Abstract
mRNA localization is a powerful mechanism for targeting factors to different regions of the cell and is used in Drosophila to pattern the early embryo. During oogenesis of the wasp Nasonia, mRNA localization is used extensively to replace the function of the Drosophila bicoid gene for the initiation of patterning along the antero-posterior axis. Nasonia localizes both caudal and nanos to the posterior pole, whereas giant mRNA is localized to the anterior pole of the oocyte; orthodenticle1 (otd1) is localized to both the anterior and posterior poles. The abundance of differentially localized mRNAs during Nasonia oogenesis provided a unique opportunity to study the different mechanisms involved in mRNA localization. Through pharmacological disruption of the microtubule network, we found that both anterior otd1 and giant, as well as posterior caudal mRNA localization was microtubule-dependent. Conversely, posterior otd1 and nanos mRNA localized correctly to the posterior upon microtubule disruption. However, actin is important in anchoring these two posteriorly localized mRNAs to the oosome, the structure containing the pole plasm. Moreover, we find that knocking down the functions of the genes tudor and Bicaudal-D mimics disruption of microtubules, suggesting that tudor's function in Nasonia is different from flies, where it is involved in formation of the pole plasm.
Keywords: Nasonia, mRNA localization, nanos, caudal, giant, tudor, otd1, Bicaudal, microtubules, actin
Introduction
The establishment of the antero-posterior and dorso-ventral axes in the early embryo is a long standing problem in developmental biology. Investigations in Drosophila have elucidated a variety of mechanisms used to establish axes, including the formation of morphogenetic gradients that pattern the segmented body (Ashe and Briscoe, 2006). The initial polarization of the Drosophila embryo, however, is achieved during early oogenesis, prior to the establishment of these morphogenetic gradients, when the cytoskeleton becomes polarized and various maternal mRNAs are specifically localized to opposite poles of the developing oocyte (St Johnston, 2005). mRNA localization is a common strategy for creating asymmetry within a cell by targeting proteins to particular areas within a cell or tissue (St Johnston, 2005). A variety of methods are employed by the cell to localize mRNAs. This includes active transport along cytoskeletal elements, diffusion and anchoring, as well as degradation and protection of mRNA. Most of these processes are mediated by elements located in the 3' and/or 5' untranslated regions (UTR) of the mRNA. Among these mechanisms, active transport via plus-end or minus-end directed motors along the microtubule network has been studied in detail in the Drosophila ovary, where many maternal mRNAs are localized to opposite poles to establish the initial antero-posterior axis of the oocyte (St Johnston, 2005).
In recent years, it has become clear that several aspects of Drosophila development are not conserved in all insects, in particular during early embryogenesis. This has led researchers to seek common and distinct developmental features of insect embryogenesis. In Drosophila, bicoid (bcd) is the critical factor that patterns the anterior-posterior axis. bcd maternal mRNA is localized at the anterior pole of the Drosophila embryo through transport along the cytoskeletal tracks via a minus-end directed motor. Upon fertilization, translation of bcd mRNA generates a morphogenetic gradient of the Bicoid protein (Steinhauer and Kalderon, 2006; Struhl et al., 1989) (Driever and Nusslein-Volhard, 1988a) (Driever and Nusslein-Volhard, 1988b). Despite the various important functions of Bcd in Drosophila segmentation, including transcriptional activation and positioning of gap genes, as well as translational repression of the posterior factor caudal, bcd is a recent addition in higher dipterans to the genetic network responsible for the formation of the body plan and is not found outside this order (Niessing et al., 2002; Niessing et al., 1999; Niessing et al., 2000; Rivera-Pomar and Jackle, 1996; Rivera-Pomar et al., 1996) (Stauber et al., 1999).
The other critical mRNA that is localized in Drosophila for antero-posterior axis formation is nanos (nos). nos takes advantage of the specification system that generates the germline and is localized to the posterior of the embryo where it generates a gradient that functions in repressing anterior development (Gavis and Lehmann, 1992). In contrast to bcd mRNA localization, nos localization is achieved through diffusion and subsequent trapping of the nos message. nos is released into the oocyte during nurse cell dumping and allowed to diffuse throughout the oocyte. This diffusion is enhanced by microtubule-facilitated cytoplasmic movements. nos mRNA is then trapped in the posterior cortex along with the germ plasm through actin dependent anchoring (Forrest and Gavis, 2003).
Much has been learned about the conserved and derived aspects of insect segmentation as a result of investigations using several distantly related insect model systems. However, a number of these studies have used the beetle Tribolium, the grasshopper Schistocerca or the milk weed bug Oncopeltus, all of which undergo a different mode of embryogenesis than Drosophila, making comparisons of the early genetic network between these insects and Drosophila difficult (Liu and Kaufman, 2005).These species undergo short germ embryogenesis, in which the bulk of the egg is composed of extraembryonic membranes, while the germ rudiment develops in the posteriormost region of the egg. As a result, only anterior structures are patterned in a syncytial environment, whereas abdominal and posterior structures form later in the posterior of the embryo from a cellularized “growth zone”. Drosophila instead undergoes long germ development where the embryo occupies the entire egg length and lacks a growth zone. Moreover, all segments of the embryo are patterned together within a syncytial environment (Davis and Patel, 2002).
We have focused our attention on Nasonia vitripennis, a parasitoid wasp (Hymenoptera) that undergoes long germ development similar to that of Drosophila, yet is evolutionarily very distant from flies (>200MY) and lacks bcd (reviewed in: (Pultz and Leaf, 2003). We have uncovered a number of striking features that differ with the highly derived system of axis formation in Drosophila. Notably, we have shown that Nasonia localizes a number of maternal mRNAs that are not localized in Drosophila, such as orthodenticle-1 (otd-1), caudal (cad) and giant (gt), as a primary step in oocyte axis specification and early embryonic patterning. This, at least in part, compensates for the absence of bcd (Lynch et al., 2006)(Olesnicky et al., 2006)(Brent et al., 2007). otd-1 mRNA is expressed maternally in the Nasonia ovary and is localized to both the anterior and posterior poles of the developing oocyte (Lynch et al., 2006). caudal (cad) mRNA, which later forms a posterior to anterior mRNA gradient in the early embryo, is transiently localized to the posterior of the oocyte during oogenesis (Olesnicky et al., 2006). As in flies and in most insect species tested, nos is strictly localized to the posterior of the oocyte during oogenesis and remains localized to the oosome in early embryos (J. Lynch and C.D. submitted). The oosome is a structure that contains the germ plasm that will later give rise to the germ line. During early Nasonia embryogenesis, the oosome migrates to the posterior pole of the embryo and buds off the posterior pole to give rise to the pole cells. Moreover, we find that giant (gt) mRNA is expressed maternally and localized to the anterior of the oocyte in a manner reminiscent of bcd in Drosophila (Brent et al., 2007).
