Abstract
Oral candidiasis is a common opportunistic infection, with Candida albicans being the most prevalent etiologic agent and Candida glabrata emerging as an important pathogen. C. glabrata is frequently co-isolated with C. albicans from oral lesions. Although C. albicans has been shown to trigger significant cytokine responses and cell damage, C. glabrata has not been systematically studied yet. The purpose of this study was to characterize the ability of C. glabrata to induce proinflammatory cytokine responses and host damage as a single infecting organism and in combination with C. albicans, using in vitro models of the oral mucosa. In monolayer oral epithelial cell cultures, C. glabrata failed to induce a significant interleukin-1α and interleukin-8 cytokine response and showed lower cytotoxicity, compared to C. albicans. However, C. glabrata triggered a significantly higher granulocyte macrophage colony stimulating factor response than C. albicans. C. glabrata strains showed a strain-dependent tissue damaging ability and a superficial invasion of the mucosal compartment in a 3-dimensional (3-D) in vitro model of the human oral mucosa and submucosa. In the 3-D system, co-infection failed to promote host damage beyond the levels of infection with C. albicans alone. These studies indicate that C. glabrata induces cytokines in human oral epithelium in a strain-specific manner, but its tissue/cell damaging ability, compared to C. albicans, is low. Synergy between C. glabrata and C. albicans in cytokine induction and host damage was not observed with the strains tested.
Keywords: Candida glabrata, Cytokines, Cytotoxicity, Oral Epithelium
1. Introduction
In recent years C. glabrata is emerging as an important pathogen in humans, being the second or third leading agent of candidiasis at all sites, including the oropharyngeal and esophageal mucosa [1]. The actual prevalence of C. glabrata-induced oropharyngeal candidiasis (OPC) is difficult to decipher from epidemiological studies, due to the fact that this organism is rarely the only Candida species isolated from the mucosal lesions [2]. C. glabrata is often co-isolated with C. albicans, however, reports of C. glabrata as the lone detectable species from oral lesions have also been rising steadily [3, 4, 5]. This is particularly important since unlike other Candida species, C. glabrata isolated from oral lesions is much more resistant to standard anti-fungal treatment than C. albicans [3, 4].
Although the pathogenicity of C. albicans in the oral mucosa has been well established (for review see [6]), little is known about the outcome of the interaction between C. glabrata and oral mucosal cells. C. albicans has been shown to trigger a multitude of proinflammatory cytokines, as well as significant cell/tissue damage in several in vitro model systems of the human oral mucosa (for review see [6]). However, reports on C. glabrata are currently scarce and not centered on this microorganism [7]. Therefore, the purpose of this work was to study the ability of C. glabrata to trigger a proinflammatory cytokine response and cell damage as a single infecting organism and in combination with C. albicans, using well established in vitro models of the oral mucosa [8-13]. The underlying hypothesis in this study is that although in a monoinfection model C. glabrata may trigger reduced cell destruction and may elicit a lower proinflammatory cytokine response compared to C. albicans, in a co-infection model with C. albicans, it may synergistically augment the oral mucosal cell inflammatory response to infection and tissue destruction. Because of its increasing importance as a pathogen, its high frequency of anti-fungal drug resistance, and the paucity of information on its specific role in the pathogenesis of OPC, new information on the ability of this organism to trigger an inflammatory response and destroy the oral mucosa is of paramount importance.
2. Results
2.1 Compared with C. albicans, C. glabrata triggered a different inflammatory cytokine profile in oral epithelial cells
We first analyzed the network of cytokines produced by oral epithelial cells OKF6/TERT-2 challenged with two oroesophageal C. glabrata strains using a cytokine protein array, in comparison with C. albicans SC5314, used as a reference strain. As shown in Table 1, both C. glabrata strains triggered a significant GM-CSF response above basal. However, only one of the two strains (MRL2302) triggered a pronounced growth-regulated oncogene (GRO) response, similar to the C. albicans reference strain. No significant induction of other proinflammatory and antiinflammatory cytokines, including IL-8, IL-6, IL-10, TNF-α and IL-1α, was observed in C. glabrata-challenged OKF6/TERT-2 cells. Compared with C. glabrata, C. albicans SC5314 triggered a more pronounced IL-1α and IL-8, but a weaker GM-CSF response in OKF6/TERT-2 cells (Table 1).
Table 1.
Cytokine profile of oral epithelial cells following challenge with C. glabrata.
| Fold protein induction compared with unchallenged cells | |||
|---|---|---|---|
| Cytokine | |||
| C. glabrata 94-11 | C. glabrata MRL2302 | C. albicans SC5314 | |
| IL-6 | 1.2±0.3 | 1.1±0.2 | 1.9±0.4 |
| IL-10 | 0.9±0.2 | 0.9±0.1 | 1.2±0.2 |
| GRO | 1.4±0.3 | 4.6±0.6* | 4.2±0.3* |
| GM-CSF | 3.6±0.2* | 5.7±0.1* | 1.8±0.1 |
| IL-1α | 1.7±0.4 | 0.9±0.2 | 2.4±0.1* |
| IL-8 | 1.6±0.5 | 1.8±0.6 | 2.2±0.2* |
| TNF-α | 1.1±0.2 | 1.2±0.5 | 1.2±0.2 |
1×106 OKF6/TERT-2 cells were challenged with an equal number of C. glabrata cells (strains 94-11 and MRL2302), as well as C. albicans SC5314 (positive control) for 24 h. Supernatants were tested for the presence of cytokines using a cytokine protein array. The results are expressed as fold induction above basal (uninfected) levels (means ± SD). Samples from two independent experiments were analyzed.
P < 0.05 for a comparison with uninfected control.
