Abstract
Haemophilus influenzae transits between niches within the human host that are predicted to differ in oxygen levels. The ArcAB two-component signal transduction system controls gene expression in response to respiratory conditions of growth and has been implicated in bacterial pathogenesis, yet the mechanism is not understood. We undertook a genome-scale study to identify genes of the H. influenzae ArcA regulon. Deletion of arcA resulted in increased anaerobic expression of genes of the respiratory chain and of H. influenzae's partial tricarboxylic acid cycle, and decreased anaerobic expression levels of genes of polyamine metabolism, and iron sequestration. Deletion of arcA also conferred a susceptibility to transient exposure to hydrogen peroxide that was greater following anaerobic growth than after aerobic growth. Array data revealed that the dps gene, not previously assigned to the ArcA modulon in bacteria, exhibited decreased expression in the arcA mutant. Deletion of dps resulted in hydrogen peroxide sensitivity and complementation restored resistance, providing insight into the previously uncharacterized mechanism of arcA-mediated H2O2 resistance. The results indicate a role for H. influenzae arcA and dps in pre-emptive defence against transitions from growth in low oxygen environments to aerobic exposure to hydrogen peroxide, an antibacterial oxidant produced by phagocytes during infection.
Introduction
Haemophilus influenzae has no identified natural niche outside of the human host where it primarily colonizes the nasopharyngeal mucosa. It can disseminate to other anatomical sites making it a common cause of otitis media, upper and lower respiratory tract infections, septicaemia and meningitis in children (Klein, 1997; Moxon and Murphy, 2000). H. influenzae also frequently colonizes the respiratory tract of individuals with chronic obstructive pulmonary disease (Sethi and Murphy, 2001; Murphy and Sethi, 2002; Murphy et al., 2004) and cystic fibrosis (Gilligan, 1991; Moller et al., 1995). The incidence of H. influenzae meningitis has dramatically declined in populations immunized with an effective vaccine against the type b capsular polysaccharide [Centers for Disease Control and Prevetion (CDC), 2002], a major factor promoting bloodstream survival by strains of this serotype. However, the vaccine has not affected the incidence of infection with non-typeable strains (NTHi), which lack the capsule. Although NTHi predominantly cause respiratory tract infections and otitis media, they have been isolated from patients with invasive disease such as meningitis in rare cases, raising the possibility that genes conferring varying degrees of bloodstream persistence could be distributed among NTHi strains (Nizet et al., 1996; Cuthill et al., 1999; O'Neill et al., 2003; Erwin et al., 2005).
We postulate that modulation of gene expression in response to environmental conditions is required by H. influenzae to express the repertoire of genes needed for survival during pathogenesis. H. influenzae likely encounters varying oxygen levels in diverse environments in the host such as growth in biofilm structures on mucosal surfaces or after invasion into the bloodstream. Signal transduction in response to varied oxygen levels represents a mechanism by which H. influenzae could co-ordinate gene expression profiles needed for efficient colonization and pathogenesis in different environments encountered within the host. In Escherichia coli, a two-component signal transduction system designated ArcAB (for anoxic redox control) responds to respiratory conditions of growth to modulate expression of genes/operons of the tricarboxylic acid (TCA) cycle and genes involved in other aspects of respiratory or fermentative metabolism (Lynch and Lin, 1996a). The ArcB sensor kinase autophosphorylates and transfers a phosphoryl group to ArcA, a DNA binding protein that can act as either repressor or activator depending on the configuration of the target promoter (Lynch and Lin, 1996a,b). The ArcAB system is most active under low oxygen conditions and least active under high oxygen conditions. Recent evidence indicates that this response is likely not via direct sensing of oxygen but that ArcB senses the oxidation or reduction (redox) status of the membrane-bound quinones, central electron carriers of respiration (Georgellis et al., 2001a; Malpica et al., 2004). The ArcAB system of H. influenzae possesses similar biochemical and regulatory functions to those of its counterpart in E. coli in modulating gene expression in response to redox conditions of growth. Of note, expression of H. influenzae ArcB in an E. coli arcB mutant can restore the response of at least two ArcAB-regulated promoters, sdh (succinate dehydrogenase) and lldP (l-lactate permease), to respiratory conditions of growth (Manukhov et al., 2000; Georgellis et al., 2001b). Several genes or proteins that are repressed by ArcA in E. coli have been identified as ArcA-regulated in H. influenzae, including lldD (l-lactate dehydrogenase) and certain subunits of formate dehydrogenase and fumarate reductase (Georgellis et al., 2001b; De Souza-Hart et al., 2003). ArcA is a global regulator in E. coli, however, the extent of the ArcA regulon of H. influenzae is unknown.
ArcA has been implicated in pathogenesis as arcA mutants of both H. influenzae and Vibrio cholerae, a diarrhoeal pathogen, exhibit reduced lethality compared with wild type in mouse mortality studies (De Souza-Hart et al., 2003; Sengupta et al., 2003). Despite extensive information concerning ArcA-mediated control of genes of respiratory pathways and enzymes of the TCA cycle in E. coli, the mechanism by which this gene regulation could alter virulence in H. influenzae is not well understood. ArcA mutants of H. influenzae type b were more sensitive than wild type to killing by human serum, however, ArcA-regulated genes encoding cell-surface structures as potential targets of humoral immune components in serum, such as complement, have yet to be identified. V. cholerae ArcA influences production of cholera toxin which is essential for virulence, yet H. influenzae produces no exotoxins implicated in pathogenesis. In Salmonella enterica serovar Enteritidis, ArcA has been implicated in resistance to reactive oxygen and nitrogen intermediates (ROI/RNI) (Lu et al., 2002). A role in oxidative stress resistance for a regulator such as ArcA, which is active under low oxygen conditions, appears to be paradoxical, and the mechanism and role of ArcA-regulated genes in this resistance profile has not been determined.
In the current study, we have extended our analysis of the H. influenzae ArcAB system to understand mechanisms by which this signalling system can influence H. influenzae pathogenesis. We analysed the global expression profile of the H. influenzae arcA mutant grown under anaerobic conditions to identify genes comprising the ArcA regulon in this organism. By microarray analysis, we identified a set of genes whose expression pattern was influenced by the arcA mutation and restored by complementation. Northern hybridizations confirmed ArcA-mediated control of all of the genes that were evaluated by this method. In addition to detecting genes encoding respiratory metabolic enzymes known to be ArcA-regulated in E. coli, this analysis identified and validated ArcA-dependent modulation of genes not previously recognized to be within the ArcA regulon. One of these genes is a putative homologue of Dps proteins in other species that participate in oxidative stress resistance yet have not been previously linked to ArcA-mediated phenotypes. Thus, the microarray results gave us insight into physiological characteristics of the H. influenzae arcA mutant that can account for its oxidative stress sensitivity. Mutational analysis of ArcA controlled genes including dps provided insight into the mechanism of ArcA-mediated resistance to hydrogen peroxide, and provides support for a model in which ArcA promotes survival of cells shifted from low oxygen conditions to oxidative stress exposure, a transition H. influenzae is likely to experience in the host.
Results
Global expression profiling of the H. influenzae arcA mutant
To investigate the role of the ArcAB signal transduction system in H. influenzae pathogenesis, the ArcA regulon in H. influenzae Rd was investigated by DNA microarray analysis. Genomic expression profiles were measured in four independent samples each of the parent strain, RdV, the arcA deletion mutant, RAA6V (ΔarcA), and the complemented strain, RAA6C (ΔarcA, arcA+), grown anaerobically. The complete set of data from these experiments is provided in the web supplement (Table S1 and S2). Of the 1697 H. influenzae protein coding genes represented on the array, expression of 19 genes was increased by greater than or equal to twofold (P-value ≤ 0.0001) in the ΔarcA mutant, RAA6V, compared with its parent, RdV (Table 1, RAA6V/RdV column). Expression of these genes was restored to levels similar to that of RdV in the complemented strain, RAA6C (Table 1, RAA6V/RAA6C column) providing a high degree of confidence in this set of candidate ArcA modulated genes. Six genes that showed increased expression in the ΔarcA mutant include genes encoding putative homologues of E. coli dehydrogenases of the respiratory chain (fdxH, fdxI, ndh and lldD) or the TCA cycle (sucA and sucB). The ΔarcA mutant also exhibited increased expression of a putative homologue of lipA, which is involved in the biosynthesis of lipoate, a cofactor used by several enzyme complexes involved in oxidative metabolism such as pyruvate dehydrogenase and α-ketoglutarate dehydrogenase (Vanden Boom et al., 1991). Two other genes whose expression was negatively controlled by ArcA are similar to E. coli genes, lldP and fdhE, that encode lactate permease and a protein involved in assembly of formate dehydrogenase, respectively (Dong et al., 1993; Abaibou et al., 1995; Nunez et al., 2002). Expression levels of five genes that were decreased in the ΔarcA mutant compared with its parent, RdV, were restored in the complemented strain, generating a list of ‘high confidence’ candidate arcA activated genes (Table 2). One of the five genes was arcA itself as expected because the gene was deleted; three are genomically linked in organization, HI0590 (potE), HI0591 (speF) and HI0592. The fifth gene, HI1349, encodes a predicted protein with sequence similarity to Dps proteins that function in protection against oxidative stress (Almiron et al., 1992; Ilari et al., 2000; Pulliainen et al., 2005).
Table 1.
