Abstract
Members of the yeast polymerase-associated factor 1 (Paf1) complex, which is composed of at least five components (Paf1, Rtf1, Cdc73, Leo1 and Ctr9), are conserved from yeast to humans. Although these proteins have been implicated in RNA polymerase II-mediated transcription, their roles in vertebrate development have not been explained. Here, we show that a zebrafish mutant with a somite segmentation defect is deficient in rtf1. In addition, embryos deficient in rtf1 or ctr9 show abnormal development of the heart, ears and neural crest cells. rtf1 is required for correct RNA levels of the Notch-regulated genes her1, her7 and deltaC, and also for Notch-induced her1 expression in the presomitic mesoderm. Furthermore, the phenotype observed in rtf1-deficient mutants is enhanced by an additional deficiency in mind bomb, which encodes an effector of Notch signalling. Therefore, zebrafish homologues of the yeast Paf1 complex seem to preferentially affect a subset of genes, including Notch-regulated genes, during embryogenesis.
Keywords: Notch, Paf1 complex, segmentation, somite, transcription
Introduction
In addition to transcriptional activators and repressors—which function together with the mediator complex and general transcription factors in transcriptional initiation—factors participating in many of the steps of transcription seem to influence transcription. One such factor is the yeast polymerase-associated factor 1 (Paf1) complex. This complex has been implicated in various processes, such as transcription initiation and elongation, histone modification, phosphorylation of RNA polymerase II (pol II), and RNA processing and export (Sims et al, 2004). The components of the Paf1 complex are evolutionarily conserved from yeast to Drosophila and humans, although the Rtf1 proteins in human and Drosophila are not stably associated with the other members of the Paf1 complex (Zhu et al, 2005; Adelman et al, 2006).
Although this complex seems to be associated with pol II at all transcriptionally active genes, the loss of each component of the yeast Paf1 complex either reduces or enhances the transcription of a small subset of genes (Penheiter et al, 2005), suggesting that the yeast Paf1 complex preferentially regulates a particular subset of genes in vivo. Similarly, Drosophila homologues of the yeast Paf1 complex seem to have preferential roles during development. For example, knockdown of Drosophila rtf1 increases the hypomorphic severity of a Notch mutant phenotype during wing development (Tenney et al, 2006); however, it is not clear whether Rtf1 preferentially affects transcription of a specific subset of genes involved in Notch signalling.
One aspect of development in which Notch signalling is important is the segmentation of somites (Pourquié, 2003). Periodical segmentation is coupled to an internal oscillator, referred to as the ‘segmentation clock', as shown by the cyclic expression of genes in the presomitic mesoderm (PSM) (Palmeirim et al, 1997). Interestingly, many genes that show cyclic expression are regulated by Notch signalling in the PSM. Notch signalling is also involved in other aspects of segmentation, synchronization of the oscillating phase between cells in the PSM (Horikawa et al, 2006) and establishment of the rostral–caudal compartment inside a somite (Takahashi et al, 2000).
In previous studies, we identified numerous ethylnitrosourea (ENU)-mutagenized zebrafish mutants with altered somite morphogenesis (Koshida et al, 2005). In contrast to previously identified Notch signalling mutants, in which the striped expression of Notch target genes is perturbed to form salt-and-pepper patterns, one of the mutants obtained in our screens, kt641, showed reduced but still striped expression of the Notch target genes in the PSM. In the present study, we found that the gene responsible for this phenotype is a zebrafish homologue of yeast rtf1.
Results And Discussion
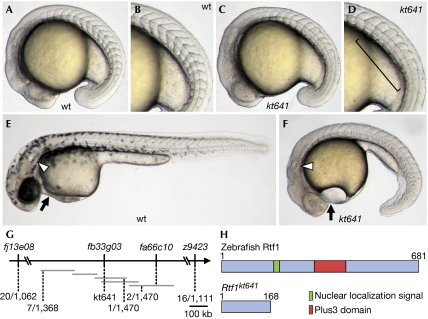
In initial experiments, we found that kt641 homozygous embryos showed a distinct phenotype: in addition to the segmentation defect (Fig 1A–D), they showed decreased pigmentation of melanophores, abnormal cardiogenesis, small ears, shortened and curled tails in later stages (Fig 1E,F; supplementary Fig S1A–D online), and embryonic lethality. In addition to these defects, in situ hybridization analysis showed abnormal neural development (supplementary Fig S1E–H online).
