toxin [28]. However, it is unknown if EF is capable of cellular entry in the absence of PA. While it has been shown that transient expression of EF from a plasmid resulted in a manifestation of some cytopathological effects congruent with exposure to Edtx [28, 29], EF contain binding sites only for PA and not for the receptors [3]. Our observation might be explained by the interactions of EF with PA on the two receptors. These interactions could result in the activation or repression of different signaling pathways through some conformational change in ANTXR and the co-receptor low-density lipoprotein receptor-related protein 6 (LRP6) [30]. Alternatively, once delivered to cytosol, the intracellular effect of EF could result in the activation or repression of different pathways that regulate ANTXR gene transcription. In evaluating our data with the current understanding of the signaling effects of anthrax toxin, a question emerges: is the regulation of transcription of ANTXR due to some conformational change or signal conferred by ANTXR and LRP6 resulting from the event of receptor binding and translocating EF into the cytosol, or is it the result of the enzymatically active moieties of EF exerting their influence on downstream signaling? Future studies could help resolve this issue.
Current and future structural understanding of conserved domains may prove useful. The two anthrax toxin receptors share 40% overall amino acid identity and 60% identity within their VWA regions. There is complete conservation within their MIDAS motif, a metal-ion dependent adhesion site often involved in ligand binding [13]. VWA domains are often sites for protein-protein interactions in cell adhesion proteins such as integrins. There is accumulated evidence from studies of the α-integrin VWA domain demonstrating its existence in an open and closed conformation which display high and low affinities for ligand, respectively [13]. It would be interesting to see if an open/closed conformation in ANTXR also correlated with signaling involved in regulation of transcription of its own gene.
This work was also undertaken to more fully characterize the expression of known anthrax toxin receptors, ANTXR1 (TEM8) and ANTXR2 (CMG2). Both receptors are conserved between diverse species, including zebrafish, humans, mouse, and rat; this suggests that they fulfill an important and possibly unknown physiological function [31]. CMG2 was originally discovered to be differentially regulated during endothelial cell morphogenesis. It was subsequently shown to be localized to the endoplasmic reticulum, and a recombinant form of CMG2 was demonstrated to bind collagen type IV and laminin, suggesting a connection with basement matrix synthesis and assembly. However, RT-PCR in that work was only able to detect expression of CMG2 in human placenta [32]. More recent work has shown that CMG2 is widely expressed in human tissues [13], and mutations within CMG2 have been shown to cause two allelic disorders, juvenile hyaline fibromatosis (JHF) and infantile systemic hyalinosis (ISH) [33]. Here, we show that in the mouse, ANTXR2 (CMG2) mRNA is expressed in all tissues examined and has the highest level in the lung. While ANTXR1 (TEM8) has been shown to be overexpressed in tumor vessels, supporting a possible role in angiogenesis [34], the general expression pattern shown in this work as well as in previous work [12] suggests that a complete description of the physiological role this receptor plays remains to be determined. Nonetheless, we have demonstrated that infection with spores affects the expression of ANTXR1 in more tissues than ANTXR2. In turn, regulation of ANTXR2 appears to play a significant role in the lung upon challenge with either spores or an anthrax toxin-based vaccine. Although both receptors are widely expressed, ANTXR2 always outnumbers ANTXR1 in all mouse tissues examined. We acknowledge the significance of the finding that the lung has the highest ANTXR mRNA expression level and its relevance to the severity of inhalational anthrax warrants to be further explored.
4. Materials and methods
4.1. Materials
Recombinant PA and EF were purchased from List Biologicals, Inc.(Campbell, CA). B. anthracis Sterne strain spores were from the Anthrax Spore Vaccine (U.S. Vet license #188) which is a viable suspension of the Sterne Strain 34F2 spores in saponin (Colorado Serum Company, Denver, CO). BioThrax™ vaccine (AVA) was produced by BioPort Corporation (Lansing, MI). Mouse monocyte macrophage cell line J774A.1 was purchased from the American Type Culture Collection (Manassas, VA). TRI RNA/DNA/Protein Isolation Reagent and 1-bromo-3-chloropropane (BCP) were from Molecular Research Center, Inc. (Cincinnati, OH). One-step Real-time RT-PCR and QuantiTect Sybr Green RT-PCR Kits were purchased from Qiagen (Valencia, CA).