Genes increased in expression in the H. influenzae arcA deletion mutant compared with parent strain.
| RAA6V/RdV | RAA6V/RAA6C | ||||
|---|---|---|---|---|---|
| Gene ID | Function | Fold change | P-value | Fold change | P-value |
| HI1218 | l-lactate permease (lldP) | 44 | 1.20E-08 | 26.8 | 1.69E-08 |
| HI0747 | NADH dehydrogenase (ndh) | 11 | 4.06E-11 | 9.6 | 5.86E-11 |
| HI0009 | FdhE protein (fdhE) | 9.7 | 8.39E-10 | 9.4 | 1.53E-09 |
| HI0008 | Formate dehydrogenase, gamma subunit (fdxI) | 9.7 | 5.42E-09 | 12 | 3.81E-09 |
| HI0007 | Formate dehydrogenase, beta subunit (fdxH) | 7.8 | 1.12E-09 | 9.2 | 7.77E-10 |
| HI1731 | Conserved hypothetical protein | 5.9 | 2.52E-09 | 6.2 | 4.67E-09 |
| HI1444 | 5,10 methylenetetrahydrofolate reductase (metF) | 5.6 | 4.91E-06 | 4.1 | 2.57E-05 |
| HI0608 | Conserved hypothetical protein | 5.5 | 3.65E-10 | 8.3 | 1.81E-10 |
| HI1727 | Argininosuccinate synthetase (argG) | 4.8 | 5.02E-07 | 4.8 | 3.56E-07 |
| HI1728 | Conserved hypothetical protein | 3.5 | 1.54E-06 | 3.1 | 2.58E-05 |
| HI1739.1 | l-lactate dehydrogenase (lldD) | 3.2 | 1.79E-10 | 4.5 | 1.07E-11 |
| HI1730 | Conserved hypothetical protein | 3.0 | 1.48E-06 | 7.0 | 3.32E-08 |
| HI1661 | 2-oxoglutarate dehydrogenase E2 component (sucB) | 2.8 | 5.33E-07 | 3.0 | 1.14E-06 |
| HI0026 | Lipoate biosynthesis protein A (lipA) | 2.6 | 4.08E-07 | 2.2 | 9.63E-06 |
| HI1662 | 2-oxoglutarate dehydrogenase E1 component (sucA) | 2.6 | 6.71E-07 | 3.5 | 1.07E-07 |
| HI0018 | Uracil DNA glycosylase (ung) | 2.5 | 1.01E-05 | 2.2 | 5.68E-05 |
| HI0890 | Dephospho-CoA kinase (coaE) | 2.5 | 4.83E-05 | 2.4 | 2.91E-04 |
| HI0889 | Serine hydroxymethyltransferase (serine methylase) (glyA) | 2.4 | 4.11E-06 | 2.5 | 6.13E-06 |
| HI0174 | Conserved hypothetical protein | 2.3 | 1.75E-05 | 2.1 | 1.92E-04 |
List contains genes whose expression levels were increased in the ΔarcA mutant, RAA6V compared with the parent strain, RdV (column RAA6V/RdV) and were restored close to parental levels in the complemented strain, RAA6C (column RAA6V/RAA6C). Fold differences are ≥ 2.0 with P ≤ 0.0001.
Table 2.
Genes decreased in expression in the H. influenzae arcA deletion mutant compared with parent strain.
| RAA6V/RdV | RAA6V/RAA6C | ||||
|---|---|---|---|---|---|
| Gene ID | Function | Fold change | P-value | Fold change | P-value |
| HI0884 | Aerobic respiration control protein ArcA (arcA) | −654.8 | 2.72E-12 | −776.7 | 9.02E-10 |
| HI0592 | Conserved hypothetical protein | −4.4 | 4.62E-08 | −3.8 | 6.36E-05 |
| HI0591 | Ornithine decarboxylase (speF) | −4.0 | 2.54E-08 | −2.7 | 2.49E-05 |
| HI1349 | Conserved hypothetical protein, similar to dps protein family | −3.4 | 1.43E-07 | −3.0 | 5.97E-06 |
| HI0590 | Putrescine-ornithine antiporter (potE) | −2.5 | 3.06E-08 | −2.8 | 5.27E-07 |
List contains genes whose expression levels were decreased in the ΔarcA mutant, RAA6V compared with the parent strain, RdV (column RAA6V/RdV) and were restored in the complemented strain, RAA6C (column RAA6V/RAA6C). Fold differences are ≥ 2.0 with P ≤ 0.0001.
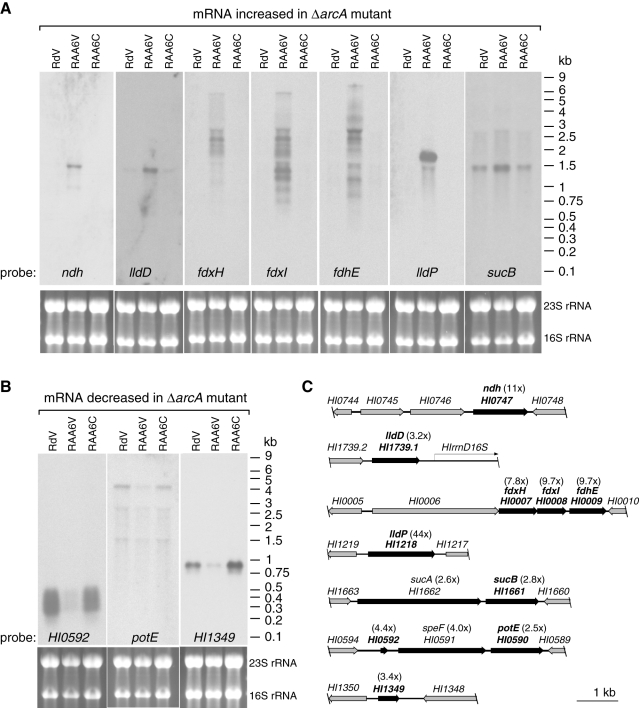
To verify the microarray results, we performed Northern blot hybridizations with genes whose expression as detected on the arrays either increased in the ΔarcA mutant (ndh, lldD, fdxH, fdxI, fdhE, lldP and sucB) or decreased in the ΔarcA mutant (HI0592, potE and HI1349) (Tables 1 and 2; Fig. 1). Northern blots containing RNA from the parent strain, RdV, the ΔarcA mutant, RAA6V, and the complemented strain, RAA6C, grown anaerobically were analysed with probes specific to these genes (Experimental procedures). Levels of each of the ndh, lldD, fdxH, fdxI, fdhE, lldP and sucB specific transcripts were higher in the ΔarcA mutant compared with its parent and complementation restored negative control of these genes (Fig. 1A and C). Levels of the HI0592, potE and HI1349 specific transcripts were decreased in the ΔarcA mutant compared with its parent and transcript abundance was restored with complementation (Fig. 1B). Transcript sizes for each of the loci appear to be generally consistent with the gene lengths annotated by The Institute for Genomic Research (TIGR) (Fig. 1C). The mRNAs of multiple sizes that were detected with the fdxH, fdxI and fdhE specific probes are likely to represent polycistronic transcripts with the largest ∼3 kb transcript potentially spanning fdxH, fdxI and fdhE. The large mRNA species (∼4 kb) detected with the potE probe is likely to represent a polycistronic transcript spanning speF-potE or HI0592-speF-potE. Based on our subsequent results implicating HI1349 in ArcA-mediated phenotypes (see below), this gene's expression pattern was additionally verified by reverse transcription quantitative polymerase chain reaction (RT-qPCR) and found to be decreased by 4.6-fold in the ΔarcA mutant compared with its parent and by 2.7-fold in comparison to the complemented strain. These results are in agreement with the microarray data and confirm ArcA-dependent modulation of these genes in H. influenzae.
Fig. 1.
Differential transcript abundance in the H. influenzae arcA mutant versus parental strain. Northern blots containing 6 μg of total RNA from anaerobically grown Rd parental strain, RdV, arcA deletion mutant, RAA6V (ΔarcA) and complemented strain, RAA6C hybridized with probes corresponding to genes increased (A) or decreased (B) in expression in the ΔarcA mutant (Tables 1 and 2 respectively). Ethidium bromide stained gel is directly below each blot. 16S and 23S are the ribosomal RNAs.
C. Schematic representation of the genomic organization of genes modulated in the ΔarcA mutant (black open reading frames) of which the genes in bold were evaluated on Northern blots in panels A and B. Numbers in parentheses near each gene represent the fold change in differential gene expression in the ΔarcA mutant versus parent comparison (Tables 1 and 2).
Sensitivity of the H. influenzae arcA mutant to oxidative stress
The microarray data suggested that the H. influenzaeΔarcA mutant might exhibit sensitivity to oxidative stress, a phenotype of relevance to the role of arcA in bacterial survival in the host. Our expression analysis detected putative homologues of eight genes encoding subunits or assembly factors of respiratory chain dehydrogenases (fdxH, fdxI, fdxE, ndh and lldD), a dehydrogenase substrate transporter (lldP), or TCA cycle enzymes (sucA and sucB) that were increased in expression in the H. influenzaeΔarcA mutant. An increase in respiratory activity mediated by these genes in the H. influenzaeΔarcA mutant could produce elevated levels of ROI during a transition from anaerobiosis to an oxygenated microenvironment (see Discussion).