2.2 The proinflammatory cytokine response to C. glabrata was strain-dependent
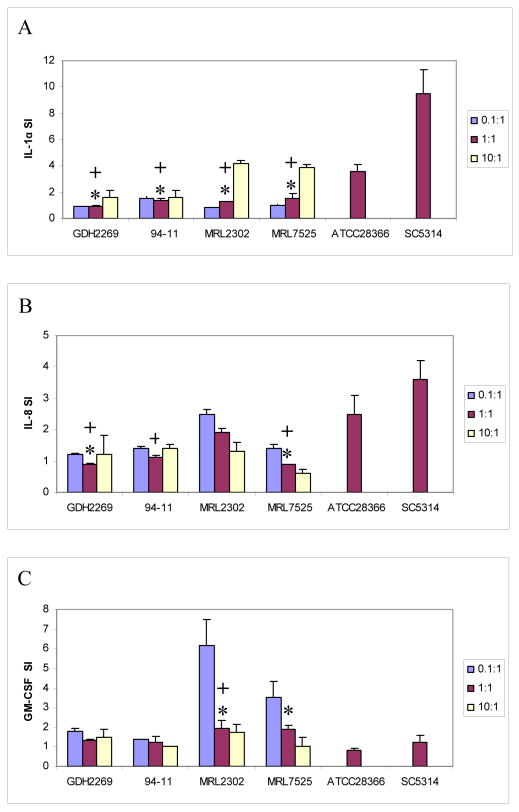
To confirm findings with the cytokine protein array and further characterize the proinflammatory cytokine response to C. glabrata in oral epithelial cells, we challenged OKF6/TERT-2 cells with four C. glabrata strains at various yeast to epithelial cell ratios (0.1:1, 1:1 and 10:1) for up to 24 h, and quantified the levels of cytokine mRNAs in cell lysates by real-time RT-PCR as well as the levels of cytokine proteins in culture supernatants by ELISA. For comparison, two C. albicans strains were also used at 1:1 yeast to epithelial cell ratio. This ratio was previously documented to trigger maximal cytokine responses by C. albicans in this system [8, 9]. Because the most pronounced difference observed between the two species related to GM-CSF, IL-1α and IL-8 induction (Table 1), we focused on these cytokines. Compared to both C. albicans strains, all C. glabrata strains triggered a lower IL-1α response when added to the epithelial cell monolayers at the 1:1 challenge ratio, at the protein (Fig. 1A) and mRNA (Table 2) levels. In addition, regardless of the challenge ratio, all C. glabrata strains triggered a low IL-8 protein response (less than 3 fold above basal) in OKF6/TERT-2 cells. With the exception of strain MRL2302, this response was significantly lower than that induced by the oral C. albicans strain ATCC28366, when added to cultures at the same infectivity ratio (Fig. 1B). Consistent with previous findings, C. albicans strain SC5314 triggered a significantly higher IL-8 response than ATCC28366 [8] and all C. glabrata strains tested (Fig. 1B). IL-8 protein findings correlated well with mRNA findings in these cultures with the exception of strain 94-11 which showed significant induction of IL-8 mRNA but not protein (Table 2 and Fig. 1B), possibly due to deficient post-transcriptional processing.
Figure 1.
Cytokine responses of oral epithelial cells to C. glabrata. 4×105 OKF6/TERT-2 cells were challenged with C. glabrata at 0.1:1, 1:1 and 10:1 or C. albicans at 1:1 fungal cell to epithelial cell ratio for 24 h. The presence of IL-1α (A), IL-8 (B) and GM-CSF (C) were examined by ELISA of culture supernatants. Results are expressed as the mean stimulation index (SI, stimulated over basal secretion levels). Mean SI values were obtained by analysis of two experiments with each condition set up in triplicate. The bars represent SD. * P < 0.05 for a comparison with strain ATCC28366; + P < 0.05 for a comparison with strain SC5314.
Table 2.
Comparison of cytokine mRNA expression in C. glabrata- or C. albicans-challenged oral epithelial cells.
| Cytokine | Duration of challenge | Fungal cell to epithelial cell ratio | Strains | mRNA fold increase over uninfected cells |
|---|---|---|---|---|
| IL-1α | 8 hours | 1:1 | GDH2269 | 0.8±0.1* |
| 1:1 | 94-11 | 1.6±0.2* | ||
| 1:1 | ATCC28366 | 2.5±0.2 | ||
| 1:1 | SC5314 | 3.3±0.1 | ||
| IL-8 | 8 hours | 1:1 | GDH2269 | 1.4±0.3* |
| 1:1 | 94-11 | 5.6±0.3* | ||
| 1:1 | ATCC28366 | 4.1±0.6* | ||
| 1:1 | SC5314 | 12.3±0.4 | ||
| GM-CSF | 24 hours | 0.1:1 | MRL2302 | 14.5±2.6* |
| 0.1:1 | SC5314 | 6.0±1.4 |
4×105 OKF6/TERT-2 cells were challenged with C. glabrata or C. albicans at 0.1:1 or 1:1 fungal cell to epithelial cell ratio. IL-1α, IL-8 and GM-CSF mRNA in OKF6/TERT-2 cells were quantified by real-time RT PCR. Data represent two separate experiments and are expressed as ratio ±SD over uninfected.
P < 0.05 for a comparison with strain SC5314 (positive control).
We have previously reported a weak, strain-dependent GM-CSF protein induction in oral keratinocytes by C. albicans, only at lower infectivity ratios [11]. Consistent with prior findings, we noted a lower than two-fold induction of GM-CSF protein above basal triggered by C. albicans SC5314 and ATCC28366, which did not differ significantly from the two oral C. glabrata strains (Fig. 1C). Interestingly, the GM-CSF response to C. glabrata esophageal isolates MRL2302 and MRL7525 at the 1:1 challenge ratio was significantly higher than that to C. albicans oral strain ATCC28366 at the same ratio (Fig. 1C). In comparison with the highly virulent C. albicans strain SC5314 used in the same ratio, C. glabrata strain MRL2302 induced a significantly higher GM-CSF response in challenged OKF6/TERT-2 cells (Fig. 1C). These findings were confirmed at the mRNA level (Table 2). When used at the lowest challenge ratio, these C. glabrata strains triggered the most pronounced GM-CSF response (4-6 fold above basal) in OKF6/TERT-2 cells (Fig. 1C). Overall, the lowest challenge ratio (0.1:1) was consistently more effective in triggering a GM-CSF response by all C. glabrata strains tested. Findings for all cytokines in OKF6/TERT-2 cells were confirmed in two primary oral keratinocyte cultures (not shown).