4.2. Cell culture and sample collection
24-well tissue culture plates were seeded with 1×105 mouse macrophage cells J774A.1 per well in 1 ml of Dulbecco’s Modified Eagles’s medium (DMEM) supplemented with 10% FBS (Hyclone), 2 g/l sodium bicarbonate from Sigma (St. Louis, MO), 100 U/ml penicillin, and 100 μg/ml streptomycin (GIBCO, Carlsbad, CA, USA), and were grown until approximately 80% confluent. The cells were either treated with individual PA (0–1μg/ml), EF (0–1 μg/ml), or with the combination (PA+EF) for 6 h in humidified air with 5% CO2 at 37°C. After the media was removed, 0.2 ml TRI RNA/DNA/Protein Isolation Reagent was added into each well and the plates were incubated for 10 min at room temperature. Then the samples were frozen at −80°C until RNA isolation.
4.3. Animals, treatments, and tissue sample collection
Female A/J mice at 6–8 weeks old were purchased from Jackson Laboratory (Bar Harbor, ME) and housed under biosafety level 2 (BSL2) pathogen-free conditions in the animal facility at the University of Rochester Medical Center. Mice were grouped to 4 per cage and maintained in a controlled environment (22 ± 2°C; 12 h light/12 h dark cycles). The animals were provided Laboratory Rodent Diet 5001 with ad libitum access to food and water. The animal research herein reported was conducted in facilities with programs accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care.
4.3.1. Experiment 1
Eight mice were sacrificed to collect tissue samples of lung, liver, heart, spleen, kidney, stomach, intestine (jejunum), ventral skin, and brain (thalamus).
4.3.2. Experiment 2
16 mice were equally divided into either a treatment or control group. The treatment group was subcutaneously injected with 100×LD50 of B. anthracis Sterne strain spores as previously described [17], while the control group was subcutaneously injected with an identical volume of 0.9% NaCl solution. Four animals in each group were sacrificed for tissue sampling at 24 and 48 h post-injection when no symptom of anthrax was observed.
4.3.3. Experiment 3
8 mice were allotted into a treatment and a control group. The treatment group was intranasally administered with 30 μl BioThrax™ Vaccine. The control group was intranasally administered the same volume of 0.9% NaCl solution. Four mice in each group were sacrificed for samples collection of lung tissues at 24 and 48 h after the treatments.
4.4. RNA isolation
Total RNA from J774A.1 cells or mouse tissues were isolated by using TRI reagent according to the manufacturer’s protocol. Briefly, cells were harvested from each well of 24-well plates and lysed in 0.2 ml TRI Reagent; 50–100 mg tissue samples were homogenized in 1.0 ml TRI Reagent. DNA, protein, and RNA were separated by centrifugation after addition of 1-bromo-3-chloropropane (BCP). RNA was precipitated from the aqueous phase with isopropanol, washed with ethanol, and solubilized in diethylpyrocarbonate (DEPC)-treated water. DNA was removed from the RNA preparation by using a DNA-free kit (Ambion) according to the manufacturer’s protocol. DNase I-treated RNA samples were purified by passing through a Qiagen RNeasy column. After the purity of RNA was assessed by 260- and 280-nm spectrophotometer reading, the RNA samples were aliquotted and stored at −80°C.
4.5. Real-time quantitative RT-PCR
The relative abundance of mouse ANTXR1 and 2 mRNA to 18S rRNA was assessed by quantitative real-time RT-PCR using the one-step QuantiTect SYBR Green RT-PCR kit (Qiagen) and the ABI Prism 7900 sequence detection system (Applied Biosystems). The nucleotide sequences of the primers and the real-time RT-PCR conditions are shown in Table 1. ANTXR1 and 2 sequences were analyzed through a BLAST algorithm to ensure that the primers generated were unique to each sequence in order to avoid cross-reaction. 18S rRNA was used as an internal quantitative standard. Each sample was run in duplicate and repeated twice. Controls were analyzed in parallel to verify the absence of DNA in the RNA preparation as well as the absence of primer dimers in control samples without template RNA. In addition, RT-PCR products were analyzed by gel electrophoresis, and in all cases, a single product was observed at the appropriate base pair size. Amounts of ANTXR mRNA in different samples were normalized relative to 18S rRNA. A mouse liver total RNA sample purchased from Ambion, Inc. (Austin, TX) was used as a calibrator during each experiment with mouse tissue samples.
