Abstract
Syringolides are water-soluble, low-molecular-weight elicitors that trigger defense responses in soybean cultivars carrying the Rpg4 disease-resistance gene but not in rpg4 cultivars. 125I-syringolide 1 previously was shown to bind to a soluble protein(s) in extracts from soybean leaves. A 34-kDa protein that accounted for 125I-syringolide 1 binding activity was isolated with a syringolide affinity-gel column. Partial sequences of internal peptides of the 34-kDa protein were identical to P34, a previously described soybean seed allergen. In soybean seeds, P34 is processed from a 46-kDa precursor protein and was shown to have homology with thiol proteases. P34 is a moderately abundant protein in soybean seeds and cotyledons but its level in leaves is low. cDNAs encoding 46-, 34-, and 32-kDa forms of the soybean protein were cloned into the baculovirus vector, pVL1392, and expressed in insect cells. The resulting 32- and 34-kDa proteins, but not the 46-kDa protein, exhibited ligand-specific 125I-syringolide binding activity. These results suggest that P34 may be the receptor that mediates syringolide signaling.
Plant pathogens carrying particular avirulence (avr) genes are recognized by plants expressing matching disease-resistance genes, thereby invoking active plant defense responses (1–3). The matching genes in plant and pathogen have been collectively called “gene-for-gene” systems. The defense responses that they mediate have been termed the “hypersensitive response,” because of the occurrence of variable degrees of plant cell necrosis at the site of pathogen invasion. The hypersensitive response generally involves recognition of a pathogen as foreign followed by the production of chemical agents and/or structural re-enforcements to arrest pathogen spread. Although a general understanding is emerging as to how this defense system functions, recognition events occurring soon after pathogen infection are not well understood (4–6).
The elicitor-receptor hypothesis appears to explain plant recognition of pathogens in many gene-for-gene systems (1, 7–10). Elicitors are defined as signal molecules produced directly or indirectly by pathogens that are recognized by plants and thereby activate defense responses. Two types of elicitors generally are recognized: nonspecific elicitors, which do not exhibit differences in cultivar responses within a plant species, and specific elicitors, which cause specific responses only in cultivars carrying matching disease-resistance genes. Nonspecific elicitors include substances associated with basic pathogen metabolism, such as cell wall glucans, chitin oligomers, and glycopeptides. Specific elicitors are signal molecules, including proteins and peptides encoded by some avirulence genes, as well as small nonprotein compounds produced by pathogens under the direction of avirulence-gene encoded enzymes. There is substantial evidence for the occurrence of plant receptors to both type of elicitors (10–14), but they have not been well characterized.
Syringolides are unusual acyl glycosides produced in Escherichia coli or several other Gram-negative bacteria expressing avirulence gene D (avrD). avrD originally was cloned from Pseudomonas syringae pv. tomato (15, 16) and occurs in several P. syringae isolates. The purified syringolides or bacteria expressing avrD elicit the hypersensitive response specifically in soybean cultivars carrying the Rpg4 disease resistance gene (15). The syringolide elicitors are at least 1,000 times more active in Rpg4 than in rpg4 cultivars. It is possible that this specificity is determined at the level of a primary syringolide binding site (receptor). Based on this presumption, we radio-labeled syringolide 1 with 125I and synthesized several syringolide derivatives with altered biological activities (17, 18). We demonstrated that the soluble fractions of extracts from both Rpg4 and rpg4 soybean leaves contain a ligand-specific, saturable binding site(s) for the syringolide elicitor (18). The observed syringolide binding site(s) showed high ligand selectivity and a strong correlation was observed between competitive binding activity and elicitor activity for all compounds tested. Therefore, the identified binding site(s) is a candidate for the physiologically active syringolide receptor. In this study we report the isolation and characterization of a 34-kDa soybean protein that binds the syringolide elicitors.
MATERIALS AND METHODS
Biological Materials.
