Abstract
Whereas vertebrates possess only two thioredoxin genes, higher plants present a much greater diversity of thioredoxins. For example, Arabidopsis thaliana has five cytoplasmic thioredoxins (type h) and at least as many chloroplastic thioredoxins. The abundance of plant thioredoxins leads to the question whether the various plant thioredoxins play a similar role or have specific functions. Because most of these proteins display very similar activities on artificial or biological substrates in vitro, we developed an in vivo approach to answer this question. The disruption of both of the two Saccharomyces cerevisiae thioredoxin genes leads to pleiotropic effects including methionine auxotrophy, H2O2 hypersensitivity, altered cell cycle characteristics, and a limited ability to use methionine sulfoxide as source of methionine. We expressed eight plant thioredoxins (six cytoplasmic and two chloroplastic) in yeast trx1, trx2 double mutant cells and analyzed the different phenotypes. Arabidopsis type h thioredoxin 2 efficiently restored sulfate assimilation whereas Arabidopsis type h thioredoxin 3 conferred H2O2 tolerance. All thioredoxins tested could complement for reduction of methionine sulfoxide, whereas only type h thioredoxins were able to complement the cell cycle defect. These findings clearly indicate that specific interactions between plant thioredoxins and their targets occur in vivo.
Thioredoxins are small oxidoreductases (molecular mass ≈12 kDa) that all contain two redox-active half-cystine residues in an exposed active center, having the amino acid sequence WCXPC (1). In its reduced form, thioredoxin can function as hydrogen donor for a variety of target proteins. The oxidized form of thioredoxin, having an intramolecular disulfide bridge between the two cysteine residues from the catalytic center, is generally reduced by a thioredoxin reductase. Thioredoxins are ubiquitous and have been described in prokaryotes as well as in eukaryotes, including, fungi, plants, invertebrates, and vertebrates (2–4). Higher plants are known to possess at least three types of thioredoxins: thioredoxin f and m, which are both encoded by the nuclear genome but are located within the chloroplast, and thioredoxin h, which is a cytosolic protein. The chloroplastic thioredoxins f and m are thought to provide a functional link between the light-absorbing pigments and several key metabolic enzymes such as fructose-1,6-bisphosphatase, phosphoribulose kinase, and malate dehydrogenase (5–7). Both oxidized thioredoxin f and thioredoxin m are reduced by a ferredoxin-dependent thioredoxin reductase (8–10). In contrast, the cytosolic thioredoxins h are reduced by an NADPH-dependent thioredoxin reductase (11, 12). Recent progress in the systematic sequencing of plant genomes provides evidence that the diversity of thioredoxins is much greater than previously expected, each class of thioredoxin being encoded by multigene families. For instance, Arabidopsis thaliana has been shown to express at least two thioredoxins f (L.V. and Y.M., unpublished results), five thioredoxins m (D. Mestres and Y.M., unpublished results), and five thioredoxins h (13). Clearly, this is in contrast to genes from vertebrates, which express only two different thioredoxins, one located in the cytosol and the other in mitochondria. Because thioredoxins have been implicated in multiple redox signal transduction pathways, the diversity of plant thioredoxin h could be interpreted as allowing a great versatility in their interaction with various target proteins. However, little progress has been made supporting this hypothesis because, in vitro, most thioredoxins h display very similar activity as donors of reducing equivalents to artificial substrates such as 5,5′-dithiobis(2-nitrobenzoate) (DTNB) and insulin. Moreover, thioredoxins h were demonstrated to be as efficient as the chloroplastic thioredoxins f and m for the in vitro reduction of chloroplastic malate dehydrogenase (14). To obtain insights into the functional specificity of thioredoxins h from A. thaliana, we devised a set of experiments that rely on the heterologous expression of these plant thioredoxins in the yeast Saccharomyces cerevisiae. Indeed, the general approach of expressing heterologous proteins in this lower eukaryote has been widely used and proven to facilitate investigations on metabolic mechanisms. Wild-type S. cerevisiae cells express two highly related thioredoxins that are encoded by two unlinked genes, TRX1 and TRX2 (15). Whereas the inactivation of each of the two genes does not significantly alter the growth of the cells, simultaneous disruption of these genes profoundly affects cell life and leads to a set of phenotypic consequences including organic sulfur auxotrophy, increased sensitivity to hydrogen peroxide, and modification of the cell cycle (16). These results thus suggest that the yeast thioredoxins are involved in very different cellular mechanisms, and we reasoned that information about the functional specificity of the plant thioredoxins would be gained by systematically testing those capable of complementing the different phenotypes of the yeast trx1, trx2 double-mutant cells.
MATERIALS AND METHODS
Strains and Media.
