Abstract
Artemisinin- and artesunate-resistant Plasmodium chabaudi mutants, AS-ART and AS-ATN, were previously selected from chloroquine-resistant clones AS-30CQ and AS-15CQ respectively. Now, a genetic cross between AS-ART and the artemisinin-sensitive clone AJ has been analysed by Linkage Group Selection. A genetic linkage group on chromosome 2 was selected under artemisinin treatment. Within this locus, we identified two different mutations in a gene encoding a deubiquitinating enzyme. A distinct mutation occurred in each of the clones AS-30CQ and AS-ATN, relative to their respective progenitors in the AS lineage. The mutations occurred independently in different clones under drug selection with chloroquine (high concentration) or artesunate. Each mutation maps to a critical residue in a homologous human deubiquitinating protein structure. Although one mutation could theoretically account for the resistance of AS-ATN to artemisinin derivates, the other cannot account solely for the resistance of AS-ART, relative to the responses of its sensitive progenitor AS-30CQ. Two lines of Plasmodium falciparum with decreased susceptibility to artemisinin were also selected. Their drug-response phenotype was not genetically stable. No mutations in the UBP-1 gene encoding the P. falciparum orthologue of the deubiquitinating enzyme were observed. The possible significance of these mutations in parasite responses to chloroquine or artemisinin is discussed.
Introduction
Malaria is estimated to cause the death of over one million people annually, mainly children in Africa. Moreover, despite intensive research, the overall disease burden arising from infections by the malaria parasite Plasmodium falciparum is increasing. One major factor has been the emergence and spread of malaria parasites which are resistant to antimalarial drugs such as chloroquine or pyrimethamine/sulphadoxine. Consequently, many countries have now introduced artemisinin (ART) derivatives as their first-line therapy, in combination with other drugs (such as mefloquine, amodiaquine, piperaquine, pyrimethamine/sulphadoxine or lumefantrine) (World Health Organization, 2006). These artemisinin combination therapies (ACTs) present favourable pharmacokinetics and are thought to reduce the probability of mutations that underlie resistance and treatment failure emerging in parasite populations (White, 1999). Artemisinin has a short half-life but acts extremely quickly in reducing parasite densities and symptoms. The activation, mechanisms of action and targets of artemisinin derivatives have been vigorously investigated and debated (Olliaro et al., 2001; Meshnick, 2002; Krishna et al., 2006). For instance, the activation of the endoperoxide group might produce a carbon-centred radical either after activation by Fe(II) in reduced haem (Meshnick et al., 1991) or as a result of interactions with iron-sulphur proteins in the mitochondrial electron-transport chain (Li et al., 2005). Candidate targets have included haem itself (Robert et al., 2005) and the SERCA-type Ca2+-ATPase (Eckstein-Ludwig et al., 2003). There is also debate regarding the extent to which chloroquine or artemisinin derivatives aggravate the oxidative stress normally incurred by parasites, for example during haemoglobin digestion and subsequent haem processing (oxidation of Fe2+-protoporphyrin IX to Fe3+-protoporphyrin IX in the food vacuole, with a concomitant release of reactive oxygen species) (reviewed in Tilley et al., 2001; Becker et al., 2004).
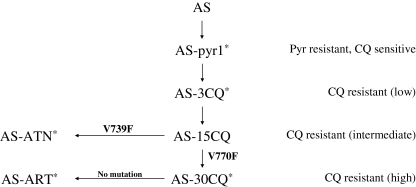
Afonso et al. (2006) recently reported the selection of two artemisinin-resistant mutant clones, AS-ART and AS-ATN (Fig. 1) in the rodent malaria parasite Plasmodium chabaudi. They were derived from chloroquine-resistant clones by serial passage in the presence of increasing but subcurative doses of artemisinin or artesunate (ATN) respectively. After cloning, both showed improved growth in the presence of either ART or ATN, relative to their sensitive progenitors, AS-30CQ and AS-15CQ. Minimum curative doses of artemisinin increased 15-fold in AS-ART and 26-fold in AS-ATN (relative to their sensitive progenitors) while those of artesunate increased five- to sixfold in both clones. These lines exhibited stable drug response phenotypes after cloning, passage in untreated mice and transmission through the mosquito vector.
Fig. 1.
Drug-resistant mutants – the AS lineage. Mutants of P. chabaudi cloned isolate AS have been selected by passage in the presence of pyrimethamine, low, intermediate and high chloroquine concentrations, artesunate and artemisinin, to give lines and clones AS-Pyr (Walliker et al., 1975), AS-3CQ (Rosario, 1976; Carlton et al., 1998), AS-15CQ and AS-30CQ (Padua, 1981), AS-ATN and AS-ART (Afonso et al., 2006) respectively. An asterisk denotes that these clones have been used to generate genetic crosses with the cloned isolate, AJ. The genetic cross investigated in this study used AS-ART and AJ as parental clones. The mutations shown, V739F and V770F, refer to those in UBP-1, described in this study.
What resistance mechanisms are possible and what are their genetic bases? Initial sequence analysis of these clones (and their sensitive progenitors) began with genes for which published evidence suggests a potential involvement in modulating parasite responses to artemisinin derivatives. These genes were the P. chabaudi orthologues of pfatp6, encoding the SERCA-type Ca2+-ATPase (Eckstein-Ludwig et al., 2003; Jambou et al., 2005; Uhlemann et al., 2005), pfcrt (Sidhu et al., 2002), pfmdr1 (Reed et al., 2000; Ferrer-Rodríguez et al., 2004; Price et al., 2004; Sidhu et al., 2005) and pftctp (encoding translationally controlled tumour protein, TCTP) (Bhisutthibhan et al., 1998; Walker et al., 2000). Sequencing of AS-ART and AS-ATN and their progenitors (AS-30CQ and AS-15CQ) showed that there were no mutations or copy number changes in these genes (Afonso et al., 2006).
Plasmodium chabaudi is a robust model system for identifying genetic loci, candidate genes and individual mutations underlying drug resistance (Carlton et al., 2001). Classical genetic studies such as traditional linkage analysis (Carlton et al., 1998; Hayton et al., 2002; Cravo et al., 2003; Hunt et al., 2004a,b) or Linkage Group Selection (LGS) (Culleton et al., 2005), both of which analyse linkage between genotype and phenotype (Carter et al., 2007), have been employed to this end. LGS, used in this current investigation, characterizes the uncloned progeny of a genetic cross (between a resistant and sensitive parasite) by measuring the proportion of parental polymorphic markers at genome-wide loci, before and after drug treatment. Markers from the sensitive parent that are linked to the gene underlying the resistance phenotype are reduced in proportion or intensity after drug treatment, forming a ‘selection valley’. We have previously used amplified fragment length polymorphisms (AFLP) that distinguish the two parental lines in the genetic cross (Culleton et al., 2005; Martinelli et al., 2005a). A genetic linkage map (Martinelli et al., 2005b), a genome sequence database and a complete syntenic map (Kooij et al., 2006) allow us to map genes and markers genetically and physically, thus allowing identification of loci under selection. Mutations in candidate genes within the locus can be identified by comparative sequencing of genes from both the resistant mutant and its sensitive progenitor.
Here we describe the analysis of a genetic cross between AS-ART and AJ and the identification of a selection valley on P. chabaudi chromosome 2 detected by LGS analysis after artemisinin treatment. Subsequently we have identified two independent non-synonymous mutations in a gene encoding a deubiquitinating enzyme within this locus. One mutation appears in AS-ATN derived from AS-15CQ after artesunate selection and the other in AS-30CQ (also derived from AS-15CQ) after chloroquine selection.