Both the Drosophila and Nasonia ovariole are meroistic, meaning that the nurse cells and oocyte are both of germ cell descent and originate from the same primordium, but differentiate during subsequent cell divisions. As each ovarian follicle develops and is positioned more distally along the ovariole, the nurse cells remain attached to one another and to the oocyte through ring canals, which arise from incomplete cleavage during cell division. The 16 sister cells that make up each germline cyst result from four of these incomplete divisions. An egg chamber forms comprising of 15 nurse cells and the oocyte, surrounded by the somatic follicle cells, which form an epithelial layer around the oocyte. Nurse cells produce metabolites and other factors that transit through the ring canals to accumulate in the oocyte (King, 1969)(Figure 1).
Figure 1. Dynamic cytoskeletal changes during Nasonia oogenesis.
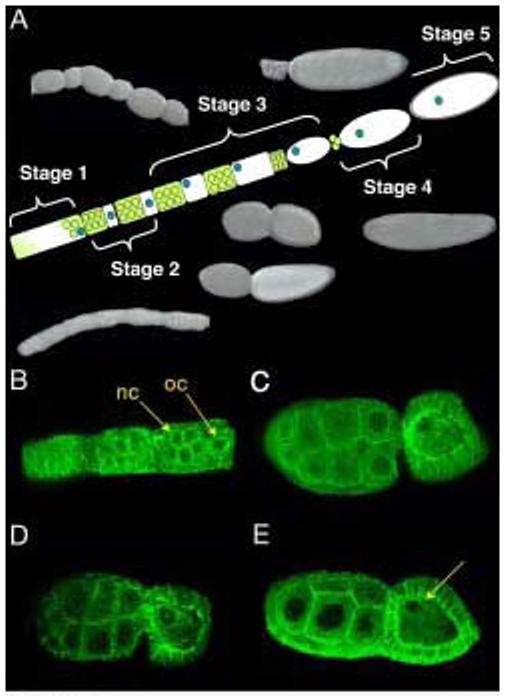
The 5 stages of oogenesis as described by King & Richards (1969). Yellow and white represents oogonium, nurse cells are in yellow, oocyte in white, oocyte nucleus in blue and vitelline membrane in grey. Photographs of fixed wild type follicles taken using Nomarsky optics. In early stage one follicles, the oocyte and nurse cells are indistinguishable. Later stage one follicles begin to distinguish the nurse cells from the oocyte which lies at the posterior of each follicle. Stage 2 follicles have the nurse cells and a smaller oocyte separated from one another with a constriction. In early stage 3 follicles, the oocyte and nurse cells are comparable in size. In later stage 3 follicles, however, the oocyte is larger than its accompanying nurse cells. Stage 4 follicles empty the nurse cell contents into the oocyte. In stage 5, a vitelline membrane is laid around the oocyte (A). Early stage 2 follicles stained using alpha tubulin antibody and analyzed using confocal microscopy. nc, nurse cells, oc, oocyte (B). Later stage 3 follicles show a complex microtubule network throughout the oocyte and nurse cells, and highly enriched at the oocyte nucleus (C,D). Late Stage 3 follicles show a breakdown of the microtubule cytoskeleton along the anterior-posterior axis, but retain a hairpin like organization surrounding the oocyte nucleus (arrowhead) (E).
The Drosophila oocyte is specified early during oogenesis as a result of the asymmetric segregation of the fusome, an organelle that connects the 16 cells. Once the oocyte has been specified, the polarity of the oocyte microtubule network becomes extremely dynamic and undergoes a major reorganization resulting from communication between the oocyte and follicle cells. This reorganization is essential to localize maternal mRNAs that will generate the axes of the embryo. At first, microtubule minus ends extend from the nurse cells into the oocyte toward a microtubule organizing center (MTOC) localized at the posterior pole of the oocyte, near its nucleus. Later, however, the posterior MTOC disassembles while multiple MTOCs form toward the anterior of the growing oocyte. At this stage, the microtubules are therefore pointing from the plus end at the posterior of the oocyte to the minus end at the anterior. mRNAs and the oocyte nucleus utilize the polarity of the microtubules to localize to the anterior or posterior pole (Steinhauer and Kalderon, 2006).
Nasonia oogenesis presents striking similarities to that of Drosophila. It is divided into five morphologically distinct stages (King, 1969)(Figure 1A). In stage 1, the nurse cells and oocyte are indistinguishable until they begin to segregate, with the oocyte lying towards the posterior of the follicle. By stage 2, the nurse cells and a smaller oocyte are clearly distinguishable, as a constriction forms between the oocyte and its supporting nurse cells. At this stage, the oocyte nucleus is positioned in the center of the cell. The oocyte continues to grow throughout stage 3 until it becomes larger than its accompanying nurse cells. Concomitantly, the oocyte nucleus migrates to the dorsal anterior cortex of the developing oocyte, as in Drosophila. Later, during stage 4, the nurse cells begin to degenerate as they empty all their contents into the oocyte. In the final stage (5), a vitelline membrane is constructed around the embryo (King, 1969)(Figure 1A).
Here, we show that the localization of four maternal mRNAs is achieved using at least 2 distinct mechanisms. We show that, during Nasonia oogenesis, microtubules play a major role in oocyte polarity and in the control of anterior localization of otd1 and gt mRNA and the posterior localization of cad mRNA. In contrast, the actin cytoskeleton is important for anchoring the oosome and is therefore essential for the localization of nanos and otd1 mRNA to the posterior pole of the oocyte.
Materials and Methods
Fixation and in situ hybridization
Nasonia wild type stocks were kept at 28°C. Ovaries were fixed in 4% Formaldehyde 0.1% Tween 20 in 1X PBS for 20 minutes and dehydrated in methanol. In situ hybridization was performed as previously described (Brent AE, 2003).
Microtubule detection and Immunohistochemistry
Microtubules were detected using the method previously described by Januschke et al, 2006 to preserve the cytoskeleton in ovaries. Monoclonal anti-α-tubulin (DM1A) FITC Conjugate (Sigma) was incubated at 1:1000 overnight at 4°C in PBS 0.1% Tween containing 2% bovine serum albumin (BSA).
Bicaudal and tudor sequence information
The following 941 base pair Bicaudal fragment and tudor 810 base pair fragment were cloned from a Nasonia cDNA library generated from ovaries. Primers were designed based on sequence similarity between the Drosophila melanogaster tud and BicD genes. The known Drosophila sequences were aligned with raw sequence data from the Nasonia genome sequence using BLAST.