Figure 1.
Isolation and molecular characterization of a zebrafish rtf1kt641 mutant defective in somite formation. (A–F) Lateral views of wild-type (wt) (A,B,E) and kt641 homozygotes (C,D,F) at the 17-somite stage (A–D) and at 36 h post-fertilization (E,F). Panels (B) and (D) show views of panels (A) and (C) respectively, at higher magnification. Somite boundaries are disrupted in the posterior trunk of the kt641 mutant (square bracket in (D)). At later stages, the kt641 mutation causes reduced pigmentation, limited tail growth and abnormal heart (arrow) and ear (arrowhead) development (F). (G) Meiotic and physical mapping of the kt641 mutation. Horizontal grey bars represent contiguous sequences deposited in linkage group 13. (H) Schematic diagrams of zebrafish Rtf1 proteins encoded by wild-type and kt641 alleles.
To identify the gene responsible for this phenotype, we mapped its chromosomal position (Fig 1G). The kt641 mutation was mapped to the region between fj13e08 (1.9 cM from the mutation) and fa66c10 (0.14 cM from the mutation) on linkage group (LG) 13, and a polymorphism within a contiguous sequence, BX284673, was found to be near the mutation (1/1,470 recombinants/meioses). Interestingly, a zebrafish homologue of rtf1 had been located near the suspected mutation. Sequence analysis of rtf1 in the kt641 mutant showed a point mutation—C to T—at codon 169, which co-segregated with the phenotype. This mutation produces a termination codon—CAG to TAG—which yielded a truncated version of the Rtf1 protein (Fig 1H). Injection of two individual antisense morpholino oligonucleotides (MOs) specific for rtf1 also resulted in morphological defects similar to those of the kt641 phenotype (supplementary Table S1 online). Injection of rtf1 MO1 into kt641 mutants did not enhance the phenotype, indicating that this is a null mutation (data not shown). Furthermore, injection of wild-type rtf1 messenger RNA at the one-cell stage rescued the phenotype, whereas injection of kt641 mutant mRNA did not (supplementary Table S2 online). Therefore, we conclude that the gene responsible for the kt641 phenotype is rtf1. In situ hybridization analysis indicated that the rtf1 transcript was present in all blastomeres at cleavage stages and in the entire embryo during early developmental stages (data not shown). Maternal expression of the transcript was also detected (data not shown).
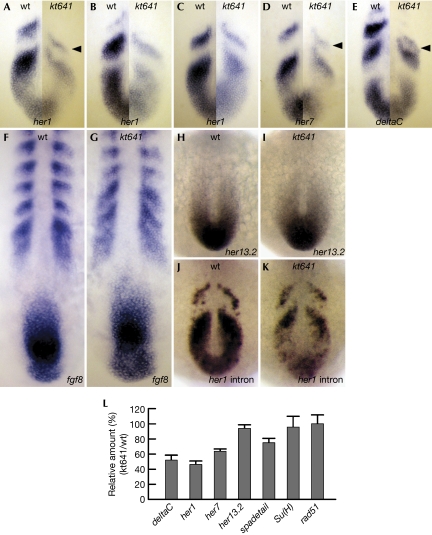
So far, posterior segmentation defects in zebrafish similar to those observed in the rtf1 mutant have been reported to be caused by abnormalities in one of two distinct signalling pathways: Notch or fibroblast growth factor (FGF). Several mutants in Notch signalling components—including deltaC, deltaD and notch1a, and also mib in E3 ubiquitin ligase for Notch ligands—lack somite boundaries in the posterior region (Holley & Takeda, 2002; Itoh et al, 2003; Jülich et al, 2005). Zebrafish embryos defective in her13.2, which is a downstream target of FGF signalling, also show a similar phenotype, probably resulting from abnormal regulation of her1, a prominent target of Notch signalling (Kawamura et al, 2005). To examine whether rtf1 is involved in either of these signalling pathways, we analysed the expression of genes involved in Notch or FGF signalling in somite segmentation at early somite stages, when tail elongation is not yet defected in rtf1 homozygous embryos. Striped expression of her1, her7 and deltaC, all of which are regulated by Notch signalling (Holley et al, 2000; Jiang et al, 2000; Oates & Ho, 2002), was strongly