Soybean (Glycine max L.) cvs. Harosoy (Rpg4/Rpg4) and Merit (rpg4/rpg4) were grown in soil overlaid with vermiculite in a growth chamber at 22°C on a 14-hr photoperiod. Tomato (Lycopersicon esculentum) cv. Rio Grande 76 (S/R), Arabidopsis thaliana (ecotype Columbia), and tobacco (Nicotiana tabacum) plants were grown under the same conditions.
Syringolide Affinity Gel Chromatography.
Syringolide 1, 4′-succinyl syringolide 1, 3-O-methyl-syringolide 1, and 125I-syringolide 1 were purified and synthesized as previously described (17, 18). Proteinase K, pepstatin A, leupeptin, Norit-A charcoal (Sigma), dextran T70 (Pharmacia), and Affi-Gel 102 (Bio-Rad) were purchased. The following steps were followed for preparation of the Affi-gel 102 affinity matrix. Activation of the carboxyl group on 4′-succinyl syringolide 1 and displacement by the nucleophile (R-NH2), releasing EDAC [(1-ethyl-3-(3-dimethyl-aminopropyl) carbodiimide HCl] as a soluble urea derivative were carried out as described by the manufacturer. The activated carboxyl group on 4′-succinyl syringolide 1 was coupled to the amino group on the Affi-Gel 102 matrix. The Affi-Gel 102 gel (6 ml) was transferred to a sintered glass filter (30 μm), washed with 6 ml of dimethyl sulfoxide (DMSO) three times, and dried under vacuum. The gel then was suspended in 6 ml of DMSO and transferred to a 30-ml flask. To the gel suspension, 30 mg of 4′-succinyl syringolide 1 was added. The pH was adjusted immediately to 5–6 with 1 M HCl. EDAC coupling reagent (5 mg) was added to the gel suspension, and the pH was again adjusted immediately to 5–6 with 1 M HCl. The crosslinking reaction proceeded for 5 hr at room temperature with gentle stirring. The resulting syringolide affinity gel was packed into a 10-ml glass econo-column (Bio-Rad), washed with 15 ml of DMSO, and kept at 4°C until use.
Preparation of Protein Extracts.
Soybean leaves of 2-week-old plants were harvested and homogenized at 4°C in extraction buffer [30 mM sodium phosphate, pH 7.2, containing 4 mM DTT, 0.04% (vol/vol) β-mercaptoethanol, 5% glycerol, 1 mM phenylmethylsulfonyl fluroide, 2 μg/ml of pepstatin A, and 2 μg/ml of leupeptin] in a blender. The homogenate was filtered through four layers of cheese cloth, and the filtrate was centrifuged at 100,000 × g for 1.5 hr at 4°C. The resulting supernatant was dialyzed against the extraction buffer at 4°C for 12 hr, concentrated on a Centricon-10 device (Amicon) to 10 mg/ml, and stored at −80°C until use. The soluble protein extracts from imbibed soybean seeds and cotyledons were prepared according to the methods described previously (19). Protein content was determined with the Protein Assay Kit (Bio-Rad) using bovine gamma globulin as a standard.
Purification of the Syringolide Binding Protein(s).
The syringolide affinity-gel column was equilibrated with sodium phosphate buffer (30 mM, pH 7.2, 50 mM KCl, 4 mM DTT, 0.04% β-mercaptoethanol), and the protein sample (10 mg in 1 ml) was loaded. The column was washed with 4 bed volumes of the phosphate buffer. A salt gradient of 0 to 1 M NaCl in sodium phosphate buffer (30 mM, pH 7.2) was applied. Fractions of 1.5 ml were collected, and the protein content of each fraction was measured with a spectrophotometer at 280 nm. Each fraction was dialyzed against sodium phosphate buffer (30 mM, pH 7.2, 4 mM DTT) at 4°C for 6 hr.
The binding activity of 125I-syringolide 1 in each fraction was examined according to methods described previously (18). The protein/125I-syringolide 1 complex also was examined by 10% (wt/vol) PAGE under native conditions (18, 20).