2.3 C. glabrata demonstrated lower cytotoxic and tissue damaging potential in comparison to C. albicans
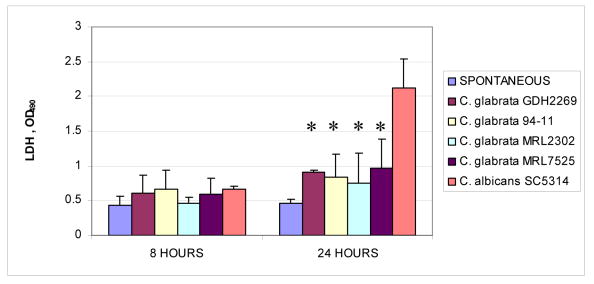
We next performed a series of experiments to characterize the cell and tissue damaging potential of C. glabrata in comparison to C. albicans. Initially the cytotoxicity of these organisms was tested in monolayer cultures of OKF6/TERT-2 cells by measuring LDH released in culture supernatants during infection. As shown in figure 2, C. albicans SC5314 was significantly more cytotoxic than the C. glabrata oropharyngeal strains when tested at the same infectivity ratio (Fig. 2). In fact, the cytotoxic potential of all four C. glabrata strains was very low, as LDH released in the presence of these organisms was not significantly higher than the spontaneous release, even after 24 h of infection (Fig. 2). Higher doses of these strains did not significantly affect the cytotoxicity levels (not shown).
Figure 2.
Comparison of cytotoxic potential of C. glabrata and C. albicans. 4×105 OKF6/TERT-2 cells were co-cultured with an equal number of C. glabrata cells (strains GDH2269, 94-11, MRL2302 and MRL7525), and C. albicans SC5314 for 8 h and 24 h. Cell supernatants were analyzed for LDH presence. * indicates that the P value is < 0.05 for a comparison with strain SC5314. Values were obtained by analysis of two separate experiments with conditions set up in duplicate. The bars represent SD.
To confirm findings with culture monolayers and compare the tissue damaging potential of these two Candida species, we infected an in-house 3-D tissue model of the oral mucosa with two C. glabrata strains and two C. albicans strains. Compared with C. albicans (Fig.3, D and E), both C. glabrata strains formed a thinner mucosal biofilm (Fig.3, B and C). Compared to the uninfected tissue models (Fig.3, A), minimal loss of the normal mucosal architecture with only a mild edema in the uppermost keratinocyte layers was observed with C. glabrata strain GDH2269 throughout the 48 h infection period (Fig.3, B), which was consistent with a prior report testing a single oral candidiasis C. glabrata isolate in a commercially available 3-D model system [7]. C. glabrata strain MRL2302 showed a superficial intraepithelial invasion accompanied by extensive intercellular edema and loss of cellular junctions in the keratinized and granular layers (Fig.3, C). In contrast, both C. albicans strains invaded past the epithelial layer into the lamina propria in this model. Degradation of the uppermost layer of epithelial cells and acantholysis was seen after 24 h of infection with C. albicans (not shown). Spongiosis, extensive cellular necrosis and loss of cellular junctions in the stratum basale was evident after 48 h (Fig.3, D and E). Consistent with the histological findings, the LDH release triggered by C. glabrata GDH2269 was significantly lower than that triggered by C. albicans ATCC28366 and SC5314 (P<0.05), and did not differ significantly from uninfected cultures (P>0.1) (Fig.3B). In contrast, the LDH release triggered by C. glabrata MRL2302 was significantly higher than that in uninfected cultures (P<0.05), but lower than both C. albicans strains (Fig.3C).
Figure 3.
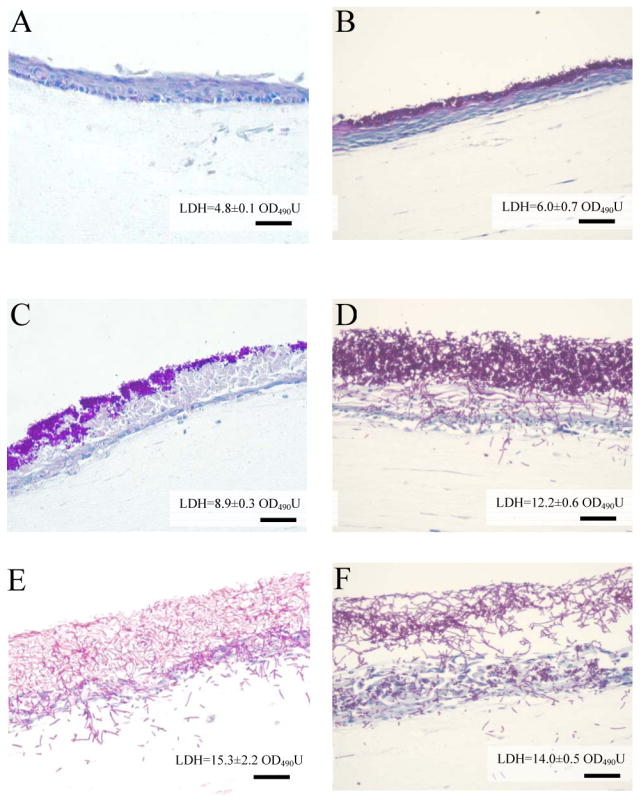
Infection of the 3-D model of oral mucosa with C. glabrata (B, GDH2269; C, MRL2302); C. albicans (D, ATCC28366; E, SC5314) and combination of GDH2269 and ATCC28366 (F) for 48 h. Tissues were infected with 1×106 fungal cells in single infections. In co-infections, 1×106 yeast cells of one strain from each species were used. The 3-D model without infection (A) was shown as a control. PAS-stained paraffin sections are shown at 20x magnifications. Shown here is one of the two representative experiments. Bars=60 μm.
2.4 Co-infection with C. glabrata and C. albicans did not enhance the host proinflammatory response or trigger increased tissue damage
Co-infection of OKF6/TERT-2 cells in monolayers using the same numbers of C. glabrata (94-11strain) cells and C. albicans (ATCC28366 or SC5314 strain) cells did not trigger a synergistic upregulation of GM-CSF, IL-1α or IL-8, since the response to co-infection we observed was, at best, additive (Table 3). Findings were similar when co-infection with the two species was carried out using lower number of yeasts (not shown). Also, when we tested cytokine responses during co-infection by adding half the dose of each species so that the final infectivity dose is kept constant in each well, we detected no significant differences between the responses to a single Candida species compared to mixed species infection (not shown).
Table 3.