Results
Genetic crosses and LGS
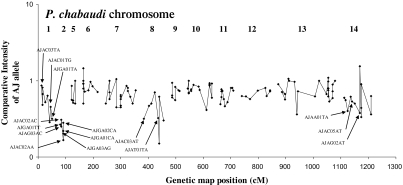
We performed three independent genetic crosses between the artemisinin-resistant clone, AS-ART (Afonso et al., 2006) and the genetically distinct sensitive parasite clone AJ. Uncloned progeny from these crosses were pooled in equal proportions and passaged through two groups of mice. One group was treated with 25 mg artemisinin/kg/day for five consecutive days [day 0 to day 4 post infection (pi)]. The other group was untreated. Parasites were harvested on days 10 pi and 8 pi, respectively, and DNA samples from these groups were analysed by AFLP. The comparative intensities (CIs) for AFLP bands specific to the sensitive parent AJ, and their location within the P. chabaudi genetic linkage map are shown in Fig. 2.
Fig. 2.
LGS – artemisin treatment of AS-ART × AJ genetic cross. The selection on a set of AJ-specific genome-wide AFLP markers (Grech et al., 2002) in the ‘treated’ group relative to the ‘untreated’ group is represented by Comparative Intensity (Martinelli et al., 2004) (logarithmic scale) of AFLP markers (vertical axis) and their position (revised, see Table 1) in a genetic linkage map (horizontal axis), assigned to specific chromosomes. AFLP markers mapped by sequence analysis (Fig. 3) are indicated.
Seven highly linked AFLP markers (mapping to chromosome 2) showed the lowest CIs. CIs of other AFLPs in three other linkage groups (mapping to chromosomes 1, 8 and 14) also appeared to be lower than those shown across the genome as a whole, suggesting possible selection or experimental variation at these loci.
Mapping of AFLP markers
All of the AFLP bands showing reduced CIs had previously been placed on a genetic linkage map (Martinelli et al., 2005b). We confirmed the physical location of these AFLP markers (see Experimental procedures). The data for a number of AFLP markers in the four linkage groups under possible selection are shown in Table 1. Markers AJGA01TA, AJAC01TG and AJAC03TA all mapped to loci on P. falciparum chromosome 6 that are syntenic to P. chabaudi chromosome 1. AJGA01CA, AJAC02AA, AJGA02CA and AJGA01TT mapped to P. falciparum chromosome 1 or 7 at loci syntenic to P. chabaudi chromosome 2. AJAC03AT and AJAT01TA mapped to loci on P. falciparum chromosome 9 that are syntenic to P. chabaudi chromosome 8. Finally, AJAG02AT, AJAC05AT and AJAA01TA mapped to P. falciparum chromosome 13 or 12 at loci that are syntenic with P. chabaudi chromosome 14 (Table 1). Other markers were not mapped, either because of poor sequence quality or because marker sequence did not identify a P. chabaudi contig unambiguously.
Table 1.
Physical and Genetic mapping of AFLP markers with low CI.
| Nearest gene | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Nearest gene | ||||||||||
| AFLP marker | Comparative intensity | P. chabaudi linkage group according to Martinelli et al. (2005b) | Contig ID | Length of contig (nt) | Approximate position of AFLP on contig | Gene | Gene annotation | Gene ID | P. falciparum genomic locus | P. chabaudi chromosome according to synteny Kooij et al. (2006) |
| AJGA01TA | 0.32 | Group 38 | 452.1 | 161 536 | 50 850 | PFF0175c | Hypothetical | 3885788 | pf06–0144 | 1 |
| AJAC01TG | 0.45 | Group 38 | 452.1 | 161 536 | 36 900 | PFF0185c | Hypothetical | 3885790 | pf06–0160 | 1 |
| AJAC03TA | 0.48 | Group 38 | 1290 | 95 398 | 92 500 | PFF0480w | Hypothetical | 3885877 | pf06–0419 | 1 |
| AJGA01CA | 0.20 | Chr 10 | 462 | 60 484 | 59 400 | MAL1P1.71 | Hypothetical | 813167 | pf01–0132 | 2 |
| AJAC02AA | 0.17 | Chr 10 | 462 | 60 484 | 34 300 | MAL7P1.147 | Ubiquitin C-terminal hydrolase | 2655023 | pf07–1029 | 2 |
| AJGA03AG | 0.22 | Chr 10 | 462 | 60 484 | 33 650 | MAL7P1.147 | Ubiquitin C-terminal hydrolase | 2655023 | pf07–1030 | 2 |
| AJGA02CA | 0.28 | Chr 10 | 462 | 60 484 | 21 700 | PF07_0108 | Hypothetical | 2655161 | pf07–1045 | 2 |
| AJAC02AC | 0.31 | Chr 10 | Insufficient sequence for analysis | na | na | |||||
| AJAG03AC | 0.25 | Chr 10 | Insufficient sequence for analysis | na | na | |||||
| AJGA01TT | 0.27 | Chr 10 | 462 | 60 484 | 12 810 | MAL7P1.149 | Hypothetical | 2655024 | pf07–1053 | 2 |
| AJAC03AT | 0.32 | Chr 8 | 936.0 | 61 964 | 13 400 | PFI0460w | Hypothetical | 813372 | pf09–0431 | 8 |
| AJAT01TA | 0.32 | Chr 8 | 1489 | 7 907 | 2 150 | PF1520w | Hypothetical | 813584 | pf09–1261 | 8 |
| AJAG02AT | 0.33 | Group 2 | 408 | 63 353 | 12 000 | PF13–0069 | Translation initiation factor-2, putative | 814047 | pf13–0482 | 14 |
| AJAC05AT | 0.50 | Group 2 | 1214 | 11 563 | 2 950 | PFL0615w | Hypothetical | 811176 | pf12–0549 | 14 |
| AJAA01TA | 0.40 | Group 2 | 882 | 61 223 | 49 600 | PFL0805w | Hypothetical | 811214 | pf12–0665 | 14 |
The AJ-specific AFLP bands that show reduced CIs are shown with a physical location (P. falciparum genomic locus) of homologous positions in P. falciparum (pfxx-yyyy denotes locus yyyy kb along chromosome xx). This locus has been mapped to P. chabaudi chromosomes using detailed syntenic relationships (Kooij et al., 2006). The previously reported linkage groups (Martinelli et al., 2005b) are shown. Those markers previously allocated to ‘linkage group 38’ had not been reported by Martinelli et al. (2005b) because it contained less than eight markers. They have now been assigned to P. falciparum chromosome 1 on the basis of physical mapping described in the text. Similarly, markers previously allocated to ‘group 2’ have now been assigned to chromosome 14. Markers now assigned to chromosome 2 had previously been assigned (incorrectly) to chromosome 10. This assignment was based upon presumed linkage of these AFLP markers to a physically mapped RFLP marker, vacuolar ATPase subunit B (Karcz et al., 1994), for which the inheritance data were only available for nine (out of 28 possible) recombinant clones (Carlton et al., 1998). However, the inheritance data for another RFLP marker, Ca2+-dependent ATPase (Murakami et al., 1990), assigned by PFGE analysis to chromosome 2, were also only available for nine (out of 28 possible) clones (Carlton et al., 1998). Inspection of data showed that the group of AFLP markers, formerly assigned to chromosome 10 could also be linked to the Ca2+-dependent ATPase. Although this may be confirmed by characterization of the inheritance patterns of RFLPs Ca2+-dependent ATPase and vacuolar ATPase, subunit B, the data presented here demonstrates physical linking of the AFLP markers to chromosome 2.