The S. cerevisiae strains used in this work were derived from W303 (17) and EMY54 (18). EMY60 is the standard wild-type strain (Mata, ade2–1, ade3-100, his3-11, leu2-3, lys2-801, trp1-1, ura3-1). EMY63 is the standard thioredoxin double mutant and is isogenic with EMY60 except at the thioredoxin loci (Mata, ade2-1, ade3-100, his3-11, leu2-3, lys2-801, trp1-1, ura3-1, trx1∷TRP1, trx2∷LEU2). Standard complete and minimal yeast media were as described by Sherman et al. (19). The B medium is a sulfur-less medium and was made according to Cherest and Surdin-Kerjan (20). The YMGE medium is a synthetic medium containing 0.2% (wt/vol) glucose, 3% (vol/vol) glycerol, and 2% (vol/vol) ethanol as carbon sources. The YMGal medium is a synthetic medium containing 2% galactose, 3% glycerol, and 2% ethanol. S. cerevisiae was transformed after lithium chloride treatment as described by Ito et al. (21).
Plant Thioredoxin Expression.
To express the plant thioredoxin in yeast cells, we used the shuttle vector Ycp2, an autonomously replicating sequence (ars) centromeric plasmid (22) containing the URA3 gene as a selectable marker and the GAL1 promoter. The different plant thioredoxin-encoding ORFs were cloned downstream from the GAL1 promoter region by using the two unique cloning sites present in Ycp2, MluI and BamHI. Each plant ORF was amplified from the cDNA by PCRs using specific oligonucleotides designed to introduce a MluI site upstream from the ATG codon and a BamHI site downstream from the termination codon. As a control, an equivalent procedure was applied to clone the yeast TRX1 gene under the control of the GAL1 promoter on the Ycp2 plasmid. To induce thioredoxin expression, the transformed cells were first grown in YMGE medium to relieve glucose repression and then either plated or diluted in YMGal medium to induce the transcription of the thioredoxin gene from the GAL1 promoter.
Immunoblot Analysis.
For the different transformant cells, total extracts were prepared from exponentially growing cells in appropriate YMGal medium according to a protocol described by Kuras et al. (23). Western blot analysis was performed as described by Meyer et al. (24) with protein from each extract (5 μg as quantitated by the Bradford method). We generally used antiserum directed against thioredoxins at a 1:10,000 dilution. Antibodies were visualized with ECL Western blotting detection reagents (Amersham) as described by the manufacturer. The activity of each serum was tested on the corresponding recombinant protein and the five sera have roughly the same detection limits in our Western blots—i.e., ≈2.5 ng of proteins per 1 μl of extract.
H2O2 Tolerance Analysis and Flow Cytometry.
To test H2O2 tolerance, the transformed cells were first grown in YMGE medium to a density of about 107 cells per ml and then diluted in YMGal medium to OD600 = 0.2. Then 15 μl of each culture was plated on YMGal agar medium containing H2O2 at various concentrations, and plates were incubated 3 days at 30°C. To measure DNA content of the transformant cells by a flow cytometry technique, 10 ml of cells were grown in YMGal medium to OD600 = 0.5, centrifuged, and washed in 10 ml of 50 mM Tris⋅HCl, pH 8. Cells were then fixed in 70% (vol/vol) ethanol for 1 h at room temperature, centrifuged, and resuspended in 1 ml of 50m mM Tris⋅HCl, pH 8, containing 1 mg/ml RNase A and incubated 2 h at 37°C. After sedimentation in a clinical centrifuge, the cell pellet was resuspended in 500 μl of propidium iodide solution (50 μg/ml) and stained in the dark at 4°C overnight. Fluorescence was analyzed by using a Bruker ACR 1000 flow cytometer. Data were collected with fried software, which was used to estimate the proportion of cells in G1 phase.
RESULTS
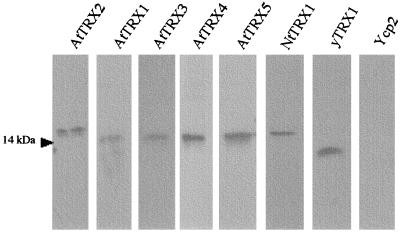
To address the question of whether the different plant thioredoxins may be functionally distinguished in vivo, we expressed several of these proteins in a heterologous cellular context consisting of S. cerevisiae cells that do not express endogenous thioredoxin. For this purpose, a set of seven thioredoxin-encoding ORFs from A. thaliana were cloned under the control of a galactose-inducible promoter on the Ycp2 plasmid, a yeast replicating vector containing the URA3 gene as a selectable marker. These series comprised five type h thioredoxins (hereafter called AtTRX1 to AtTRX5), one type m thioredoxin (AtTRXm), and one type f thioredoxin (AtTRXf). We also cloned one thioredoxin of type h from Nicotiana tabacum (NtTRX1) in the same vector (see Materials and Methods). All of these constructs were used to transform EMY63 cells (ura3, trx1∷TRP1, trx2∷LEU2), and transformants were selected for their ability to grow in the absence of uracil. As controls, EMY63 cells were transformed by the empty Ycp2 plasmid and by a plasmid expressing the yeast thioredoxin 1 (yTRX1) under the control of the GAL1 promoter. To ensure a maximal level of expression of the thioredoxin genes, the same growth procedure was applied in all the following experiments. The transformed cells were first grown in the YMGE medium to a density of about 107 cells per ml, allowing the glucose repression of the GAL1 promoter to be relieved, and then diluted in YMGal medium, a synthetic minimal medium containing 2% galactose (see Materials and Methods). As shown in Fig. 1, immunoblot analysis performed with total cellular extracts of the different transformed cells revealed that this growth procedure resulted in an equivalent level of expression of all the plant thioredoxins. In addition, it should be noted that, as expected, the EMY63 cells did not express any detectable thioredoxin.