Table 1.
Nucleotide sequences of primers and RT-PCR conditions for analysis of ANTXR mRNA expression
| GeneBank Access | Expected Product Size | Primer sequences | RT-PCR reaction conditions | |
|---|---|---|---|---|
| ANTXR1 | NM054041 | 231 bp (975–1205) | F: 5′-CTTTCAAGTGGTCGTAAGAG-3′
R: 5′-GTGATGATGACAAGAACTGGA-3′ |
RT: 50°C 30 min.
94°C 15 min. 1 cycle |
| ANTXR2 | NM 33738 | 242 bp (1088–1330) | F: 5′-TCCTCCAAGTGTCTGTGTGTAG-3′
R: 5′-GGCTGTGATTGTTAAGGATC-3′ |
PCR: 94 °C 15 s
55 °C 30 s 72 °C 30 s 40 cycles |
| 18S rRNA | AY248756 | 101 bp (950–1050) | F: 5′-CGCCGCTAGAGGTGAAATTCT-3′
R: 5′-CGAACCTCCGACTTTCGTTCT-3′ |
4.6. Statistical analysis
ANOVA and T-test were performed to analyze the results and are expressed as mean ± SE. Data presented are representatives of experiments run in duplicates and repeated twice. A paired comparison was done between the control groups and the experimental groups under different treatment conditions, or between tissues. All statistical analysis was performed using Statistica 6.0 software from StatSoft, Inc. (Tulsa, OK).
Acknowledgments
This work was supported by National Institutes of Health grant AI053598 to M. Z. The US Department of Defense provided BioThrax™ (AVA) for our research. The authors are grateful to Michael E. Pichichero, Sarah Van Cor-Hosmer, and Danielle C. Alcena for careful reviewing of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Baillie L, Hibbs S, Tsai P, Cao GL, Rosen GM. Role of superoxide in the germination of Bacillus anthracis endospores. FEMS Microbiol Lett. 2005;245(1):33–8. doi: 10.1016/j.femsle.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 2.Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Friedlander AM, et al. Anthrax as a biological weapon: medical and public health management. Working Group on Civilian Biodefense. Jama. 1999;281(18):1735–45. doi: 10.1001/jama.281.18.1735. [DOI] [PubMed] [Google Scholar]
- 3.Bradley KA, Young JA. Anthrax toxin receptor proteins. Biochem Pharmacol. 2003;65(3):309–14. doi: 10.1016/s0006-2952(02)01455-7. [DOI] [PubMed] [Google Scholar]
- 4.Leppla SH, Robbins JB, Schneerson R, Shiloach J. Development of an improved vaccine for anthrax. J Clin Invest. 2002;110(2):141–4. doi: 10.1172/JCI16204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makino S, Uchida I, Terakado N, Sasakawa C, Yoshikawa M. Molecular characterization and protein analysis of the cap region, which is essential for encapsulation in Bacillus anthracis. J Bacteriol. 1989;171(2):722–30. doi: 10.1128/jb.171.2.722-730.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guidi-Rontani C. The alveolar macrophage: the Trojan horse of Bacillus anthracis. Trends Microbiol. 2002;10(9):405–9. doi: 10.1016/s0966-842x(02)02422-8. [DOI] [PubMed] [Google Scholar]
- 7.Lacy DB, Wigelsworth DJ, Scobie HM, Young JA, Collier RJ. Crystal structure of the von Willebrand factor A domain of human capillary morphogenesis protein 2: an anthrax toxin receptor. Proc Natl Acad Sci U S A. 2004;101(17):6367–72. doi: 10.1073/pnas.0401506101. Epub 2004 Apr 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brossier F, Mock M. Toxins of Bacillus anthracis. Toxicon. 2001;39(11):1747–55. doi: 10.1016/s0041-0101(01)00161-1. [DOI] [PubMed] [Google Scholar]