Conversely, the candidate ArcA activated genes likely promote resistance to oxidative stress. Two genes whose expression levels were lower in the arcA mutant have potential roles, based on sequence similarity to their E. coli counterparts, in the biosynthesis of the polyamine putrescine (speF), or transport of putrescine (potE) (Table 2). The putative H. influenzae SpeF and PotE show 65% and 77% amino acid identity to E. coli SpeF and PotE respectively. Polyamines are present in all organisms and are associated with a variety of vital biological processes such as replication, transcription and cell growth (Pegg, 1988; Tabor and Tabor, 1985). Addition of exogenous polyamines can protect polyamine deficient E. coli from the toxicity of the reactive oxygen species H2O2 (Tkachenko et al., 2001; Chattopadhyay et al., 2003; Jung and Kim, 2003). Moreover, HI1349, encoding a Dps-like protein, was also downregulated in the ΔarcA mutant (Table 2). HI1349 has approximately 18% identity and 20% similarity to E. coli Dps and contains a conserved iron-binding motif found in Dps homologues (see Discussion). Members of the ferritin-like Dps protein family function in iron storage/detoxification (Chiancone et al., 2004). Dps was first discovered in E. coli as a DNA-binding protein that protects DNA from hydrogen peroxide-mediated oxidative damage (Almiron et al., 1992). Nearly half (11/23) of the detected ArcA controlled genes in H. influenzae have potential roles in ROI generation or resistance to oxidative stress. Therefore, in addition to its apparent role in optimizing metabolic flux under anaerobic conditions, it is possible that ArcA plays a role in pre-emptive protection against exposure of anaerobically grown cells to oxidants. Potentially consistent with this hypothesis, the ArcA of S. enterica serovar Enteritidis has been implicated in resistance to reactive oxygen and nitrogen intermediates by an unknown mechanism (Lu et al., 2002).
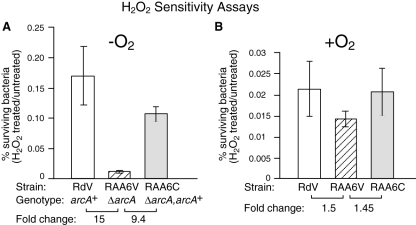
To determine if arcA has a role in protection of H. influenzae during a transition from anaerobic growth to aerobic oxidative stress conditions, we tested the effect of transient H2O2 exposure on the viability of the H. influenzaeΔarcA mutant grown under anaerobic versus aerobic conditions. We could detect no appreciable differences in growth rates of the ΔarcA mutant, the parental strain and the complemented strain under these conditions alone (Experimental procedures). Anaerobic or aerobic cultures of the parent strain, RdV, the ΔarcA mutant and the complemented strain were incubated aerobically for 10 min in the absence and presence of 0.5 mM H2O2 followed by quenching of the H2O2 with sodium pyruvate before plating to enumerate survivors (Fig. 2). Consistent with greater activity of ArcA under low oxygen conditions, anaerobically grown ΔarcA mutant exhibited an approximate 15-fold decrease in the number of survivors compared with its parent after H2O2 treatment and complementation restored the survival phenotype to near the parental level (Fig. 2A). Similar fold differences in survival were obtained with exposure of anaerobic cultures to 0.25 mM H2O2 (10.5 ± 0.9% survival of the parental strain, 0.5 ± 0.3% survival of the ΔarcA mutant, and 10.1 ± 1.1% survival of the complemented strain). Growth in an anaerobic chamber prior to H2O2 exposure at either concentration yielded similar fold differences in survival compared with the sealed tube condition (data not shown). In contrast, the aerobically grown ΔarcA mutant showed only a 1.5-fold decrease relative to the parental strain in the number of survivors after exposure to H2O2 and complementation restored survival similar to the parental level (Fig. 2B).
Fig. 2.
Sensitivity of the H. influenzae arcA mutant to hydrogen peroxide. The parental strain, RdV, arcA deletion mutant, RAA6V (ΔarcA) and complemented strain, RAA6C (ΔarcA, arcA+) from anaerobically (A) and aerobically (B) grown cultures were treated with 0.5 mM H2O2 for 10 min (Experimental procedures). The survival ratios are plotted as the percentage of cfu obtained from H2O2 treated/untreated samples. Values represent the mean of three independent cultures of each strain tested, and the error bars represent the standard deviations. Statistically significant differences between the arcA mutant and parent (P < 0.01) and between the arcA mutant and complemented strain (P < 0.05) were observed (one-way anova with Bonferroni's multiple comparison test) for cultures grown anaerobically. The arcA mutant exhibits a 15-fold and 1.5-fold increase in sensitivity to H2O2 challenge compared with parent when grown anaerobically and aerobically respectively.
To evaluate whether the speF, potE and HI0592 genes participate in the H2O2 sensitivity phenotype as observed in the arcA mutant, we created a strain, RputV, which contains a deletion of all three loci. Anaerobic growth of RputV, followed by challenge with 0.5 mM H2O2 resulted in a slight reduction (∼14%) in the number of survivors in the mutant compared with the parental strain that did not reach statistical significance (data not shown).
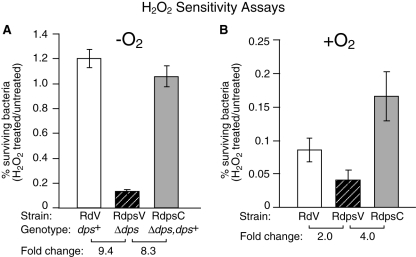
Next, we examined the role of the putative dps homologue, HI1349 in resistance to H2O2. We observed that deletion of this locus renders the mutant, RdpsV, more sensitive to H2O2 challenge compared with the parent when grown anaerobically, as it showed approximately ninefold reduction in the number of survivors (Fig. 3A). In contrast, H2O2 treatment of the aerobically grown RdpsV resulted in only a twofold decrease in the number of survivors compared with the parent, RdV (Fig. 3B). Under either condition, complementation restored the survival phenotypes of the mutant to levels at or above those of the parental strain. Based on sequence and motif similarity to putative homologues in other species, together with its role in H2O2 resistance demonstrated here, HI1349 is referred to as dps in this report.
Fig. 3.
Sensitivity of the H. influenzae HI1349 (dps) mutant to hydrogen peroxide. H2O2 sensitivity assays with the parental strain, RdV, dps deletion mutant, RdpsV (Δdps) and complemented strain, RdpsC (Δdps, dps+), from anaerobically (A) and aerobically (B) grown cultures were performed as described in Fig. 2. Statistical differences: parent versus dps mutant (P < 0.001 for anaerobic growth, P > 0.05 for aerobic growth); complemented strain versus dps mutant (P < 0.001 for anaerobic and aerobic growth) (one-way anova with Bonferroni's multiple comparison test). The dps mutant exhibits a 9.4-fold and twofold increase in sensitivity to H2O2 challenge compared with parent when grown anaerobically and aerobically respectively.
These results indicate that the H. influenzae arcA mutant has an increased sensitivity to H2O2 following anaerobic growth. In addition, dps, a gene identified as a target of ArcA control in these experiments, plays a role in resistance to H2O2 hypersensitivity providing evidence for a mechanism by which ArcA mediates protection from ROI in this species.
Effect of arcA on survival of H. influenzae in a murine model of bacteraemia
Intravascular colonization is a well-established virulence trait of H. influenzae. Bacteraemia is primarily associated with encapsulated strains, which are of declining clinical significance in countries with adequate Hib vaccination programs. Nevertheless, recent evidence indicates that a subset of NTHi strains, which are non-encapsulated, also has the capacity to infect the bloodstream (Nizet et al., 1996; Cuthill et al., 1999; O'Neill et al., 2003). Bloodstream colonization by NTHi is less efficient and persistent than is observed with encapsulated strains, yet it is possible that a limited ability to infect the mammalian bloodstream and resist immunological clearance is a general feature of H. influenzae that varies quantitatively between diverse isolates. For these studies we used a nonencapsulated Rd strain clonally related to KW20, whose genome sequence has been available for many years and has provided a reference point for genetic analysis of H. influenzae (Fleischmann et al., 1995). Rd strains are efficiently transformable, unlike typical NTHi clinical isolates. Despite exhibiting less virulence than encapsulated strains, Rd derivatives generate transient infections in animal models of infection and have been useful for characterization of pathogenic properties of H. influenzae (Weiser et al., 1995; Daines et al., 2003). The recently determined genome sequence of NTHi strain 86–028NP contains putative homologues of ∼97% of Rd genes indicating that genes implicated in infection related phenotypes with Rd are potentially present in other H. influenzae strains, although some unique genes present in clinical NTHi isolates are absent from Rd (Erwin et al., 2005; Harrison et al., 2005).