Figure 1.
Western blot analysis of protein extracts from yeast double mutant transformed cells. They were grown in YMGal medium to induce transcription of the thioredoxin gene from the GAL1 promoter. Each lane represents the separation of 5 μg of total soluble protein extracts blotted to nitrocellulose and probed with corresponding antibodies.
Expression of Either AtTRX2 or NtTRX1 Restores Sulfate Assimilation.
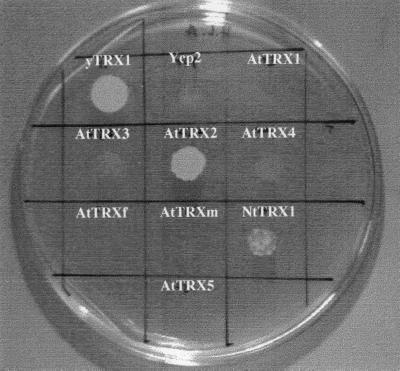
The most obvious phenotype exhibited by the trx1, trx2 double disrupted yeast cells is their methionine requirement. Such an inability to use sulfate ions in a glucose-based medium for the biosynthesis of the sulfur amino acids is accounted for by the fact that thioredoxins are needed to provide reducing equivalents to an enzyme implicated in the sulfate assimilation pathway (2). Indeed, the involvement of thioredoxin in the reduction of 3′-phosphoadenosine 5′-phosphosulfate (PAPS) into sulfite has been discussed in some detail during the past few years, in Escherichia coli as well as in S. cerevisiae, at least in vitro (25, 26). Fig. 2 shows that, as expected, on the galactose-based medium, the same phenotype was observed for the trx1, trx2 mutated cells transformed by the Ycp2 plasmid, whereas the expression of the yeast TRX1 gene from the GAL1 promoter led to methionine-prototrophic cells. Among the cells expressing the plant thioredoxins, only those expressing the AtTRX2 thioredoxin from A. thaliana were capable of vigorous growth in the absence of methionine. A partial functional complementation was also observed when thioredoxin NtTrx1 from N. tabacum was expressed. The corresponding transformed cells indeed displayed slow but sustained growth in the absence of methionine. In all the other cases, the transformed cells were completely unable to grow in the absence of methionine.
Figure 2.
Sulfate assimilation of trx1, trx2 cells expressing transgenic thioredoxins. Cells were first grown in YMGE medium to a density of about 107 cells per ml. Cultures were then diluted to OD600 = 0.2 and 15 μl of each one was plated on YMGal agar medium free of methionine. Plates were incubated 6 days at 30°C.
Expression of Either AtTRX3 or AtTRX4 Confers Tolerance to H2O2.
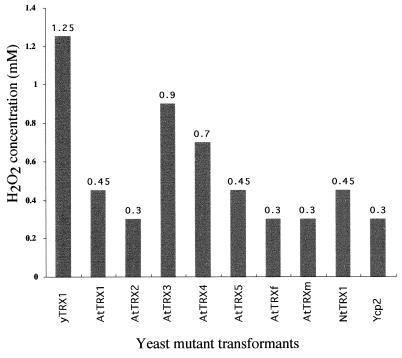
Evidence has accumulated suggesting that thioredoxins protect living organisms from oxidative stress by scavenging reactive oxygen species as well as by regenerating proteins inactivated by radical molecules (27–29). In yeast, Chae and coworkers (30) described a 25-kDa antioxidant enzyme that is able to reduce H2O2 and alkyl hydroperoxides with the use of hydrogens provided by thioredoxin, thioredoxin reductase, and NADPH. Kuge and Jones (31) showed that yeast cells overexpressing yeast thioredoxin 2 are resistant to H2O2 and to tBOOH (t-butyl hydroperoxide). Such results suggest that yeast cells that do not express functional thioredoxin would be sensitive to H2O2. In an attempt to verify this hypothesis, the trx1, trx2 double mutant cells transformed by the Ycp2 plasmid as well as those expressing the yeast thioredoxin 1 from the GAL1 promoter were plated on galactose-containing media in the presence of 0.01 mM l-methionine and different concentrations of H2O2. As reported in Fig. 3, the trx1, trx2 double mutant cells transformed by the Ycp2 plasmid could not grow in the presence of H2O2 at concentrations higher than 0.3 mM, whereas the trx1, trx2 cells expressing the yeast yTRX1 from the GAL1 promoter are capable of growing in the presence of 1.2 mM H2O2. This result prompted us to test whether the expression of plant thioredoxins might preserve the trx1, trx2 mutant cells from the detrimental effects of high concentrations of H2O2. The transformed cells expressing the different plant thioredoxins were thus plated on the media described above and their growth capacities in the presence of different concentrations of H2O2, ranging from 0.3 to 1.4 mM, were analyzed. As reported in Fig. 3, the expression of two A. thaliana thioredoxins of type h (AtTRX3 and AtTRX4) led to cells capable of growing in the presence of high concentrations of H2O2 (0.9 mM and 0.7 mM, respectively), whereas the expression of the other plant thioredoxins resulted in cells whose tolerance to H2O2 did not differ significantly from the parental trx1, trx2 cells.