reduced in rtf1 homozygous embryos at early somite stages (Fig 2A–E,L) and at later stages (supplementary Fig S2A and B online). The most anterior expression of these genes also shifted in the posterior direction, although the reason for this shift was uncertain (Fig 2A,D,E, arrowheads). By contrast, the expression of fgf8 and her13.2 in the PSM was normal in rtf1 homozygous embryos (Fig 2F–I,L), suggesting that Rtf1 is preferentially required for the correct expression of Notch-regulated genes. As de novo mRNA synthesis, as monitored by a her1 intron probe, was also decreased in rtf1 mutants (Fig 2J,K), the reduction of Notch-regulated genes apparently occurs at the transcriptional level. By contrast, expression of Su(H), a mediator of Notch signalling, was not significantly altered in the mutants (data not shown; Fig 2L), indicating that the decrease in Notch-regulated genes was not due to a reduction in this Notch signalling mediator. Notably, in contrast to previously identified Notch signalling mutants, expression of the Notch-regulated genes remained periodic in rtf1 homozygous embryos. Therefore, reduction, but not perturbed patterning, of the expression of these genes might produce defective segmentation of somites in rtf1 mutants.
Figure 2.
The rtf1kt641 mutation reduces the expression of genes involved in somite segmentation. (A–E) A comparative representation of expression patterns of her1 (A–C), her7 (D) and deltaC (E) messenger RNAs between wild-type siblings (wt; left halves) and rtf1kt641 mutants (kt641; right halves) indicates a reduction of these genes in rtf1kt641 mutants. Each half panel represents the cyclic expression patterns of wild-type siblings and rtf1kt641 mutants at comparative oscillation phases. Three consecutive expression phases of her1 mRNA are represented (A–C). Arrowheads indicate the anterior-most stripes in rtf1kt641 mutants, which are shifted in the posterior direction. (F–I) Expression patterns of fgf8 (F,G) and her13.2 (H,I) mRNA in wild-type siblings (F,H) and rtf1kt641 mutants (G,I). (J,K) her1 nascent transcript hybridized with intron probe in wild-type siblings (J) and rtf1kt641 mutants (K). Expression of her1 is reduced at the transcriptional level in rtf1kt641 mutants. Embryos are at the 10- or 12-somite stage and the anterior is top in (A–K). (L) Reverse transcription–PCR analysis of transcript abundance for genes involved in somitogenesis. Data are presented as means+s.d. from triplicate assays.
It has been proposed that the segmentation process in the posterior PSM is subsequently translated into a rostro–caudal pattern within a segmental unit in the anterior PSM (Pourquié, 2004). In rtf1 homozygous mutants, the expression patterns of genes that are normally expressed in a segmental fashion in the anterior PSM and/or somites are disrupted. Anterior compartment-specific expression of papc and mesp-b was broadened and became less localized (supplementary Fig S2C–F online). Furthermore, expression of mesp-a, which is specifically expressed in the anterior compartment within a segmental unit of wild-type embryos, was markedly reduced in rtf1 mutants (supplementary Fig S2G and H online). Caudal-specific expression of myoD and rostral-specific expression of fgf8 in the somite was broadened in rtf1 embryos (Fig 2F and G; supplementary Fig S2I and J online) probably as a result of abnormal rostro–caudal compartmentation. Thus, rtf1 is required in the anterior PSM for correct rostro–caudal patterning, and this patterning defect seems to result in morphological defects in somites.