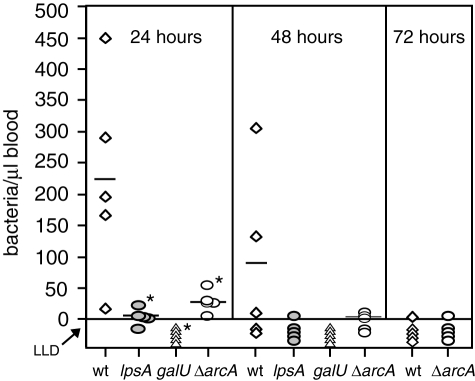
As expected for non-encapsulated H. influenzae, Rd produced a transient infection in mice. Bacteria were recovered at an average density of 224 colony-forming units (cfu) μl−1 of blood at 24 h after intraperitoneal (IP) inoculation, began to decline by 48 h, and were fully cleared by 72 h post inoculation (Fig. 4). The ability of this model to detect H. influenzae virulence properties is demonstrated by comparison of the Rd strain and isogenic mutants deficient in lpsA or galU, lipooligosaccharide (LOS) biosynthesis genes essential for bloodstream colonization by H. influenzae type b in an infant rat model (Hood et al., 1996). The lpsA gene is the glycosyltransferase required for glucosyl addition and extension of the third heptosyl residue of the LOS inner core, and galU encodes the UDP-glucose pyrophosphorylase required for production of the UDP-glucose precursor essential for all hexose additions to the LOS. No viable bacteria were recovered at any time point from mice infected with the galU mutant. Likewise, mutation of lpsA yielded a similar level of attenuation to that of the galU mutation. Therefore, this model is capable of detecting predicted phenotypes associated with the classical virulence factors of H. influenzae.
Fig. 4.
Persistence defect of H. influenzae mutants in a mouse model of bacteraemia. H. influenzae Rd wild-type (wt), Rcp5 (lpsA) and Rcp19 (galU), and arcA deletion mutant (ΔarcA) were inoculated IP into five C57BL/6 mice per strain and blood was sampled daily for cfu determination (Experimental procedures). Bars represent the average cfu μl−1 (224 and 28 for wt and ΔarcA mutant, respectively, at 24 h post inoculation). Asterisks denote differences from wild type that were statistically significant (P < 0.01). lower limit of detection (LLD) = 2 cfu μl−1.
If signalling in response to redox conditions is important for adaptation to niches encountered by H. influenzae during infection, then arcA may be expected to play a role in some aspect of survival or persistence in the host. A mutant containing a non-polar deletion of the arcA gene was detected at an average density of 28 cfu μl−1 of blood at 24 h post inoculation. This represents an approximately eightfold reduction in cfu relative to the parent strain at 24 h post inoculation (Fig. 4). At 48 h, the ΔarcA mutant remains attenuated, although the difference at this later time did not reach statistical significance. Consistent with these results, De Souza-Hart et al. have shown that an arcA mutation confers an increase in LD50 with H. influenzae type b after IP inoculation of BALB/c mice (De Souza-Hart et al., 2003). This virulence defect was attributed to the increased serum sensitivity of the arcA mutant that was observed. We evaluated the Rd ΔarcA mutant versus parental strain for serum sensitivity after anaerobic growth. Both strains were sensitive to treatment with human serum with an approximate IC50 of 2% (Experimental procedures) and did not differ detectably in serum sensitivity in three independent experiments: survival ratio in 2% serum: Rd (parent) = 0.57, standard deviation (SD) = 0.1; RAA6 ( ΔarcA) = 0.56, SD = 0.04. It therefore seems likely that the role of ArcA in serum resistance differs between strains. Nevertheless, the more rapid clearance of the arcA mutant from the blood raises the possibility that an impaired ability to maintain high numbers of bacteria in the blood contributes to the previously observed reduction in lethality of the arcA mutant versus the parental type b strain. Furthermore, decreased ability of the mutant to survive in the mammalian bloodstream suggests that ArcA is active in controlling genes during a critical stage of pathogenesis.
Because deletion of the ArcA-regulated dps gene resulted in an increased sensitivity to H2O2 (Fig. 3), we evaluated whether this gene could contribute to H. influenzae's ability to persist in the host bloodstream. We coinoculated 6-week-old C57BL/6 mice by the IP route at a dose of 2 × 108 cfu of H. influenzae Rd carrying the lacZ gene (strain RdlacZ) and an equal number of either the parent (strain RdV), Δdps mutant (RdpsV), or complemented dps strain (RdpsC). After 24 h, the ratios and standard deviations of the coinoculated strains recovered from the blood were not significantly different: RdV/RdlacZ = 1.32, SD = 0.73, n = 4; RdpsV/RdlacZ = 2.27, SD = 0.74, n = 4; RdpsC/RdlacZ = 0.97, SD = 0.22, n = 5. The results indicate that the Δdps mutant does not appear to exhibit a survival defect compared with the parental strain in the bloodstream at 24 h post inoculation. The H. influenzae ArcA regulates multiple genes as indicated by expression profiling and it is likely that the combined effect of more than one gene expressed at inappropriately high or low levels in the ArcA mutant confers the in vivo defect.
Discussion
Overview
We report the role of H. influenzae ArcA in resistance to hydrogen peroxide during transitions between redox conditions of growth and enhanced survival in a murine model of bloodstream clearance. Application of DNA microarray-based expression profiling led to the identification of an ArcA controlled gene, dps which encodes a putative homologue of the ferritin-like Dps protein and has not previously recognized to be within bacterial ArcA regulons. The H. influenzae dps gene was implicated in ArcA-mediated resistance to hydrogen peroxide, providing evidence for a molecular mechanism of ArcA-mediated ROI resistance. It will be of interest to determine whether ArcA controls genes fulfilling similar functions in other species, or whether ArcA-mediated regulation of dps is unique to this inhabitant of the human respiratory tract.
Of the 1697 protein coding genes represented on the microarray, we found 23 (∼1.3% of the 1740 genes in the genome) that were differentially expressed in the arcA mutant when compared with its parent strain and complemented arcA strain (Tables 1 and 2). The H. influenzae arcA mutant exhibited increased anaerobic expression of genes of the respiratory chain and TCA cycle, and decreased anaerobic expression levels of genes with putative functions in polyamine biosynthesis/transport and oxidative stress resistance. In this mutant, a potential increase in cellular respiration could lead to an increase in intracellular generation of ROI, whereas a concurrent decrease in the uptake/biosynthesis of polyamines and in the levels of the putative Dps protein could lower resistance to ROI.
This model based on the expression data suggested a mechanism by which the H. influenzae ArcA could promote oxidative stress resistance. We found that mutants containing non-polar deletions of either the H. influenzae arcA or HI1349 (a putative dps homologue), a gene shown in this study to be positively regulated by arcA, possess enhanced sensitivity to H2O2 following a shift from growth in low oxygen to a transient aerobic exposure to this oxidant. We postulate that as H. influenzae transits from low to high oxygen environments within the host, ArcAB protects the cell against oxidative stress, contributing to the ability of H. influenzae Rd to resist clearance in a mouse model of intravascular infection as we report here.
Resistance to oxidative stress is a major feature of bacterial pathogens that must survive encounters with defences of the host innate immune system (Nathan and Shiloh, 2000). The importance of this trait in H. influenzae is supported by the observation that the sodA gene encoding superoxide dismutase, which was not detected by our microarray analysis as an ArcA-regulated gene in H. influenzae, is required for oxidative stress defence in vitro and for optimal nasopharyngeal colonization in infant rats (D'Mello et al., 1997). Our results suggest that co-ordinate regulation of additional oxidative stress resistance pathways could represent a mechanism by which ArcA promotes survival of H. influenzae during bloodstream infection.
ArcA regulon diversity
Compared with the relatively small number of ArcA targets in H. influenzae identified in the microarray data, global expression profiling experiments conducted with E. coli arcA mutants have indicated that transcript levels of 372 genes (Liu and De Wulf, 2004) or as many as 1139 genes (Salmon et al., 2005) (∼9% and ∼26%, respectively, of the E. coli genome) are controlled directly or indirectly by ArcA anaerobically, while 110 genes were found to be ArcA-regulated under an aerobic growth condition (Oshima et al., 2002). Use of an ArcA-P recognition weight matrix from footprinting data for 10 known ArcA-regulated genes identified approximately 50 additional E. coli operons as probable direct targets of ArcA (Liu and De Wulf, 2004). The difference in the number of ArcA targets between H. influenzae and E. coli is likely related, in part, to their difference in genome size (the E. coli genome is ∼2.3-fold larger) and host environment. H. influenzae inhabits a more restricted environment (primarily the human nasopharynx) compared with E. coli. In addition, some ArcA-regulated promoters in E. coli are subject to complex combinatorial control by other regulators such as FNR, which is considered a direct sensor of oxygen and is active under a more restricted range of low oxygen conditions than ArcA (Lynch and Lin, 1996a; Kiley and Beinert, 1998). It is likely that under diverse environmental conditions, different subsets of ArcA-regulated genes may be detected as a result of the presence or absence of contributions from other regulatory systems.
Of the 23 ArcA modulated genes on our list (Tables 1 and 2, columns RAA6V/RdV), all but HI0592 have putative homologues in E. coli with 10 showing agreement in anaerobic expression ratios in arcA versus parental strains in the two species suggesting similarities and differences in their respective arcA regulons (Liu and De Wulf, 2004; Salmon et al., 2005) (Supplementary material, Table S3). These results are consistent with regulon diversity among putative ArcA homologues in other species. Of note, Gralnick and coworkers (Gralnick et al., 2005) applied the E. coli ArcA-P recognition weight matrix to the genome of Shewanella oneidensis and experimentally validated several candidates detected by this method. In S. oneidensis, the dmsEFAB genes, encoding dimethyl sulphoxide reductase, were positively controlled by ArcA and the cydAB genes, encoding cytochrome d oxidase, were negatively controlled. In contrast, dms genes in E. coli are not known to be ArcA-regulated and cydAB of E. coli is ArcA activated (Lynch and Lin, 1996b). Each of these three bacterial species inhabits distinctly different environments in nature and it is likely that their respective ArcA regulons are each uniquely adapted to growth and survival in these settings.
