Abstract
Objective
To analyze the comparison, through their results, of two distinct approaches applied to aligning two representations of anatomy.
Materials
Both approaches use a combination of lexical and structural techniques. In addition, the first approach takes advantage of domain knowledge, while the second approach treats alignment as a special case of schema matching. The same versions of FMA and GALEN were aligned by each approach. 2199 concept matches were obtained by both approaches.
Methods and results
For matches identified by one approach only (337 and 336 respectively), we analyzed the reasons that caused the other approach to fail.
Conclusions
The first approach could be improved by addressing partial lexical matches and identifying matches based solely on structural similarity. The second approach may be improved by taking into account synonyms in FMA and identifying semantic mismatches. However, only 33% of the possible one-to-one matches among anatomical concepts were identified by the two approaches together. New directions need to be explored in order to handle more complex matches.
Keywords: ontology, anatomy, Foundational Model of Anatomy (FMA), GALEN, ontology alignment
1. Introduction
Anatomy is central to the biomedical domain. While macroscopic anatomy is required for the representation of diseases and procedures, subcellular anatomy has become increasingly important for molecular biology. Not only is a sound representation of anatomy fundamental to biomedicine, but the various representations of anatomy currently available also need to be aligned in order to ensure interoperability. This need inspired two groups of researchers to take up the challenge of aligning two sizeable representations of anatomy: the Foundational Model of Anatomy (FMA) and the GALEN common reference model.
The first effort in aligning these two systems occurred at the U.S. National Library of Medicine (NLM). In parallel, but unrelated to it, another alignment was performed at Microsoft Research. Both approaches use a combination of lexical and structural techniques. In addition, the first approach takes advantage of domain knowledge, while the second approach is domain-independent and thus can be applied to other domains.
The contribution of this study is a comparison and analysis of the results of the two alignments in an effort to determine the strengths and weaknesses of each approach. This analysis illustrates how each approach can be improved based on the results of the other.
2. Background
2.1. Approaches to aligning ontologies
Ontology alignment is an active field of research. The objective of aligning ontologies is to identify correspondence among entities (i.e., concepts and relationships) across ontologies with overlapping content. Some ontology systems essentially rely on manual curation for their alignment. In Cyc, for example, several ontologies of varying complexity were aligned with Cyc’s large commonsense knowledge base through manually written term mapping predicates [1]. Among the many automatic and semi-automatic methods developed for merging and aligning ontologies, some are specific to this task, while others treat ontology alignment as a specific example of a more general problem. A brief overview of these methods is presented next.
Specific approaches
Specifically developed for aligning and merging ontologies are the interactive tools PROMPT [2] and Chimaera [3], which make suggestions to users based on the similarity between terms, relationships, instances and slot constraints identified across ontologies. The ONION system semi-automatically generates articulation rules to represent the semantic implication between terms across ontologies based on a graph-oriented model extended with some algebraic operators [4]. The bottom-up FCA-MERGE approach offers a structural description of the global merging process under a mathematical framework including the computation of the pruned concept lattice [5].
What distinguishes the first alignment in this study from other specific approaches is the use of domain knowledge. Implicit knowledge embedded in concept names and combination of relations is made explicit to facilitate the alignment. Semantic constraints are used to ensure that the concepts aligned belong to the same domain.
Generic approaches
The problem of aligning two ontologies can be seen as an example of the problem of schema matching, which has been a subject of database research for many years. Approaches to schema matching are surveyed in [6] which categorizes approaches based on the type of information used to compute the match result. Such information may include linguistic information about the names of elements, constraint information such as keys and is-a relationships, and structural information such as the set of component elements of a given element.
Most schema matching algorithms work by computing a similarity matrix, whose rows and columns denote elements of the two schemas to be matched. The value of each cell of the matrix is a real number in the range [0, 1] which denotes the degree of similarity of the row and column elements. Usually, two or more matching techniques are combined to produce the similarity matrix. For example, the Cupid algorithm [7] uses a first phase that computes a linguistic match and then a second phase to incorporate structural information. The COMA system offers a platform where matching techniques can be flexibly combined in different ways [8].
After the matrix is computed, a mapping between the two schemas is constructed, e.g., for each row, one can select the column with largest similarity value provided that the corresponding cell exceeds a given threshold. Techniques for computing the mapping are discussed in [9]. The second alignment in this study uses schema matching algorithm and, more specifically, relies on the Cupid algorithm.
Like many approaches, both alignments studied in this paper compare concepts based on lexical information (i.e., concept names) and structural information (i.e., relationships to other concepts). Other algorithms also exploit instance information (e.g., [6,10]). In our study, the instances of anatomical classes correspond to the organs, tissues and cells of individual persons (e.g., this author’s liver). Ontologies of anatomy do not typically record information about instances, but only about classes. For this reason, approaches based on instance information were not considered in this study.