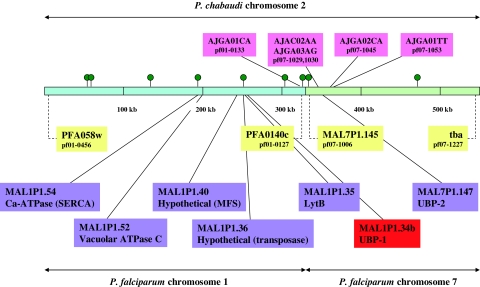
Plasmodium chabaudi chromosome 2 is syntenic with two blocks of the P. falciparum genome, one each on chromosomes 1 and 7 (Kooij et al., 2006) (Fig. 3). These regions define approximately 551 kb of syntenic sequence in P. falciparum, encoding approximately 130 genes.
Fig. 3.
P. chabaudi chromosome 2 map – markers, genes and synteny. P. chabaudi chromosome 2 (blue + green bar; double-headed arrow, top) and its syntenic blocks relative to P. falciparum chromosome 1 (blue) and 7 (green) (double-headed arrows, bottom) are shown. The genes at the extremities of these blocks are shown with their chromosomal loci (pfxx-yyyy, where xx is the chromosome and yyyy the position in kb, yellow). Note that the syntenic block from P. falciparum chromosome 1 is reversed in direction. The position of AFLPs that were physically mapped (blast) and homologous positions in P. falciparum are shown (pink). Genes (not mutated) sequenced during current study are shown (purple). Mutated gene (UBP-1) is shown (red). The scale (100 kb intervals) indicates distances in P. falciparum. The positions of SNP allele quantification assays (pyrosequencing) are shown (green circles); these are, left to right, pf01–0406, pf01–0399, pf01–0323, pf01–0265, pf01–0197, pf01–0150, pf01–0129, pf07–1006 and pf07–1151.
Confirmation of selection by quantitative SNP analysis (pyrosequencing)
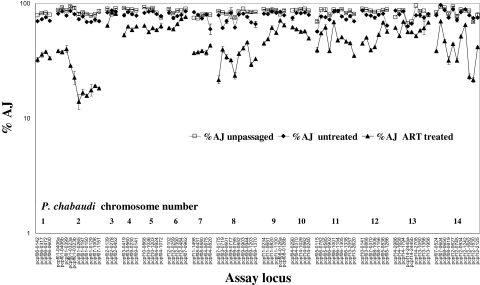
We wished to confirm independently the proportions of AJ alleles of genes in parasite populations after artemisinin treatment. We therefore measured proportions of AS and AJ SNPs at 93 loci distributed evenly across the genome, using pyrosequencing (Cheesman et al., 2007) which can measure the proportions of SNPs in a mixed population with greater precision and accuracy than proportional AFLP. The data (Fig. 4) show that the proportions of genome-wide AJ alleles after artemisinin treatment are generally reduced (relative to the untreated group) as expected, probably because of the loss of parental drug-sensitive AJ parasites during drug treatment. However, there were particularly large and consistent reductions at loci on chromosome 2 and, to a lesser extent, at loci on chromosomes 1, 8 and 14, giving further evidence of selection at these loci. Using pyrosequencing, we also estimated SNP proportions at loci on chromosomes 2, 8 and 14 in DNA derived from the three independent crosses between AS-ART and AJ. We observed reduced AJ proportions on chromosomes 2, 8 and 14, while loci on chromosome 6 (control) showed no selection in artemisinin-selected populations (Supplementary material).
Fig. 4.
LGS – pyrosequencing analysis. The proportions of AJ alleles in the uncloned progeny prior to passage (‘unpassaged’), and after passage with (‘ART treated’) or without (‘untreated’) treatment with artemisinin are shown for 92 loci genome-wide. The loci are named (x-axis) after the positions of the P. falciparum orthologues using the same nomenclature as Table 1 and Fig. 3. Four loci (pf01–0406, pf01–0323, pf08–0126, pf14–2445) have two SNPs in the same assay and therefore give two sets of data, referred to as ‘a’ or ‘b’. Data shown are means, with standard errors indicated (for ‘treated’ and ‘untreated’, 3 < n < 8). Oligonucleotide primers for amplification and sequencing of loci are given in Supplementary material.
Identification of ubp-1 mutation on P. chabaudi chromosome 2
Within P. chabaudi chromosome 2, we sequenced a number of genes (Fig. 3) from AS-ART and AS-ATN to search for mutations that might account for selection of this locus during LGS. One gene, the orthologue of P. falciparum MAL1P1.34b, encodes a putative deubiquitinating protease (DUB), here termed ubiquitin-specific protease-1 (UBP-1). Within this gene, we identified two G to T mutations that are predicted to generate two non-synonymous substitutions in AS-ATN and AS-30CQ, respectively, namely V739F and V770F (Fig. 1). However, there was no mutation in AS-ART relative to AS-30CQ. Mutations that underlie the resistance of AS-ART to artemisinin (and presumably, artesunate), relative to those of AS-30CQ have therefore not been identified because the V770F mutation was already present in the AS-30CQ clone before ART selection. In order to evaluate the significance of the two mutations in the P. chabaudi homologue of MAL1P1.34b, especially regarding their possible role in P. falciparum, we aligned the predicted amino-acid sequences of the orthologous P. falciparum and the P. chabaudi proteins (Fig. 5). Extensive identity between the two sequences occurs close to the N-terminus and in a larger section in the C-terminus of the proteins. Both mutations map to the highly conserved C-terminal part of this DUB, suggesting that the mutations are located in functionally significant parts of the enzyme.
Fig. 5.
Alignment of P. falciparum (MAL1P1.34b) and its P. chabaudi orthologue UBP-1. P. chabaudi AS-30CQ mutated UBP-1 and the P. falciparum homologue MAL1P1.34b alignments are shown. The position of the mutation V770F (and the V739F mutation in AS-ATN) are indicated in rectangular boxes.
Because AS-ART markers on chromosome 2 appear to be positively selected by artemisinin treatment, we also sequenced, from AS-ART, another DUB homologue (orthologous to MAL71P.147) which also lies on the predicted locus of P. chabaudi chromosome 2 (Fig. 3). No mutation was identified in this gene (termed here as ubp-2) in the AS-ART clone. In order to discount the possibility that ubp-1 mutations arose by chance during a burst of mutation activity around this locus (genomic region), we sequenced four other genes nearby, namely the P. chabaudi orthologues of MAL1.P1.35, MAL1P1.36, MAL1P1.40 and MAL1P1.52. None of these showed mutations in their predicted coding sequences or deviation from the P. chabaudi AS genome database entries.
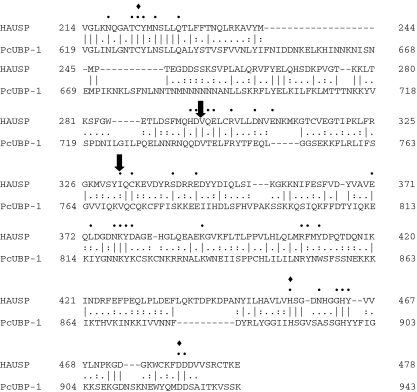
Mapping UBP-1 mutations in a crystal structure
We used the highly conserved predicted C-terminal P. chabaudi amino acid sequence (aa 595–963) of UBP-1 to interrogate a protein structure database by blast search. We identified the core domain of a human homologue (herpes virus-associated ubiquitin-specific protease, HAUSP), for which the crystal structure has been solved (Hu et al., 2002). The alignment between the P. chabaudi UBP-1 and the HAUSP predicted protein sequences is shown (Fig. 6). Twenty-eight per cent of the P. chabaudi UBP-1 amino acids are identical to those of HAUSP, 23% are conservative substitutions and 49% are not conserved. We investigated whether those residues identified as having important structural roles (e.g. catalytic involvement, binding of ubiquitin) were particularly conserved between the two proteins. Out of 35 residues identified as having such roles (Hu et al., 2002) 25 (72%) were identical in the P. chabaudi UBP-1 sequence, five (14%) were conservative substitutions and only five (14%) residues showed non-conservative substitutions (Fig. 6). We therefore concluded that these homologues are likely to share many structural and functional similarities. The V739F mutation (equivalent to HAUSP V296) maps to an end of alpha helix 5 close to the proposed catalytic cysteine, C628 (equivalent to HAUSP C223). The other mutation, V770F (equivalent to HAUSP I332), maps to a hydrophobic pocket required for ubiquitin binding.