To determine whether Notch-dependent transcription was preferentially affected in the PSM of rtf1 mutants, we examined her1 expression in embryos in which Notch signalling was strongly activated by expressing the intracellular domain of Notch1a. Although her1 expression was elevated in the entire PSM of wild-type embryos (Fig 3A), its expression was reduced in rtf1 mutant embryos (Fig 3B), indicating that Rtf1 is required for transcriptional activation by Notch signalling. In addition, rtf1 mutation enhanced the phenotype of mib, which encodes the E3 ubiquitin ligase that ubiquitinates Notch ligands. Compared with rtf1 and mib mutants, compound homozygotes for rtf1 and mib showed more severe phenotypes (Fig 3C–M): specifically, a lack of morphological somites (Fig 3C–G) and shortened tails (Figs 1F, 3H,I) and also lower her1 expression (Figure 3J–M). These results strongly indicate that Rtf1 cooperatively participates in transcription with Notch signalling during somite development. By contrast, overall suppression of pol II-mediated transcription by α-amanitin resulted in abnormal development, but with a phenotype distinct from that of the rtf1 mutant (supplementary Fig S3 and Table S4 online). Furthermore, in addition to her13.2, which is induced by FGF, expression of the Wnt signalling target, cdx4, was almost unchanged in the posterior region of rtf1 homozygous embryos (supplementary Fig S2K and L online). Therefore, we conclude that Rtf1 is preferentially required for Notch-mediated transcription in somite development.
Figure 3.
Functional cooperation between Rtf1 and Notch signalling. (A,B) her1 expression in wild-type (wt; A) and kt641 mutant (B) embryos injected with synthetic messenger RNA encoding Notch1a intracellular domain (NICD). In kt641 mutant embryos, the elevated expression is reduced, although the pattern of her1 expression is still abnormal. (C–F) Lateral views of 12-somite-stage embryos from crossing of rtf1kt641/+;mibta52b/+ fish. Wild type (C), kt641 mutant (D), mib mutant (E) and kt641;mib compound mutant (com; F) were generated at a ratio (57.1:18.1:19.3:5.5%; n=590) close to the expected mendelian frequency (56.3:18.8:18.8:6.3%). Square brackets in (D–F) indicate abnormally segmented somites. The kt641;mib double mutation enhances the somite segmentation defect. (G) Number of fully segmented somites in kt641 (n=47), mib (n=59) and kt641;mib (com; n=15) at 18 h post-fertilization. Data are presented as means+s.d. P<0.001 for differences between single mutants (kt641 or mib) and compound mutants (kt641;mib) by Student's t-test are represented. (H,I) In addition to the somite defect, kt641;mib mutants (I) show more severe defects in the eye, pigmentation and tail elongation than kt641 (Fig 1F) and mib single mutants (H) at 36 h post-fertilization. (J–M) In contrast to wild-type siblings (J), kt641 mutants, which show cyclic but reduced her1 expression (K), and mib mutants, which lack the striped expression of her1 (L), kt641;mib compound mutants show markedly reduced her1 expression without stripes (M) at the 12-somite stage.
Next, we examined whether zebrafish homologues of other components of the yeast Paf1 complex are required for expression of Notch-regulated genes and somite segmentation. Injection of two individual MOs specific for zebrafish ctr9 resulted in morphological defects similar to the rtf1 mutant phenotype (Fig 4A,B; supplementary Table S1 online). The ctr9 MO also enhanced somite and neural crest defects, and reduced her1 expression in rtf1 mutants (Fig 4C–J; supplementary Fig S5A–D online). These findings indicate that Rtf1 associates with another member of the Paf1 complex in zebrafish embryos. As the yeast Paf1 complex is involved in transcription elongation, we examined the genetic interaction between rtf1 and genes encoding two factors involved in transcriptional elongation, spt5 and spt6. Injection of an rtf1-specific antisense MO enhanced the defects in somite segmentation and neural crest formation, and reduced her1 expression in foggy/spt5 homozygous embryos (Keegan et al, 2002), suggesting a genetic interaction between these genes (supplementary Figs S4 and S5 online). We also observed genetic interaction between rtf1 and pandra/spt6 (Keegan et al, 2002; data not shown). These results indicate that Rtf1 functions together with Spt5 and Spt6 during the elongation process in the transcription of her1 in the PSM.