2.2. Approaches to comparing alignments
In the process of aligning ontologies, the alignment itself only represents a first step. Comparing several approaches to aligning ontologies or several alignments requires that the information resulting from the alignment be represented uniformly. Several formalisms have been designed for representing alignments, including a RDF format and the corresponding ontology alignment API [11], an extension to the OWL language [12], and a framework for defining formal languages for specifying alignments and their associated semantics [13]. A uniform formalism not only facilitates the comparison of alignments, but also enables various operations to be performed on the alignments, such as transformation, derivation of new alignments, as well as reasoning about the alignments.
In this study, our objective is to compare the concepts and relationships aligned by each approach, as well as the complexity of the two alignment processes. Therefore, the comparison performed in this study is simple. No particular formal representation of alignments is used.
3. Materials
3.1. FMA and GALEN
The Foundational Model of Anatomy1 (FMA) [July 2, 2002 version] is an evolving ontology that has been under development at the University of Washington since 1994 [14,15]. Its objective is to conceptualize the physical objects and spaces that constitute the human body. The underlying data model for FMA is a frame-based structure implemented with Protégé2. 58,957 concepts cover the entire range of macroscopic, microscopic and subcellular canonical anatomy. Concept names in FMA are pre-coordinated, and, in addition to preferred terms (one per concept), 28,499 synonyms are provided (up to 6 per concept). For example, there is a concept named Uterine tube, which has two synonyms: Oviduct and Fallopian tube.
The Generalized Architecture for Languages, Encyclopedias and Nomenclatures in medicine3 (GALEN) [v. 4] has been developed as a European Union AIM project led by the University of Manchester since 1991 [16,17]. The GALEN common reference model is a clinical terminology represented using GRAIL [18], a formal language based on description logics. GALEN contains 23,428 concepts and intends to represent the biomedical domain, of which canonical anatomy is only one part. Concept names in GALEN are post-coordinated, and only one name is provided for each non-anonymous concept (e.g., Lobe of thyroid gland). There are 2,960 anonymous concepts (e.g., SolidStructure which < isPairedOrUnpaired leftRight-Paired >).
Both FMA and GALEN are modeled by is-a relationship. Additionally, FMA uses two kinds of partitive relationships (part of and general part of) and GALEN 26, including isStructuralComponentOf and IsDivisionOf. The hierarchy of associative relationships is also more extensive in GALEN than in FMA There are 514 relationship types in GALEN (e.g., IsSpecificallyNonPartitivelyContainedIn) and 54 in FMA (e.g., nerve supply). In addition to inter-concept relationships, there are 85 slots in FMA describing atomic properties of concepts, whose types are Boolean, Integer, Symbol, String and Instance. Examples of such slots include has dimension (Boolean), laterality (Symbol) and definition (String).
3.2. The UMLS®
An additional resource used in the alignment is the Unified Medical Language System® (UMLS®)4 developed by NLM. The UMLS Metathesaurus® is organized by concept or meaning. A concept is defined as a cluster of terms representing the same meaning (synonyms). The 14th edition (2003AA) of the Metathesaurus contains over 1.75 million unique English terms drawn from more than sixty families of medical vocabularies, and organized in some 875,000 concepts. In the Metathesaurus, each concept is categorized by at least one semantic type from the UMLS Semantic Network. A subset of these semantic types is used to define the domain of anatomy. Also part of the UMLS distribution is the SPECIALIST Lexicon, a large syntactic lexicon of both general and medical English.
4. Methods
4.1. Alignment 1
Alignment 1 first compares the concepts between FMA and GALEN in two steps: lexical alignment and structural alignment [19]. Then, based on the matching concepts identified, Alignment 1 compares the associative relationships across systems [20].
The lexical alignment identifies shared concepts across systems lexically through exact match and after normalization. Concepts exhibiting similarity at the lexical level across systems are called anchors, as they are going to be used as reference concepts in the structural alignment and for comparing associative relationship. Additional anchors are identified through UMLS synonymy. Two concepts across systems are considered anchors if their names are synonymous in the UMLS Metathesaurus (i.e., if they name the same concept) and if the corresponding concept is in the anatomy domain (i.e., has a semantic type related to Anatomy). For FMA, both preferred concept names and synonyms were used in the lexical alignment process. For GALEN, only non-anonymous concept names were used. For example, the concepts Cardiac valve in FMA and Valve in heart in GALEN are identified as anchor concepts because Cardiac valve has Valve of heart as a synonym in FMA and Valve in heart matches Valve of heart after normalization.
The structural alignment first consists of acquiring the semantic relations explicitly represented within systems. Inter-concept relationships are generally represented by semantic relations <concept1, relationship, concept2>, where relationship links concept1 to concept2. For the purpose of aligning the two ontologies, we considered as only one PART-OF relationship the various subtypes of partitive relationships present in FMA (e.g., part of, general part of) and in GALEN (e.g., IsStructuralComponentOf,IsDivisionOf). Only hierarchical relationships were considered at this step, i.e., is-a, inverse-is-a, part-of, and has-part. Implicit semantic relations are then extracted from concept names and various combinations of hierarchical relations. Augmentation and inference are the two main techniques used to acquire implicit knowledge from FMA and GALEN.