A gel filtration column (1.2 × 60 cm, polyacrylamide P-30, Bio-Gel) was used for purification of proteins from fractions after the affinity-gel column. This filtration column was equilibrated with a sodium phosphate buffer (pH 7.0, 30 mM, 0.1 M NaCl) and run with 150 ml of the equilibrium buffer at a flow rate of 0.15 ml/min. The protein content in each fraction (30 drops/each) was monitored as described above.
Microsequencing of Internal Tryptic Peptides.
Proteins in the active fractions eluted from the syringolide affinity gel column were concentrated with a Centricon-10 device (Amicon) and separated by 12% SDS/PAGE. The proteins then were immobilized on a ProBlot membrane (Applied Biosystems) according to the manufacturer’s instructions. After Coomassie blue R-250 staining, the protein bands were excised and sent to the Harvard Microchemistry Facility (Harvard University, Cambridge, MA) for amino acid composition and sequence analyses. For sequence analysis, proteins were subjected to in situ digestion with trypsin, and the resulting peptides were separated by reverse-phase HPLC. The peptides from well-resolved peak fractions were collected and sequenced.
Subcloning and Expression of the Cloned P34 Gene.
The P34 cDNA of 1,350 bp was cloned into pBluescript SK(−) (Stratagene) (21, 22). The full coding sequence of the 46-kDa precursor protein and two partial coding sequences corresponding to the 34- and 32-kDa processed protein products were amplified, respectively, by PCR with the following primers. M1: 5′-TT TTT TTT CAT ATG GGT TTC CTT GTG TTG C-3′; M2: 5′-CAG CAA ATC CAT ATG GCC AAC AAG AAA ATG-3′; M3: 5′-GG AAA AAA GGT CAT ATG ACC CAA GTA AAG T-3′. M1 introduced a NdeI site at the first start codon without altering the nucleotide sequence of the coding region. M2 and M3 introduced a NdeI site at bp 359 and bp 437, respectively, without altering the nucleotide sequence of the remaining coding region. The M13/pUC sequencing primer (5′-GTAAAACGACGGCCAGT-3′), which is located on the cloning vector was used to facilitate amplification of the complete 3′ end of the gene. The PCR products of M1, M2, and M3 were designated 46M1, 34M2, and 32M3. Each of them was digested with NdeI and EcoRI and cloned into the corresponding sites of MTL22 (23). Insert fragments then were released with NdeI and XhoI and recloned into the corresponding sites of pET19b (Novagen). Each of the constructs was digested with XbaI–SmaI and the resulting fragments containing His-tag fusion sequences finally were cloned into the corresponding sites of the baculovirus transfer vector, pVL1392 (Invitrogen). All the above constructs were confirmed by sequencing. Constructs were expressed in insect cells and purified by nickel affinity gel chromatography following the manufacturer’s instructions (Novagen).
Antibody Production and Western Blotting.
MTL22-P34M3 was digested with BamHI and the resulting 0.6-kb fragment, designated M25, comprising the carboxy terminal amino acids of P34 was cloned in-frame as a carboxyl fusion to the lacZ gene of pUR292 (24). This plasmid, pUR292-M25, was expressed in E. coli strain RB791, and the fusion protein was purified on thio-galactoside columns (Sigma). The fusion protein was used to prepare rabbit polyclonal antiserum against P34. Antibodies were isolated from sera by using an ImmunoPure IgG Purification Kit (Pierce) following the manufacturer’s instructions. Western blotting was performed according to the manufacturer’s (Bio-Rad) instructions.
RESULTS
Identification of a Syringolide Binding Protein.