Cytokine responses in oral epithelial cells co-infected with C. glabrata and C. albicans.
| Cultures | Infections | Cytokines (pg/ml) | ||
|---|---|---|---|---|
| IL-1α | IL-8 | GM-CSF | ||
| Monolayer | Unchallenged | 145±63 | 1717±753 | 236±25 |
| C. glabrata 94-11 | 246±28 | 2104±818 | 574±93 * | |
| C. albicans ATCC28366 | 301±35 * | 2187±141 | 189±40 | |
| C. albicans SC5314 | 978±122 * | 2079±225 | 278±12 | |
| C. glabrata 94-11+C. albicans ATCC28366 | 589±95 * | 2410±77 | 259±1 | |
| C. glabrata94-11 +C. albicans SC5314 | 1195±207* | 2781±133 | 241±23 | |
| 3-D model | Unchallenged | 115±31 | 7450±70 | 58±3 |
| C. glabrata GDH2269 | 932±1129* | 15200±8626 | 145±29 * | |
| C. albicans ATCC28366 | 913±165 * | 17500±7495 | 99±24 | |
| C. glabrata GDH2269+C. albicans ATCC28366 | 930±382 * | 16500±5445 | 164±18* | |
In monolayer cultures, 4×105 OKF6/TERT-2 cells were challenged with an equal number of C. glabrata 94-11, C. albicans ATCC28366 or C. albicans SC5314 cells, and supernatants were collected 24 h after challenge. In co-infections, 4×105 yeast cells of one strain from each species were used. In the 3-D models, tissues were infected with 1×106 C. glabrata GDH2269 or 1×106 C. albicans ATCC28366 cells, and supernatants were collected 48 h postinfection. In co-infections, 1×106 yeast cells of one strain from each species were used. Data were obtained by analysis of two separate experiments and shown as mean ± SD.
P<0.05 compared to unchallenged.
Since C. glabrata GDH2269 triggered a weak cytokine response in OKF6/TERT-2 cells (Fig.1) and a minimal tissue damage (Fig. 3B) in the tissue model system, this strain was used along with oral C. albicans strain ATCC28366 to co-infect the 3-D cultures in order to examine how these strains would perform in a co-infection setting. Consistent with the findings in monolayer cultures, co-infection of the 3-D model of oral mucosa with C. glabrata GDH2269 and C. albicans ATCC28366 did not trigger a synergistic upregulation of GM-CSF, IL-1α or IL-8 (Table 3). The LDH release triggered by co-infection with strains GDH2269 and ATCC28366 was not significantly different from that triggered by strain ATCC28366 alone (P>0.1). Interestingly, co-infection with the two species produced a soft tissue biofilm with lower cell density, but no reduction in thickness, compared to C. albicans alone (Fig. 3B, D, F).
3. Discussion
A number of new Candida species are emerging as pathogens causing oral infections with significantly increasing morbidity in immunocompromised patient cohorts [5]. C. glabrata is frequently isolated from the oral cavity of patients receiving radiation treatment of head and neck tumors [3], the elderly [15], as well as patients with advanced extra-oral cancer [16]. Infection in these patients is frequently associated with more than one co-isolated Candida species [3, 4]. Clinical reports suggest that oral infection with mixed C. albicans and C. glabrata may be more severe and is more difficult to treat [5], therefore insight in the pathogenesis of this mixed infection is critical.
In this study we showed that C. glabrata, with the exception of GM-CSF, failed to induce a proinflammatory cytokine response in oral epithelial cells that is comparable to C. albicans. These findings are in agreement with the only other report known, regarding oral mucosal cytokine responses to this species [7]. Schaller and coworkers reported similar findings in a commercially available 3-D oral mucosa system, infected with a single oral C. glabrata isolate. These findings are also consistent with a prior report in endothelial cells, which demonstrated that while chemotactic cytokines are induced in response to C. albicans, this is not the case with C. glabrata [17]. Because chemotactic cytokines such as IL-8 also promote the release of azurophilic granules by neutrophils, which enhance Candidacidal activity [18], a lack of such a response by C. glabrata may contribute to its reduced clearance and persistence in the oral mucosa. Ours is the first systematic attempt to examine the proinflammatory cytokine responses of several oral epithelial cell systems to multiple strains of this species.
A significant finding of our study is the ability of C. glabrata to trigger high amounts of GM-CSF at the lowest infectivity ratio. The protective role of GM-CSF in oral candidiasis has been suggested in clinical reports using this proinflammatory cytokine as an adjunct to antifungal treatment [19, 20]. By enhancing the proliferation, activation as well as fungicidal activity of immunoeffector cells such as neutrophils and monocytes, GM-CSF may play a major role in local control of Candida and prevention of invasive infection [21, 22]. At higher infectivity doses GM-CSF production by oral epithelial cells returned to almost baseline levels. Although the mechanism for this high dose-associated inhibition of GM-CSF induction is unknown, it is possible that C. glabrata, like other fungal species [23], produces fatty acid metabolites, which at higher concentrations may potentially inhibit GM-CSF synthesis by epithelial cells [24]. Alternatively, higher doses of C. glabrata may trigger the generation of prostaglandins or lipoxins by oral epithelial cells, which acting in an autocrine manner may inhibit GM-CSF synthesis [25, 26]. This effect of high C. glabrata infectious loads on oral epithelium may be analogous to the cytokine-mediated “cell paralysis” which takes place when immune cells face high antigenic loads [27], and may be another one of the innate immune evasion strategies of this organism in the oral mucosa.
We also showed that C. glabrata was not capable of inflicting significant oral epithelial cell monolayer damage and did not invade past the basal epithelial cell layers in a 3-D model of the oral mucosa. Secretion of phospholipases and proteases can play a role in disrupting epithelial cell membranes and these enzymes have been postulated to be essential in invasive Candida infections [26]. Most studies examining a large number of C. glabrata strains or clinical isolates have reported that only a small fraction or none of these are able to produce phospholipases [27, 28, 29]. In addition several studies failed to detect protease activity in C. glabrata [29, 30]. Our discrepant findings in tissue invasion and damage with C. glabrata GDH2269 and MRL2302 suggest differences in the expression of such extracellular hydrolytic enzymes in these strains.