Augmentation attempts to represent with relations knowledge that is otherwise embedded in the concept names. Augmentation based on reified part-of relationships consists of creating a relation <P,part-of, W> between concepts P (the part) and W (the whole) from a relation <P,is-a, Part of W>, where the concept Part of W reifies, i.e., embeds in its name, the part-of relationships to W. For example, <Neck of femur,part-of, Joint> was added from the relation <Neck of femur,is-a, Component of joint>, where the concept Component of joint reifies a specialized part-of relationship. Examples of augmentation based on other linguistic phenomena include <Prostate gland,is-a, Gland> (from the concept name Prostate gland) and <Extensor muscle of leg,part-of, Leg> (from the concept name Extensor muscle of leg).
Inference generates additional semantic relations by applying inference rules to the existing relations. These inference rules, specific to this alignment, represent limited reasoning along the part-of hierarchy, generating a partitive relation between a specialized part and the whole or between a part and a more generic whole. For example, <First tarsometatarsal joint,part-of, Foot> was inferred based on the relations <First tarsometatarsal joint,is-a, Joint of foot> and <Joint of foot,part-of, Foot>.
With these explicit and implicit semantic relations, the structural alignment identifies structural similarity and conflicts among anchors across systems. Structural similarity, used as positive structural evidence, is defined by the presence of common hierarchical relations among anchors across systems, e.g., <c1,part-of, c2> in one system and <c1’,part-of, c2’> in another where {c1, c1’} and {c2, c2’} are anchors across systems. The anchor concepts Cardiac valve in FMA and Valve in heart in GALEN, presented earlier, received positive structural evidence because they share hierarchical links to some of the other anchors across systems. For example, Cardiac valve is related to Heart(part-of), to Tricuspid valve(inverse-is-a) and to Mitral valve(inverse-is-a).
Conflicts, on the other hand, are used as negative structural evidence. The first type of conflict is defined by the existence of opposite hierarchical relationships between the same anchors across systems, e.g., <c1,part-of, c2> in one system and <c1’,has-part, c2’> in another. The second type of conflict is based on the disjointedness of top-level categories across systems. For example, Nail in FMA is a kind of Skin appendage which is an Anatomical structure, while Nail in GALEN is a Surgical fixation device which is an Inert solid structure. Anatomical structure and Inert solid structure being disjoint top-level categories, the two concepts of Nail across systems are semantically distinct, although they share the exact same name.
Based on the anchors (except those receiving negative structural evidence), associative relationships are compared across systems. The most frequent matches indicate a correspondence between an associative relationship in one system and one relationship (hierarchical or associative) or combination thereof in the other. For example, from Heart -contained in→ Middle mediastinum -part-of→ Mediastinum in FMA and Heart -boundsSpace→ Mediastinum in GALEN, the relationship match {FMA:contained in - part-of, GALEN:boundsSpace} can be extracted.
4.2. Alignment 2
The second alignment also includes a lexical phase and a structural phase, followed by a hierarchical match phase [21]. For each phase, generic schema matching algorithms were adapted to 1) cope with the number of concepts present and 2) handle the more expressive modeling environments (Protégé and GRAIL). Summarizing from [21], the second alignment proceeds as follows.
The lexical phase identifies concepts whose names are similar. Each concept name from FMA and GALEN is first mapped to the UMLS Metathesaurus after normalization and reduced to a set of UMLS concept identifiers. Each concept identifier is further annotated with part-of-speech information identified using the SPECIALIST Lexicon. The similarity between two concepts from FMA and GALEN depends on the ratio of shared UMLS concepts to the total number of UMLS concept mapped to. Part-of-speech information is further used to distinguish between roots (nouns and verbs) and modifiers (adjectives and adverbs) [7].
For example, Valve in heart from GALEN is first normalized to heart valve and mapped to two UMLS concepts. Cardiac valve from FMA is normalized to cardiac valve and mapped to three UMLS concepts, two of which being shared with the mappings of Valve in heart. Based on this, the similarity between Valve in heart and Cardiac valve was assigned a score of .8 (where 0 indicates no similarity and 1.0 indicates a perfect match).
The structural phase attempts to identify concepts (and relationships) that are used similarly in both systems. The first step is to reify every relation present in FMA or GALEN, thereby creating new, artificial concepts. For example, one such concept is created from the relation <Cardiac valve, part-of, Heart>. Similarity scores can then be assigned to matches among these artificial concepts, corresponding to relation matches. The similarity of two relations in a match is estimated to be the average similarity of the concepts and relationships involved in the relations. This process makes it possible to identify the similarity of relations, not only concepts. For example, this is how we identified that both FMA and GALEN assert that cardiac valves are part of the heart.
Moreover, the similarity between relations can be back-propagated to improve the similarity of the corresponding concepts and relationships. Whenever two concepts (or relationships) are mentioned in similar relations, the similarity between those concepts is increased. This back-propagation detects similarity of use, especially between relationships. For example, the similarity between isBranchOf and branch of increases from .28 to .98 using back-propagation.