Crude, soluble extracts from soybean leaves were passed through a syringolide affinity column, and, after extensive washing, eluted with a gradient of 0–1 M NaCl. Several fractions (12 through 18) contained specific syringolide binding activity. Fig. 1A demonstrates the specific binding activity of proteins in each fraction to 125I-syringolide 1. Maximal binding activity was observed in the 15th fraction. The binding activity in this fraction was 1.97 fmol/μg, which was about 50 times greater than observed in the soluble crude extract (0.04 fmol/μg). The binding activity in each fraction was further examined with the native gel binding assay developed previously (Fig. 1B) (18). Syringolide binding with crude extracts occurred at a position higher in the native gel (Fig. 1B, lane 1) than with the affinity-purified fractions (Fig. 1B, lanes 2–10). Proteins corresponding to syringolide binding in the native gel were eluted and examined by SDS/PAGE. Six proteins, including a 34-kDa protein, were observed (data not shown). This finding suggests that the syringolide binding protein(s) may be complexed with other components in the crude extracts.
Figure 1.

Isolation of syringolide binding protein(s) by affinity chromatography. (A) Specific binding activity of 125I-syringolide 1 to soybean leaf soluble proteins recovered from a syringolide affinity column. (B) Detection of syringolide binding proteins using the native gel binding assay as described in Materials and Methods, with specific bound radioactivity shown by the arrows. CR denotes crude extracts. Unbound 125I-syringolide 1 occurs at the bottom of the gel.
The active fractions from the affinity column contained three major proteins, as detected by SDS/PAGE (Fig. 2). These proteins were ca. 34, 31, and 28 kDa, respectively. After further purification by gel filtration, the isolated 31-kDa protein had weak binding activity and the 28-kDa protein had no detectable syringolide binding activity (Fig. 3B). However, the 34-kDa protein bound the radioactive probe with a specific binding activity of 1.84 fmol/μg and accordingly was concluded to be the major syringolide binding protein in the affinity fractions.
Figure 2.

12% SDS acrylamide gel of proteins in fractions from a syringolide affinity column. Lane 1, protein size markers; lane 2, crude soybean leaf extract; lane 3, fraction 6; lane 4, fraction 12; lane 5, fraction 15; lane 6, fraction 16; lane 7, fraction 18. Arrows denote 34-, 31-, and 28-kDa proteins eluted.
Figure 3.

Purification of syringolide binding protein. (A) Gel filtration purified 28-, 31-, and 34-kDa proteins were electrophoresed on a 12% SDS acrylamide gel. Lane 1, fraction 15 from the affinity column run in Fig. 1; lane 2, 28-kDa protein; lane 3, 31-kDa protein; lane 4, 34-kDa protein. (B) Syringolide binding activity of purified 28-, 31-, and 34-kDa proteins. Lanes are as in A.
The three proteins were eluted from SDS gels and submitted for tryptic digestion and peptide sequencing (Table 1). The N termini of internal peptides of the 31- and 28-kDa proteins had 100% homology to vegetative storage proteins previously isolated from soybean leaves (25). The amino acid sequences of two internal fragments of the 34-kDa protein disclosed 100% agreement with the published sequence of the P34 protein isolated from soybean seeds (21, 22).
Table 1.
Partial amino acid sequences of 28-, 31-, and 34-kDa proteins
| Proteins
|
Amino acid sequences
|
|---|---|
| 28 kDa | LAVEAHNIFGFETIPEECVEATK |
| 31 kDa | ARTPEVKCASWRLAV |
| 34 kDa | 1) ASWDWRKKGVIT |
| 2) LVSLSEQELV |
Presence and Processing of the P34 Protein in Soybean Plants.