- 9.Bradley KA, Mogridge J, Mourez M, Collier RJ, Young JA. Identification of the cellular receptor for anthrax toxin. Nature. 2001;414(6860):225–9. doi: 10.1038/n35101999. [DOI] [PubMed] [Google Scholar]
- 10.Carson-Walter EB, Watkins DN, Nanda A, Vogelstein B, Kinzler KW, St Croix B. Cell surface tumor endothelial markers are conserved in mice and humans. Cancer Res. 2001;61(18):6649–55. [PubMed] [Google Scholar]
- 11.Hotchkiss KA, Basile CM, Spring SC, Bonuccelli G, Lisanti MP, Terman BI. TEM8 expression stimulates endothelial cell adhesion and migration by regulating cell-matrix interactions on collagen. Exp Cell Res. 2005;305(1):133–44. doi: 10.1016/j.yexcr.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 12.Bonuccelli G, Sotgia F, Frank PG, Williams TM, de Almeida CJ, Tanowitz HB, et al. ATR/TEM8 is highly expressed in epithelial cells lining Bacillus anthracis’ three sites of entry: implications for the pathogenesis of anthrax infection. Am J Physiol Cell Physiol. 2005;288(6):C1402–10. doi: 10.1152/ajpcell.00582.2004. Epub 2005 Feb 2. [DOI] [PubMed] [Google Scholar]
- 13.Scobie HM, Rainey GJ, Bradley KA, Young JA. Human capillary morphogenesis protein 2 functions as an anthrax toxin receptor. Proc Natl Acad Sci U S A. 2003;100(9):5170–4. doi: 10.1073/pnas.0431098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banks DJ, Barnajian M, Maldonado-Arocho FJ, Sanchez AM, Bradley KA. Anthrax toxin receptor 2 mediates Bacillus anthracis killing of macrophages following spore challenge. Cell Microbiol. 2005;7(8):1173–85. doi: 10.1111/j.1462-5822.2005.00545.x. [DOI] [PubMed] [Google Scholar]
- 15.Premanandan C, Lairmore MD, Fernandez S, Phipps AJ. Quantitative measurement of anthrax toxin receptor messenger RNA in primary mononuclear phagocytes. Microb Pathog. 2006;41(4–5):193–8. doi: 10.1016/j.micpath.2006.05.003. Epub 2006 Jul 18. [DOI] [PubMed] [Google Scholar]
- 16.Welkos SL, Keener TJ, Gibbs PH. Differences in susceptibility of inbred mice to Bacillus anthracis. Infect Immun. 1986;51(3):795–800. doi: 10.1128/iai.51.3.795-800.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng M, Xu Q, Hesek ED, Pichichero ME. N-fragment of edema factor as a candidate antigen for immunization against anthrax. Vaccine. 2006 Jan 30;24(5):662–70. doi: 10.1016/j.vaccine.2005.08.056. Epub 2005 Aug 26. [DOI] [PubMed] [Google Scholar]
- 18.Anthrax Vaccine Adsorbed (BioThrax) Product Insert. Lansing, Michigan: BioPort Corporation; 2002. January 31, [Google Scholar]
- 19.Reuveny S, White MD, Adar YY, Kafri Y, Altboum Z, Gozes Y, et al. Search for correlates of protective immunity conferred by anthrax vaccine. Infect Immun. 2001;69(5):2888–93. doi: 10.1128/IAI.69.5.2888-2893.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kudva IT, Griffin RW, Garren JM, Calderwood SB, John M. Identification of a protein subset of the anthrax spore immunome in humans immunized with the anthrax vaccine adsorbed preparation. Infect Immun. 2005;73(9):5685–96. doi: 10.1128/IAI.73.9.5685-5696.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedlander AM, Pittman PR, Parker GW. Anthrax vaccine: evidence for safety and efficacy against inhalational anthrax. Jama. 1999;282(22):2104–6. doi: 10.1001/jama.282.22.2104. [DOI] [PubMed] [Google Scholar]
- 22.Collier RJ, Young JA. Anthrax toxin. Annu Rev Cell Dev Biol. 2003;19:45–70. doi: 10.1146/annurev.cellbio.19.111301.140655. [DOI] [PubMed] [Google Scholar]