Negative control of genes of respiration and the TCA cycle by arcA
The H. influenzae arcA mutant exhibits increased expression relative to the parental strain of a set of genes (lldD, lldP, fdxH, fdxI, fdhE, sucA, sucB and ndh) encoding putative respiratory dehydrogenases, substrate transporters for these dehydrogenases, or enzymes involved in dehydrogenase assembly. These enzymes include α-ketoglutarate dehydrogenase, formate dehydrogenase and l-lactate dehydrogenase (LldD). Consistent with these results, LldD and a subunit of formate dehydrogenase (FdxG) have been detected as ArcA-regulated proteins in type b H. influenzae (De Souza-Hart et al., 2003). Substrates for these enzymes are created endogenously or available in the host. For example, l-lactate is produced at significant levels in the host with persistent levels in the blood ranging between 0 and 1.5 mM in resting, healthy individuals and levels as high as 4 mM in sepsis (Levraut et al., 1998). Expression levels of these enzymes correlate with their activity levels in E. coli (Iuchi and Lin, 1988), and this expression pattern is likely to signify increased cellular respiration in the mutant.
The observed anaerobic increase in the expression of a number of genes involved in energy metabolism in the H. influenzae arcA mutant is consistent with the reported role of ArcA in E. coli as a negative regulator of genes of the respiratory pathway and TCA cycle (Lynch and Lin, 1996a; Patschkowski et al., 2000; Oshima et al., 2002; Liu and De Wulf, 2004; Salmon et al., 2005). In addition, we detected ArcA regulation of ndh. The E. coli ndh encodes a non-proton-translocating NADH dehydrogenase (Matsushita et al., 1987). A second NADH dehydrogenase in E. coli, encoded by the nuo operon, is coupled to the generation of proton motive force (Weidner et al., 1993) and has no apparent homologue in H. influenzae. Although the nuo operon is repressed by ArcA in E. coli (Bongaerts et al., 1995), regulation of the E. coli ndh gene involves the oxygen-responsive regulator, FNR (Green and Guest, 1994; Meng et al., 1997). To our knowledge, transcriptional regulation of ndh by ArcA as detected in this study has not been reported previously. Together, these results demonstrate increased expression of multiple genes of cellular respiration in the ArcA mutant.
In the presence of oxygen, respiration generates endogenous ROI. E. coli arcA mutants exhibit increased rates of respiration (Nystrom et al., 1996; Vemuri et al., 2006). Potentially consistent with these results, a S. enterica serovar Enteritidis arcA mutant has been shown to be more susceptible to H2O2 compared with wild type (Lu et al., 2002). It is likely that increased levels of respiration are generated in the ArcA mutant, contributing to its hydrogen peroxide sensitivity.
Positive control of oxidative stress resistance genes by ArcA
Deletion of arcA resulted in decreased expression of a set of genes with probable roles in oxidative stress resistance including dps (HI1349), encoding a putative homologue of ferritin-like iron storage proteins in other bacteria, and a putative operon containing genes similar to those mediating polyamine biosynthesis and transport in other organisms. A non-polar deletion of dps resulted in H2O2 sensitivity similar to that of the ArcA mutant under equivalent conditions, consistent with an important role for ArcA-mediated activation of this gene in H2O2 resistance.
The dps gene is conserved in all of the sequenced isolates of H. influenzae (Fleischmann et al., 1995; Harrison et al., 2005). Dps proteins bind iron and oxidize Fe(II) with H2O2 to form a stable ferric oxide mineral core within the cavity of the protein, thereby avoiding generation of toxic hydroxyl radicals mediated by Fenton chemistry (Zhao et al., 2002). Since the discovery that Dps in E. coli plays a role in protection against DNA damage from oxidative stress mediated by H2O2, homologues have been found to confer resistance to H2O2 in diverse bacteria (Chen and Helmann, 1995; Martinez and Kolter, 1997; Cooksley et al., 2003; Ishikawa et al., 2003; Ueshima et al., 2003; Halsey et al., 2004; Loprasert et al., 2004; Olsen et al., 2005; Pulliainen et al., 2005). HI1349 has approximately 18% identity and 20% similarity to E. coli Dps. Furthermore, HI1349 contains a conserved amino acid motif comprised of residues His38, His50, Glu54, Asp65 and Glu69 that are located at equivalent positions in the Listera innocua Dps and were implicated as sites for iron-binding based on structural analysis (Ilari et al., 2000). The Glu54 residue in the putative H. influenzae Dps represents a conserved variation of Asp at this equivalent position in other Dps proteins. Functional evidence of iron-binding capabilities of the corresponding residues has been obtained with the Streptococcus suis Dps protein (Pulliainen et al., 2005).
Our results indicate that the H. influenzae dps-like protein also functions in conferring resistance to H2O2, with the protection against transient exposure to H2O2 being more pronounced in anaerobically grown cells and lower in aerobically grown cells. The dps genes have been shown to be regulated by OxyR in the presence of exogenous H2O2 in multiple species including E. coli (Zheng et al., 2001), Bacteroides (Rocha et al., 2000), Archaea (Wiedenheft et al., 2005) and OxyR-dependant regulation of the dps-like gene of H. influenzae was recently observed (Harrison et al., 2006). In addition, dps mRNA and protein levels in Borrelia burgdorferi were higher in cultures grown microaerobically versus anaerobically, indicating redox regulation of this gene in this species (Seshu et al., 2004). To our knowledge, a role for ArcA in regulation of dps has not been demonstrated previously in any species. The role we have detected for ArcA in anaerobic expression of the putative H. influenzae Dps homologue, and the H2O2 sensitivity of the dps mutant versus the parental strain after anaerobic growth, provide evidence that a Dps protein can operate under a physiological condition not previously recognized to induce its upregulation. Because the H. influenzae dps-like gene was more critical for resistance to H2O2 in cells that were grown anaerobically versus aerobically prior to oxidant exposure, it is likely that cells grown under aerobic conditions rely more heavily on other systems including catalase, superoxide dismutase and peroxiredoxin-glutaredoxin (Bishai et al., 1994; D'Mello et al., 1997; Vergauwen et al., 2003), for resistance subsequent to exposure to H2O2. It is also likely that aerobic dps transcription, activated by OxyR in response to H2O2 (Harrison et al., 2006), and subsequent translation of sufficient amounts of Dps protein to afford protection require more time to confer protection than the transient exposure period used here. If cells are able to survive the initial exposure to H2O2, then Dps could accumulate, and we would predict that it could contribute to the multiple mechanisms of resistance to H2O2 present in H. influenzae growing aerobically.
In the ΔarcA mutant, we also observed decreased expression relative to the parent of a locus similar to the speF-potE operon of E. coli which encodes an inducible ornithine decarboxylase (speF), responsible for conversion of ornithine to putrescine, and the putrescine transport protein (potE) (Kashiwagi et al., 1991). Polyamines, such as putrescine and spermidine, have roles in a wide variety of biological processes and their optimal cellular concentrations are maintained by biosynthesis, degradation and transport (Igarashi and Kashiwagi, 1999). Polyamines have also been implicated in resistance to oxidative stress in E. coli and other organisms (Tkachenko et al., 2001; Chattopadhyay et al., 2003; Jung and Kim, 2003). The cellular amines putrescine, cadaverine and 1,3-diaminopropane are present in H. influenzae as measured by HPLC (Hamana and Nakata, 2000).
Downregulation of speF and potE in the H. influenzaeΔarcA mutant could disrupt the optimal balance of cellular polyamine levels, thereby contributing to oxidant sensitivity. However, a mutant containing a deletion of speF, potE and HI0592 did not exhibit an appreciable increase in sensitivity to H2O2 exposure under the conditions tested. This result could potentially be attributed to the presence in H. influenzae of possible alternative pathways for production/uptake of polyamines. HI0949 and HI0946.1 have been shown to express the enzymatic activities required for 1,3-diaminopropane production (Ikai and Yamamoto, 1998). H. influenzae potABCD genes, whose E. coli counterparts function as a spermidine (preferential) and putrescine uptake system (Furuchi et al., 1991) could provide a similar capability to H. influenzae.
ArcA, oxidative stress resistance and pathogenesis
Growth of H. influenzae within the human host likely requires adaptation to diverse conditions as the bacteria accumulate in biofilms on mucosal surfaces, invade the epithelium within or between cells, enter the bloodstream, or encounter oxidative defences of phagocytic cells. Decreased survival of the arcA mutant in the mouse model suggests that ArcA-mediated regulation is required during infection. Our results implicate ArcA in repression of genes of respiratory catabolism and activation of an oxidative stress resistance gene, dps, in addition to genes of polyamine metabolism. These regulatory effects of ArcA could protect cells growing under relatively low oxygen levels, a condition they likely encounter in venous blood or in biofilms on mucosal surfaces, against sudden oxidative stresses such as exposure to the oxidative defences of phagocytic cells. Consistent with this hypothesis, both arcA and dps exerted greater effects on hydrogen peroxide resistance in cells grown anaerobically than in cells grown aerobically prior to exposure. A pre-emptive role of certain oxidative stress resistance mechanisms has been proposed previously, such as anaerobic expression of an iron-dependant superoxide dismutase of E. coli (Kargalioglu and Imlay, 1994), but this aspect of resistance has received relatively little attention compared to defence mechanisms induced upon exposure to oxidants. Our results indicating arcA-dependant anaerobic activation of dps expression and the role of these genes in hydrogen peroxide resistance in H. influenzae demonstrate an additional mechanism by which bacteria can prepare for rapid transitions from low oxygen conditions to exposure to oxidative stress, conditions they likely encounter within the mammalian host.