The P34 protein is a highly processed protein of 379 amino acids and has homology to thiol proteases (21). The 34-kDa protein is produced from a 46-kDa precursor protein after seed germination. To quantify P34 in soybean leaves, we produced polyclonal antiP34 antibodies using an internal P34 fragment fused to lacZ (see Materials and Methods). This antiserum (designated β-gal/M25) is active at a 1:9,000 dilution in Western blots of P34 from soluble protein extracts of soybean cotyledons (data not shown). The 34-kDa protein was present in leaf soluble extracts from both Harosoy (Rpg4/Rpg4) and Merit (rpg4/rpg4) cultivars (Fig. 4). However, a few additional bands that reacted with β-gal/M25 were observed in the extracts from seeds and cotyledons. These bands had apparent molecular masses of 67, 46, 34, and 32 kDa. As shown in Fig. 4, levels of P34 were low in leaves and higher in seeds and cotyledons.
Figure 4.

Detection of P34 in soluble protein fractions of soybean seeds, cotyledons, and leaves. Equal amounts of protein (20 μg) were loaded in each lane.
Baculovirus Expression of P34 and Syringolide Binding Activity.
The PCR products of 46M1, 34M2, and 32M3 were cloned into the baculovirus vector pVL1392 as described in Materials and Methods. The resulting constructs were designated pVL1392–46M1, pVL1392–34M2, and pVL1392–32M3. They were expressed in insect cells and purified by Ni affinity gel chromatography (Fig. 5). When pVL1392–46M1 encoding the full-length 46-kDa protein was expressed in insect cells, a Western blot of the cell fraction showed a protein at 46 kDa as well as several lower weight bands (data not shown). This finding suggests that processing or random proteolysis might occur in insect cells, but a protein size of 34 kDa was not detected. 34M2 and 32M3 led to 34- and 32-kDa proteins of the expected size in Western blots and after purification (Fig. 5). The 125I-syringolide binding activity in insect cells expressing pVL1392 was negligible, but significant binding activities were observed in cells expressing all three constructs (Table 2). The purified 46-, 34-, and 32-kDa proteins all were active in the native gel 125I-syringolide binding assay (Fig. 6). However, the 46-kDa protein had reduced specific syringolide binding (Fig. 6B) in the presence of excess unlabeled ligand. The specific binding activity detected in insect cells infected with pVL1392–46M1 probably was contributed by proteolytic fragments of the 46-kDa protein. The purified 34- and 32-kDa proteins both exhibited ligand-specific syringolide binding activity. This finding suggests that neither deletion of the 10 terminal amino acids of the 34-kDa protein nor the polyhistidine N-terminal extension significantly affects specific syringolide binding. In addition, syringolide binding to cotyledon and imbibed seed protein extracts was higher than that of leaves (Table 2). Interference was observed in specific syringolide binding to the protein extracts of cotyledons and imbibed seeds because of high lipid content (data not shown).
Figure 5.
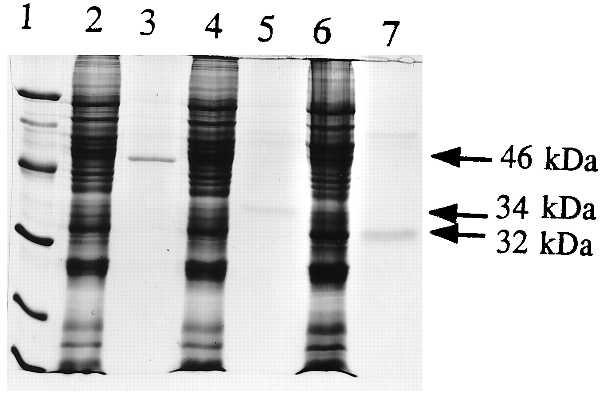
Expression of His-tagged 46-, 34-, and 32-kDa soybean proteins in baculovirus expression vector pVL1392. Lane 1, protein size markers; lane 2, protein extract from cells expressing pVL1392–46M1; lane 3, Ni-purified 46-kDa protein; lane 4, protein extract from cells expressing pVL1392–34M2; lane 5, Ni-purified 34-kDa protein; lane 6, protein extract from cells expressing pVL1392–32M3; lane 7, Ni-purified 32-kDa protein.
Table 2.