Nasonia tudor
TGAAGAAATAACTACGGTACGCTTTATCGACTATGGTAACACTGATGTAA TCCACAATAATGTAAGCAAGATCAAACAATTGCCTGATAAATGGAAGGCC ATAAAGAAGTACGCTATTGAGTGTAAACTGGATGTACTCCCGGTTAATGC TGACGACTGGAGCGACGAAGCCAGCGCCAAATTGAGTGAACTGGTTACAA CTGAAGACCCTATCCACGCTTTGATAATCGCCGATAAAGCTCCAATGCGT ATAGATCTCTATAGCAAAGGTGATAGTATCTGTAAGATGCTGATCGACGA GAAGCTCGCTACGTACGTTCAAAGTTCCGAAGATCTGAATGAGGAGATTG TTGAAGAAGTCCAGTTAGATCCACGATCTGCCTTCGTCAGTAACATTATA TCAGTAGATCAATTTTGGGTACAGGAAGAAAAATGGATTAACGATTTGGA AATGATAGAGGATAGACTTGTGATGGCTCCCATGTTCCCACAAGTACCAG AAATCGAAGAAGGACTCGTCTGCATTGCTCATTTTCCTGATGACAACTTG TACTACAGAGCTATCATTTTGTCACACACCGATGAAGGAACAAAGGTTCG TTATATCGATTATGGCAATTCAGCCATCACAAAGGACCTTAAAACGATTC CAGGGGACTTGGCCCAAATTCGTCCGCTGTCGAGAAAATGCTGTTTGGCA AAACCAGATGGGATCGAACAGTGGCCTGATGGAATTCATGATGAATTTGT CACCCTAGCGGCGTCTGGTGCAACAGTATTTCTTCTAGACGTAATCGAGA GAGCGAAACT
Nasonia Bicaudal
CTGCGCCATGAGTTGGAACGTGTTCAATCTGAGCGTGATCACGCTCTACA AGAAAGAGAAGACGTAGGCAAGGACCATTTGTTAGTGGAGTCTGAACGGA GAAGCTTGCGTACAGAGCTTAAAGAGAGCAGATTTCGCGAGACAAGACTT CTGCAAGACTACACTGAACTCGAGGATGAAAACATTTCCTTGCAGAAGCA AGTATCCACTTTGCGGTCTAGTCAGGTGGAATTTGAAGGTGCCAAACATG AAATACGTAGACTGACTGAGGAGGTTGAGCTTCTGAACAGTCAAGTAGAAGAGTTGTCTAATTTAAAGAAAATAGCTGAGAAGCAGATGGAAGAAGCATT GGAGTCGTTGCAAGCCGAAAGGGAGGCAAAATATGTCCTCAAGAAGGAAC TGGATCAGAGAATGAACAGCGAATCTATCTACAACCTCAGTAATCTTGCA CTTTCCATTCGCGGTATTACGGATGACCAAACGATGTGCAGCGATGGTGA GGATGATTCTCCAGCACTAAGAAGAATAGAGGATGATCTGAAGACTCAAG AACCTGGCACCTCATCGCCAAACAAGCAGGTCGACCTGTTCTCGGAGATT CATTTGAACGAACTGAAAAAGCTTGAAAAGCAACTTGAAATGGCTGAAAC TGAAAAGGCAGTACTGACGCAGAACCTGAAGGAATCACAGTGTGCTGTTG ACAAGAGCCAAGCGGAATTGCAGTCGTTCATTGCGCGCATTGTACAATTA GCTGCCCATGTCCAGTCTCTGCAGCACATTCACTCGAAGCTACCAGAGAA GCAAAGCGATGAAACCACCCTGGACAAGCTGAATTTGGCCATTATTCAAT ATCACCAATGGGAGTACATTGTCAGCCTCAAGAAGTTCCACCAACTACCA AAAGACTTGGCTGAATTGGATAAGGGTCTGACGATTTCAGA
RNA interference
parental RNAi was performed as described in Lynch and Desplan (2006). To generate templates via PCR for transcription of double stranded RNA the following primers were used (T7 promoters in bold.): For Bicaudal dsRNA transcription templates: forward primer (5' TAATACGACTCACTATAGGGAGACCACGCTGCGCCATGAGTTGGAACG 3') and reverse primer (5'TAATACGACTCACTATAGGGAGACCACCTTGTTACGAAGATTCGTAAG 3') were used. For tudor dsRNA transcription templates: forward primer (5' TAATACGACTCACTATAGGGAGACCACCGGTACGCTTTATCGACTATGG 3') and reverse primer (5' TAATACGACTCACTATAGGGAGACCACGTTTCGCTCTCTCGATTACGTC 3') were used. Female pupae were injected with dsRNA and kept at 28°C until eclosion. Upon eclosion, females were fed sugar water for 2 days at 25°C. Ovaries were subsequently dissected and fixed for analysis by in situ hybridization.
Chemical treatments
One day after eclosion, wild type females were fed 65μg/mL colchicine in 20% sucrose dissolved in dH2O for 15 hours, ovaries were subsequently fixed and examined for defects in mRNA localization and oogenesis.
As cytochalasin D is lethal when fed to Nasonia for long periods of time, Nasonia ovaries were instead cultured in the presence of cytochalasin D. Females were reared at 28°C and fed 10% sugar water for 2 days after eclosion at 25°C. Ovaries were then dissected in 500 μl Schneider's Insect Medium (Sigma-Aldrich) supplemented with 20% fetal bovine serum. 1μl/ml ethanol was added to control ovaries, whereas experimental ovaries were supplemented with 1μg/ml cytochalasin D. Ovaries were incubated on a nutator for 1 hour at room temperature and subsequently fixed and analyzed by in situ hybridization.
Results and Discussion
Microtubules function in oocyte specification and polarity
A comparison of early developmental processes in Drosophila and Nasonia has highlighted the critical role of mRNA localization in Nasonia. Of particular interest is the fact that various maternal mRNAs appear to exhibit distinct localization profiles in Nasonia. For instance, while cad mRNA is not localized in flies, it is initially localized to the posterior in Nasonia, but this is rapidly lost, leading to the formation of an mRNA gradient in the embryo (Olesnicky et al., 2006). In contrast, nos and posterior otd1 remain very tightly localized. Anterior otd1 and gt are also more loosely associated with the anterior pole. We therefore investigated the mechanisms involved in mRNA localization, focusing first on the mechanisms that are known to play critical roles in Drosophila.
We first investigated the role of microtubules. Microtubules are crucial for oocyte specification in Drosophila, as their disruption often results in the formation of germline cysts with 16 nurse cells and no oocyte. Moreover, active transport along the microtubule network is important for the localization of various mRNAs during Drosophila oogenesis. For example, the minus-end directed motor dynein is involved in transporting the mRNAs of bcd and gurken (grk) to the anterior of the oocyte along cytoskeletal tracks. In contrast, oskar (osk), a posteriorly localized mRNA, is likely transported along microtubules by the plus-end directed motor kinesin (St Johnston, 2005).