Experimental procedures
Media and H. influenzae growth conditions
The non-encapsulated Rd derivative of H. influenzae type d (BA042), termed Rd in this report (Akerley et al., 2002) was grown at 35°C ± 1.5°C in Brain Heart Infusion supplemented with 10 μg ml−1 nicotinamide adenine dinucleotide and 10 μg ml−1 haemin (sBHI) on agar plates or in sBHI broth cultures shaken at 250 r.p.m. as indicated. DNA was transformed into naturally competent H. influenzae prepared as previously described (Barcak et al., 1991). Kanamycin (Km), gentamicin (Gm) and tetracycline (Tet) were added to sBHI at 20 μg ml−1, 10 μg ml−1 and 8 μg ml−1 respectively. H. influenzae cultures grown in unaerated culture containers filled and sealed to exclude air to approximate anaerobiosis were termed ‘anaerobic’ for simplicity. H. influenzae is a haem auxotroph and requires haem for aerobic growth, yet grows to equivalent levels in these sealed containers in the presence and absence of exogenous haem, suggesting oxygen levels are very low or absent in sealed tubes (data not shown). Aerobically grown H. influenzae are cultured in 10 ml of sBHI in 500 ml Erlenmeyer flasks. The generation times for parent strain (RdV), arcA deletion mutant (RAA6V) and the complemented strain (RAA6C) were 29 ± 3 min (n = 3), 31 ± 1.6 min (n = 3), and 31 ± 5 min (n = 3), respectively, for aerobic growth; 67 ± 3 min (n = 3), 71 ± 8 min (n = 3), and 69 ± 3 min (n = 3), respectively, for anaerobic growth.
Plasmid and H. influenzae strain construction
Standard molecular biology methods were used for plasmid construction and Northern blot analysis (Ausubel et al., 1995). Strain RdV was generated by transforming Rd with vector pXT10 and selection with tetracycline for homologous recombination at the xyl locus (Wong and Akerley, 2003). Strain RAA6 contains a non-polar, in-frame deletion of the arcA protein coding sequences (Georgellis et al., 2001b). Vector pXT10 was introduced into RAA6 at the xyl locus to generate RAA6V. RAA6C contains the deletion of arcA with a wild-type copy of arcA introduced in trans at the xyl locus as described previously (Georgellis et al., 2001b). Strains Rcp5 and Rcp19 containing transposon insertion mutations in lpsA and galU, respectively, were generated by in vitro transposon mutagenesis with the Himar1 derivative magellan1 as previously described (Akerley et al., 2002), and mapped by DNA sequence analysis of PCR-amplified transposon junctions. Rcp5 and Rcp19 contain magellan1 insertions in their coding sequences at nucleotide positions 635 in lpsA and 264 of galU respectively. H. influenzae Rd carrying lacZ was made by cloning a promoterless E. coli lacZ into the SapI sites of pXT10 and introduced into Rd at the xyl locus.
A non-polar, in-frame deletion of HI1349 (dps) was created by replacement of the protein coding sequences with the aacC1 gentamicin resistance cassette to create Rdps by PCR ‘stitching’ as follows: A 1172 bp PCR product containing the 5′ flanking region of HI1349 was amplified from Rd with primers 1350-3 (5′-TTACAAAGAATAATACTCTAATTCTAC) and 1349-5outgent (5′-ATTCGAGAATTGACGCGTAATAATTTCCTTTTTCTAGTTGAA). A 1881 bp PCR product containing the 3′ flanking region of HI1349 was amplified from Rd with primers 1349-3outgent3 (5′-GTTCAAGCCGAGATCTGAATAAATTTCAACGCTAACGAA) and 1348-5upout (5′-TCAAGATGTTTTCTATTTTTCTCG). A 791 bp fragment containing the aacC1 gentamicin resistance cassette was amplified with primers gentMluI5 (5′-ACGCGTCAATTCTCGAATTGACAT) and gent-3′ (5′-GATCTCGGCTTGAACGAATTGTTA) from pBSL182 (Alexeyev and Shokolenko, 1995). The 1172 bp, 1881 bp and 791 bp products were stitched in a PCR reaction with primers 1350-3 and 1348-5upout. The resultant 3.84 kb product was introduced into Rd and GmR transformants were selected on sBHI agar containing Gm to create strain Rdps.
To generate RdpsV, the vector pXT10 was introduced into the xyl locus of Rdps. RdpsC was created as follows: A 333 bp fragment containing the putative hel (HI0693) promoter was PCR-amplified with primers 692–5ATGoutSap2 (5′-AACTGCAGATCTGCTCTTCAATGCATTTGAAACATATCCCAAGT) and hel5′ATGout2 (5′-CAGGGTATAGTAAGTCTTTCTGA) from Rd. A 525 bp product containing the HI1349 gene was amplified from Rd with primers 1349SDATG (5′-ACTTACTATACCCTGTAGAAAAAGGAAATTATTATGTCA) and 1349-3Sap (AAAGATCTGCAGGCTCTTCTTTAATTATGGCAAGTTTGGCAAGC). These two products were stitched in a PCR reaction with primers 692-5ATGoutSap2 and 1349-3Sap. The resultant 858 bp PCR product was digested with SapI and cloned into the SapI sites of pXT10. This plasmid was introduced into the xyl locus of strain Rdps and TetR transformants were selected on sBHI agar containing Tet to create strain RdpsC.
A non-polar, in-frame deletion of HI0592, speF and potE was created as follows: A 2086 bp PCR product containing the 5′ flanking region of HI0592 was amplified from Rd with primers 594 5′ORF (5′ATGGATGCATCCAAAAAG) and 592out + kan (5′-TTGAATATGGCTCATAGGAAAAATCCCTCTTTCTATCTA). A 1054 bp PCR product containing the 3′ flanking region of HI0590 was amplified from Rd with primers potE 3 + kan (5′-GATGAGTTTTTCTAAAAAAGAAACGCCTACATCTTAATG) and 588–5int (5′-AAAGAGGATTATTAATTGAGTTAC). A 818 bp fragment containing the kanamycin resistance gene, aphI from Tn903 was PCR-amplified with primers kan5 + ATG (5′-ATGAGCCATATTCAACGGGAAAC) and kan3′ + TAA (5′-TTAGAAAAACTCATCGAGCATCAAA). The three resultant products were used as template in PCR to amplify a 3921 bp ‘stitched’ product with primers 594-5′ORF and 588–5int. The 3921 bp product was introduced into RdV and KmR transformants were selected on sBHI agar containing Km to create strain RputV. All transformants were verified by PCR analysis.
Murine bacteraemia model
Haemophilus influenzae were grown to logarithmic phase (OD600 = 0.3) as 20 ml of cultures in 50 ml shake flasks at 35°C. Five- to six-week-old C57BL6 mice were inoculated by the IP route at a dose of 2–8 × 108 cfu. Blood (5 μl) was collected daily by tail vein sampling and serially diluted in BHI for cfu determination and results compared via the one-way anova with the Bonferroni correction. Experiments were conducted with approval and in accordance with guidelines of the University of Massachusetts Institutional Animal Use and Care Committee.
Serum bacteriocidal assays
Serum bactericidal testing was performed as described previously (McQuillen et al., 1994). Briefly ∼2000 cfu of bacteria grown anaerobically to the mid-log phase were incubated with or without normal human serum (NHS) (concentration of NHS specified for each experiment) in a final reaction mixture volume of 150 μl. Aliquots of NHS treated versus untreated samples were plated on sBHI agar plates at 0 and 30 min. In all cases, the number of bacteria recovered from treated and untreated samples at 0 min were equivalent. Survival was calculated as the ratio of the number of cfu recovered at 30 min relative to cfu recovered from untreated samples. An initial study with a range of serum concentrations (1, 2, 3, 4, 5 and 10% NHS) revealed that both Rd and RAA6 were affected similarly at each dose with 4% NHS or above, killing greater than 99% of cells of either strain and both strains had an approximate IC50 of 2%. Therefore, three replicate experiments were conducted with 1% and 2% NHS as indicated in the text.