Comparison of specific 125I-syringolide binding activities of soluble protein extracts from soybean leaves, cotyledons, and seeds and from insect cells expressing 46-, 34-, and 32-kDa cDNA fragments (46M1, 34M2, and 32M3, respectively)
| Sample | Specific binding activity, fmol/mg |
|---|---|
| Leaves | 37 ± 6.1 |
| Cotyledons | 72 ± 8.9 |
| Seeds | 69 ± 11.3 |
| Insect cells expressing pVL1392 | 1.9 ± 1.7 |
| pVL1392-46M1 | 49 ± 15.5 |
| pVL1392-34M2 | 78 ± 12.9 |
| pVL1392-32M3 | 73 ± 13.6 |
Figure 6.

125I-syringolide 1 binding activity of soybean protein produced as His-tagged fusions (Fig. 5). (A) Binding activity in the native gel assay. Four micrograms of protein was loaded in each lane. (B) Quantitation of A of the protein–125I-syringolide complexes by scintillation counting. −, absence of competitive cold syringolide 1; +, presence of 1,500-fold cold syringolide 1.
DISCUSSION
We have demonstrated that the previously reported soybean allergen P34 (19, 26) is a specific syringolide binding protein. Although its cellular localization in leaves has not been investigated, presence of the P34 protein in the soluble fraction is not surprising because of the amphipathic nature of the syringolides that allows their diffusion across cell membranes. The 34-kDa protein is a posttranslationally processed protein of 379 amino acids. The 122 N-terminal amino acids are deleted from the preprotein via a single cleavage at the carboxyl side of an Asn residue, presumably mediated by a thiol protease present in developing seeds (22). In addition, P34 can be partially processed further to a 32-kDa protein after seed germination (Fig. 4; refs. 19, 21, and 22). Interestingly, the level of P34 is high in soybean seeds and cotyledons but very low in soybean leaves. This finding mirrors the pattern with legume lectins, which are produced in large amounts in seeds, apparently as storage proteins, but in very small amounts in vegetative tissues. Such lectins have been shown to be important determinants of recognitional specificity of Rhizobium strains by plant roots (27). However, P34 may differ from lectins in terms of posttranslational processing. Why the P34 protein undergoes processing remains to be determined. It is likely that the P34 protein has multiple functions such as involvement in senescence (22) as well as in signal perception (our studies), and its differential processing may influence its compartmentalization and functionality. For example, the 122 terminal amino acids on the 46-kDa preprotein might play a role in protein folding and therefore affect syringolide binding specificity and/or its subsequent function. One intriguing finding, however, is that both the 34- and 32-kDa proteins possess syringolide binding activity. The 10 amino acids at the N terminus of the 34-kDa protein include several basic residues that are not required for binding activity.
The 34-kDa protein is present in both cultivar Harosoy, carrying the Rpg4 gene, and cultivar Merit, lacking the Rpg4 gene (Fig. 4). P34 analogs were detected in soybean roots, stems, suspension cells, and tomato leaves but not in Arabidopsis or tobacco leaves as determined by Western blotting of soluble extracts (data not shown). P34 does not contain leucine-rich repeats that are present in many cloned resistance genes (6). The resistance gene, Pto, is a protein kinase and appears to interact with elicitor directly but requires a LRR protein, Prf, for function (12, 13). These observations suggests that initial elicitor perception may not necessarily be by a resistance gene protein(s). Similar observations were reported with the avr9 peptide elicitor in tomato (11). These findings together are of considerable significance because they indicate that there is more complexity in elicitor perception than previously appreciated. Dangl (28) noted similarities between active defense in higher plants and immune systems in higher vertebrates, particularly in the major histocompatiblity complex (MHC). One especially interesting feature of the MHC is antigen presentation to T cells (29, 30). Although it is premature to speculate about functional analogies between MHC presentation of antigens and elicitor recognition by plants expressing particular disease resistance genes, it is nonetheless likely that elicitor presentation and/or display are important in plant defense.