Fig. 6.
Alignment of P. chabaudi UBP-1 and human HAUSP. Alignment of human HAUSP (core protein) and P. chabaudi UBP-1. Residues marked ‘•’ are those with presumed catalytic function, role in substrate binding or making intramolecualar contacts which contribute to the structure (Hu et al., 2002). The three residues thought to be involved in catalysis are marked ‘♦’. The positions of mutations V739F and V770F are each indicated.
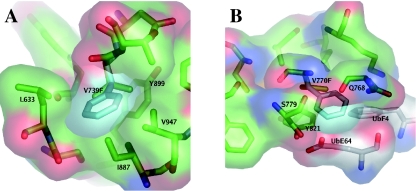
A homology model for the P. chabaudi UBP-1 amino acid sequence was produced using the catalytic core domain of HAUSP as a template. The model was checked manually and side-chain positions adjusted to minimize steric clashes. The wild-type P. chabaudi UBP-1 is likely to form a very similar structure to that found in HAUSP. For both the V739F and V770F mutations the large size increase of the side chain is likely to have a significant effect. In the HAUSP structure the valine 296 (V739F in P. chabaudi) side-chain points into a small hydrophobic pocket principally defined by Y465 and L454. In order for the V739F mutation to be accommodated, some movement of these side chains would be needed (Fig. 7A). It is most likely that the phenylalanine side chain would point into a small pocket produced by V947, I887 and L633. However, the V739F side chain comes too close to these residues to be accommodated without further movement. These movements are likely to affect the catalytic cysteine, C628, found on the same helix as L633. It is also likely that ubiquitin binding will be affected, because Y465 interacts directly with ubiquitin.
Fig. 7. Structural consequences of UBP-1 mutations.
A. Residue environment around V739F. Side chains are shown as sticks with carbons coloured green for the P. chabaudi UBP-1 sequence.
The mutation to F739 in the most likely orientation is represented in stick form with carbons coloured cyan. A surface representation is shown to indicate the tightness of fit around the phenylalanine.
B. Residue environment around V770F. Side chains are shown as sticks with carbons coloured green for the P. chabaudi UBP-1 sequence and white for ubiquitin (UbF4, UbE64). The mutation to F770 in the most likely orientation is represented in stick form with carbons coloured cyan. A surface representation is shown to indicate the tightness of fit around the phenylalanine.
The V770F mutation also involves a large size increase in the side chain. The valine side-chain points into a small hydrophobic pocket that is principally defined by Y821 (UBP-1) and F4 (ubiquitin). The large increase in the size of the V770F mutation results in dramatic protein–protein clashes in side-chain orientations. A very small pocket is formed by Q768, S779 (UBP-1) and E64 (ubiquitin) and is shown in Fig. 7B. Even in this orientation the phenylalanine side chain comes too close to all these residues and a large amount of protein movement would be needed to accommodate it. It is therefore likely that the V770F mutation would disrupt ubiquitin binding in this region.
Phenotypic characterization of ART-resistant P. falciparum lines
ART-resistant parasite lines of the NF54 and 7G8 genetic backgrounds were selected in vitro by exposing parasites to stepwise increases in ART concentrations. Selected drug-resistant lines were designated ART-RNF54 and ART-R7G8. The degree of ART resistance was determined by [3H]-hypoxanthine incorporation assays performed in parallel with their parental control lines. ART IC50 values for ART-RNF54 and ART-R7G8 were 148 and 98 nM, respectively, as compared with IC50 values of 30 and 20 nM for control lines NF54 and 7G8 respectively. These in vitro sensitivity assays showed that selection for resistance to ART resulted in approximately fivefold decrease in susceptibility to this drug in both resistant lines. To confirm the stability of the ART-resistant phenotype, frozen ART-RNF54 and ART-R7G8 parasite lines were thawed and after growing in the absence of drug pressure for 2 weeks, ART IC50 values were retested. These assays revealed ART IC50 values of 33 nM and 25 nM for ART-RNF54 and ART-R7G8 lines, respectively, which were comparable to their respective parental controls. This loss of the ART resistance phenotype in the drug-selected parasites indicates that ART-RNF54 and ART-R7G8 parasite lines acquired a transient drug-induced ART resistance phenotype.
Sequence analysis of the pfcrt, pfmdr1 and pfubp-1 genes
To study the molecular basis of (transient) ART resistance, we polymerase chain reaction (PCR)-amplified and sequenced the pfcrt, pfmdr1 and pfubp-1 genes of ART-resistant and wild-type parasite lines. We selected these genetic determinants based on the observation that mutations in these genes modulate susceptibility to ART and other antimalarials. There were no sequence changes observed at the polymorphic positions of the pfcrt (position 76) and pfmdr1 (positions 86, 1034, 1042 and 1246) genes of ART-RNF54 and ART-R7G8 lines grown in the presence of drug. Sequencing of the complete open reading frame of the pfubp-1 gene of ART-resistant lines also did not reveal any sequence change compared with the parental lines, although two novel SNPs were detected in the 7G8 line at V260A and E265K. These investigations suggest that the transient resistance phenotye of the ART-RNF54 and ART-R7G8 lines was due to reversible modulation of gene expression.
Discussion
We have used LGS to analyse the uncloned progeny of a genetic cross between the artemisinin- (and artesunate-) resistant clone, AS-ART and the sensitive clone AJ, before and after treatment with artemisinin, using AFLP marker intensities and genome-wide pyrosequencing. Using the pooled uncloned progeny from three independent genetic crosses, we obtained a selection valley on chromosome 2, and also evidence of selection on chromosomes 1, 8 and 14. An analysis of the three individual component genetic crosses by pyrosequencing (supplementary data) for loci on chromosomes 2, 6 (control), 8 and 14 confirms possible selection signatures on chromosome 2, 8 and 14. The similarity of our quantitative AFLP and pyrosequencing data confirms that genome-wide pyrosequencing is a useful tool for the de novo mapping of genes underlying selectable phenotypes in LGS-type experiments, as previously suggested (Cheesman et al., 2007).
Because pcatp6 (encoding SERCA Ca2+-ATPase) is located within chromosome 2 (Fig. 3) and because its involvement in artemisinin activation and resistance has been proposed (Jambou et al., 2005; Uhlemann et al., 2005), we wished to investigate whether there were pcatp6 mutations which might affect the activity or expression of this protein. However, there were no mutations in AS-ART coding region, over 4 kb of 5′-UT or over 1 kb of 3′-UT (Afonso et al., 2006).
Instead, within P. chabaudi chromosome 2, we identified two mutations in a gene encoding a DUB enzyme (UBP-1); V739F in the artesunate-resistant clone AS-ATN (relative to its progenitor AS-15CQ), and V770F in the high chloroquine-resistant clone AS-30CQ (relative to AS-15CQ). These two mutations within the same gene occurred independently in two different lines under selection with artesunate and chloroquine respectively. Each mutation produces an identical substitution (V to F), and occurs in one of only two regions that are highly conserved relative to the orthologous P. falciparum gene.
An analysis of the structure of a human homologue (HAUSP) of the mutated P. chabaudi protein, showed that residues underlying structural, intermolecular or catalytic roles in HAUSP are highly conserved in the P. chabaudi DUB. Both mutations increase the size of the side group and map onto residues that are likely to be critical to the molecule's activity.