Because C. glabrata is most often co-isolated with C. albicans we postulated that interactions between the two species play a role in the pathogenesis of mixed oral infection. C. albicans has been shown to efficiently adhere to a preformed C. glabrata biofilm in a catheter model, suggesting that coaggregation of the two species is possible [31]. Ours is the first study to examine the host-pathogen interactions of C. albicans and C. glabrata in a co-infection tissue model. We found that, contrary to our original hypothesis, C. glabrata did not synergize with C. albicans to potentiate the cell damage and the inflammatory response in our in vitro systems. These findings would suggest that C. glabrata is frequently co-isolated from oral lesions largely due to its remarkable resistance to antifungal medications [4], salivary innate antifungal molecules such as histatins [32] or oral epithelial cell derived anti-fungal β-defensins [33, 34], which allow it to overgrow in the oral mucosa and not due to its active role in the disease process. However, additional co-infection studies with clinical isolates from oroesophageal lesions are needed to reach this conclusion.
Strain differences were noted in the ability of C. glabrata to trigger a proinflammatory cytokine response. We have previously shown that adhesion of Candida to oral epithelial cells is an important step in cytokine induction [12]. The EPA family of cell wall proteins is responsible for Ca++-dependent adhesion of C. glabrata to mucosal epithelial cells [35]. C. glabrata adherence to oral epithelial cells is strain specific, with strain GDH2269 having the lowest capacity to adhere, among a number of strains and species examined in a prior study [19]. This may explain the strain variations in cytokine responses and the lack of a rigorous cytokine response to strain GDH2269 in our study.
The lesional C. glabrata isolate MRL2302 triggered a stronger proinflammatory cytokine response than all other C. glabrata strains tested in this study. In previous studies we showed that C. albicans hyphal transformation enhances the interaction of C. albicans with oral epithelial cells, by increasing the ability of the organism to adhere to, and trigger a potent proinflammatory response by these cells [8, 9, 12]. Upon microscopic examination strain MRL2302 appeared to have a greater propensity to form pseudohyphae when co-cultured with oral epithelial strains (not shown) than the other C. glabrata strains, which appeared almost exclusively as fully separated yeast cells. It has been shown that disruption of the virulence moderating gene ACE2 in C. glabrata, which encodes for a transcriptional activator needed for mother-daughter separation, results in defective cell separation and a strikingly pronounced proinflammatory cytokine response in a murine model of disseminated candidiasis. Upon infection with the ace2 mutant the mice quickly succumbed to severe septic shock indicated by a sharp rise in circulating inflammatory cytokines [36]. Further investigation is needed to conclusively address whether pseudohyphal formation due to incomplete cell separation during budding is one of the virulence attributes of certain clinical strains isolated from the oral mucosal lesions.
In light of our findings, we conclude that human oral epithelial cells respond to C. glabrata infection with a pronounced GM-CSF synthesis and weak or undetectable other synthesized proinflammatory cytokines in vitro. C. glabrata oroesophageal strains demonstrated variable tissue damaging and invasive potential, which was overall lower than that of C. albicans. The ability of C. glabrata to trigger a host inflammatory response and invade oral epithelium suggests the active participation of this species in orchestrating the host immune and inflammatory response in the oral mucosa and its contribution to pathogenesis.
4. Materials and methods
4.1 Organisms
C. glabrata oral strains GDH2269 and 94-11 were obtained by the American Type Culture Collection (ATCC, Rockville, MD). Two C. glabrata clinical isolates from esophageal candidiasis, MRL2302 and MRL7525, were kindly provided by Dr. M. Ghannoum (Case Western Reserve University). These isolates were confirmed by the germ tube test and the API 20C system (Biomerieux Vitek, Hazelwood, MO, USA). For comparison to C. glabrata as well as in co-infection studies, two C. albicans strains were used: C. albicans strain SC5314 (kindly provided by Dr. A. Mitchell, Columbia University), originally isolated from a patient with disseminated candidiasis [37], with the ability to trigger high levels of proinflammatory cytokines and cell damage [8, 9, 13]; and C. albicans oral strain 28366 (ATCC), which exhibits moderate ability to do so in oral mucosal models in vitro [10, 12]. All of the strains used in this study showed similar growth rates in the inoculation media used for infection (see below), as determined by direct cell counts of yeast cells grown in these media, or by the XTT assay when germinated organisms were tested. The organisms were routinely propagated in YPD or Sabouraud agar (Difco Laboratorie, Detroit, MI), at 25°C.
4.2 Cell cultures
Because epithelial cells are the major cell type encountered by Candida in the oral cavity, cytotoxicity and proinflammatory responses were characterized in these cells. Epithelial cells included primary oral keratinocytes, isolated as described previously from discarded gingival tissues [8, 9], as well as the oral keratinocyte line OKF6/TERT-2. This cell line represents normal oral mucosal epithelium (floor of the mouth) and was immortalized by overexpression of telomerase and deletion of the p16INK4a regulatory protein [38]. We [8, 9] and others [39] have shown that responses of this cell line to infection closely resemble primary oral keratinocyte responses. Keratinocytes were maintained in Keratinocyte Serum Free Media (KSFM, Invitrogen, Carlsbad, CA), supplemented with 0.4 mM CaCl2, 0.1 ng/ml EGF, 50 μg/ml bovine pituitary extract (Invitrogen, Carlsbad, CA) and antibiotics (Penicillin/Streptomycin, 100 U/ml and 100 μg/ml, respectively).
4.3 Coculture of Candida with epithelial cell monolayers
Stationary phase yeast were prepared by growth for 18 h at room temperature in YPD (Difco Laboratories, Detroit, MI), supplemented with 2% (wt/vol) glucose. The fungal cells were harvested by centrifugation and washed in PBS. Subsequently yeast were counted in a hemacytometer and adjusted to the final concentration in complete KSFM before adding to epithelial cells.
Oral keratinocytes were seeded at or near confluence in 6- or 12-well polystyrene plates (Corning Incorporated, Corning, NY) and were incubated overnight in complete KSFM media at 37°C in 5% CO2 atmosphere. Preliminary experiments based on cell counts of viable cells showed that the number of cells seeded was similar to viable cell number just prior to infection challenge. The following day the media were discarded and the cells were challenged with suspensions of stationary phase viable organisms at varying fungal cell to host cell ratios, for up to 48 h. In co-infection studies equal numbers of C. albicans and C. glabrata yeast cells were added per well. Negative controls for these experiments included uninfected cultures and Candida alone. At the end of the experimental period supernatants and RNA were collected and stored at −70°C until assayed.