The processing requirement for formation of the 34-kDa protein by a thiol protease and the putative thiol protease activity of P34 itself are interesting because of the precedents for involvement of proteases in processing of signals after ligand binding, cleavage of transcription factors and prohormones, and in apoptosis (31–33). It will be of interest, for example, to see whether the putative thiol protease activity of P34 is important for syringolide binding and induction of the hypersensitive response, and to know what is the native substrate of P34.
It is also noteworthy that there were three proteins isolated from the syringolide affinity gel column (Fig. 2). The 28- and 31-kDa proteins are abundant in vegetative tissues and they tend to associate with other proteins. Although the 31- and 28-kDa proteins did not have significant syringolide binding affinity, this does not necessarily rule out their roles in syringolide-mediated signaling in vivo. For example, it is possible that they might interact with P34 in the presence of syringolides. The vegetative storage proteins were reported to have phosphatase activity (34), and phosphatase activity has been shown to be involved in elicitor-mediated signal transduction (35, 36). It would be interesting to find out whether the 31- and 28-kDa proteins associate with P34 and whether other components are present in the native binding complex (Fig. 1B).
Acknowledgments
We thank William Lane (Harvard Microchemistry Facility, Harvard University, Cambridge, MA) for protein sequencing, and Barbara Walter and Jolinda Traugh (Department of Biochemistry, University of California, Riverside, CA) for helping set up the baculovirus expression system. C.J. is a postdoctoral fellow in N.T.K.’s laboratory. This work was supported by U.S. Department of Agriculture Grant No. 9501093 and by Grant in Aid No. 08760038 for Scientific Research from the Ministry of Education, Science, and Culture of Japan.
References
- 1.Keen N T. Annu Rev Genet. 1990;24:447–463. doi: 10.1146/annurev.ge.24.120190.002311. [DOI] [PubMed] [Google Scholar]
- 2.Staskawicz B J, Ausubel F M, Baker B J, Ellis J G, Jones J D G. Science. 1995;268:661–667. doi: 10.1126/science.7732374. [DOI] [PubMed] [Google Scholar]
- 3.Bent A F. Plant Cell. 1996;8:1757–1771. doi: 10.1105/tpc.8.10.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamb C J. Cell. 1994;76:419–422. doi: 10.1016/0092-8674(94)90106-6. [DOI] [PubMed] [Google Scholar]
- 5.Dangl J L. Cell. 1995;80:363–366. doi: 10.1016/0092-8674(95)90485-9. [DOI] [PubMed] [Google Scholar]
- 6.Baker B, Zambryski P, Staskawicz B, Dineshkumar S P. Science. 1997;276:726–733. doi: 10.1126/science.276.5313.726. [DOI] [PubMed] [Google Scholar]
- 7.DeWit P J G M. Annu Rev Phytopathol. 1992;30:391–418. doi: 10.1146/annurev.py.30.090192.002135. [DOI] [PubMed] [Google Scholar]
- 8.Boller T. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:189–214. [Google Scholar]
- 9.Leach J G, White F F. Annu Rev Phytopathol. 1996;34:153–179. doi: 10.1146/annurev.phyto.34.1.153. [DOI] [PubMed] [Google Scholar]
- 10.Hahn M G. Annu Rev Phytopathol. 1996;34:387–412. doi: 10.1146/annurev.phyto.34.1.387. [DOI] [PubMed] [Google Scholar]