Importantly, while it is theoretically possible to argue that V739F could confer artesunate/artemisinin resistance relative to AS-15CQ, there is no corresponding mutation to explain the resistance of AS-ART relative to AS-30CQ, because mutation V770F is present in both AS-ART and AS-30CQ.
Some reports have linked protein ubiquitination/deubiquitination to DNA repair function (Huang and D'Andrea, 2006). However, we reject the hypothesis that the mutations reported here may underlie a change in a phenotype known as Accelerated Resistance to Multiple Drugs, alternatively known as a ‘mutator’ phenotype (Rathod et al., 1997). Such mutations may occur during the original selection of drug-resistant mutants (i.e. at some stage during the generation of the drug-resistant AS lineage). However, these pre-existing mutations would not be selected during LGS experiments unless, of course, they are genetically linked to other, as yet, unidentified, mutations that underlie the resistance phenotype.
We have considered other possible interpretations. The first is simply that there is a mutation in a different gene (gene X) on chromosome 2 that arose during artemisinin selection on AS-30CQ during the AS-ART mutant generation. This mutation could be responsible for the selection valley obtained during LGS, and would differentiate AS-ART and AS-30CQ. Whether or not AS-ATN also has a mutation in the same (or similar) gene X relative to AS-15CQ is an open question that can be addressed by analysing a genetic cross between AS-ATN and AJ and by comparative sequencing of AS-ATN. We are currently investigating the presence of other mutations in this linkage group by further comparative sequencing of chromosome 2 genes from the resistant clones and their sensitive progenitors. If gene X is mutated in both AS-ATN and AS-ART, and if this mutation is causing the selection of chromosome 2 in LGS, what would be our interpretation of the UBP mutations reported here? One possibility is that UBP mutations, which are all relative to the parasite AS-15CQ, may compensate for mutations selected during the acquisition of intermediate resistance to CQ in AS-15CQ from AS-3CQ (resistant to low concentrations of chloroquine) (Fig. 1). This would suggest that a mutation in AS-15CQ has compromised the parasite's normal physiological fitness and that subsequent selections with other drugs have selected a compensatory UBP mutation that abrogates the fitness costs of the AS-15CQ mutation.
If, on the other hand, gene X lies on a different chromosome (for example, chromosomes 1, 8 and 14), we must consider why chromosome 2 shows a selection valley. We would suggest that the compensation of mutations underlying intermediate chloroquine resistance in AS-15CQ by mutations in UBP-1 is required for the expression of resistance to artemisinin, i.e. that the phenotypic expression of a mutation in gene X is epistatic to the UBP mutations.
We wish to emphasize one possible implication of this interpretation. The UBP-1 mutations that may fulfil this role occurred during artesunate selection of AS-ATN and during chloroquine selection of AS-30CQ. The fact that similar mutations in the same gene were selected during long-term passage in the presence of two different drugs suggests that there may be some underlying common cellular function that might be compromised during treatment with artemisinin derivatives or chloroquine, such as regulation of oxidative stress in drug-treated parasites (Krungkrai and Yuthavong, 1987; Tilley et al., 2001; Becker et al., 2004). Indeed a DUB enzyme was a primary target of oxidative stress in mutant superoxide dismutase-1 (SOD1) transgenic mice (Poon et al., 2005). Also, the expression of its homologue was modified by oxygen stress in a human tumour cell line; upregulation and downregulation were associated with surviving and apoptotic cells, respectively (Shen et al., 2006).
We also wish to suggest that the basis of artemisinin resistance is likely to be multigenic for the following reasons. Artemisinin resistance in P. chabaudi required significant passage using subcurative doses of artemisinin, and required the use of parasites with previously generated chloroquine resistance. Stable, artemisinin resistance is difficult to obtain with P. falciparum in culture. Selection of genetic crosses with artemisinin is suggesting an important contribution of a gene on chromosome 2, and perhaps, to a lesser extent, an involvement of other genes on, for example, chromosomes 1, 8 and 14.
We have also selected two lines of P. falciparum parasites that show decreased susceptibility to artemisinin but whose phenotype is not stable. These lines do not bear mutations in pfubp-1. In contrast, the P. chabaudi parasites AS-ART and AS-ATN are both genetically stable and selected in vivo. These factors emphasize the value of the P. chabaudi rodent malaria for the generation and genetic characterization of mutations underlying drug resistance. We await with interest a characterization of DUB enzymes in genetically stable artemisinin-resistant P. falciparum mutants.
What effect might UBP-1 mutations have on malarial parasites? We cautiously expect that a partial loss of UBP-1 activity might lead to an increase in degradation (via the proteasome) of proteins that act as UBP-1 substrates. Identification of UBP-1 substrates and UBP-1 allelic exchange transfections of P. falciparum parasites in culture should allow us to address these questions, and are planned. Transfection of mutated ubp-1 genes in P. chabaudi would allow us to evaluate their role in an in vivo system. Up to now, however, successful stable transfection is not an established technique.
Previous studies have considered the ubiquitination/deubiquitination of mdr1 in human cancer cell lines and its effects upon intracellular drug accumulation and responses to drugs. For example Zhang et al. (2004) report that in a human breast cancer cell line, the mdr1 gene product, P-glycoprotein (P-gp), is constitutively ubiquitinated and that increased ubiquitination enhanced the rate of degradation of the mdr1 gene product. Enhanced ubiquitination increased drug accumulation and sensitivity to the drugs doxorubicin and vinblastine, both of which are normally modulated by mdr1 expression. Because the P. falciparum homologue, pfmdr1, has been shown to modulate the responses of malaria parasites to both artemisinin derivatives and to chloroquine, the effect of UBP-1 mutations upon possible post-translational modifications (e.g. ubiquitination or phosphorylation) of the mdr1 gene product may be worthy of further study in P. falciparum.
We acknowledge that we have yet to provide direct evidence of a role for mutations in UBP-1 in artemisinin resistance in the rodent malaria parasite P. chabaudi. Nevertheless, we believe that the data presented here are important for two main reasons. First, they suggest new opportunities for genetic and biochemical investigations into drug resistance in malaria, especially in the vital area of resistance to artemisinin based therapies. Second, we believe that the demonstration of mutations in UBP and its deep involvement in central cellular processes such as responses to oxidative damage may encourage conceptual developments in our understanding of drug resistance, its genetic basis and its evolution.
Experimental procedures
Mice, parasites (cloned isolates, drug-resistant mutants), mosquitoes
Four- to six-week-old female CBA-inbred mice were used for all parasite infections. They had permanent access to 41B mouse maintenance diet (Harlon, UK) and drinking water supplemented with 0.05% p-amino-benzoic acid (pABA).
The Plasmodium chabaudi chabaudi (referred to here as P. chabaudi) parasites investigated here (Fig. 1) include the drug-resistant parasites AS-15CQ, AS-30CQ (Padua, 1981), AS-ART and AS-ATN (Afonso et al., 2006), which are derived from the drug-sensitive cloned isolate AS obtained from thicket rats, Thamnomys rutilans, in the Central African Republic (Landau, 1965; Landau and Chabaud, 1965). AJ is a genetically different drug-sensitive cloned isolate. All parasite clones were available as cryopreserved stabilates, deep-frozen in liquid nitrogen.
Anopheles stephensi mosquitoes were maintained in a dedicated insectary as previously described (Martinelli et al., 2005a).