4.4 Cytokine detection
4.4.1 Real-time RT-PCR
Total RNA was extracted using Tri-reagent (Molecular Research Center, Cincinnati, OH), following the manufacturer's instructions. Expression of IL-1α, IL-8 and GM-CSF transcripts by OKF6/TERT-2 monolayers was quantitatively assessed by real-time RT-PCR and data were analyzed using the iCycle iQ system software (BioRad, Hercules, CA). Briefly, 3 μg total RNA of each sample was reverse transcribed into cDNA using random primer oligonucleotides (Invitrogen, Carisbad, CA) and BD sprint™ PowerScript™ (BD Biosciences, Palo Alto, CA). Real-time PCR was performed on 96-well optical reaction plates (BioRad, Hercules, CA). All PCR reaction mixtures contained 7.5μl of iQ™ SYBR® Green Supermix (BioRad, Hercules, CA), 0.5μl of forward primer (10μM), 0.5μl of reverse primer (10μM), 4μl of each diluted RT product, and 2.5μl of distilled water per well. The sequence of primers used for real-time PCR were as follows: human IL-1α (forward), 5′-GTCTCTGAATCAGAAATCCTTCTATC-3′ and (reverse) 5′-CATGTCAAATTTCACTGCTTCATCC-3′; human IL-8 (forward), 5′-CAAGAGCCAGGAAGAAACCAC-3′ and (reverse) 5′-GGCATCTTCACTGATTCTTGG-3′; human GM-CSF (forward) 5′-ATGTGAATGCCATCCAGGAG-3′ and (reverse) 5′-AATCTGGGTTGCACAGGAAG-3′. PCR amplification of the housekeeping gene GAPDH was performed to control for sample loading and normalize samples. The specificity of each primer pair was confirmed by agarose gel electrophoresis. Water controls were included for specificity control. Negative controls also included RNA extracted from C. albicans or C. glabrata incubated under the same conditions, in the absence of oral epithelial cells. The PCR reaction was performed at 95°C for 3 minutes followed by 50 cycles of 30 s at 95 °C, 60 °C and 72 °C. The GAPDH-normalized data were expressed as fold induction of cytokine gene expression in infected compared to uninfected cultures. Using these procedures we were unable to amplify a GAPDH signal from wells containing C. albicans or C. glabrata alone.
4.4.2 ELISA
To confirm the findings with QRT-PCR and quantify IL-1α, IL-8 and GM-CSF protein levels, culture supernatants obtained from duplicate or triplicate experiments, were analyzed by ELISA. In each experiment supernatants from 2 replicate wells were pooled and assayed by duplicate sandwich enzyme-linked immunosorbent assays (ELISA) using commercially available antibody pairs (Endogen MiniKit, Pierce, Rockford, IL), as previously described [8-11]. Absorbance values and corresponding cytokine concentrations were determined with an Opsys MR Microplate reader (Dynex Technologies Inc., Chantilly, VA) using the Revelation QuickLink® software (Thermo Labsystems, Chantilly, VA). The sensitivity of these assays ranged between 8 and 16 pg/ml.
4.4.3 Cytokine protein array
Cytokines in culture supernatants were detected using an enzyme-linked immunosorbent assay (ELISA)-based cytokine protein array (Ray Bio cytokine array; RayBiotech, Norcross, GA). Briefly, after membrane blocking, 1 ml of supernatant was added and incubated for 2h, followed by addition of biotinylated detection antibodies at a dilution of 4:1,000. The membranes were developed by addition of horseradish peroxidase-conjugated streptavidin for 2h and subsequent addition of an enhanced chemiluminescence-type solution. The membranes were exposed to X-ray film (Kodak BioMax film; Kodak, Rochester, NY) for 1 min and processed by autoradiography. The intensity of signals was determined by densitometry analysis using the Chemilmager software (Alpha Innotech Corporation, San Leandro, CA)
4.5 Assessment of cell and tissue damage
4.5.1 Monolayer Cultures
Since Candida infection can cause extensive epithelial cell necrosis, manifesting as ulceration of the oral mucosa in vivo, we set out to compare the ability of different strains and species to damage oral epithelial cells in vitro, alone or in combination. The ability of Candida to injure oral epithelial cells was assessed by the CytoTox-96® assay (Promega, Madison, WI), which measures release of lactate dehydrogenase (LDH) from dying cells. In these experiments OKF6/TERT-2 cells were cocultured with increasing doses of each strain, or a combination of equal doses of one strain from each species, for 8h or 24h. The LDH released from this coculture system was quantified by spectrophotometry, as described previously [13]. Spontaneous release of LDH by uninfected cultures, or by Candida alone, incubated under identical conditions was included as a negative control in each experiment.
4.5.2 Three-dimensional Model of the Human Oral Mucosa
To further characterize the tissue damaging and invasive properties of these strains, we used a three-dimensional (3-D) model system of the oral mucosa, as described previously [14]. Briefly, this system is composed of 3T3 fibroblasts embedded in a biomatrix of collagen type I, overlaid by a multilayer of OKF6/TERT-2 cells. In order to develop the oral mucosal model, fibroblasts were suspended in pre-cooled gel medium DMEM (4° C; 2 X Dulbecco's Modified Eagle Medium), containing 100mM HEPES at a final concentration of 3 × 105 cells/ml. The cell suspension was mixed with an equal volume of acidic collagen solution (Collagen Type I from rat tails, Sigma, 4mg/ml in 0.1% acetic acid), and 200 μl of this solution was placed in the upper compartment of a Millicell Culture Plate Insert (Millicell-MA, 13 mm diameter, Millipore, Bedford, MA). The gel was allowed to solidify for 15 minutes at 37°C in 5% CO2. Each insert was transferred to a well of a 24-well plate containing 0.5 ml of DMEM-10% FBS (Mediatech, Herndon, VA). Two days later 200μl of KSFM (Invitrogen, Carlsbad, CA) containing 1 × 106 keratinocytes were added into each insert. The culture medium was changed every other day, using Keratinocyte-SFM inside the inserts and DMEM-10% FBS basolaterally. After 6 days under submerged conditions, the inserts were transferred into 6-well plates and the cells were exposed to air. Airlift media (Keratinocyte-SFM containing 5% FBS, 1.88 mM CaCl2 and 0.025mM glucose) were added basolaterally only. The cultures were grown for 14 additional days with daily change of media until a multilayer of 10-15 keratinocyte layers was formed.