Using the fixation technique described in Januschke et al (2006) to preserve the cytoskeleton, we stained for microtubules in Nasonia ovaries using an anti-alpha-tubulin antibody (Januschke et al., 2006). Microtubules are evident throughout the Nasonia nurse cells and oocyte in late stage 1 and stage 2 follicles (Figure 1B). In stage 3, after the nucleus has migrated to the dorsal anterior cortex, they are enriched around the nucleus and extend along both the dorso-ventral and antero-posterior axes throughout the oocyte (Figure 1C,D). At the end of stage 3, much of the microtubule network breaks down along the anterior-posterior axis. One population, however, remains surrounding the nucleus, forming a hairpin-like structure that opens toward the center of the oocyte. At this stage, the nucleus is located at the dorsal anterior cortex of the oocyte (Figure 1E see arrow). Therefore, an intricate microtubule network exists in the Nasonia follicle, similar to the network described in Drosophila. Moreover, as is seen in Drosophila, we find that microtubules are enriched at the oocyte nucleus, suggesting that microtubule nucleation might be mediated in a manner dependent on the oocyte nucleus (Januschke et al., 2006).
Role of microtubules in formation of ovarian cysts
We next sought to disrupt the microtubule cytoskeleton pharmacologically using colchicine, which acts by depolymerizing microtubules. One day after eclosion, females were fed 65μg/ml colchicine dissolved in sugar water for 15 hours; ovaries were subsequently fixed and examined for defects in mRNA localization and oogenesis. We find this the most efficient approach to disrupt microtubules in a consistent way since cultured ovaries do not survive long enough to exhibit phenotypes. The morphology and organization of developing follicles is affected after this prolonged treatment in a way that is related to what has been observed in flies. Additionally, the overall shape of the nurse cells is elongated and antero-posterior polarity is often affected within the developing follicle chain, with some follicles exhibiting inverted polarity, a phenotype that has not been previously reported in Drosophila in response to microtubule disruption (Figure 2A,B). Colchicine treatment also results in altered number of nurse cells. Although we do not find egg chambers composed solely of nurse cells, as has been reported in Drosophila in response to similar drug treatment (reviewed by (Huynh and St Johnston, 2004),we do find egg chambers with twice the number of nurse cells. We interpret these as resulting from 2 follicles that have fused during oogenesis, but where only 1 oocyte was specified (i.e. 31 nurse cells and 1 oocyte; see Figure 2C). It might also be possible that this phenotype is the result of an extra round of mitosis in the germline, as has been previously reported in Drosophila encore mutant egg chambers, which contain 31 nurse cells and 1 oocyte. encore has been shown to be required for regulating mitotis entry/exit and oocyte differentiation during oogenesis (Hawkins et al., 1996). Additionally, as a result of microtubule drug inhibition, some follicles often contain a double oocyte with a furrow in the center, surrounded on each side by a set of nurse cells with opposite polarity. This phenotype probably results from the fusion of 2 cysts, one of which has inverted polarity (Figure 2B).
Figure 2. nos mRNA localization is microtubule independent.
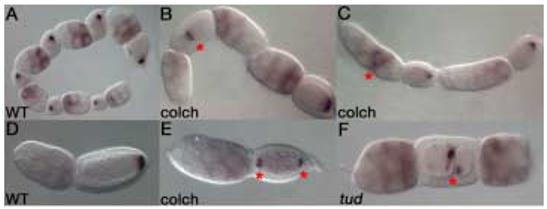
Wild type nos mRNA localization (A,D). Inverted, fused follicle, marked by an asterisk, is shown on the left with posterior nos localization. The adjacent follicle on the right shows the normal orientation of the follicular chain (B). Follicles often display extra nurse cells, with nos mRNA accumulating in central nurse cells (marked by an asterisk) in response to colchicine-treatment (C). A bipolar follicle shows localization of nos to both poles of the oocyte (E). Bipolar localization is marked by asterisks. tud RNAi results in dicephalic follicles with abnormal nurse cell numbers (F). The furrow within the oocyte is marked by an asterisk. Nonetheless, tud RNAi affected follicles show normal posterior nos mRNA localization (F).
These results indicate that microtubules play a major role in oocyte specification and overall polarity of developing follicles. This major role for the microtubule network in oogenesis is consistent with previous studies in hymenopterans using colchicine to inhibit the microtubule network (Bilinski and Jaglarz, 1999). Although microtubules do play an important role in oocyte specification and transport of factors from the nurse cells to the oocyte in Drosophila, microtubule inhibition in Nasonia results in strong oogenesis phenotypes that have not previously been reported in Drosophila despite use of similar drug inhibition treatments. Results from such experiments indicate that microtubules play a greater role in the wasp than in Drosophila in maintaining the general morphology of the nurse cells, positioning nurse cell nuclei, as well as in transferring maternal factors to the oocyte (Bilinski and Jaglarz, 1999).
Microtubules are necessary for localization of giant and caudal mRNAs
Our results show that colchicine treatment efficiently disrupts the microtubule network in Nasonia follicles as oogenesis phenotypes similar to those reported for Drosophila are obtained. We next examined the effects of microtubule disruption on localization of various maternal mRNAs.
giant (gt) is expressed maternally and its mRNA is localized to the anterior of the early Nasonia oocyte in a perinuclear manner (Figure 3A; Brent et al. 2007). This localization breaks down once the embryo is deposited, resulting in an mRNA gradient in freshly laid embryos. gt mRNA localization is microtubule dependent, as microtubule depolymerization prevents its localization to the anterior of the oocyte. Instead, gt mRNA is homogenously dispersed, resulting in a faint, even signal throughout the oocyte (Figure 3B). Interestingly, a population of microtubules surrounds the oocyte nucleus in developing wild type follicles in a pattern similar to the pattern observed during gt localization in stage 3 of oogenesis (Figure 3G,H). Thus it is possible that a specific population of microtubules that surrounds the nucleus is responsible for maintaining the perinuclear localization of gt mRNA. Similarly, in Drosophila it is believed that different microtubule populations are responsible for directing transport of specific mRNAs to different regions of the oocyte (St Johnston, 2005).
Figure 3. gt mRNA anterior localization and maintenance is microtubule-dependent.
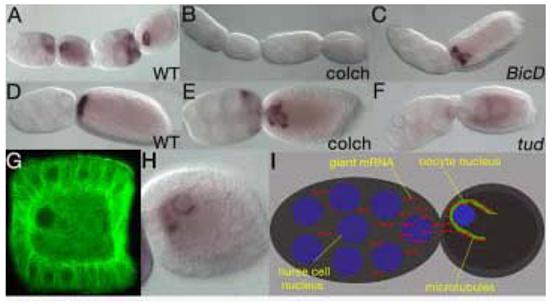
gt mRNA localizes in a perinuclear manner in early wild type follicles (A). This early localization is microtubule dependent since colchicine treated females fail to localize gt mRNA to the anterior pole (B). gt localizes to the anterior pole of the oocyte in later wild type follicles (D). Colchicine treatment disrupts the maintenance of gt mRNA localization in older follicles, where gt mRNA is streaming towards the center of the oocyte and is no longer tightly associated with the anterior pole (E). Similarly, BicD (C) and tud (F) mutant follicles show a failure to localize (F) and a failure to maintain normal (C) gt anterior localization. tud mutant follicles also display migration of nurse cell nuclei into the oocyte (F). Microtubules surround the oocyte nucleus in a manner similar to gt mRNA localization as evidenced by alpha tubulin staining (G). gt mRNA is localized to the anterior of the oocyte in a perinuclear manner (H). A model for gt mRNA localization using a microtubule dependent mechanism for transport and maintenance of localization (I).