The final hierarchical phase attempts to identify concepts with similar descendants. Similarity scores across leaf concepts were established during the previous phases, but few higher-level correspondences were identified. In this final phase, the similarity between two concepts is increased if there are many descendants that match. In theory, similarity is pushed up the inheritance hierarchy from the leaves, but [22] notes that few matches were found in this manner.
4.3. Comparing Alignment 1 and 2
Alignment 1 identified a set of concept matches across systems with an indication of the presence of structural evidence and relationship matches with their frequency. A concept match is supported by Alignment 1 if it receives positive structural evidence; not supported otherwise.
Alignment 2 identified a set of matches for both concepts and relationships, each match being qualified by similarity score. A match is supported by Alignment 2 if its similarity score is higher than or equal to a pre-specified threshold; not supported otherwise. The threshold selected in this study is .83, determined heuristically by examining the validity of a subset of matches.
We compared the concept matches obtained by Alignment 1 and 2 by classifying them into four categories: 1) matches supported by both alignments, 2) matches supported by Alignment 1 but not supported or identified by Alignment 2, 3) matches supported by Alignment 2 but not supported or identified by Alignment 1, and 4) matches ignored by both alignments. We then used a similar approach to compare the relationship matches obtained by the two alignments.
5. Results
The matches obtained in Alignment 1 and 2 are first presented separately. Then, we analyze the results of their comparison. These results are summarized in Table 1 (concept matches).
Table 1.
Concept matches in Alignment 1 and 2
| Alignment 2 | |||||
|---|---|---|---|---|---|
| Identified | Not identified | ||||
| Similarity ≥ .83 | Similarity < .83 | ||||
| Alignment 1 | Identified | Positive evidence | 2,199 | 42 | 295 |
| No evidence | 168 | 3 | 29 | ||
| Negative evidence | 36 | 0 | 4 | ||
| Not identified | 132 | 1,074 | |||
5.1. Matches in Alignment 1
2,410 pairs of matching concepts across systems were identified by lexical alignment between FMA and GALEN. Through UMLS synonyms, 366 additional pairs of matching concepts were found across systems, resulting in totally 2,776 concept matches in Alignment 1.
By structural alignment, 2,536 (91.4%) of the 2,776 matches received positive evidence, 40 (1.4%) negative evidence and 200 (7.2%) no evidence. The concept Pancreas, which has the same name in FMA and in GALEN, exemplifies a match with positive evidence as this concept is in has-part relationship to three anchors across systems: Head of pancreas, Tail of pancreas and Neck of pancreas. By contrast, Pectoral girdle (synonym: Shoulder girdle) in FMA and Shoulder girdle in GALEN, although matching lexically, were identified to be a mismatch from the conflicting relationships these concepts have across systems, i.e., <Pectoral girdle,has-part, Shoulder> in FMA and <Shoulder girdle, PART-OF, Shoulder> in GALEN. Finally, although linked to anchors including Cardiovascular system(part-of) and Body Part(is-a) in GALEN, Carotid body does not have any hierarchical links to these or other anchors in FMA, and therefore receives no structural evidence.
The alignment of associative relationships resulted in 182 relationship matches. Matches with high frequency include {FMA:branch of, GALEN:isBranchOf} and {FMA:tributary of, GALEN:isBranchOf}.
In summary, a total of 2,958 matches (2,776 for concepts and 182 for relationships) were identified between FMA and GALEN by Alignment 1.
5.2. Matches in Alignment 2
A total of 3,780 matches were identified by Alignment 2, 3,503 of them in the lexical phase, 64 in the structural phase, and 213 in the hierarchical phase. 2,583 (68.3%) of the 3,780 matches were assigned similarity scores above the threshold of .83. As a matter of fact, 2,539 of these matches have the similarity score of 1.0 (e.g., {FMA: Pancreas, GALEN: Pancreas}). 1,197 (31.7%) of the 3,780 matches have a similarity score lower than .83 and were ignored (e.g., {FMA: Upper lobe of lung, GALEN: Lobe of left lung} has a similarity of .5).
Among the 3,780 matches, there are 3,654 concept matches and 22 relationship matches (e.g., {FMA:part-of, GALEN:IsDivisionOf} has a similarity of 1.0). The remaining 104 matches associate things other than two concepts or two relationships. In 102 cases, a concept in one system matches a relationship in the other (e.g., {FMA:insertion, GALEN: Insertion point}). Finally, two FMA Boolean-typed slots match GALEN relationships (e.g., has dimension in FMA and hasDimension in GALEN).
5.3. Concept matches supported by both alignments
2,776 concept matches were identified by Alignment 1 and 3,654 by Alignment 2. Among them, 2,199 both received positive structural evidence and had a similarity score above the threshold of .83, as shown in the upper left part of Table 1. These matches are supported by both alignments. For example, the match {FMA: Cardiac valve, GALEN: Valve in heart}, presented earlier, received positive evidence in Alignment 1, and its similarity score is .88 in Alignment 2.