- 23.Wigelsworth DJ, Krantz BA, Christensen KA, Lacy DB, Juris SJ, Collier RJ. Binding stoichiometry and kinetics of the interaction of a human anthrax toxin receptor, CMG2, with protective antigen. J Biol Chem. 2004;279(22):23349–56. doi: 10.1074/jbc.M401292200. Epub 2004 Mar 24. [DOI] [PubMed] [Google Scholar]
- 24.Scobie HM, Thomas D, Marlett JM, Destito G, Wigelsworth DJ, Collier RJ, et al. A soluble receptor decoy protects rats against anthrax lethal toxin challenge. J Infect Dis. 2005;192(6):1047–51. doi: 10.1086/432731. Epub 2005 Aug 10. [DOI] [PubMed] [Google Scholar]
- 25.Maldonado-Arocho FJ, Fulcher JA, Lee B, Bradley KA. Anthrax oedema toxin induces anthrax toxin receptor expression in monocyte-derived cells. Mol Microbiol. 2006;61(2):324–37. doi: 10.1111/j.1365-2958.2006.05232.x. [DOI] [PubMed] [Google Scholar]
- 26.Voth DE, Hamm EE, Nguyen LG, Tucker AE, Salles II, Ortiz-Leduc W, et al. Bacillus anthracis oedema toxin as a cause of tissue necrosis and cell type-specific cytotoxicity. Cell Microbiol. 2005;7(8):1139–49. doi: 10.1111/j.1462-5822.2005.00539.x. [DOI] [PubMed] [Google Scholar]
- 27.Firoved AM, Miller GF, Moayeri M, Kakkar R, Shen Y, Wiggins JF, et al. Bacillus anthracis edema toxin causes extensive tissue lesions and rapid lethality in mice. Am J Pathol. 2005;167(5):1309–20. doi: 10.1016/S0002-9440(10)61218-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kushner N, Zhang D, Touzjian N, Essex M, Lieberman J, Lu Y. A fragment of anthrax lethal factor delivers proteins to the cytosol without requiring protective antigen. Proc Natl Acad Sci U S A. 2003;100(11):6652–7. doi: 10.1073/pnas.1131930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong J, Beeler J, Zhukovskaya NL, He W, Tang WJ, Rosner MR. Anthrax edema factor potency depends on mode of cell entry. Biochem Biophys Res Commun. 2005;335(3):850–7. doi: 10.1016/j.bbrc.2005.07.132. [DOI] [PubMed] [Google Scholar]
- 30.Wei W, Lu Q, Chaudry GJ, Leppla SH, Cohen SN. The LDL receptor-related protein LRP6 mediates internalization and lethality of anthrax toxin. Cell. 2006;124(6):1141–54. doi: 10.1016/j.cell.2005.12.045. [DOI] [PubMed] [Google Scholar]
- 31.Scobie HM, Young JA. Interactions between anthrax toxin receptors and protective antigen. Curr Opin Microbiol. 2005;8(1):106–12. doi: 10.1016/j.mib.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 32.Bell SE, Mavila A, Salazar R, Bayless KJ, Kanagala S, Maxwell SA, et al. Differential gene expression during capillary morphogenesis in 3D collagen matrices: regulated expression of genes involved in basement membrane matrix assembly, cell cycle progression, cellular differentiation and G-protein signaling. J Cell Sci. 2001;114(Pt 15):2755–73. doi: 10.1242/jcs.114.15.2755. [DOI] [PubMed] [Google Scholar]
- 33.Hanks S, Adams S, Douglas J, Arbour L, Atherton DJ, Balci S, et al. Mutations in the gene encoding capillary morphogenesis protein 2 cause juvenile hyaline fibromatosis and infantile systemic hyalinosis. Am J Hum Genet. 2003;73(4):791–800. doi: 10.1086/378418. Epub 2003 Aug 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nanda A, St Croix B. Tumor endothelial markers: new targets for cancer therapy. Curr Opin Oncol. 2004;16(1):44–9. doi: 10.1097/00001622-200401000-00009. [DOI] [PubMed] [Google Scholar]