As our method for colchicine treatment employs feeding females over relatively long periods of time and females ingest different quantities of microtubule inhibiting drugs, follicles are affected at varied stages of oogenesis. Additionally, we observe a range of phenotypes and varied strength within the affected follicles. Follicles treated with colchicine, presumably later during the development of the follicle than those that fail to localize gt mRNA in early stage 2, exhibit a failure to maintain gt localization in later stage 3 of oogenesis. In these follicles, “streams” of gt mRNA are seen detaching from the anterior pole, stretching towards the center of the oocyte (Figure 3E). Microtubules are thus important for both the initial localization of gt mRNA and subsequently for its maintenance.
We next examined the effect of microtubule depolymerization on the localization of caudal (cad) mRNA. cad mRNA is expressed throughout oogenesis in nurse cells and is localized to the posterior of the oocyte (Olesnicky et al., 2006) (Figure 4A). During stage 4, however, cad mRNA localization begins to break down such that once the embryo is laid, a cad mRNA gradient is formed. cad mRNA localization is also microtubule dependent as colchicine treatment results in a failure to localize to the posterior. Instead, cad mRNA is homogenously dispersed throughout the oocyte (Figure 4 compare A & C). Therefore, although at opposite poles, the mRNA localization of both gt and cad appears to exhibit similar features: Both mRNAs are transiently localized and begin to diffuse as the oocyte reaches stage 5, leading to the formation of mRNA gradients in the early embryo. They both appear to rely on a similar microtubule-dependent mechanism to achieve their initial mRNA localization. Later, gt mRNA also relies on microtubules to maintain its localization, whereas cad mRNA localization breaks down quickly in the wild type and has no need for maintaining localization.
Figure 4. Anterior otd1 and posterior cad mRNAs are localized in a microtubule-dependent manner.
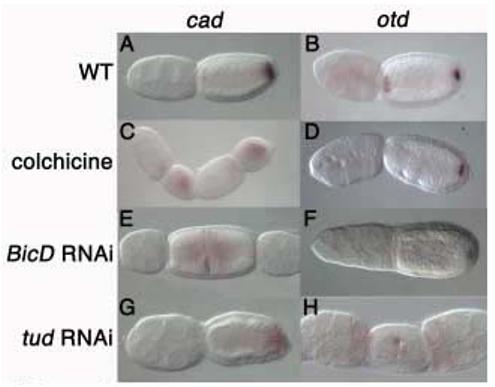
Wild type cad (A) and otd1 (B) localization. cad localization is lost in colchicine-treated follicles (C). Anterior otd1 localization is abolished in colchicine-treated follicles, while posterior otd1 remains localized (D). cad mRNA is homogenously expressed in inverted and fused follicles derived from BicD RNAi females (E). BicD mutant follicles show loss of only anterior otd1 localization, while posterior otd1 localization is reduced (F). cad mRNA fails to localize to the posterior of tud mutant follicles (G). tud RNAi follicles show fusion and inverted polarity phenotypes with aberrant anterior otd1 localization (H).
In Drosophila, a number of transiently localized mRNAs, including Bicaudal-D and orb, have been previously characterized. These mRNAs are also sensitive to microtubule inhibiting drug treatment, which results in uniform distribution of mRNA throughout the oocyte (Pokrywka, 1995; Pokrywka and Stephenson, 1991) (Pokrywka and Stephenson, 1995). Our results in Nasonia suggest that microtubules might play a conserved role in the transport of mRNA. Furthermore, the conserved sensitivity of transiently localized mRNAs to microtubule inhibitors among different species might reflect a common general mechanism for achieving this temporary localization.
Nasonia utilizes two different mechanisms to localize otd1 mRNA
otd1 mRNA is localized to both the anterior and posterior poles of the oocyte where it plays a major role in patterning the embryo. In particular, the anteriorly localized otd1 functions in a manner reminiscent of bcd in Drosophila (Lynch et al., 2006). During oogenesis, otd1 is first localized to the posterior of the Nasonia oocyte and closely resembles the tight localization seen for nos mRNA (see below; J. Lynch and C.D. submitted). Later, otd1 mRNA is also localized to the anterior of the oocyte. Anterior otd1 mRNA is loosely localized in comparison to the tight posterior otd1 mRNA localization, suggesting that the localization of this mRNA to opposite poles relies on 2 different mechanisms. Moreover anterior otd1 differs from gt anterior localization, as it is not perinuclear. Interestingly, in response to microtubule depolymerization, anterior otd1 mRNA localization is abolished, whereas posterior localization is maintained (Figure 4 compare B & D). Therefore, the anterior and posterior localization of otd1 mRNA depend on two different mechanisms: Anterior localization is microtubule-dependent while posterior localization is not.
Microtubules are not essential for nanos localization
The posterior otd1 localization is reminiscent of the tight localization of nos. This suggests that both mRNAs rely on a common mechanism for localization. As in Drosophila, nos mRNA is maternally expressed in Nasonia (J. Lynch and C.D., submitted). Using fluorescent in situ hybridization techniques, early nos mRNA can be seen localized in a perinuclear manner in oocytes (J. Lynch and C.D. submitted). Later, nos is tightly localized to the posterior pole of the developing oocyte (Figure 2A,D; Lynch and C.D., submitted). Depolymerization of microtubules using colchicine does not affect nos posterior localization within the egg chamber, even in conditions where severe oogenesis and polarity defects described above are fully apparent and cad and gt mRNAs are completely delocalized. In fused egg chambers, each set of nurse cells is oppositely oriented, resulting in nos mRNA localizing to the ‘posterior’ of each apposed oocyte (Figure 2B). We also observe nos localization to both poles of the oocyte in a small percentage of follicles examined, likely resulting from the transport of nos mRNA from 2 sets of inverted nurse cells (“bicaudal oocytes” Figure 2E). Therefore, although microtubules are necessary for oocyte specification and overall polarity of egg chambers throughout oogenesis, they are only required to transport some mRNAs and do not appear to be necessary for the localization of nos. This is consistent with previous reports in Drosophila, where nos mRNA is dispersed throughout the oocyte and is localized to the posterior by being trapped and anchored in an actin dependent manner to the pole plasm (Forrest and Gavis, 2003).