Figure 3.
H2O2 tolerance of trx1, trx2 cells expressing transgenic thioredoxins. Cells were plated on YMGal agar medium containing 0.1 mM methionine and H2O2 at various concentrations. Plates were incubated 3 days at 30°C. Numbers above data bars indicate the average H2O2 concentration between the maximum concentration at which the yeast could grow and the lowest concentration that completely inhibited growth. These data have been obtained from three independent experiments.
Restoration of in Vivo Methionine Sulfoxide Reduction by Plant Thioredoxins.
The work of Brot and Weissbach (32) has established the widespread occurrence of enzymatic systems dedicated to the reduction of methionine sulfoxide entities. Methionine is indeed particularly susceptible to oxidative damage, and both prokaryotic and eukaryotic organisms possess enzymes capable of specifically reducing methionine sulfoxide found either as free amino acid or as residues within polypeptide chains. The thioredoxin from E. coli was demonstrated to serve in vitro as an efficient hydrogen donor to the methionine sulfoxide reductase from this organism (2, 25). The involvement of thioredoxin during the reduction of methionine sulfoxide was further sustained by the fact that E. coli cells whose thioredoxin- and glutaredoxin-encoding genes were both inactivated are unable to use methionine sulfoxide as a source of methionine (33). In yeast, methionine sulfoxide was recently shown to be used as an efficient sulfur source, and this metabolic utilization of methionine sulfoxide was postulated to involve its reduction by methionine sulfoxide reductase (34). We thus decided to test whether yeast cells that do not express endogenous thioredoxin were capable of growing on methionine sulfoxide as a sulfur source. The assay relies on the use of a particular synthetic medium (B medium) that is completely devoid of both mineral and organic sulfur elements. In such a medium, the growth of the yeast cells is strictly dependent on the added methionine (or methionine sulfoxide). As shown in Fig. 4, trx1, trx2 double mutant cells transformed by the Ycp2 plasmid showed only residual growth on methionine sulfoxide (mean generation time of about 23 h), whereas the cells expressing yTRX1 from the GAL1 promoter grew with a mean generation time of about 3 h. In contrast, both strains grew well in the presence of methionine as sulfur source with the same mean generation time (data not shown). These results thus confirm that the yeast thioredoxins are indeed involved in the in vivo reduction of methionine sulfoxide, in agreement with the first biochemical characterization of the methionine sulfoxide reductase from S. cerevisiae. The limited ability of the trx1, trx2 double mutant cells to grow in the presence of methionine sulfoxide used as a sulfur source allowed us to determine whether the expression of the plant thioredoxins might lead to a functional complementation of this growth defect. The growth in the presence of methionine sulfoxide of the trx1, trx2 double mutant cells expressing each of the seven plant thioredoxins was thus monitored. As reported in Fig. 4, expression of all the plant thioredoxins confers the capability of growing in the presence of methionine sulfoxide to the trx1, trx2 mutant cells. However, the observed complementations varied from one plant thioredoxin to another and could be grouped into three classes according to the measured growth characteristics. The first class comprises four type h thioredoxins, including thioredoxins AtTRX2, AtTRX3, and AtTRX4 from A. thaliana and NtTRX1 from N. tabacum. The trx1, trx2 cells expressing these thioredoxins exhibited both a mean generation time and a growth yield equivalent to those of cells expressing the yeast thioredoxin 1 from the GAL1 promoter. The second class comprises the two type f and m thioredoxins as well as one type h thioredoxin (AtTRX1). The expression of the three thioredoxins allowed the trx1, trx2 cells to grow on methionine sulfoxide but with a mean generation time of between 9 and 15 h and a growth yield half of that measured for the cells expressing the yeast thioredoxin 1. In addition to the above classified thioredoxins, the expression of AtTRX5 led to an intermediary phenotype. The trx1, trx2 cells expressing AtTRX5 displayed a growth yield equivalent to that measured with the cells expressing the thioredoxin of the first class, but the stationary phase was reached with a mean generation time of 9 h.