Northern and RT-qPCR
Total RNA from H. influenzae Rd was obtained from cultures grown anaerobically in sBHI to OD600 = 0.3–0.4 as described above. RNA was isolated using TRIzol Reagent (Invitrogen), treated with DNase I (Ambion) and phenol extracted. For Northern blotting, 6 μg of total RNA was separated by electrophoresis on a 1.5% agarose gel containing 1.1% formaldehyde and transferred to a Nytran nylon membrane (Amersham Pharmacia Biotech). Probes were generated by amplification from Rd using 5′ and 3′ primer pairs for ndh (HI0747) (5′-ATGAAAAACGTCGTGATC and 5′-ATGCAATTTTAATCTTGGTTTTAAATAAC), lldD (HI1739.1) (5′-ATGATTATTTCATCAGCTAG and 5′-AAGTTTACTTAGATCAACC), fdxH (HI0007) (5′-ATGGCTGGAACTGCTCAAGGCG and 5′-GAAACACGATCTACACAAAGAG), fdxI (HI0008) (5′-ATGAGTAAAATTGAAATTAGCAAC and 5′-AGATACCAGTGAATAACATAAAAG), fdhE (HI0009) (5′-ATGAGTATCAAAATCTTATC and 5′-TGCTTCTTCTGCAGGAAAAATAAATG), lldP (HI1218) (5′-ATGCTGTCTTTTATTCTAAG and 5′-TAGATTATAAAATAAAGGTAC), sucB (HI1661) (5′-ATGGCAATCGAAATTCTTG and 5′-GATTTCTAATAACAATCTTG), HI0592 (5′-ATGCTATTTCGTACATATATAC and 5′-GAGAGCCCTGTTGGATG), potE (HI0590) (5′-ATGAGTGCTAAAAGCAATAAAATTG and 5′-TTTTTTAAGATCAAATTTGTAAG) and HI1349 (5′-AACTGCAGATCTGCTCTTCAATGTCAAAAACATCAATCGGACTA and 5′-AAAGATCTGCAGGCTCTTCTTTAATTATGGCAAGTTTGGCAAGC). PCR products were labelled with the Gene Images AlkPhos Direct Labeling Kit and signals visualized with CDP-Star chemiluminescent detection system (Amersham Pharmacia Biotech). The lldD Northern blot was performed by stripping of the lldP probe from the membrane followed by hybridization with the lldD probe. Washing and hybridization conditions were according to the manufacturer's instructions.
Quantification of mRNA expression of the HI1349 (dps) and HI0802 (rpoA) from strains RdV, RAA6V and RAA6C with RNA samples from four independent cultures used in the microarray analysis was performed using iQ SYBR Green Supermix (Bio-Rad Laboratories) in quantitative real-time PCR measured with the DNA Engine Opticon II System (MJ Research). Briefly, 2.5 μg of DNase I-treated total RNA from the above strains was used as template in cDNA synthesis using random primers (New England Biolabs) and SuperScript II reverse transcriptase (Invitrogen). One-tenth of the reverse transcriptase reactions was used as template in qPCR for amplification using 5′ and 3′ primer pairs for dps (5′-AACTGCAGATCTGCTCTTCAATGTCAAAAACATCAATCGGACTA and 5′-ACATTCTTGTGCCTCACTTACTGC) and rpoA (5′-GTAGAAATTGATGGCGTATTG and 5′-TCACCATCATAGGTAATGTCC). Real-time cycler conditions were as follows: 95°C for 3 min, followed by 39 cycles of 96°C for 20 s, 58°C for 30 s and 72°C for 30 s, followed by one cycle of 72°C for 7 min. Fluorescence was read at 72, 74, 76 and 78°C and normalized to the housekeeping gene, rpoA, which encodes the alpha subunit of RNA polymerase. Control reactions were performed in parallel with mock cDNA reactions generated without reverse transcriptase to verify specific amplification. Product sizes were confirmed by agarose gel electrophoresis.
Microarray analysis
Total RNA from four independent cultures of H. influenzae Rd grown anaerobically to OD600 = 0.3–0.5 was obtained and treated with DNase I as described above. Thirty micrograms of RNA from each quadruplicate culture was used as template for generation of probes by reverse-transcription in the presence of random primers, essentially as described previously (Wong and Akerley, 2005) except that biotinylated nucleotides (Mergen, San Leandro, CA) were used in the reverse-transcription. Biotinylated cDNAs were hybridized to glass slide microarrays containing 45-mer oligonucleotides representing a total of 1697 genes of the H. influenzae KW20 genome in addition to negative controls specific for human genes. Array printing, fluorescent labelling, hybridization to microarrays, and array scanning was conducted by Mergen (San Leandro, CA). The total signal intensity for every gene represented on the array was corrected by subtracting the local background and normalized by dividing by the mean of the values for all of the genes represented on the array. Bacterial cultures, RNA isolation, labelling and hybridizations were conducted independently to obtain four experimental replicates for each strain. The corrected signal intensity for each gene represents the mean of separate hybridization experiments conducted with labelled cDNAs from each of the four independent cultures of each strain. Expression ratio data were generated by comparing the corrected mean signal intensity values from arrays hybridized to cDNAs generated from the parent strain versus the arcA mutant or complemented arcA strain versus the arcA mutant. Statistical analysis of the expression data were performed using the Cyber-T Bayesian statistics framework (Baldi and Long, 2001) available as a web interface from the Institute for Genomics and Bioinformatics at the University of California, Irvine (http://visitor.ics.uci.edu/genex/cybert). Genes whose expression ratios had Bayesian P-values based on the regularized t-test ≤ 0.0001 and showed ≥ 2.0-fold differential gene expression levels were considered to exhibit significant differences. Mean signal intensities greater than or equal to twofold above the mean local background averaged over all the spots on all of the arrays were considered to have reached the threshold value for significant gene expression.
Hydrogen peroxide sensitivity assays
Survival after exposure to H2O2 was evaluated for anaerobically or aerobically grown H. influenzae. Aerobically grown H. influenzae was prepared by diluting overnight cultures 1:200 in sBHI in standard culture tubes and growing at 35°C with shaking at 250 r.p.m. to an OD600 = 0.1–0.2. Triplicate cultures of each strain were inoculated at 0.005 OD600 units ml−1 into 10 ml of sBHI in 500 ml Erlenmeyer flasks and grown at 35°C with shaking at 250 r.p.m. to an OD600 = 0.3–0.4. Anaerobic cultures were prepared by diluting overnight standing cultures to 0.005 OD600 units ml−1 in sBHI (triplicates per strain) in glass vials filled to exclude air and grown at 35°C with shaking at 250 r.p.m. to an OD600 = 0.3–0.4. Cells from 1 ml of each culture were pelleted and resuspended in 1 ml of MIc medium (Barcak et al., 1991). The H2O2 sensitivity assay was conducted in Costar 24-well cell culture plates seeded with 100 μl per well. An equal volume of MIc medium with and without 1 mM H2O2 was added (final concentration of 0.5 mM) and the plate was shaken at 250 r.p.m. at 35°C for 10 min followed by addition of 10 mM sodium pyruvate to quench H2O2 (Maciver and Hansen, 1996) to all samples. H2O2 treated and untreated cells were diluted and plated onto sBHI agar plates for cfu determination. Differences were compared via the one-way anova with Bonferroni post-tests.
Acknowledgments
We thank Drs John Leong and John Trawick for helpful comments on the manuscript. We thank TIGR, the Institute for Genomics and Bioinformatics at UC Irvine, and the Kanehisa Laboratory of Kyoto University Bioinformatics Center for web-based bioinformatics resources. This work was supported in part by grants from the American Heart Association, Philip Morris, USA and Philip Morris Intl., and NIH NIAID 1RO1-AI49437 to B.J.A.; a grant from the Cystic Fibrosis Foundation to S.M.W.; and NIH RO1 AI054544 to S.R.
Supplementary material
The following supplementary material is available for this article:
Microarray data of parent vs. ΔarcA mutant in H. influenzae Rd. Microarray data was analyzed using the Cyber T statistics program. Input for the Cyber T program is the corrected signal intensity values from 1697 genes representing the genome of H. influenzae and control probes (see Col. 1 definitions in table) hybridized with cDNA from quadruplicate cultures of the RdV parental strain, ΔarcA mutant, RAA6V, and complemented strain, RAA6C grown anaerobically. In this table, the Cyber T analysis represents the comparison between the parent strain and ΔarcA mutant. The column heading definitions are listed at the end of the data file.
Microarray data of arcA complemented strain vs. ΔarcA mutant in H. influenzae Rd. Microarray data was analyzed using the Cyber T statistics program. Input for the Cyber T program is the corrected signal intensity values from 1697 genes representing the genome of H. influenzae and control probes (see Col. 1 definitions in table) hybridized with cDNA from quadruplicate cultures of the RdV parental strain, ΔarcA mutant, RAA6V, and complemented strain, RAA6C grown anaerobically. In this table, the Cyber T analysis represents the comparison between the complemented strain and ΔarcA mutant. The column heading definitions are listed at the end of the data file.
Comparsion of arcA microarray expression data between H. influenzae and E. coli. Similarities and differences across species in putative ArcA regulons. H. influenzae arcA microarray expression data (from Tables 1 and 2, columns RAA6V/RdV) was compared to E. coli arcA microarray expression data of Salmon et al. (Salmon, K.A., et al. 2005. J. Biol. Chem. 280: 15084–15096) and Liu and DeWulf (Liu, X. and P. De Wulf. 2004. J. Biol. Chem. 279: 12588–12597). Putative E. coli homologs in bold have expression data from either microarray sets of Salmon et al. or Liu and De Wulf that show agreement with the H. influenzae array data in anaerobic expression ratios in the ΔarcA mutant, RAA6V vs. the parent strain, RdV. Fold change values represent an increase (positive) or decrease (negative) in gene expression in the ΔarcA mutant vs. parent strain, RdV. E. coli homologs with blank entries are genes whose expression levels did not meet the authors′ criteria for minimum signal detection on the array.
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1365-2958.2007.05747.x
(This link will take you to the article abstract).