5.4. Concept matches supported by Alignment 1 only
As shown in the upper right part of Table 1, 42 concept matches received similarity scores lower than the threshold by Alignment 2, and 295 were not identified by Alignment 2. However, these 337 matches were supported by positive structural evidence of Alignment 1.
167 are FMA synonyms matching GALEN concept names in Alignment 1. Alignment 2 failed to identify or to select these matches in the lexical phase because it did not use synonyms in FMA. For example, Prostate in FMA was matched to Prostate gland in GALEN by Alignment 1 because the former has a synonym Prostate gland in FMA. The positive structural evidence for this match includes their sharing is-a link to Gland and has-part link to Lobe of prostate across systems.
158 were obtained through UMLS synonyms in Alignment 1. One such match is {FMA: First tarsometatarsal joint, GALEN: First tarso metatarsal joint}. This match received positive structural evidence from the shared hierarchical links to other anchors such as Foot(part-of) and Joint of foot5(is-a) across systems. It was not obtained by Alignment 2 because the two alignments used slightly different matching criteria for mapping to UMLS concepts.
12 are FMA preferred concept names matching GALEN concept names in Alignment 1, e.g., {FMA: Immunoglobulin M, GALEN: Immunoglobulin M}, which shared hierarchical links to anchors such as Immunoglobulin(is-a) and Protein(is-a) across systems. The reasons why these matches were not obtained by Alignment 2 were investigated and found to be essentially unimportant.
5.5. Concept matches supported by Alignment 2 only
The lower left part of Table 1 shows the concept matches with similarity scores above the threshold by Alignment 2 but not supported or identified by Alignment 1.
168 received no structural evidence by Alignment 1, e.g., {FMA: Carotid body, GALEN: Carotid body}, presented earlier. Although its similarity score is 1.0 by Alignment 2, this match was not supported by Alignment 1 because no structural evidence could be found (in this case, because of a lack of relations being represented in FMA for this concept).
36 received negative structural evidence by Alignment 1. Both {FMA: Nail, GALEN: Nail} and {FMA: Pectoral girdle, GALEN: Shoulder girdle}, with negative evidence in Alignment 1 as presented earlier, received the similarity score of 1.0 by Alignment 2. These 36 matches were inappropriately supported by Alignment 2 because, unlike Alignment 1, this approach does not attempt to identify semantic mismatches.
- 132 were only identified by Alignment 2.
- 78 could have been obtained by Alignment 1 through UMLS synonymy. They were filtered out by Alignment 1 because they caused two different concepts in one system to be synonymous. In the UMLS Metathesaurus, the terms Prostate, Prostate gland and Prostatic gland are synonymous. In FMA, Prostate refers to the organ while Prostatic gland is subdivision of the organ. Being different concepts in FMA, their matching to the same UMLS synonym was rejected. Therefore, Alignment 1 did not get the match {FMA: Prostatic gland, GALEN: Prostate gland} while Alignment 2 did.
- 18 were rejected by Alignment 1 through the UMLS Semantic Network filter for Anatomy, e.g., {FMA: Flatulence, GALEN: Flatus} (similarity = 1.0). Neither Flatulence norFlatus is related to Anatomy in UMLS and this match was rejected by Alignment 1 for this reason.
- 36 were not identified by Alignment 1 because at least one of the concept names did not match any UMLS synonyms. For example, Alignment 1 missed {FMA: Colic flexure, GALEN: Colonic flexure} (similarity = 1.0) through UMLS because Colonic flexure in GALEN does not match any UMLS synonyms. Some of these matches of Alignment 2 were determined to be valid by a domain expert.
5.6. Concept matches ignored by both alignments
The lower right part of Table 1 shows the concept matches ignored by both alignments. These matches are either not identified by one alignment and not supported by the other or identified but not supported by either alignment.
1,074 were only identified by Alignment 2 but their similarity scores are lower than the threshold. 72 are FMA concepts matching GALEN anonymous concepts, purposely ignored by Alignment 1. 1,002 are FMA concepts matching GALEN non-anonymous concepts. Most of these matches correspond to partial matches, not addressed by Alignment 1 (e.g., {FMA: Ligament of knee joint, GALEN: Ligament of knee}, with a similarity score of .35).
32 received no structural evidence by Alignment 1, of which 3 of them had similarity scores lower than the threshold and 29 were not identified by Alignment 2.
4 received negative structural evidence by Alignment 1 and were not identified by Alignment 2.
5.7. Relationship matches
182 relationship matches were identified in Alignment 1. Alignment 2 identified 22 matches, of which 17 were supported by a similarity score above .83. Seven relationship matches were identified by both alignments (e.g., {FMA: nerve supply, GALEN:IsServedBy}). Seven were supported by Alignment 2 only (e.g., {FMA:lymphatic drainage, GALEN:IsServedBy}). Alignment 1, relying on the concepts already aligned, failed to identify these matches, because these relationships occurred among concepts that have not been aligned. Finally, in three cases, the match identified by Alignment 2 corresponded to a match created manually in Alignment 1 between the subtypes of part-of relationships (e.g., {FMA:part-of, GALEN:IsDivisionOf}).