Figure 4.
Growth of transformed trx1, trx2 mutant cells in B medium with methionine sulfoxide (0.1 mM) as sole sulfur source. Cells were first grown in YMGE medium lacking sulfur and then diluted in YMGal medium also completely devoid of sulfur. Absorbance was measured at 600 nm in a Uvikon 930 spectrophotometer. Data reported in the key of the graph represent generation times (g.t.) calculated from OD600 between 0.1 and 1.
Plant Type h Thioredoxins Complement the Cell Cycle Defect of the trx1, trx2 Yeast Mutant Cells.
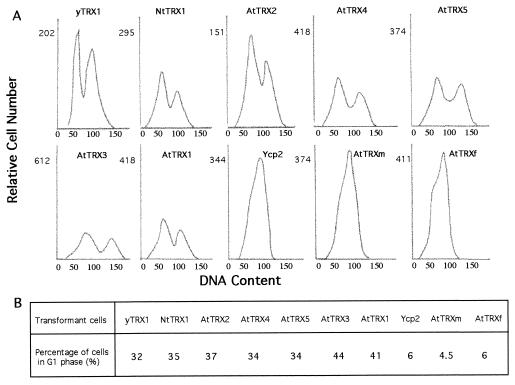
In addition to the phenotypes described above, the simultaneous inactivation of the thioredoxin-encoding genes in S. cerevisiae also results in a marked modification of the cell cycle. Muller demonstrated that the trx1, trx2 double mutant cells displayed a modified cell cycle with an considerably lengthened S phase (16). In asynchronous cultures, this change could be observed by flow cytometry and resulted in the case of the trx1, trx2 double mutant in a marked decrease of cells having a G1 content of DNA. As expected, we were able to repeat these results by analyzing by flow cytometry the DNA content of trx1, trx2 mutated cells transformed by the Ycp2 plasmid. In the presence of galactose (Fig. 5) only 6% of the observed cells are in the G1 phase as estimated by the fried peak deconvolution. In the glucose medium used by Muller the generation time of the mutant is longer (130 min) than that of the wild type (90 min). In the galactose/methionine medium growth is slower (180 min) and no difference can be seen between wild-type and mutant cells. As predicted, the expression of yeast thioredoxin 1 in mutant cells restored a normal cell distribution with about 32% of cells in G1 phase. We thus asked whether the expression of plant thioredoxins might restore a normal cell cycle to the yeast trx1, trx2 double mutant. The results reported in Fig. 5 clearly demonstrate that the expression of all of the tested plant thioredoxins of type h indeed led to a normal cell cycle. In contrast, the expression of the plant thioredoxins of both type f and m could not complement the cell cycle defect of the trx1, trx2 mutant cells. In these two cases, flow cytometry analysis show that only about 6% of the cells had a G1 DNA content, which is very close to that observed with yeast cells that do not express their endogenous thioredoxins. The generation time of the complemented cells is the same as for the wild-type and mutant cells.
Figure 5.
DNA histograms of asynchronous cultures of transformed trx1, trx2 mutant cells. Cells were grown in YMGal medium to exponential phase and prepared for flow cytometry. In A the relative fluorescence intensity is plotted against the relative cell number. The two peaks observed for yTRX1, AtTRX1, … , AtTRX5, and NtTRX1 represent 1n and 2n DNA contents. For each sample, between 10,000 and 20,000 fluorescent events were measured. The proportion of cells in G1 phase estimated by fried software analysis is indicated in B.
DISCUSSION
As shown by Muller (16), the yeast thioredoxin double mutant (EMY63) presents a phenotype with multiple aspects, including methionine auxotrophy and a perturbed cell cycle.
In addition, recent studies suggest the implication of thioredoxin in the control of the cellular redox state. Indeed, Muller (35) reported that, in S. cerevisiae, mutants that lack glutathione reductase require one of two thioredoxins for growth, and mutants deficient for the two thioredoxins require glutathione reductase. Furthermore, Kuge and Jones (31) reported that yTRX2 plays a unique role, since its promoter presents a H2O2-activable YAP1 element. In fact, our experiments indicate that yTRX1 also confers H2O2 tolerance to trx1, trx2 mutant cells. This result suggests that both proteins encoded by the thioredoxin genes allow growth in the presence of high concentrations of H2O2, and the particular role assigned to the yTRX2 gene is likely due to its activable YAP1 promoter element. Moreover, methionine is an easily oxidizable amino acid leading to methionine sulfoxide. In vitro, thioredoxin constitutes a good hydrogen donor for methionine sulfoxide reductase. This prompted us to test the ability of trx1, trx2 mutant cells to grow on methionine sulfoxide as sole sulfur source. Our results clearly indicate that thioredoxin plays a central role in the reduction of methionine sulfoxide in vivo as previously shown for E. coli (33).