In vitro selection of artemisinin-resistant parasite lines and antimalarial drug assays
Plasmodium falciparum NF54 (chloroquine-sensitive) and 7G8 (chloroquine-resistant) strains were used for the selection of artemisinin (ART) resistance. Drug resistance selection experiments were started with approximately 5 × 1010 mixed stage parasites, which were exposed to 10 nM of ART for 10 days. Cultures were maintained carefully by feeding two to three times daily with ART-containing complete media. Cultures were smeared every 2–3 days and healthy asexual stage parasites were observed by examining Giemsa-stained thin blood smears. After initial drug exposure, ART drug concentration was increased to 25 nM for both parasite lines and kept at this level for the following 12 days. At this drug concentration dying asexual- and gametocyte-stage parasites were observed by microscopic examination of Giemsa-stained smears. The ART drug selection level was then increased to 50 nM that apparently eliminated asexual stage parasites, and cultures were further maintained at this drug level for following 4 weeks. During the entire selection process 30–40% red blood cells were replaced with freshly washed cells once a week. For both parasite lines, ring stage parasites growing in the presence of ART at 50 nM concentration were observed by day 45. Once the parasitaemias reached 2–3%, frozen stocks of ART-selected parasites were prepared using Glycerolyte 81. Parasites were phenotypically characterized for their ART response profiles using [3H]-hypoxanthine incorporation assays as described previously (Desjardins et al., 1979).
Genetic cross
For each genetic cross, six mosquito cages (approximately 20 cm cubes) were set up each containing approximately 200 female A. stephensi mosquitoes, 5–7 days old. Glucose was removed from cages 24 h before mosquito feeds. Four mice were inoculated with both AJ and AS-ART (1 × 106 parasites of each) and two mice were inoculated with either one of the parental clones. All mice were monitored for the presence of gametocytes. On day 6 post infection, mice with mixed infections or single infections of the parental parasites were each laid on top of a mosquito cage for 30 min. Glucose and water solution were then replaced and non-blood fed mosquitoes removed. After 7–9 days, 10 mosquitoes from each cage were removed and their midguts examined for the presence of oocysts. After 14 days following feeding, the salivary glands were removed from the mosquitoes and placed in 50% Ringer's solution and 50% heat-inactivated calf serum. The glands were gently disrupted using a pestle and mortar to release the sporozoites. This suspension was kept on ice, and injected intraperitoneally into a group of mice in 0.1 ml aliquots.
Drugs and selection of cross-progeny (LGS)
When the sporozoite-induced infections (‘unpassaged’) reached parasitaemias of between 10% and 15%, the parasites were harvested, pooled and inoculated (1 × 107 parasites) into two groups of mice, one treated with artemisinin (‘treated’) and the other left untreated (‘untreated’). Artemisinin was dissolved in dimethylsulphoxide and administered orally at a dose of 25 mg kg−1 of mouse body weight daily at 24 h intervals for 5 days, starting 3 h after parasite challenge. The treated and the untreated blood-stage cross progeny were allowed to grow until parasitaemias reached 30%−40% for untreated, and 15%−20% for treated, when parasites were harvested (days 6 and 12 pi respectively).
Parasite harvesting
Mouse lymphocytes and other nucleated cells were removed from blood by filtration (twice) through 5 ml columns of powdered cellulose (CF11, Sigma) washed with citrate saline. Blood was then further filtered through Plasmodipur™ filters (Euro-Diagnostica) twice. The filtrate was centrifuged for 5 min at 3000 r.p.m. and the supernatant removed, leaving a pellet of packed cells. The pellet was resuspended in two volumes of 0.15% saponin in phosphate-buffered saline (PBS). After lysis of erythrocytes, PBS was added in excess to prevent parasite lysis. This solution was then centrifuged at 4000 r.p.m. for 5 min and washed twice in PBS. Supernatant was discarded and pellets stored at −70°C.
DNA extraction, AFLP amplification, analysis and nomenclature
DNA from ‘unpassaged’‘treated’ and ‘untreated’ groups was extracted, prepared for AFLP analysis, and amplified using ‘non-selective’ and ‘selective’ oligonucleotide primers, as previously described (Grech et al., 2002). Polymorphic markers (between AS-ART and AJ) were named to denote specificity of the polymorphic band, the size of the band (relative to other polymorphic bands in the same gel lane) and the selective bases used. Thus, AJAG02CT denotes the second largest AJ-specific band obtained using EcoRI primers with AG as additional ‘selective’ 3′-nucleotides, and CT as ‘selective’ nucleotides on the MseI primers. Genomic DNA from P. falciparum drug-selected and parental lines was prepared using DNeasy Blood and Tissue Kit (Qiagen).
Determination of CIs of AJ-specific bands
Marker band intensities were measured with PhosphorImager and IMAGEQUANT software (Molecular Dynamics), and the values converted to relative intensity indices (RIIs) which estimate the intensity of a polymorphic marker in a mixture, relative to its intensity in the parental clone (Martinelli et al., 2004). CIs of polymorphic markers are defined as the RII of an AFLP marker in the cross progeny selected in ‘treated’ mice (RIIt), divided by the RII of the marker of the cross progeny grown in a parallel ‘untreated’ group of mice (RIIut), and expressed as a percentage, i.e. CI = (RIIt/RIIut) × 100.
Mapping of AFLP markers
Markers with low CIs were identified and their position on the genetic linkage map (Martinelli et al., 2005b) noted. AFLP bands that appeared to be under selection were sequenced as previously described (Hunt et al., 2004b). The positions of corresponding sequences in the P. falciparum genome were determined as previously described (Hunt et al., 2004b). Briefly, P. chabaudi contig sequences containing highly similar AFLP sequences were identified by blast search. These contigs (usually containing a number of genes) were used to identify loci in P. falciparum containing orthologous genes, by blast search. These loci were expressed in the form pfxx-yyyy, where xx denotes the P. falciparum chromosome and where yyyy denotes the position in kb. The corresponding loci in P. chabaudi were predicted using a genome-wide syntenic map (Kooij et al., 2006).
Sequencing, sequence analysis and SNP identification
Polymerase chain reactions were sequenced using the ABI PRISM Big DyeTM Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems) according to manufacturer's instructions. Sequencing reaction products were purified by precipitation with sodium acetate and 95% ethanol, and washed in 70% ethanol, before analysis on an ABI 3700 sequencer. All fragments were sequenced in both forward and reverse directions. Sequencing results were analysed using SeqED v1.0.3 software (Applied Biosystems, 1992) and CHROMAS 2.31 (Technelysium Pty). To determine the sequence at the pfcrt polymorphic position 76, a 600 bp fragment was amplified by using the primers CF5C and BB84. The 5′ region of the pfmdr1 gene encompassing the polymorphic site 86 was amplified as a 1.0 kb fragment using primers P285 and P423. For sequencing of the pfmdr1 polymorphic positions 1034, 1042 and 1246, a 2.0 kb fragment was PCR-amplified with primers P213 and P215. The full-length 4.0 kb PfUBP-1 gene was PCR-amplified with primers P1595 and P1597 and fully sequenced on both strands using internal primers.
Oligonucleotide primers
Oligonucleotide primers used for amplification and sequencing of candidate genes, and for pyrosequencing analysis are detailed in Supplementary material.
Pyrosequencing
Assays to estimate proportions of SNPs were designed using Assay design software v1.0.6 (Biotage AB, Sweden). This defines two oligonucleotides as PCR primers, one of which is biotinylated, and a sequencing primer close to the SNP. The primers used for each of the assays are given in Supplementary material. Pyrosequencing assays were performed on a Biotage Pyrosequencer HS96A, according to manufacturer's procedures.
Identification of DUB structural homologue and structural analysis
The sequence for P. chabaudi was entered into Phyre (Protein Homology/analogy Recognition Engine) software, which identified the catalytic core domain of HAUSP as a template (pdb id 1NBF) and created an initial protein model. The program Coot (Emsley and Cowtan, 2004) was used to manually examine the resultant model and adjust side chains to minimize steric clashes. The modelled mutations were created using Coot and also manually adjusted to minimize steric clashes. The structural figures were created using the program PyMOL (Delano, 2002).