6. Discussion
6.1. Improving the alignments
In fact, the philosophy behind each approach is different. Alignment 1 takes advantage of domain knowledge. It requires lexical matches to be supported by structural matches, at the cost of inaccurately rejecting some valid matches. Therefore, it favors precision over recall. On the other hand, Alignment 2 relies on generic algorithms and, by imposing no penalty for lack of structural matches, favors recall over precision. Theoretically, the two approaches could be combined. In practice, however, despite their differences, their results are surprisingly close and any improvement would only be marginal at best.
Nevertheless, each approach can be improved based on the results of the other. Alignment 1 would benefit from addressing partial lexical alignment and identifying matches based solely on structural similarity. Alignment 2 could be improved by taking into account synonyms in FMA and identifying semantic mismatches.
Of particular interest are the 875 relation matches obtained by Alignment 2 in the structural phase for the purpose of increasing the similarity scores of the corresponding concepts and relationships. In addition to increasing the chances of identifying matches, these relation matches could be used for themselves. For example, the match by {FMA: <Lung,contained in, Thoracic cavity>, GALEN: <Lung,IsSpecificallyNonPartitivelyContainedIn, Pleural membrane>} whose similarity score is .33, captured the difference the two ontologies have in representing the knowledge about equivalent concepts.
6.2. Validating the alignments
The validation of the results of the alignment has been an issue for both groups. Anatomy is a vast domain and, in addition to domain knowledge, the experts are also required to have some knowledge of the two systems under investigation. No group has achieved a comprehensive evaluation of its results. One interest of disposing of two alignments is that there is the possibility of a cross-validation. In fact, while the matches of Alignment 1 can certainly validate those of Alignment 2, the contrary is not necessarily true. In Alignment 1, a lexical match is required to be supported by some structural evidence. Conversely, in Alignment 2, lexical matches get the highest score possible and structural evidence, if any, is only used to increase the score of partial lexical matches. However, matches from Alignment 2 supported by structural evidence could be used to validate the results of Alignment 1. Unfortunately, the similarity score used in Alignment 2 to indicate the quality of the match does not strictly reflect the presence of structural evidence.
6.3. Challenges
Evaluating completeness
Neither alignment identified enough matches. A total of 3,982 concept matches were identified by the two alignments together, only accounting for about 7% of all FMA concepts and 17% of all GALEN concepts. Arguably, these proportions represent a conservative estimate of completeness for the alignment. While the coverage of FMA is restricted to canonical anatomy, GALEN includes categories from biomedical subdomains other than anatomy (e.g., Non-normal phenomenon, Basic drug form, Clinical process and Food). These concepts and their descendants do not belong to the anatomical domain and, therefore, are not expected to have any matches in FMA. Examples of such concepts include Supernumerary thumb, Tetanus vaccine, Cardiac valvotomy, and Diary product. 11,384 non-anatomical concepts were identified in GALEN, accounting for 49% of the 23,428 concepts in GALEN. In other words, only 12,044 concepts in GALEN can be the target of a match for FMA concepts. This indicates that there is a maximum of 12,044 one-to-one concept matches between FMA and GALEN. By this measure, the two alignments together have identified 33% of all possible concept matches, i.e., 3,982 out of 12,044.
Identifying complex matches
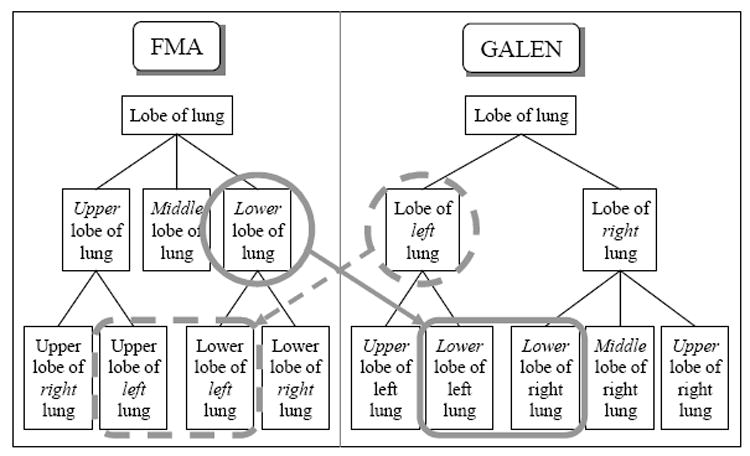
By design, all concept matches identified by the two alignments are one-to-one matches. However, there are more complex cases where a single entity in one ontology may match a group of entities in the other [22]. For example, the information about arterial and nerve supply and venous and lymphatic drainage is represented by four distinct relationships in FMA (arterial supply,venous drainage,nerve supply and lymphatic drainage), while GALEN uses a single relationship (isServedBy). A simple way to address this difference in approach is to establish a one-to-many match that relates the single relationship type in GALEN to the four relationship types in FMA. Groups of concepts may also match across ontologies. For example, as illustrated in Figure 1, along the IS-A hierarchy of FMA, Lobe of lung is first modeled by upper/lower positions (i.e., Upper lobe of lung and Lower lobe of lung) and then by laterality (e.g., for Upper lobe of lung: Upper lobe of left lung and Upper lobe of right lung). By contrast, in GALEN, Lobe of Lung is first modeled by laterality and then by upper/lower positions. Although one-to-one matches were identified for fine-grained concepts such as Upper lobe of left lung, because of these modeling differences, no single match can be found in the other system for concepts such as Lobe of Left Lung in GALEN and Lower lobe of lung in FMA. One possibility would be for such concepts to be associated not with one concept in the other ontology, but with several concepts (e.g., Lobe of left lung in GALEN with Upper lobe of left lung and Lower lobe of left lung in FMA; and, Lower lobe of lung in FMA with Lower lobe of left lung and Lower lobe of right lung in GALEN). Additional alignment techniques need to be explored to handle such complex cases.