Figure 4.
Rtf1 cooperates with the Paf1 complex during somitogenesis. (A,B) Knockdown of ctr9 results in defects that are similar to but weaker than those of kt641 mutants. Embryos injected with ctr9 MO1 (A), but not 5-mismatched MO1 (B), show reduced pigmentation, abnormal cardiogenesis (arrow), small ears (arrowhead), slightly shortened tails and disorganized somite boundaries in the tail (square bracket) at 36 h post-fertilization. Two independent ctr9 MOs cause similar morphological defects. (B) 5-mispaired ctr9 MO1 hardly impairs development. (C–E) At the 12-somite stage, no marked defects were observed in ctr9 MO-injected embryos (C). 5-mispaired control MO1 had no effect on the kt641 phenotype (D), whereas injection of ctr9 MO1 into kt641 mutant embryos resulted in an enhanced segmentation defect with few segmentation boundaries (E). The square brackets in (D) and (E) indicate the posterior somites with obscure boundaries. (F) Number of segmented somites in ctr9 MO1-injected wild-type embryos (n=82), ctr9 5-mismatched MO1-injected kt641 mutants (n=18) and ctr9 MO1-injected kt641 mutants (n=23) at 24 h post-fertilization. Error bars represent the s.d. P<0.001 between single mutants (kt641<5-mismatched MO1 or wt<ctr9 MO1) and double mutants (kt641<ctr9 MO1) are represented. (G–J) her1 expression in 5-mismatched control MO1-injected (G,I), ctr9 MO1-injected (H,J) wild-type (G,H) or kt641 mutant (I,J) embryos. In comparison with wild-type siblings injected with 5-mismatched control MO1 (G), those injected with the ctr9 MO1 show reduced her1 expression (H). This reduction is enhanced in kt641 mutant embryos injected with ctr9 MO1 (J). MO, morpholino oligonucleotides.
Our results indicate a developmental role of the Paf1 complex homologues in vertebrates. As in yeast (Penheiter et al, 2005), zebrafish Rtf1 and Ctr9 function preferentially in the transcription of a specific subset of genes, including Notch-regulated genes. Ligand stimulation of Notch signalling causes proteolytic cleavage of the Notch receptor and translocation of its intracellular fragment (NotchICD) to the nucleus. NotchICD binds to a transcription factor of the CBF1/Su(H)/Lag-1 family (CBF1 in vertebrates) and functions as its co-activator (Lubman et al, 2004). As rtf1-defective PSM cells show reduced NotchICD-induced her1 expression, Rtf1 seems to function downstream of NotchICD. This suggests that Rtf1 functions in Notch-specific gene expression in association with NotchICD or CBF1.
In contrast to somite development, we have not yet determined whether abnormal development of other tissues in rtf1 homozygous embryos is due to a decrease in Notch signalling. At present, it is uncertain whether other signals cooperate with Rtf1 in the development of these tissues. The rtf1 mutation did not affect the expression of genes affected by other signals, including foxd3, a Wnt target in neural crest development, or her9, prdm1/blimp1 and gata5, the expression patterns of which are affected by bone morphogenetic protein signalling in neural, neural crest and cardiac development (data not shown). Conversely, zebrafish Rtf1 might also function in transcriptional elongation. Our results showed that defective translation of the transcriptional elongation factors fog/spt5 and pan/spt6 also enhanced the phenotype of rtf1 mutants. This result suggests another possibility: that the transcription of the Notch-regulated genes preferentially involves cooperative interaction between Rtf1 and these elongation factors.