Acknowledgments
This research was supported by Medical Research Council research Grant G0400476 (71003), the European Community FP5, Project Grant RESMALCHIP (QLK2-CT-2001-01503) and by FCT/MCTES of Portugal (POCTI/BME/46526/2002). AA was supported by Portuguese Foundation for Science and Technology PhD fellowship SFRH/BD/8913/2002. DAF gratefully acknowledges the support of the Burroughs Wellcome Fund for their Investigator of Pathogenesis in Infectious Diseases award, which provided support for his contribution. We thank Kathryn Degnan, Richard Fawcett and Les Steven for technical assistance, Axel Martinelli and Sittiporn Pattaradilokrat for assistance with AFLP, and the University of Edinburgh, School of Biological Science Sequencing Service for excellent sequencing facilities. The assistance of sequence database resources and blast search facilities at the Pathogen Sequencing Unit at the Wellcome Trust Sanger Institute is gratefully acknowledged. We thank African Artemisia for the gift of artemisinin. We also acknowledge the contribution of Taco Kooij (Oxford, UK) for making the rodent/human malaria synteny map available to us, before its publication.
Supplementary material
The following supplementary material is available for this article:
Pyrosequencing data at 14 loci (chromosomes 2, 6, 8 and 14) in the 3 individual crosses (AS-ART × AJ) which were components of the pooled crosses analysed by genome-wide pyrosequencing in the main text of the paper.
Primers for amplifying and sequencing candidate genes in P. chabaudi.
Primers for amplifying and sequencing candidate genes in P. falciparum.
Primers for genome-wide pyrosequencing (amplification and sequencing).
This material is available as part of the online article from:
http://www.blackwell-synergy.com/doi/abs/10.1111/j.1365-2958.2007.05753.x
(This link will take you to the article abstract).
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Afonso A, Hunt P, Cheesman S, Alves CA, Cunha CV, do Rosario V, Cravo P. Malaria parasites can develop stable resistance to artemisinin but lack mutations in candidate genes atp6 (encoding the sarcoplasmic and endoplasmic reticulum Ca2+-ATPase), tctp, mdr1, and cg10. Antimicrob Agents Chemother. 2006;50:480–489. doi: 10.1128/AAC.50.2.480-489.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K, Tilley L, Vennerstrom JL, Roberts D, Rogerson S, Ginsburg H. Oxidative stress in malaria parasite-infected erythrocytes: host–parasite interactions. Int J Parasitol. 2004;34:163–189. doi: 10.1016/j.ijpara.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Bhisutthibhan J, Pan XQ, Hossler PA, Walker DJ, Yowel CA, Carlton J, et al. The Plasmodium falciparum translationally controlled tumor protein homolog and its reaction with the antimalarial drug artemisinin. J Biol Chem. 1998;273:16192–16198. doi: 10.1074/jbc.273.26.16192. [DOI] [PubMed] [Google Scholar]
- Carlton J, Mackinnon M, Walliker D. A chloroquine-resistance locus in the rodent malaria parasite Plasmodium chabaudi. Mol Biochem Parasitol. 1998;93:57–72. doi: 10.1016/s0166-6851(98)00021-8. [DOI] [PubMed] [Google Scholar]
- Carlton J, Hayton K, Cravo P, Walliker D. of mice and malaria mutants: unravelling the genetics of drug resistance using rodent malaria models. Trends Parasitol. 2001;17:236–242. doi: 10.1016/s1471-4922(01)01899-2. [DOI] [PubMed] [Google Scholar]
- Carter R, Hunt P, Cheesman S. Linkage Group Selection – a fast approach to the genetic analysis of malaria parasites. Int J Parasitol. 2007;37:285–293. doi: 10.1016/j.ijpara.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Cheesman S, Creasey A, Degnan K, Afonso A, Cravo P, Hunt P. Validation of Pyrosequencing for accurate and high throughput estimation of allele frequencies in malaria parasites. Mol Biochem Parasitol. 2007;152:213–219. doi: 10.1016/j.molbiopara.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Cravo PVL, Carlton JM-R, Hunt P, Bisoni L, Padua RA, Walliker D. Genetics of mefloquine resistance in the rodent malaria Plasmodium chabaudi. Antimicrob Agents Chemother. 2003;47:709–718. doi: 10.1128/AAC.47.2.709-718.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culleton R, Martinelli A, Hunt P, Carter R. Linkage group selection: rapid gene discovery in malaria parasites. Genome Res. 2005;15:92–97. doi: 10.1101/gr.2866205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delano WL. 2002. The PyMOL molecular graphics system. [WWW document]. URL http://pymd.sourceforge.net.
- Desjardins RE, Canfield CJ, Haynes JD, Chulay JD. Quantitative assessment of antimalarial activity in vitro by a semiautomated microdilution technique. Antimicrob Agents Chemother. 1979;16:710–718. doi: 10.1128/aac.16.6.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckstein-Ludwig U, Webb RJ, Van Goethem ID, East JM, Lee AG, Kimura M, et al. Artemisinins target the SERCA of Plasmodium falciparum. Nature. 2003;424:957–961. doi: 10.1038/nature01813. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Ferrer-Rodríguez I, Pérez-Rosado J, Gervais GW, Peters W, Robinson BL, Serrano AE. Plasmodium yoelii: identification and partial characterization of an mdr1 gene in an artemisinin-resistant line. J Parasitol. 2004;90:152–160. doi: 10.1645/GE-3225. [DOI] [PubMed] [Google Scholar]
- Grech K, Martinelli A, Pathirana S, Walliker D, Hunt P, Carter R. Numerous, robust genetic markers for Plasmodium chabaudi by the method of amplified fragment length polymorphism. Mol Biochem Parasitol. 2002;123:95–104. doi: 10.1016/s0166-6851(02)00142-1. [DOI] [PubMed] [Google Scholar]
- Hayton K, Ranford-Cartwright L, Walliker D. Sulfadoxine-pyrimethamine resistance in the rodent malaria parasite Plasmodium chabaudi. Antimicrob Agents Chemother. 2002;46:2482–2489. doi: 10.1128/AAC.46.8.2482-2489.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Li P, Li M, Li W, Yao T, Wu J-W, et al. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- Huang T, D'Andrea A. Regulation of DNA repair by ubiquitylation. Nat Rev Mol Cell Biol. 2006;7:323–334. doi: 10.1038/nrm1908. [DOI] [PubMed] [Google Scholar]
- Hunt P, Cravo P, Donleavy P, Carlton J, Walliker D. Chloroquine resistance in Plasmodium chabaudi: are chloroquine-resistance transporter (crt) and multi-drug resistance (mdr1) orthologues involved? Mol Biochem Parasitol. 2004a;133:27–35. doi: 10.1016/j.molbiopara.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Hunt P, Martinelli A, Fawcett R, Carlton J, Carter R, Walliker D. Gene synteny and chloroquine resistance in Plasmodium chabaudi. Mol Biochem Parasitol. 2004b;136:157–164. doi: 10.1016/j.molbiopara.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Jambou R, Legrand E, Niang M, Khim N, Lim P, Volney B, et al. Resistance of Plasmodium falciparum field isolates to in-vitro artemether and point mutations of the SERCA-type PfATPase6. Lancet. 2005;366:1960–1963. doi: 10.1016/S0140-6736(05)67787-2. [DOI] [PubMed] [Google Scholar]
- Karcz S, Hermann V, Trottein F, Cowman A. Cloning and characterisation of the vacuolar ATPase B subunit from Plasmodium falciparum. Mol Biochem Parasitol. 1994;65:123–133. doi: 10.1016/0166-6851(94)90121-x. [DOI] [PubMed] [Google Scholar]