Figure 1.
Example of complex concept matches between FMA and GALEN
Representing the alignment formally
In this study, no particular formalism was used to represent the simple, one-to-one matches identified across ontologies. However, for more complex matches, the result of the alignment would benefit from being represented formally. One possible solution is to construct a mediating ontology. This is called a mapping in [22], which shows how it can help align FMA and GALEN. In the case of the supply and drainage relationships mentioned earlier, for example, the mapping contains all five relationship types (four from FMA and one from GALEN) and states explicitly that isServedBy in GALEN subsumes the four relationship types in FMA. Expressing the mapping as a mediating ontology allows one to address more subtle situations including differences in granularity. In GALEN, the Fibrous trigone is a division of the Heart. In FMA, there is an additional level of indirection: the Fibrous trigone is part of the Fibrous skeleton, which is part of the Heart. Thus, GALEN contains a single assertion relating the heart and fibrous trigone, whereas FMA contains two assertions. One way to align these assertions is to place all three assertions in the mapping, which states that there is a partitive relationship between Fibrous trigone and Heart. Moreover, this relationship is composed of two sub-relationships that link the fibrous trigone to the heart via the fibrous skeleton. The mapping makes it possible to demonstrate that the two assertions contained in FMA refine the assertion in GALEN. Note that the transitive closure of the hierarchical relations used in Alignment 1 already identified the equivalence of the relations between Fibrous trigone and Heart in the two systems.
7. Conclusion
We have compared two approaches to aligning two representations of anatomy. Common to the two approaches is the use of a combination of lexical and structural techniques. However, the approaches differ in that one takes advantage of domain knowledge (and is therefore specific to the domain under investigation), while the other draws on a generic schema matching approach (and is therefore applicable to an arbitrary domain). Having aligned the same versions of FMA and GALEN allowed us to cross-validate our results. The alignments obtained by the two approaches were surprisingly close, but each approach identified a limited number of valid matches that the other approach failed to identify. A detailed analysis of the differences in the results helped reveal the strengths and weaknesses of each approach and suggested possible improvements to them. Complex matches, where one entity in one ontology corresponds to several entities in the other, were beyond the reach of these approaches. Further research is needed to identify these complex matches.
Acknowledgments
This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM), and by the Natural Science Foundation of China (No.60496324), the National Key Research and Development Program of China (Grant No. 2002CB312004), the Knowledge Innovation Program of the Chinese Academy of Sciences, MADIS of the Chinese Academy of Sciences, and Key Laboratory of Multimedia and Intelligent Software at Beijing University of Technology. This work was done in part while Songmao Zhang was a visiting scholar at the Lister Hill National Center for Biomedical Communications, NLM, NIH. Support for Peter Mork’s work was provided in part by NLM training grant T15LM07442.
Thanks for their support and encouragement to Cornelius Rosse for FMA, Alan Rector for GALEN, as well as their collaborators.