In Nasonia, both nos and posterior otd1 mRNA remain tightly localized to the posterior of the oocyte throughout oogenesis. Later, once the embryo is laid, nos and otd1 mRNA co-localize with the oosome, a structure that will give rise to the pole cells in the developing embryo (J. Lynch and C.D. submitted;(Lynch et al., 2006)). This suggests that a conserved mechanism involving pole plasm is utilized in both Nasonia and Drosophila to localize nos mRNA in both species, and posterior otd1 in Nasonia. However, since Drosophila has pole plasm structures but lacks an oosome, there may be some differences in the role of the pole plasm in RNA localization.
The actin cytoskeleton is required for anchoring nos and posterior otd1
To test whether actin might be required for the localization of those mRNAs that do not depend on microtubules, Nasonia ovaries were cultured with 1μg cytochalasin D for 1 hour, to disrupt the actin cytoskeleton, fixed and analyzed by in situ hybridization. Treatment with the drug is lethal at doses that affect oogenesis. Actin disruption results in nos detaching from the posterior pole of the oocyte (Figure 5D). Although detached from the posterior cortex, nos remains aggregated in a ring-like structure, which may represent the remains of the oosome. It has been shown that Drosophila nos mRNA localizes to the posterior of the oocyte through trapping and actin dependent anchoring of diffuse mRNA. Moreover, nos is associated with the pole plasm in Drosophila, although this structure is more diffuse and not as clearly defined as the Nasonia oosome (Forrest and Gavis, 2003). Our results suggest that a similar actin-dependent mechanism is responsible for anchoring nos to the posterior and is likely dependent on proper germ plasm assembly.
Figure 5. nos and posterior otd1 mRNA require actin for anchoring.
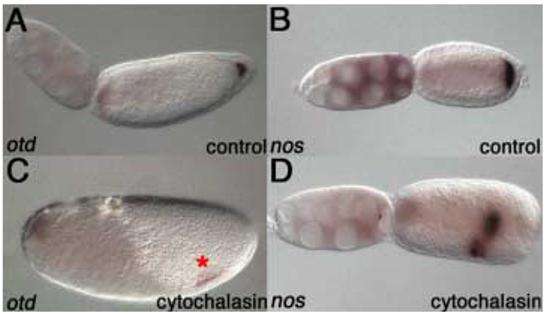
Wild type follicles stained for otd1 (A) and nos (B) mRNA after 60 mins of culturing at room temperature. Culturing for 60 mins with cytochalasin D results in detachment of posterior otd1 mRNA from the posterior pole of the oocyte (marked by an asterisk). Anterior otd1 mRNA localization remains unaffected (C). nos mRNA is also delocalized from the posterior in response to actin disruption. Most of nos mRNA, however, remains localized to the oosome (D).
We also examined otd1 localization in cytochalasin D treated ovaries. While anterior otd1 localization is unaffected, posterior otd1 becomes delocalized from the posterior pole of the oocyte (Figure 5C). The disruption of posterior otd1 localization is similar to that of nos, where the mRNA becomes detached from the posterior cortex but remains mostly aggregated within the oosome. This suggests that whereas anterior otd1 mRNA relies on microtubules for localization, posterior otd1 mRNA localizes to the posterior cortex of the oocyte in an actin-dependent manner, similar to nos mRNA. We also tested whether actin disruption affects the localization of cad or gt mRNA. However, cad posterior localization and gt anterior localization, like anterior otd1 mRNA localization, are not affected by cytochalasin D treatment (data not shown), indicating that actin does not play a role in driving or maintaining localization of these mRNAs. Instead, these mRNAs appear to utilize the microtubule network for proper localization.
Bicaudal-D and tudor RNAi phenocopy colchicine treatments
We next sought to identify genes that might be involved in microtubule organization and mRNA localization. In Drosophila, when egg chambers are depleted of Bicaudal D (BicD) during mid-oogenesis, follicles sometimes have double or half the number of nurse cells. Apposed egg chambers are also seen where the oocyte lies between two sets of nurse cells and, often, nurse cell nuclei protrude into the oocyte (Swan and Suter, 1996) (Oh and Steward, 2001). BicD loss of function egg chambers also often fail to specify an oocyte and result in egg chambers consisting of 16 nurse cells (Oh and Steward, 2001) (Suter and Steward, 1991). These phenotypes are similar to the phenotypes that result from prolonged colchicine treatment described in flies and here, in Nasonia follicles (see above). Indeed, in Drosophila BicD has been shown to play a role in the formation of the microtubule network (Oh and Steward, 2001) (Bullock and Ish-Horowicz, 2001) (Navarro et al., 2004). In BicD and egalitarian (egl) mutant egg chambers, the microtubule network fails to polarize as evidenced through tubulin staining, and mRNAs such as osk fail to localize (Swan and Suter, 1996) (Huynh and St Johnston, 2004). Egl protein forms a complex with BicD (Mach and Lehmann, 1997) and also binds dynein light chain to facilitate microtubule dependent transport via the dynein motor complex (Bullock and Ish-Horowicz, 2001; Navarro et al., 2004). Thus, BicD appears to serve as an adaptor between its cargo and the motor complex required for active transport along the microtubule cytoskeleton. We therefore next examined the role of BicD in Nasonia oogenesis.
In Nasonia, BicD is expressed homogenously throughout the egg chamber during oogenesis. We examined the function of BicD in Nasonia oogenesis using parental RNAi (pRNAi). Female pupae were injected with double stranded BicD RNA and their ovaries were subsequently fixed for analysis. Whereas females injected with GFP dsRNA show no oogenesis defects, females injected with BicD dsRNA show a range of phenotypes, closely resembling those observed in follicles of females treated with colchicine. Nurse cell morphology is affected in 40% of BicD pRNAi follicles. Additionally, 15% of the follicles derived from females injected with BicD dsRNA exhibit aberrant nurse cell number (Figure 4E). Some follicles show half the normal number of nurse cells and often display polarity defects resulting in fused egg chambers arranged in a symmetric manner (Figure 4E). Follicles with twice the number of nurse cells are also observed, albeit to a lesser extent (5%), although these egg chambers do not show polarity defects (data not shown). The most affected BicD pRNAi egg chambers show migration of nurse cell nuclei into the oocyte (data not shown), a phenotype that is reminiscent to the loss of function BicD phenotype in Drosophila. These data suggest that BicD function in forming and polarizing the microtubule network during oogenesis is conserved between flies and Nasonia.
In Drosophila, tudor (tud) is required for polar granule assembly and germ cell formation (Amikura et al., 2001; Arkov et al., 2006). tud functions in the transport of mitochondrial large ribosomal RNA from mitochondria to the polar granules (Amikura et al., 2001) (Thomson and Lasko, 2004). Moreover, tud mutant embryos fail to maintain nos localization due to aberrant polar granule assembly and often display loss of pole cells (Thomson and Lasko, 2004).Therefore, we expected tud to participate in nos and posterior otd1 mRNA localization in Nasonia.