- Kooij TWA, Carlton JM, Bidwell SL, Hall N, Ramesar J, Janse CJ, Waters AP. A Plasmodium whole-genome synteny map: indels and synteny breakpoints as foci for species-specific genes. PLoS Pathog. 2006;1:e44. doi: 10.1371/journal.ppat.0010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna S, Woodrow CJ, Staines HM, Haynes RK, Mercereau-Puijalon O. Re-evaluation of how artemisinins work in light of emerging evidence of in vitro resistance. Trends Mol Med. 2006;12:200–205. doi: 10.1016/j.molmed.2006.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krungkrai S, Yuthavong Y. The antimalarial action on Plasmodium falciparum of qinghaosu and artesunate in combination with agents which modulate oxidant stress. Trans R Soc Trop Med Hyg. 1987;81:710–714. doi: 10.1016/0035-9203(87)90003-4. [DOI] [PubMed] [Google Scholar]
- Landau I. Description of Plasmodium chabaudi, parasite of african rodents. C R Acad Sci Hebd Seances Acad Sci D. 1965;260:3758–3761. [PubMed] [Google Scholar]
- Landau I, Chabaud AG. Natural infection by 2 plasmodia of the rodent Thamnomys rutilans in the Central African Republic. C R Acad Sci Hebd Seances Acad Sci D. 1965;261:230–232. [PubMed] [Google Scholar]
- Li W, Mo W, Shen D, Sun L, Wang J, Lu S, et al. Yeast model uncovers dual roles of mitochondria in the action of artemisinin. PLoS Genet. 2005;1:e36. doi: 10.1371/journal.pgen.0010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli A, Hunt P, Cheesman S, Carter R. Amplified fragment length polymorphism measures proportions of malaria parasites carrying specific alleles in complex genetic mixtures. Mol Biochem Parasitol. 2004;136:117–122. doi: 10.1016/j.molbiopara.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Martinelli A, Cheesman S, Hunt P, Culleton R, Raza A, Mackinnon M, Carter R. A genetic approach to the de novo identification of targets of strain-specific immunity in malaria parasites. Proc Natl Acad Sci USA. 2005a;102:814–819. doi: 10.1073/pnas.0405097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli A, Hunt P, Fawcett R, Cravo P, Walliker D, Carter R. An AFLP-based Genetic Linkage Map of Plasmodium chabaudi chabaudi. Malar J. 2005b;4:11–20. doi: 10.1186/1475-2875-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshnick SR. Artemisinin: mechanisms of action, resistance and toxicity. Int J Parasitol. 2002;32:1655–1660. doi: 10.1016/s0020-7519(02)00194-7. [DOI] [PubMed] [Google Scholar]
- Meshnick SR, Thomas A, Ranz A, Xu CM, Pan HZ. Artemisinin (qinghaosu): the role of intracellular hemin in its mechanism of antimalarial action. Mol Biochem Parasitol. 1991;49:181–189. doi: 10.1016/0166-6851(91)90062-b. [DOI] [PubMed] [Google Scholar]
- Murakami K, Tanabe K, Takada S. Structure of a Plasmodium yoelii gene-encoded protein homologous to the Ca2+-ATPase of rabbit skeletal muscle sarcoplasmic reticulum. J Cell Sci. 1990;97:487–495. doi: 10.1242/jcs.97.3.487. [DOI] [PubMed] [Google Scholar]
- Olliaro PL, Haynes RK, Meunier B, Yuthavong Y. Possible modes of action of the artemisinin-type compounds. Trends Parasitol. 2001;17:122–126. doi: 10.1016/s1471-4922(00)01838-9. [DOI] [PubMed] [Google Scholar]
- Padua RA. Plasmodium chabaudi: genetics of resistance to chloroquine. Exp Parasitol. 1981;52:419–426. doi: 10.1016/0014-4894(81)90101-6. [DOI] [PubMed] [Google Scholar]
- Poon H, Hensley K, Thongboonkerd V, Merchant M, Lynn B, Pierce W, et al. Redox proteomics analysis of oxidatively modified proteins in G93A-SOD1 transgenic mice – a model of familial amylotrophic lateral sclerosis. Free Rad Biol Med. 2005;39:453–462. doi: 10.1016/j.freeradbiomed.2005.03.030. [DOI] [PubMed] [Google Scholar]
- Price RN, Uhlemann A-C, Brockman A, McGready R, Ashley E, Phaipun L, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathod PK, McErlean T, Lee P-C. Variations in frequencies of drug resistance in Plasmodium falciparum. Proc Natl Acad Sci USA. 1997;94:9389–9393. doi: 10.1073/pnas.94.17.9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403:906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- Robert A, Benoit-Vical F, Claparols C, Meunier B. The antimalarial drug artemisinin alkylates heme in infected mice. Proc Natl Acad Sci USA. 2005;102:13676–13680. doi: 10.1073/pnas.0500972102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario VE. Genetics of chloroquine-resistance in malaria parasites. Nature. 1976;261:585–586. doi: 10.1038/261585a0. [DOI] [PubMed] [Google Scholar]
- Shen H, Sikorska M, LeBlanc J, Walker P, Liu Q. Oxidative stress regulated expression of ubiquitin carboxyl-terminal hydrolase-L1: role in cell survival. Apoptosis. 2006;11:1049–1059. doi: 10.1007/s10495-006-6303-8. [DOI] [PubMed] [Google Scholar]
- Sidhu AB, Verdier-Pinard D, Fidock DA. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science. 2002;298:210–213. doi: 10.1126/science.1074045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu AB, Valderramos SG, Fidock DA. Pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol Microbiol. 2005;57:913–926. doi: 10.1111/j.1365-2958.2005.04729.x. [DOI] [PubMed] [Google Scholar]
- Tilley L, Loria P, Foley M. Chloroquine and other quinoline antimalarials. In: Totowa RPJ, editor. Antimalarial Chemotherapy: mechanisms of action, resistance and new directions in drug discovery. Totowa, NJ: Humana Press; 2001. pp. 87–122. [Google Scholar]
- Uhlemann A-C, Cameron A, Eckstein-Ludwig U, Fischbarg J, Iserovich P, Zuniga FA, et al. A single amino acid residue can determine the sensitivity of SERCAs to artemisinins. Nat Struct Mol Biol. 2005;12:628–629. doi: 10.1038/nsmb947. [DOI] [PubMed] [Google Scholar]
- Walliker D, Carter R, Sanderson A. Genetic studies on Plasmodium chabaudi: recombination between enzyme markers. Parasitology. 1975;66:309–320. doi: 10.1017/s0031182000048824. [DOI] [PubMed] [Google Scholar]
- Walker DJ, Pitsch JL, Peng MM, Robinson BL, Peters W, Bhisutthibhan J, Meshnick SR. Mechanisms of artemisinin resistance in the rodent malaria pathogen Plasmodium yoelii. Antimicrob Agents Chemother. 2000;44:344–347. doi: 10.1128/aac.44.2.344-347.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White N. Antimalarial drug resistance and combination chemotherapy. Philos Trans R Soc Lond B Biol Sci. 1999;354:739–749. doi: 10.1098/rstb.1999.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO) Guidelines for the Treatment of Malaria. Geneva: WHO; 2006. [Google Scholar]
- Zhang Z, Wu J-Y, Hait W, Yang J-M. Regulation of the stability of P-glycoprotein by ubiquitination. Mol Pharmacol. 2004;66:395–403. doi: 10.1124/mol.104.001966. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pyrosequencing data at 14 loci (chromosomes 2, 6, 8 and 14) in the 3 individual crosses (AS-ART × AJ) which were components of the pooled crosses analysed by genome-wide pyrosequencing in the main text of the paper.
Primers for amplifying and sequencing candidate genes in P. chabaudi.
Primers for amplifying and sequencing candidate genes in P. falciparum.
Primers for genome-wide pyrosequencing (amplification and sequencing).