Footnotes
http://fma.biostr.washington.edu/ (Accessed December 9, 2006)
http://protege.stanford.edu/ (Accessed December 9, 2006)
http://www.opengalen.org/ (Accessed December 9, 2006)
http://umlsks.nlm.nih.gov/ (Accessed December 9, 2006)
The anchor is named Foot joint in GALEN.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reed SL, Lenat D. Proceedings of AAAI. MIT Press; 2002. [Accessed: December 9, 2006]. Mapping ontologies into Cyc. http://citeseer.ist.psu.edu/509238.html. [Google Scholar]
- 2.Noy NF, Musen MA. Proceedings of AAAI. MIT Press; 2000. PROMPT: algorithm and tool for automated ontology merging and alignment; pp. 450–455. [Google Scholar]
- 3.McGuinness DL, Fikes R, Rice J, Wilder S. Proceedings of AAAI. MIT Press; 2000. The Chimaera ontology environment; pp. 1123–1124. [Google Scholar]
- 4.Mitra P, Wiederhold G, Kersten M. In: Zaniolo C, Lockemann PC, Scholl MH, Grust T, editors. A graph-oriented model for articulation of ontology interdependencies; Proceedings of the conference on Advances in Database Technology (EDBT 2000); Springer; 2000. pp. 86–100. [Google Scholar]
- 5.Stumme G, Alexander M. Proceedings of IJCAI 2001. Morgan Kaufmann; 2001. FCA-MERGE: Bottom-up merging of ontologies; pp. 225–230. [Google Scholar]
- 6.Rahm E, Bernstein PA. A survey of approaches to automatic schema matching. VLDB Journal. 2001;10:334–350. [Google Scholar]
- 7.Madhavan J, Bernstein PA, Rahm E. Generic schema matching using Cupid. In: Apers PMG, Atzeni P, Ceri S, Paraboschi S, Ramamohanarao K, Snodgrass RT, editors. Proceedings of 27th International Conference on Very Large Data Bases. Morgan Kaufmann; San Francisco, CA, USA: 2001. pp. 49–58. [Google Scholar]
- 8.Do HH, Rahm E. COMA - A system for flexible combination of schema matching approaches. In: Bernstein PA, Loannidis YE, Ramakrishnan R, editors. Proceedings of 28th International Conference on Very Large Data Bases. Morgan Kaufmann; San Francisco, CA, USA: 2002. pp. 610–621. [Google Scholar]
- 9.Melnik S, Garcia-Molina H, Rahm E. Similarity flooding: A versatile graph matching algorithm and its application to schema matching. IEEE Computer Society; Proceedings of the 18th International Conference on Data Engineering; Washington, DC, USA. 2002. pp. 117–128. [Google Scholar]
- 10.Doan A, Madhavan J, Dhamankar R, Domingos P, Halevy A. Learning to match ontologies on the Semantic Web. VLDB Journal. 2003;12:303–319. [Google Scholar]
- 11.Euzenat J. An API for ontology alignment. In: McIlraith SA, Plexousakis D, van Harmelen F, editors. Proceedings; Third International Semantic Web Conference; Hiroshima, Japan. November 7-11, 2004; Berlin / Heidelberg: Springer; 2004. pp. 698–712. [Google Scholar]
- 12.Stuckenschmidt H, van Harmelen F, Bouquet P, Giunchiglia F, Serafini L. Using C-OWL for the alignment and merging of medical ontologies. In: Hahn U, editor. Electronic proceedings; Proceedings of the KR 2004 Workshop on Formal Biomedical Knowledge Representation; 2004. [Accessed: December 9, 2006]. pp. 88–101. http://ceur-ws.org/Vol-102/stuckenschmidt.pdf. [Google Scholar]
- 13.Madhavan J, Bernstein PA, Domingos P, Halevy AY. Representing and reasoning about mappings between domain models. AAAI; Proceedings of the 18th national conference on artificial intelligence; 2002. pp. 80–86. [Google Scholar]
- 14.Rosse C, Mejino JL., Jr A reference ontology for biomedical informatics: the Foundational Model of Anatomy. J Biomed Inform. 2003;36:478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Noy NF, Musen MA, Mejino JLV, Rosse C. Pushing the envelope: challenges in a frame-based representation of human anatomy. Data & Knowledge Engineering. 2004;48:335–359. [Google Scholar]
- 16.Rector AL, Bechhofer S, Goble CA, Horrocks I, Nowlan WA, Solomon WD. The GRAIL concept modelling language for medical terminology. Artif Intell Med. 1997;9:139–171. doi: 10.1016/s0933-3657(96)00369-7. [DOI] [PubMed] [Google Scholar]
- 17.Rogers J, Rector A. In: Overhage JM, editor. GALEN’s model of parts and wholes: experience and comparisons; Proceedings of the AMIA Symposium; 2000. pp. 714–718. [PMC free article] [PubMed] [Google Scholar]
- 18.Zanstra PE, van der Haring EJ, Flier F, Rogers JE, Solomon WD. Using the GRAIL language for classification management. Proceedings of the 15th International Congress of the European Federation for Medical Informatics; 1997. pp. 897–901. [PubMed] [Google Scholar]
- 19.Zhang S, Bodenreider O. Aligning representations of anatomy using lexical and structural methods. In: Musen M, editor. Proceedings of the AMIA Symposium. 2003. pp. 753–757. [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang S, Bodenreider O. Comparing associative relationships among equivalent concepts across ontologies. In: Fieschi M, Coiera E, Li Y-CJ, editors. Medinfo. Vol. 2004. IOS Press; 2004. pp. 459–466. [PMC free article] [PubMed] [Google Scholar]
- 21.Mork P, Bernstein PA. 20th International Conference on Data Engineering. IEEE; Boston, MA: 2004. Adapting a generic match algorithm to align ontologies of human anatomy; pp. 787–790. [Google Scholar]
- 22.Mork P, Pottinger R, Bernstein PA. Challenges in precisely aligning models of human anatomy using generic schema matching. In: Fieschi M, Coiera E, Li Y-CJ, editors. Medinfo. Vol. 2004. IOS Press; 2004. pp. 401–405. [PubMed] [Google Scholar]