Finally, the segmentation phenotype of the rtf1 mutant showed a unique feature in comparison with previously reported mutants. Although periodic expression of the clock genes her1, her7 and deltaC was maintained in rtf1 mutants, the expression levels of these genes were markedly decreased. This observation suggests that the periodicity of their transcription is maintained even when their expression levels are reduced, which is consistent with a theoretical analysis reported previously by Lewis (2003). However, despite periodic gene expression in the posterior PSM, segmental gene expression in the anterior PSM was disrupted. This might imply that sufficient expression levels of the clock genes in the posterior PSM are required for distinct expression boundaries between genes in the anterior PSM, as well as in somites. Conversely, it also seems possible that the segmentation process in the anterior PSM, which is also regulated by the Notch signalling, is affected by the reduction of rtf1.
Methods
Fish strains and mutant screening. All studies on wild-type zebrafish (Danio rerio) were carried out using fish with a TL2 closed-colony background (Kishimoto et al, 2004). Embryos obtained from natural crosses were maintained in egg water (0.03% artificial sea salt in water) at 23°C or 28°C. ENU-based mutagenesis was carried out as described previously (Kishimoto et al, 2004).
Genetic mapping. kt641 heterozygous fish (TL background) were mated with wild-type Tübingen fish to generate F1 families. Homozygous mutant embryos were obtained from the F1 crosses. We then carried out PCR for specific simple sequence length polymorphism or single-strand conformation polymorphism markers using their genomic DNA. The PCR was carried out for 30–40 cycles of 94°C for 30 s, 60°C for 30 s and 72°C for 30 s.
Whole-mount in situ hybridization. Whole-mount in situ hybridization was carried out as described previously (Nikaido et al, 1997). In the case of the her1 intron probe, embryos were hybridized at 55°C because of its high AT content (Gajewski et al, 2003).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank M. Ito for the mib mutant, D. Yelon for the fog and pan mutants, Y.J. Jiang, H. Takeda, A. Sawada and M. Fishman for probes, and Y. Bessho, A. Kawakami and H. Kondoh for technical advice and discussion. This work was supported by a Grant-In-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (S.T. and S.K).
References
- Adelman K, Wei W, Ardehali MB, Werner J, Zhu B, Reinberg D, Lis JT (2006) Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol Cell Biol 26: 250–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewski M, Sieger D, Alt B, Leve C, Hans S, Wolff C, Rohr KB, Tautz D (2003) Anterior and posterior waves of cyclic her1 gene expression are differentially regulated in the presomitic mesoderm of zebrafish. Development 130: 4269–4278 [DOI] [PubMed] [Google Scholar]
- Holley SA, Takeda H (2002) Catching a wave: the oscillator and wavefront that create the zebrafish somite. Semin Cell Dev Biol 13: 481–488 [DOI] [PubMed] [Google Scholar]
- Holley SA, Geisler R, Nusslein-Volhard C (2000) Control of her1 expression during zebrafish somitogenesis by a delta-dependent oscillator and an independent wave-front activity. Genes Dev 14: 1678–1690 [PMC free article] [PubMed] [Google Scholar]
- Horikawa K, Ishimatsu K, Yoshimoto E, Kondo S, Takeda H (2006) Noise-resistant and synchronized oscillation of the segmentation clock. Nature 441: 719–723 [DOI] [PubMed] [Google Scholar]
- Itoh M et al. (2003) Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell 4: 67–82 [DOI] [PubMed] [Google Scholar]
- Jiang YJ, Aerne BL, Smithers L, Haddon C, Ish-Horowicz D, Lewis J (2000) Notch signalling and the synchronization of the somite segmentation clock. Nature 408: 475–479 [DOI] [PubMed] [Google Scholar]