Nvit tud is expressed homogenously in egg chambers throughout oogenesis (data not shown). Mutant follicles resulting from knockdown of tud function via pRNAi often display migration of nurse cell nuclei into the oocyte (Figure 3F), abnormal nurse cell number and apposed egg chambers with inverted polarity (Figure 2F, 4H) at similar frequencies to that seen in follicles derived from females injected with BicD dsRNA. Surprisingly, these phenotypes closely resemble the polarity defects seen in BicD RNAi and colchicine treatments, suggesting that in Nasonia, tud functions in microtubule assembly or stabilization, a role that has not been ascribed to tud in Drosophila. Instead, this more closely resembles Xenopus tudor repeat protein (Xtr), which is expressed in oogenesis. Xtr was shown to play a role in the organization of microtubules surrounding the nucleus in early Xenopus embryos, which also show cleavage arrest and abnormal cleavage formation in blastomeres (Hiyoshi et al., 2005), suggesting that Xtr functions during early embryogenesis in microtubule organization. These findings, together with our results, suggest that tudor may have a conserved role in microtubule assembly that was not detected in Drosophila.
Novel and conserved roles for tud and BicD in mRNA localization
We next tested whether mRNA localization is affected in BicD RNAi and tud RNAi follicles. In agreement with the oogenesis phenotypes described above, phenotypes observed with pRNAi are similar to those obtained with colchicine treatment. nos mRNA localizes normally in both tud (Figure 2F) and BicD mutant egg chambers (data not shown). However, cad mRNA fails to localize and instead shows diffuse staining throughout the oocyte (Figure 4G). Similarly, gt mRNA fails to localize to the anterior (data not shown). As pRNAi generates an allelic series, we also observe weaker phenotypes that affect mRNA localization at later stages of oogenesis. In later stage 5 of oogenesis, gt fails to maintain localization in both tud and BicD pRNAi follicles (Figure 3C,F). Finally, we examined the localization pattern of otd1 mRNA in BicD and tud RNAi follicles. Anterior otd1 localization is abolished in the absence of tud and BicD function, but posterior otd1 remains localized, albeit at a lower level (Figure 4F,H). These results suggest that, in addition to oocyte specification, BicD plays a fundamental role in mRNA localization, similar to Drosophila. Moreover, BicD has been shown to play a role in MTOC formation in the Drosophila ovary. As the mRNA localization phenotypes in BicD RNAi follicles are very similar to those obtained in microtubule disruption, it is probable that BicD plays a conserved role in MTOC formation in Nasonia. Consistent with the function of Xtr in Xenopus, our data also suggest that tud plays a role in mRNA localization.
Conclusions
We propose that Nasonia utilizes two basic mechanisms for the localization of mRNA, a microtubule-dependent mechanism and an actin-dependent, microtubule-independent one. Anterior localization of gt and otd1 mRNA, as well as posterior localization of cad mRNA, all rely on a similar microtubule-dependent mechanism while posterior localization of otd1 and nos mRNAs rely on actin. In wild type follicles, cad and gt mRNAs are initially localized, while later in oogenesis this localization is relaxed to achieve a more graded mRNA distribution. otd1 anterior mRNA, although not graded, is also localized loosely in wild type follicles. nos mRNA localization and posteriorly localized otd1 mRNA, however, are tightly localized to the posterior in a microtubule-independent manner. Interestingly, in freshly laid embryos both posterior otd1 mRNA and nos mRNA are localized to the oosome. Maintaining localization of these two posteriorly localized mRNAs relies on the actin cytoskeleton. Additionally, actin might be required to anchor the oosome to the posterior pole of the oocyte, as well as to trap mRNA to the oosome. It is therefore likely that both mRNAs are localized to structures within the germ plasm, resulting in a tight localization that is maintained throughout oogenesis and early embryogenesis and does not rely extensively on microtubules.
Acknowledgements
We are grateful to Ava Brent, Jeremy Lynch for their helpful comments on the manuscript. We thank the Desplan and Small labs and Flynet for their constant support. This work was supported by a grant from NIH GM-64864 to C.D. E.C.O. was supported by NIH Training Grant 5 T32 HD007520. This investigation was conducted in a facility constructed with support from Research Facilities Improvement Grant C06 RR-15518-01 from NCRR, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amikura R, Hanyu K, Kashikawa M, Kobayashi S. Tudor protein is essential for the localization of mitochondrial RNAs in polar granules of Drosophila embryos. Mech Dev. 2001;107:97–104. doi: 10.1016/s0925-4773(01)00455-5. [DOI] [PubMed] [Google Scholar]
- Arkov AL, Wang JY, Ramos A, Lehmann R. The role of Tudor domains in germline development and polar granule architecture. Development. 2006;133:4053–62. doi: 10.1242/dev.02572. [DOI] [PubMed] [Google Scholar]
- Ashe HL, Briscoe J. The interpretation of morphogen gradients. Development. 2006;133:385–94. doi: 10.1242/dev.02238. [DOI] [PubMed] [Google Scholar]
- Bilinski SM, Jaglarz MK. Organization and possible functions of microtubule cytoskeleton in hymenopteran nurse cells. Cell Motil Cytoskeleton. 1999;43:213–20. doi: 10.1002/(SICI)1097-0169(1999)43:3<213::AID-CM4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Brent AE, Tabin CJ. A somitic compartment of tendon progenitors. Cell. 2003;113:235–248. doi: 10.1016/s0092-8674(03)00268-x. S. R. [DOI] [PubMed] [Google Scholar]
- Brent AE, Yucel G, Small S, Desplan C. Permissive and instructive anterior patterning functions rely on mRNA localization in the wasp embryo. Science. 2007 doi: 10.1126/science.1137528. in press. [DOI] [PubMed] [Google Scholar]
- Bullock SL, Ish-Horowicz D. Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature. 2001;414:611–6. doi: 10.1038/414611a. [DOI] [PubMed] [Google Scholar]
- Davis GK, Patel NH. Short, long, and beyond: molecular and embryological approaches to insect segmentation. Annu Rev Entomol. 2002;47:669–99. doi: 10.1146/annurev.ento.47.091201.145251. [DOI] [PubMed] [Google Scholar]