To study infection in this system 100μl of airlift medium containing 1×106Candida yeast cells were added inside the inserts. In mixed infections 1×106 yeast cells from each species were added simultaneously. Two days later all cultures were fixed with 10% Formaldehyde-PBS and embedded in paraffin. Sections (5μm thick) were stained with Periodic Acid-Schiff (PAS) stain and were evaluated under a light microscope. To assess tissue damage, supernatants from these cultures were analyzed for LDH content as above.
4.6 Statistical analyses
The statistical significance of the differences in cytokine levels, and cytotoxicity between pairs of different Candida species was determined by two-tailed t-test, assuming equal variances. Differences were statistically significant at p < 0.05.
Acknowledgments
The authors would like to thank Dr. Aaron Mitchell and Dr. Mahmoud Ghannoum for providing some of the strains used in this study. This study was supported by NIH/NIDCR grant RO1 DE13986 to ADB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fidel PL, Jr, Vazquez JA, Sobel JD. Candida glabrata: review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin Microbiol Rev. 1999;12:80–96. doi: 10.1128/cmr.12.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vazquez JA. Options for the management of mucosal candidiasis in patients with AIDS and HIV infection. Pharmacotherapy. 1999;19:76–87. doi: 10.1592/phco.19.1.76.30509. [DOI] [PubMed] [Google Scholar]
- 3.Redding SW, Zellars RC, Kirkpatrick WR, McAtee RK, Caceres MA, Fothergill Epidemiology of oropharyngeal Candida colonization and infection in patients receiving radiation for head and neck cancer. J Clin Microbiol. 1999;37:3896–900. doi: 10.1128/jcm.37.12.3896-3900.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Redding SW. The role of yeasts other than Candida albicans in oropharyngeal candidiasis. Curr Opin Infect Dis. 2001;14:673–7. doi: 10.1097/00001432-200112000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Redding SW, Kirkpatrick WR, Coco BJ, Sadkowski L, Fothergill AW, Rinaldi MG, et al. Candida glabrata oropharyngeal candidiasis in patients receiving radiation treatment for head and neck cancer. J Clin Microbiol. 2002;40:1879–81. doi: 10.1128/JCM.40.5.1879-1881.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dongari-Bagtzoglou A, Fidel PL., Jr The host cytokine responses and protective immunity in oropharyngeal candidiasis. J Dent Res. 2005;84:966–77. doi: 10.1177/154405910508401101. [DOI] [PubMed] [Google Scholar]
- 7.Schaller M, Mailhammer R, Grassl G, Sander CA, Hube B, Korting HC. Infection of human oral epithelia with Candida species induces cytokine expression correlated to the degree of virulence. J Invest Dermatol. 2002;118:652–7. doi: 10.1046/j.1523-1747.2002.01699.x. [DOI] [PubMed] [Google Scholar]
- 8.Dongari-Bagtzoglou A, Kashleva H. Candida albicans triggers interleukin-8 secretion by oral epithelial cells. Microb Pathog. 2003;34:169–77. doi: 10.1016/s0882-4010(03)00004-4. [DOI] [PubMed] [Google Scholar]
- 9.Dongari-Bagtzoglou A, Kashleva H, Villar CC. Bioactive interleukin-1alpha is cytolytically released from Candida albicans-infected oral epithelial cells. Med Mycol. 2004;2:31–41. doi: 10.1080/1369378042000193194. [DOI] [PubMed] [Google Scholar]
- 10.Dongari-Bagtzoglou A, Villar CC, Kashleva H. Candida albicans-infected oral epithelial cells augment the anti-fungal activity of human neutrophils in vitro. Med Mycol. 2005;43:545–9. doi: 10.1080/13693780500064557. [DOI] [PubMed] [Google Scholar]
- 11.Dongari-Bagtzoglou A, Kashleva H. Granulocyte-macrophage colony-stimulating factor responses of oral epithelial cells to Candida albicans. Oral Microbiol Immunol. 2003;18:165–70. doi: 10.1034/j.1399-302x.2003.00061.x. [DOI] [PubMed] [Google Scholar]
- 12.Villar CC, Kashleva H, Dongari-Bagtzoglou A. Role of Candida albicans polymorphism in interactions with oral epithelial cells. Oral Microbiol Immunol. 2004;19:262–9. doi: 10.1111/j.1399-302X.2004.00150.x. [DOI] [PubMed] [Google Scholar]
- 13.Villar CC, Kashleva H, Mitchell AP, Dongari-Bagtzoglou A. Invasive phenotype of Candida albicans affects the host proinflammatory response to infection. Infect Immun. 2005;73:4588–95. doi: 10.1128/IAI.73.8.4588-4595.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dongari-Bagtzoglou A, Kashleva H. Development of a novel three-dimensional in vitro model of oral Candida infection. Microb Pathog. 2006;40:271–8. doi: 10.1016/j.micpath.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Lockhart SR, Joly S, Vargas K, Swails-Wenger J, Enger L, Soll DR. Natural defenses against Candida colonization breakdown in the oral cavities of the elderly. J Dent Res. 1999;78:857–68. doi: 10.1177/00220345990780040601. [DOI] [PubMed] [Google Scholar]
- 16.Bagg J, Sweeney MP, Lewis MA, Jackson MS, Coleman D, Al MA, et al. High prevalence of non-albicans yeasts and detection of anti-fungal resistance in the oral flora of patients with advanced cancer. Palliat Med. 2003;17:477–81. doi: 10.1191/0269216303pm793oa. [DOI] [PubMed] [Google Scholar]
- 17.Filler SG, Pfunder AS, Spellberg BJ, Spellberg JP, Edwards JE., Jr Candida albicans stimulates cytokine production and leukocyte adhesion molecule expression by endothelial cells. Infect Immun. 1996;64:2609–17. doi: 10.1128/iai.64.7.2609-2617.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Djeu JY, Matsushima K, Oppenheim JJ, Shiotsuki K, Blanchard DK. Functional activation of human neutrophils by recombinant monocyte-derived neutrophil chemotactic factor/IL-8. J Immunol. 1990;144:2205–10. [PubMed] [Google Scholar]