- Jülich D, Hwee Lim C, Round J, Nicolaije C, Schroeder J, Davies A, Geisler R, Lewis J, Jiang YJ, Holley SA (2005) beamter/deltaC and the role of Notch ligands in the zebrafish somite segmentation, hindbrain neurogenesis and hypochord differentiation. Dev Biol 286: 391–404 [DOI] [PubMed] [Google Scholar]
- Kawamura A, Koshida S, Hijikata H, Sakaguchi T, Kondoh H, Takada S (2005) Zebrafish hairy/enhancer of split protein links FGF signaling to cyclic gene expression in the periodic segmentation of somites. Genes Dev 19: 1156–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan BR, Feldman JL, Lee DH, Koos DS, Ho RK, Stainier DY, Yelon D (2002) The elongation factors Pandora/Spt6 and Foggy/Spt5 promote transcription in the zebrafish embryo. Development 129: 1623–1632 [DOI] [PubMed] [Google Scholar]
- Kishimoto Y, Koshida S, Furutani-Seiki M, Kondoh H (2004) Zebrafish maternal-effect mutations causing cytokinesis defect without affecting mitosis or equatorial vasa deposition. Mech Dev 121: 79–89 [DOI] [PubMed] [Google Scholar]
- Koshida S, Kishimoto Y, Ustumi H, Shimizu T, Furutani-Seiki M, Kondoh H, Takada S (2005) Integrin α5-dependent fibronectin accumulation for maintenance of somite boundaries in zebrafish embryos. Dev Cell 8: 587–598 [DOI] [PubMed] [Google Scholar]
- Lewis J (2003) Autoinhibition with transcriptional delay: a simple mechanism for the zebrafish somitogenesis oscillator. Curr Biol 13: 1398–1408 [DOI] [PubMed] [Google Scholar]
- Lubman OY, Korolev SV, Kopan R (2004) Anchoring notch genetics and biochemistry; structural analysis of the ankyrin domain sheds light on existing data. Mol Cell 13: 619–626 [DOI] [PubMed] [Google Scholar]
- Nikaido M, Tada M, Saji T, Ueno N (1997) Conservation of BMP signaling in zebrafish mesoderm patterning. Mech Dev 61: 75–88 [DOI] [PubMed] [Google Scholar]
- Oates AC, Ho RK (2002) Hairy/E(spl)-related (Her) genes are central components of the segmentation oscillator and display redundancy with the Delta/Notch signaling pathway in the formation of anterior segmental boundaries in the zebrafish. Development 129: 2929–2946 [DOI] [PubMed] [Google Scholar]
- Palmeirim I, Henrique D, Ish-Horowicz D, Pourquié O (1997) Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell 91: 639–648 [DOI] [PubMed] [Google Scholar]
- Penheiter KL, Washburn TM, Porter SE, Hoffman MG, Jaehning JA (2005) A posttranscriptional role for the yeast Paf1–RNA polymerase II complex is revealed by identification of primary targets. Mol Cell 20: 213–223 [DOI] [PubMed] [Google Scholar]
- Pourquié O (2003) The segmentation clock: converting embryonic time into spatial pattern. Science 301: 328–330 [DOI] [PubMed] [Google Scholar]
- Pourquié O (2004) The chick embryo: a leading model in somitogenesis studies. Mech Dev 121: 1069–1079 [DOI] [PubMed] [Google Scholar]
- Sims RJ III, Belotserkovskaya R, Reinberg D (2004) Elongation by RNA polymerase II: the short and long of it. Genes Dev 18: 2437–2468 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Koizumi K, Takagi A, Kitajima S, Inoue T, Koseki H, Saga Y (2000) Mesp2 initiates somite segmentation through the Notch signalling pathway. Nat Genet 25: 390–396 [DOI] [PubMed] [Google Scholar]
- Tenney K, Gerber M, Ilvarsonn A, Schneider J, Gause M, Dorsett D, Eissenberg JC, Shilatifard A (2006) Drosophila Rtf1 functions in histone methylation, gene expression, and Notch signaling. Proc Natl Acad Sci USA 103: 11970–11974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Mandal SS, Pham AD, Zheng Y, Erdjument-Bromage H, Batra SK, Tempst P, Reinberg D (2005) The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev 19: 1668–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information