- 11.Kooman-Gersmann M, Honee G, Bonnema G, DeWit P J G M. Plant Cell. 1996;8:929–938. doi: 10.1105/tpc.8.5.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scofield S R, Tobias C M, Rathjen J P, Chang J H, Lavelle D T, Michelmore R W, Staskawicz B J. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- 13.Tang X, Frederick R D, Zhou J, Halterman D A, Jia Y, Martin G B. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- 14.Umemoto N, Kakitani M, Isamatsu A, Yoshikawa M, Yamaoka N, Ishida I. Proc Natl Acad Sci, USA. 1997;94:1029–1034. doi: 10.1073/pnas.94.3.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keen N T, Buzzell R I. Theor Appl Genet. 1991;81:133–138. doi: 10.1007/BF00226123. [DOI] [PubMed] [Google Scholar]
- 16.Midland S L, Keen N T, Sims J J, Midland M M, Stayton M M, Burton V, Smith M J, Mazzola E P, Graham K J, Clardy J. J Org Chem. 1993;58:2940–2945. [Google Scholar]
- 17.Tsurushima T, Midland S L, Zeng C M, Ji C, Sims J J, Keen N T. Phytochemistry. 1996;43:1219–1225. [Google Scholar]
- 18.Ji C, Okinaka Y, Takeuchi Y, Yamaoka N, Tsurushima T, Buzzell R I, Sims J J, Midland S L, Slaymaker D, Yoshikawa N, Keen N T. Plant Cell. 1997;9:1425–1433. doi: 10.1105/tpc.9.8.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herman E M, Melroy D L, Buckout R W. Plant Physiol. 1990;94:341–349. doi: 10.1104/pp.94.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji C, Kuc J. Mol Plant–Microbe Interact. 1995;8:899–905. doi: 10.1094/mpmi-8-0899. [DOI] [PubMed] [Google Scholar]
- 21.Kalinski A, Weisemann J M, Mathews B F, Herman E M. J Biol Chem. 1990;265:13843–13848. [PubMed] [Google Scholar]
- 22.Kalinski A, Melroy D L, Dwived R S, Herman E M. J Biol Chem. 1992;267:12068–12076. [PubMed] [Google Scholar]
- 23.Chambers S P, Prior S E, Barstow D A, Minton N P. Gene. 1988;68:139–149. doi: 10.1016/0378-1119(88)90606-3. [DOI] [PubMed] [Google Scholar]
- 24.Ruther U, Muller-Hill B. EMBO J. 1983;2:1791–1794. doi: 10.1002/j.1460-2075.1983.tb01659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staswick P. Plant Physiol. 1988;87:250–254. doi: 10.1104/pp.87.1.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hook V T H, Schiller M R, Azargan A V. Arch Biochem Biophys. 1996;328:107–114. doi: 10.1006/abbi.1996.0149. [DOI] [PubMed] [Google Scholar]
- 27.Spaink H P. Annu Rev Phytopathol. 1995;33:345–368. doi: 10.1146/annurev.py.33.090195.002021. [DOI] [PubMed] [Google Scholar]
- 28.Dangl J L. Plant J. 1992;2:3–11. [Google Scholar]
- 29.Brenner M, Porcelli S. Science. 1997;277:332. doi: 10.1126/science.277.5324.332. [DOI] [PubMed] [Google Scholar]
- 30.Maffei A, Papadopoulos K, Harris P E. Hum Immunol. 1997;54:91–103. doi: 10.1016/s0198-8859(97)00084-0. [DOI] [PubMed] [Google Scholar]
- 31.Hollenberg M D. Trends Pharmacol Sci. 1996;17:3–6. doi: 10.1016/0165-6147(96)81562-8. [DOI] [PubMed] [Google Scholar]
- 32.Rawlings N D, Barnett A J. Methods Enzymol. 1994;224:461–486. doi: 10.1016/0076-6879(94)44034-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogawa T, Tsumu H, Bando N, Kitamura K, Zhu Y L, Hirano H, Nishikawa K. Biosci Biotechnol Biochem. 1993;57:1030–1033. doi: 10.1271/bbb.57.1030. [DOI] [PubMed] [Google Scholar]
- 34.Staswick P, Papa C, Huang J F, Rhee Y. Plant Physiol. 1994;104:49–57. doi: 10.1104/pp.104.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conrath U, Silva H, Klessig D F. Plant J. 1997;11:747–757. [Google Scholar]
- 36.Hunter J. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]