We were interested in estimating the ability of several plant thioredoxins to complement the mutant for these different aspects of the phenotype. For this purpose, we chose a low copy plasmid to limit the production of the transgenic protein because a very high overexpression of the protein could mask specific interactions. In addition, we placed the ORF under the control of an inducible promoter to mitigate the possible toxicity of the transgenic thioredoxin. In fact, such toxicity was not observed in this study. We noted that trx1, trx2 mutant cells displayed methionine auxotrophy and a very short G1 phase in YMGal medium as described in glucose medium. In contrast, trx1, trx2 cells expressing the yeast yTRX1 from the GAL1 promoter are methionine prototrophs and show a normal cell cycle. Moreover, the plant and yeast thioredoxins are produced at approximately the same level in mutant cells. These findings demonstrate the feasibility of our approach.
Table 1 summarizes the capacity of different plant thioredoxins to complement the four aspects of the yeast double-mutant phenotype. We can observe that each plant thioredoxin displays a unique complementation profile: for example, AtTRX2 confers methionine prototrophy but no H2O2 tolerance, whereas AtTRX3 confers H2O2 tolerance but no methionine prototrophy. These observations lead us to draw several conclusions:
Table 1.
Complementation results obtained for the various thioredoxins expressed in yeast trx1, trx2 double-mutant cells
| Thioredoxin | Source | Accession no. | Sulfate assimilation | H2O2 tolerance | MetO reduction | Cell cycle defect |
|---|---|---|---|---|---|---|
| AtTRX1 | A. thaliana | Z14084 | − | − | +/−− | + |
| AtTRX2 | A. thaliana | Z35475 | + | − | + | + |
| AtTRX3 | A. thaliana | Z35474 | − | + | + | + |
| AtTRX4 | A. thaliana | Z35473 | − | + | + | + |
| AtTRX5 | A. thaliana | Z35476 | − | − | +/− | + |
| NtTRX1 | N. tabacum | X58527 | +/− | − | + | + |
| AtTRXf | A. thaliana | Z34673 | − | − | +/−− | − |
| AtTRXm | A. thaliana | Z46485 | − | − | +/−− | − |
+, Efficient complementation; +/− and +/−−, partial complementation; −, absence of complementation. Numbers reported are GenBank accession numbers of each thioredoxin used in this work. MetO, methionine sulfoxide.
First, our findings show that the pleiotropic effect of yeast thioredoxins results from the interaction with multiple targets, rather than from the reduction of one unique protein. These targets have not yet been characterized, but in vitro experiments have shown that thioredoxin is a good hydrogen donor for 3′-phosphate 5′-phosphosulfate reductase, thioredoxin-dependent peroxidases (Tpx) (30, 36), and methionine sulfoxide reductase. They are good candidates for sulfate assimilation, tolerance to H2O2, and growth on methionine sulfoxide, respectively. Nevertheless, more that one target may be implicated in each aspect of the phenotype. Presently, the component of the cell cycle that interacts with thioredoxin is not known. Although ribonucleotide reductase is one candidate, the finding of normal deoxyribonucleotide levels in the mutant lacking both thioredoxins (37) prompted consideration of alternative targets.
The second consequence is that the thioredoxin/target interaction is specific in vivo. In other words, the sequence of AtTRX2 presents a specific determinant allowing a strong interaction with the target responsible for sulfate assimilation. This determinant is lacking in other Arabidopsis thioredoxins. Similarly, the sequence of AtTRX3 presents another specific determinant allowing a strong interaction with the target responsible for H2O2 tolerance. This specific complementation also suggests that yeast thioredoxins present multiple determinants allowing interaction with the multiple targets, rather than interacting nonspecifically with every protein presenting a disulfide bridge. This is in agreement with recent findings of Gronenborn’s group, who have shown that human thioredoxin interacts with NFκB and Ref1 in different ways (38, 39). Most in vitro experiments suggest nonspecific interaction with the targets, but clearly our in vivo experiments show specific interactions. This result also strongly suggests that each thioredoxin h plays a specific function in plant cells.
The five A. thaliana thioredoxins h, as well as the tobacco thioredoxins, have very different sequences, ranging from 95% to only 50% identity (13). These differences are clearly observable in a phylogenetic tree reported previously (40). In addition, three of them have the plant-specific active site variant WCPPC instead of the classical WCGPC. Obviously the similarity throughout the molecule and the nature of the active site are not directly related to the ability to complement specific aspects of the phenotype in yeast. For example, AtTRX5 shows only 9 amino acid replacements compared with AtTRX3. Nevertheless, it does not confer H2O2 tolerance and stimulates less growth on methionine sulfoxide than AtTRX3. Mutagenesis, construction of hybrid thioredoxins, and testing of their effect on EMY63 should allow the characterization of the sequence(s) and/or conformation responsible for in vivo specificity of the yeast and plant thioredoxins.