- 19.Nicolatou-Galitis O, Dardoufas K, Markoulatos P, Sotiropoulou-Lontou A, Kyprianou K, Kolitsi G, Pissakas, et al. Oral pseudomembranous candidiasis, herpes simplex virus-1 infection, and oral mucositis in head and neck cancer patients receiving radiotherapy and granulocyte-macrophage colony-stimulating factor [GM-CSF] mouthwash. J Oral Pathol Med. 2001;30:471–80. doi: 10.1034/j.1600-0714.2001.030008471.x. [DOI] [PubMed] [Google Scholar]
- 20.Vazquez JA, Gupta S, Villanueva A. Potential utility of recombinant human GM-CSF as adjunctive treatment of refractory oropharyngeal candidiasis in AIDS patients. Eur J Clin Microbiol Infect Dis. 1998;17:781–3. doi: 10.1007/s100960050185. [DOI] [PubMed] [Google Scholar]
- 21.Kupper TS. The activated keratinocyte: a model for inducible cytokine production by non-bone marrow-derived cells in cutaneous inflammatory and immune responses. J Invest Dermatol. 1990;94:146S–150S. doi: 10.1111/1523-1747.ep12876130. [DOI] [PubMed] [Google Scholar]
- 22.Wang M, Friedman H, Djeu JY. Enhancement of human monocyte function against Candida albicans by the colony-stimulating factors [CSF]: IL-3, granulocyte-macrophage-CSF, and macrophage-CSF. J Immunol. 1989;14:671–7. [PubMed] [Google Scholar]
- 23.Noverr MC, Phare SM, Toews GB, Coffey MJ, Huffnagle GB. Pathogenic yeasts Cryptococcus neoformans and Candida albicans produce immunomodulatory prostaglandins. Infect Immun. 2001;69:2957–63. doi: 10.1128/IAI.69.5.2957-2963.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castro M, Ralston NV, Morgenthaler TI, Rohrbach MS, Limper AH. Candida albicans stimulates arachidonic acid liberation from alveolar macrophages through alphamannan and beta-glucan cell wall components. Infect Immun. 1994;62:3138–45. doi: 10.1128/iai.62.8.3138-3145.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clarke DL, Belvisi MG, Catley MC, Yacoub MH, Newton R, Giembycz MA. Identification in human airways smooth muscle cells of the prostanoid receptor and signalling pathway through which PGE2 inhibits the release of GM-CSF. Br J Pharmacol. 2004;141:1141–50. doi: 10.1038/sj.bjp.0705716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gewirtz AT, Collier-Hyams LS, Young AN, Kucharzik T, Guilford WJ, Parkinson JF, et al. Lipoxin a4 analogs attenuate induction of intestinal epithelial proinflammatory gene expression and reduce the severity of dextran sodium sulfate-induced colitis. J Immunol. 2002;168:5260–7. doi: 10.4049/jimmunol.168.10.5260. [DOI] [PubMed] [Google Scholar]
- 27.Samaranayake LP, Raeside JM, MacFarlane TW. Factors affecting the phospholipase activity of Candida species in vitro. Sabouraudia. 1984;22:201–7. [PubMed] [Google Scholar]
- 28.Collin B, Clancy CJ, Nguyen MH. Antifungal resistance in non- albicans Candida species. Drug Resist Updat. 1999;2:9–14. doi: 10.1054/drup.1998.0059. [DOI] [PubMed] [Google Scholar]
- 29.Kantarcioglu AS, Yucel A. Phospholipase and protease activities in clinical Candida isolates with reference to the sources of strains. Mycoses. 2002;45:160–5. doi: 10.1046/j.1439-0507.2002.00727.x. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto T, Nohara K, Uchida K, Yamaguchi H. Purification and characterization of secretory proteinase of Candida albicans. Microbiol Immunol. 1992;36:637–41. doi: 10.1111/j.1348-0421.1992.tb02064.x. [DOI] [PubMed] [Google Scholar]
- 31.El-Azizi MA, Starks SE, Khardori N. Interactions of Candida albicans with other Candida spp. and bacteria in the biofilms. J Appl Microbiol. 2004;96:1067–73. doi: 10.1111/j.1365-2672.2004.02213.x. [DOI] [PubMed] [Google Scholar]
- 32.Helmerhorst EJ, Venuleo C, Beri A, Oppenheim FG. Candida glabrata is unusual with respect to its resistance to cationic antifungal proteins. Yeast. 2005;22:705–14. doi: 10.1002/yea.1241. [DOI] [PubMed] [Google Scholar]
- 33.Feng Z, Jiang B, Chandra J, Ghannoum M, Nelson S, Weinberg A. Human beta-defensins: differential activity against Candidal species and regulation by Candida albicans. J Dent Res. 2005;84:445–50. doi: 10.1177/154405910508400509. [DOI] [PubMed] [Google Scholar]
- 34.Joly S, Maze C, McCray PB, Jr, Guthmiller JM. Human beta-defensins 2 and 3 demonstrate strain-selective activity against oral microorganisms. J Clin Microbiol. 2004;42:1024–9. doi: 10.1128/JCM.42.3.1024-1029.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cormack BP, Ghori N, Falkow S. An adhesin of the yeast pathogen Candida glabrata mediating adherence to human epithelial cells. Science. 1999;23:578–82. doi: 10.1126/science.285.5427.578. [DOI] [PubMed] [Google Scholar]
- 36.Kamran M, Calcagno AM, Findon H, Bignell E, Jones MD, Warn P, et al. Inactivation of transcription factor gene ACE2 in the fungal pathogen Candida glabrata results in hypervirulence. Eukaryot Cell. 2004;2:546–52. doi: 10.1128/EC.3.2.546-552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gillum AM, Tsay EY, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–82. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 38.Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA et al. Human keratinocytes that express hTERT and also bypass a p16 INK4a—enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. 2000;20:1436–47. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feucht EC, DeSanti CL, Weinberg A. Selective induction of human beta-defensin mRNAs by Actinobacillus actinomycetemcomitans in primary and immortalized oral epithelial cells. Oral Microbiol Immunol. 2003;18:359–63. doi: 10.1046/j.0902-0055.2002.00097.x. [DOI] [PubMed] [Google Scholar]