- Driever W, Nusslein-Volhard C. A gradient of bicoid protein in Drosophila embryos. Cell. 1988a;54:83–93. doi: 10.1016/0092-8674(88)90182-1. [DOI] [PubMed] [Google Scholar]
- Driever W, Nusslein-Volhard C. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell. 1988b;54:95–104. doi: 10.1016/0092-8674(88)90183-3. [DOI] [PubMed] [Google Scholar]
- Forrest KM, Gavis ER. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol. 2003;13:1159–68. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- Gavis ER, Lehmann R. Localization of nanos RNA controls embryonic polarity. Cell. 1992;71:301–13. doi: 10.1016/0092-8674(92)90358-j. [DOI] [PubMed] [Google Scholar]
- Hawkins NC, Thorpe J, Schupbach T. Encore, a gene required for the regulation of germ line mitosis and oocyte differentiation during Drosophila oogenesis. Development. 1996;122:281–90. doi: 10.1242/dev.122.1.281. [DOI] [PubMed] [Google Scholar]
- Hiyoshi M, Nakajo N, Abe S, Takamune K. Involvement of Xtr (Xenopus tudor repeat) in microtubule assembly around nucleus and karyokinesis during cleavage in Xenopus laevis. Dev Growth Differ. 2005;47:109–17. doi: 10.1111/j.1440-169x.2005.00787.x. [DOI] [PubMed] [Google Scholar]
- Huynh JR, St Johnston D. The origin of asymmetry: early polarisation of the Drosophila germline cyst and oocyte. Curr Biol. 2004;14:R438–49. doi: 10.1016/j.cub.2004.05.040. [DOI] [PubMed] [Google Scholar]
- Januschke J, Gervais L, Gillet L, Keryer G, Bornens M, Guichet A. The centrosome-nucleus complex and microtubule organization in the Drosophila oocyte. Development. 2006;133:129–39. doi: 10.1242/dev.02179. [DOI] [PubMed] [Google Scholar]
- King PE. Oogenesis in Nasonia vitripennis (Hymenoptera: Pteromalidae) Proceedings of the Entomological Society of London, Series A, General Entomology. 1969;44:143–157. [Google Scholar]
- Liu PZ, Kaufman TC. Short and long germ segmentation: unanswered questions in the evolution of a developmental mode. Evol Dev. 2005;7:629–46. doi: 10.1111/j.1525-142X.2005.05066.x. [DOI] [PubMed] [Google Scholar]
- Lynch JA, Brent AE, Leaf DS, Pultz MA, Desplan C. Localized maternal orthodenticle patterns anterior and posterior in the long germ wasp Nasonia. Nature. 2006;439:728–32. doi: 10.1038/nature04445. [DOI] [PubMed] [Google Scholar]
- Mach JM, Lehmann R. An Egalitarian-BicaudalD complex is essential for oocyte specification and axis determination in Drosophila. Genes Dev. 1997;11:423–35. doi: 10.1101/gad.11.4.423. [DOI] [PubMed] [Google Scholar]
- Navarro C, Puthalakath H, Adams JM, Strasser A, Lehmann R. Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat Cell Biol. 2004;6:427–35. doi: 10.1038/ncb1122. [DOI] [PubMed] [Google Scholar]
- Niessing D, Blanke S, Jackle H. Bicoid associates with the 5′-cap-bound complex of caudal mRNA and represses translation. Genes Dev. 2002;16:2576–82. doi: 10.1101/gad.240002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessing D, Dostatni N, Jackle H, Rivera-Pomar R. Sequence interval within the PEST motif of Bicoid is important for translational repression of caudal mRNA in the anterior region of the Drosophila embryo. Embo J. 1999;18:1966–73. doi: 10.1093/emboj/18.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessing D, Driever W, Sprenger F, Taubert H, Jackle H, Rivera-Pomar R. Homeodomain position 54 specifies transcriptional versus translational control by Bicoid. Mol Cell. 2000;5:395–401. doi: 10.1016/s1097-2765(00)80434-7. [DOI] [PubMed] [Google Scholar]
- Oh J, Steward R. Bicaudal-D is essential for egg chamber formation and cytoskeletal organization in drosophila oogenesis. Dev Biol. 2001;232:91–104. doi: 10.1006/dbio.2001.0170. [DOI] [PubMed] [Google Scholar]
- Olesnicky EC, Brent AE, Tonnes L, Walker M, Pultz MA, Leaf D, Desplan C. A caudal mRNA gradient controls posterior development in the wasp Nasonia. Development. 2006;133:3973–82. doi: 10.1242/dev.02576. [DOI] [PubMed] [Google Scholar]
- Pokrywka NJ. RNA localization and the cytoskeleton in Drosophila oocytes. Curr Top Dev Biol. 1995;31:139–66. doi: 10.1016/s0070-2153(08)60226-4. [DOI] [PubMed] [Google Scholar]
- Pokrywka NJ, Stephenson EC. Microtubules mediate the localization of bicoid RNA during Drosophila oogenesis. Development. 1991;113:55–66. doi: 10.1242/dev.113.1.55. [DOI] [PubMed] [Google Scholar]
- Pokrywka NJ, Stephenson EC. Microtubules are a general component of mRNA localization systems in Drosophila oocytes. Dev Biol. 1995;167:363–70. doi: 10.1006/dbio.1995.1030. [DOI] [PubMed] [Google Scholar]
- Pultz MA, Leaf DS. The jewel wasp Nasonia: querying the genome with haplo-diploid genetics. Genesis. 2003;35:185–91. doi: 10.1002/gene.10189. [DOI] [PubMed] [Google Scholar]
- Rivera-Pomar R, Jackle H. From gradients to stripes in Drosophila embryogenesis: filling in the gaps. Trends Genet. 1996;12:478–83. doi: 10.1016/0168-9525(96)10044-5. [DOI] [PubMed] [Google Scholar]
- Rivera-Pomar R, Niessing D, Schmidt-Ott U, Gehring WJ, Jackle H. RNA binding and translational suppression by bicoid. Nature. 1996;379:746–9. doi: 10.1038/379746a0. [DOI] [PubMed] [Google Scholar]
- St Johnston D. Moving messages: the intracellular localization of mRNAs. Nat Rev Mol Cell Biol. 2005;6:363–75. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- Stauber M, Jackle H, Schmidt-Ott U. The anterior determinant bicoid of Drosophila is a derived Hox class 3 gene. Proc Natl Acad Sci U S A. 1999;96:3786–9. doi: 10.1073/pnas.96.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer J, Kalderon D. Microtubule polarity and axis formation in the Drosophila oocyte. Dev Dyn. 2006;235:1455–68. doi: 10.1002/dvdy.20770. [DOI] [PubMed] [Google Scholar]
- Struhl G, Struhl K, Macdonald PM. The gradient morphogen bicoid is a concentration-dependent transcriptional activator. Cell. 1989;57:1259–73. doi: 10.1016/0092-8674(89)90062-7. [DOI] [PubMed] [Google Scholar]
- Suter B, Steward R. Requirement for phosphorylation and localization of the Bicaudal-D protein in Drosophila oocyte differentiation. Cell. 1991;67:917–26. doi: 10.1016/0092-8674(91)90365-6. [DOI] [PubMed] [Google Scholar]
- Swan A, Suter B. Role of Bicaudal-D in patterning the Drosophila egg chamber in mid-oogenesis. Development. 1996;122:3577–86. doi: 10.1242/dev.122.11.3577. [DOI] [PubMed] [Google Scholar]
- Thomson T, Lasko P. Drosophila tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis. 2004;40:164–70. doi: 10.1002/gene.20079. [DOI] [PubMed] [Google Scholar]