Our first intention was to use the chloroplastic thioredoxins AtTRXf and AtTRXm as negative controls because they are reactivated in the chloroplast by a ferredoxin-dependent thioredoxin reductase, an enzyme that has no homologue in the yeast genome. Surprisingly, the two sequences stimulate growth on methionine sulfoxide as sole source of sulfur. In fact, the Arabidopsis NADP:thioredoxin reductase is able to reduce pea thioredoxin m in vitro to some extent (14). Thus in this study the chloroplastic thioredoxins are probably reduced by the NADPH:thioredoxin reductase. This low complementation cannot be discussed in terms of interaction with the targets because the reduction of the chloroplastic thioredoxins is clearly a factor that limits their activity in the yeast cells. Nevertheless, this partial complementation suggests that transformation of the mutant EMY63 by a library and screening for growth on methionine sulfoxide could be a very useful way to isolate thioredoxin clones, even if these thioredoxins are reduced in the original organism by a reductase unrelated to the yeast NADPH reductase, as is the case for the thioredoxins from chloroplasts, blue green algae, and vertebrates. In a previous study, Hartman et al. (41) showed that spinach thioredoxin m inhibits DNA synthesis in Xenopus eggs. Here we show that expression of Arabidopsis thioredoxins f and m in the yeast double mutant has no effect on S phase. In fact, the thioredoxin-dependent process concerning DNA synthesis described by Hartman et al. in Xenopus (41) and Muller in S. cerevisiae (42) is probably different in the two organisms, because in Xenopus it is redox independent, whereas in S. cerevisiae it is redox dependent.
Acknowledgments
We thank Eric Muller for giving us the wild-type and mutant yeasts used in this study, Yvette Chartier for the yeast thioredoxin antibody, Richard Cooke for critical reading of the manuscript, and Yolande Surdin-Kerjan for discussions.
References
- 1.Holmgren A. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 2.Gleason F K, Holmgren A. FEMS Microbiol. 1988;4:271–297. doi: 10.1111/j.1574-6968.1988.tb02747.x. [DOI] [PubMed] [Google Scholar]
- 3.Salz H K, Flickinger T W, Mittendorf E, Pellicenapalle A, Petschek J P, Albrecht E B. Genetics. 1994;136:1075–1086. doi: 10.1093/genetics/136.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wetterauer B, Veron M, Miginiac-Maslow M, Decottignies P, Jacquot J P. Eur J Biochem. 1992;209:643–649. doi: 10.1111/j.1432-1033.1992.tb17331.x. [DOI] [PubMed] [Google Scholar]
- 5.Marcus F, Moberly L, Latshaw S P. Proc Natl Acad Sci USA. 1988;85:5375–5383. doi: 10.1073/pnas.85.15.5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandes H K, Larimer F W, Hartman F C. J Biol Chem. 1996;271:3333–3335. doi: 10.1074/jbc.271.7.3333. [DOI] [PubMed] [Google Scholar]
- 7.Decottignies P, Schmitter J M, Miginiac-Maslow M, Le Maréchal P, Jacquot J P, Gadal P. J Biol Chem. 1988;263:11780–11785. [PubMed] [Google Scholar]
- 8.Buchanan B B. Arch Biochem Biophys. 1981;33:147–162. [Google Scholar]
- 9.Droux M, Miginiac-Maslow M, Jacquot J-P, Gadal P, Crawford N, Buchanan B B. Arch Biochem Biophys. 1987;256:372–380. doi: 10.1016/0003-9861(87)90458-9. [DOI] [PubMed] [Google Scholar]
- 10.Falkenstein E, Von Schaewen A, Scheibe R. Biochim Biophys Acta. 1994;1185:252–254. doi: 10.1016/0005-2728(94)90218-6. [DOI] [PubMed] [Google Scholar]
- 11.Florencio F J, Yee B C, Johnson T C, Buchanan B B. Arch Biochem Biophys. 1988;266:496–507. doi: 10.1016/0003-9861(88)90282-2. [DOI] [PubMed] [Google Scholar]
- 12.Marcus F, Chamberlain S H, Chu C, Maziarz F R, Shin S, Yee B C, Buchanan B B. Arch Biochem Biophys. 1991;287:195–198. doi: 10.1016/0003-9861(91)90406-9. [DOI] [PubMed] [Google Scholar]
- 13.Rivera-Madrid R, Mestres D, Marinho P, Jacquot J P, Decottignies P, Miginiac-Maslow M, Meyer Y. Proc Natl Acad Sci USA. 1995;92:5620–5624. doi: 10.1073/pnas.92.12.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacquot J P, Rivera-Madrid R, Marinho P, Kollarova M, Le Marechal P, Miginiac-Maslow M, Meyer Y. J Mol Biol. 1994;235:1357–1363. doi: 10.1006/jmbi.1994.1091. [DOI] [PubMed] [Google Scholar]
- 15.Gan Z R. J Biol Chem. 1991;266:1692–1696. [PubMed] [Google Scholar]
- 16.Muller E G D. J Biol Chem. 1991;266:9194–9202. [PubMed] [Google Scholar]
- 17.Wallis J W, Chrebet G, Brodsky G, Rolfe M, Rothstein R. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- 18.Muller E G D. Yeast. 1992;8:117–120. doi: 10.1002/yea.320080206. [DOI] [PubMed] [Google Scholar]
- 19.Sherman F, Fink G R, Hicks J B. Methods in Yeast Genetics: Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1979. [Google Scholar]
- 20.Cherest H, Surdin-Kerjan Y. Genetics. 1992;130:51–58. doi: 10.1093/genetics/130.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito H, Fukuda Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cole G M, Stone D E, Reed S I. Mol Cell Biol. 1990;10:510–517. doi: 10.1128/mcb.10.2.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuras L, Thomas D. Mol Cell Biol. 1995;15:208–216. doi: 10.1128/mcb.15.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer Y, Grosset J, Chartier Y, Cleyet-Marel J C. Electrophoresis. 1988;9:704–712. doi: 10.1002/elps.1150091105. [DOI] [PubMed] [Google Scholar]
- 25.Tsang M L, Schiff J A. J Bacteriol. 1976;125:923–933. doi: 10.1128/jb.125.3.923-933.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porqué P G, Baldesten A, Reichard P. J Biol Chem. 1970;245:2371–2374. [PubMed] [Google Scholar]
- 27.Schenk H, Klein M, Erdrugger W, Droge W, Schulze-Osthoff K. Proc Natl Acad Sci USA. 1994;91:1672–1676. doi: 10.1073/pnas.91.5.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spector A, Yan G Z, Huang R R, McDermott M J, Gascoyne P R, Pigiet V. J Biol Chem. 1988;263:4984–4990. [PubMed] [Google Scholar]
- 29.Bjornstedt M, Xue J, Huang W, Akesson B, Holmgren A. J Biol Chem. 1994;269:29382–29384. [PubMed] [Google Scholar]
- 30.Chae H Z, Chung S J, Rhee S G. J Biol Chem. 1994;269:27670–27678. [PubMed] [Google Scholar]
- 31.Kuge S, Jones N. EMBO J. 1994;13:655–664. doi: 10.1002/j.1460-2075.1994.tb06304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brot N, Weissbach H. Arch Biochem Biophys. 1983;223:271–281. doi: 10.1016/0003-9861(83)90592-1. [DOI] [PubMed] [Google Scholar]
- 33.Russel M, Model P, Holmgren A. J Bacteriol. 1990;172:1923–1929. doi: 10.1128/jb.172.4.1923-1929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Isnard A D, Thomas D, Surdin-Kerjan Y. J Mol Biol. 1996;262:473–484. doi: 10.1006/jmbi.1996.0529. [DOI] [PubMed] [Google Scholar]
- 35.Muller E G D. Mol Biol Cell. 1996;7:1805–1813. doi: 10.1091/mbc.7.11.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chae H Z, Robison K, Poole L B, Church G, Storz G, Rhee S G. Proc Natl Acad Sci USA. 1994;91:7017–7021. doi: 10.1073/pnas.91.15.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muller E G D. J Biol Chem. 1994;269:24466–24471. [PubMed] [Google Scholar]
- 38.Qin J, Clore G M, Kennedy W P, Huth J R, Gronenborn A M. Structure. 1995;3:289–297. doi: 10.1016/s0969-2126(01)00159-9. [DOI] [PubMed] [Google Scholar]
- 39.Qin J, Clore G M, Kennedy W P, Kuszewski J, Gronenborn A M. Structure. 1996;4:613–620. doi: 10.1016/s0969-2126(96)00065-2. [DOI] [PubMed] [Google Scholar]
- 40.Sahrawy M, Hecht V, Lopez-Jaramillo J, Chueca A, Chartier Y, Meyer Y. J Mol Evol. 1996;42:422–431. doi: 10.1007/BF02498636. [DOI] [PubMed] [Google Scholar]
- 41.Hartman H, Wo M, Buchanan B B, Gerhart J C. Proc Natl Acad Sci USA. 1993;90:2271–2275. doi: 10.1073/pnas.90.6.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muller E G D. Arch Biochem Biophys. 1995;318:356–361. doi: 10.1006/abbi.1995.1240. [DOI] [PubMed] [Google Scholar]