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Abaibou H, Pommier J, Benoit S, Giordano G, Mandrand-Berthelot MA. Expression and characterization of the Escherichia coli fdo locus and a possible physiological role for aerobic formate dehydrogenase. J Bacteriol. 1995;177:7141–7149. doi: 10.1128/jb.177.24.7141-7149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerley BJ, Rubin EJ, Novick VL, Amaya K, Judson N, Mekalanos JJ. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc Natl Acad Sci USA. 2002;99:966–971. doi: 10.1073/pnas.012602299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeyev MF, Shokolenko IN. Mini-Tn10 transposon derivatives for insertion mutagenesis and gene delivery into the chromosome of Gram-negative bacteria. Gene. 1995;160:59–62. doi: 10.1016/0378-1119(95)00141-r. [DOI] [PubMed] [Google Scholar]
- Almiron M, Link AJ, Furlong D, Kolter R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992;6:2646–2654. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DE, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1995. [Google Scholar]
- Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- Barcak GJ, Chandler MS, Redfield RJ, Tomb JF. Genetic systems in Haemophilus influenzae. Meth Enzymol. 1991;204:321–342. doi: 10.1016/0076-6879(91)04016-h. [DOI] [PubMed] [Google Scholar]
- Bishai WR, Howard NS, Winkelstein JA, Smith HO. Characterization and virulence analysis of catalase mutants of Haemophilus influenzae. Infect Immun. 1994;62:4855–4860. doi: 10.1128/iai.62.11.4855-4860.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongaerts J, Zoske S, Weidner U, Unden G. Transcriptional regulation of the proton translocating NADH dehydrogenase genes (nuoA-N) of Escherichia coli by electron acceptors, electron donors and gene regulators. Mol Microbiol. 1995;16:521–534. doi: 10.1111/j.1365-2958.1995.tb02416.x. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) Progress towards elimination of Haemophilus influenzae type b invasive disease among infants and children – United States, 1998–2000. MMWR Morb Mortal Wkly Rep. 2002;51:234–237. [PubMed] [Google Scholar]
- Chattopadhyay MK, Tabor CW, Tabor H. Polyamines protect Escherichia coli cells from the toxic effect of oxygen. Proc Natl Acad Sci USA. 2003;100:2261–2265. doi: 10.1073/pnas.2627990100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Helmann JD. Bacillus subtilis MrgA is a Dps (PexB) homologue: evidence for metalloregulation of an oxidative-stress gene. Mol Microbiol. 1995;18:295–300. doi: 10.1111/j.1365-2958.1995.mmi_18020295.x. [DOI] [PubMed] [Google Scholar]
- Chiancone E, Ceci P, Ilari A, Ribacchi F, Stefanini S. Iron and proteins for iron storage and detoxification. Biometals. 2004;17:197–202. doi: 10.1023/b:biom.0000027692.24395.76. [DOI] [PubMed] [Google Scholar]
- Cooksley C, Jenks PJ, Green A, Cockayne A, Logan RP, Hardie KR. NapA protects Helicobacter pylori from oxidative stress damage, and its production is influenced by the ferric uptake regulator. J Med Microbiol. 2003;52:461–469. doi: 10.1099/jmm.0.05070-0. [DOI] [PubMed] [Google Scholar]
- Cuthill SL, Farley MM, Donowitz LG. Nontypable Haemophilus influenzae meningitis. Pediatr Infect Dis J. 1999;18:660–662. doi: 10.1097/00006454-199907000-00024. [DOI] [PubMed] [Google Scholar]
- D'Mello RA, Langford PR, Kroll JS. Role of bacterial Mn-cofactored superoxide dismutase in oxidative stress responses, nasopharyngeal colonization, and sustained bacteremia caused by Haemophilus influenzae type b. Infect Immun. 1997;65:2700–2706. doi: 10.1128/iai.65.7.2700-2706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daines DA, Cohn LA, Coleman HN, Kim KS, Smith AL. Haemophilus influenzae Rd KW20 has virulence properties. J Med Microbiol. 2003;52:277–282. doi: 10.1099/jmm.0.05025-0. [DOI] [PubMed] [Google Scholar]
- De Souza-Hart JA, Blackstock W, Di Modugno V, Holland IB, Kok M. Two-component systems in Haemophilus influenzae: a regulatory role for ArcA in serum resistance. Infect Immun. 2003;71:163–172. doi: 10.1128/IAI.71.1.163-172.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong JM, Taylor JS, Latour DJ, Iuchi S, Lin ECC. Three overlapping lct genes involved in 1-lactate utilization by Escherichia coli. J Bacteriol. 1993;175:6671–6678. doi: 10.1128/jb.175.20.6671-6678.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin AL, Nelson KL, Mhlanga-Mutangadura T, Bonthuis PJ, Geelhood JL, Morlin G, et al. Characterization of genetic and phenotypic diversity of invasive nontypeable Haemophilus influenzae. Infect Immun. 2005;73:5853–5863. doi: 10.1128/IAI.73.9.5853-5863.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- Furuchi T, Kashiwagi K, Kobayashi H, Igarashi K. Characteristics of the gene for a spermidine and putrescine transport system that maps at 15 min on the Escherichia coli chromosome. J Biol Chem. 1991;266:20928–20933. [PubMed] [Google Scholar]
- Georgellis D, Kwon O, Lin ECC. Quinones as the redox signal for the Arc two-component system of bacteria. Science. 2001a;292:2314–2316. doi: 10.1126/science.1059361. [DOI] [PubMed] [Google Scholar]
- Georgellis D, Kwon O, Lin ECC, Wong SM, Akerley BJ. Redox signal transduction by the ArcB sensor kinase of Haemophilus influenzae lacking the PAS domain. J Bacteriol. 2001b;183:7206–7212. doi: 10.1128/JB.183.24.7206-7212.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilligan PH. Microbiology of airway disease in patients with cystic fibrosis. Clin Microbiol Rev. 1991;4:35–51. doi: 10.1128/cmr.4.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralnick JA, Brown CT, Newman DK. Anaerobic regulation by an atypical Arc system in Shewanella oneidensis. Mol Microbiol. 2005;56:1347–1357. doi: 10.1111/j.1365-2958.2005.04628.x. [DOI] [PubMed] [Google Scholar]
- Green J, Guest JR. Regulation of transcription at the ndh promoter of Escherichia coli by FNR and novel factors. Mol Microbiol. 1994;12:433–444. doi: 10.1111/j.1365-2958.1994.tb01032.x. [DOI] [PubMed] [Google Scholar]
- Halsey TA, Vazquez-Torres A, Gravdahl DJ, Fang FC, Libby SJ. The ferritin-like Dps protein is required for Salmonella enterica serovar Typhimurium oxidative stress resistance and virulence. Infect Immun. 2004;72:1155–1158. doi: 10.1128/IAI.72.2.1155-1158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamana K, Nakata K. Distribution of diaminopropane, putrescine and cadaverine in Haemophilus and Actinobacillus. Microbios. 2000;103:43–51. [PubMed] [Google Scholar]
- Harrison A, Dyer DW, Gillaspy A, Ray WC, Mungur R, Carson MB, et al. Genomic sequence of an otitis media isolate of nontypeable Haemophilus influenzae: comparative study with H. influenzae serotype d, strain KW20. J Bacteriol. 2005;187:4627–4636. doi: 10.1128/JB.187.13.4627-4636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison A, Ray WC, Baker BD, Armbruster DW, Bakaletz LO, Munson RS., Jr J Bacteriol. 2006. The OxyR regulon in nontypeable Haemophilus influenzae. [DOI] [PMC free article] [PubMed]
- Hood DW, Deadman ME, Allen T, Masoud H, Martin A, Brisson JR, et al. Use of the complete genome sequence information of Haemophilus influenzae strain Rd to investigate lipopolysaccharide biosynthesis. Mol Microbiol. 1996;22:951–965. doi: 10.1046/j.1365-2958.1996.01545.x. [DOI] [PubMed] [Google Scholar]
- Igarashi K, Kashiwagi K. Polyamine transport in bacteria and yeast. Biochem J. 1999;344 Part 3:633–642. [PMC free article] [PubMed] [Google Scholar]
- Ikai H, Yamamoto S. Two genes involved in the 1,3-diaminopropane production pathway in Haemophilus influenzae. Biol Pharm Bull. 1998;21:170–173. doi: 10.1248/bpb.21.170. [DOI] [PubMed] [Google Scholar]
- Ilari A, Stefanini S, Chiancone E, Tsernoglou D. The dodecameric ferritin from Listeria innocua contains a novel intersubunit iron-binding site. Nat Struct Biol. 2000;7:38–43. doi: 10.1038/71236. [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Mizunoe Y, Kawabata S, Takade A, Harada M, Wai SN, Yoshida S. The iron-binding protein Dps confers hydrogen peroxide stress resistance to Campylobacter jejuni. J Bacteriol. 2003;185:1010–1017. doi: 10.1128/JB.185.3.1010-1017.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Lin ECC. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc Natl Acad Sci USA. 1988;85:1888–1892. doi: 10.1073/pnas.85.6.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung IL, Kim IG. Transcription of ahpC, katG, and katE genes in Escherichia coli is regulated by polyamines: polyamine-deficient mutant sensitive to H2O2-induced oxidative damage. Biochem Biophys Res Commun. 2003;301:915–922. doi: 10.1016/s0006-291x(03)00064-0. [DOI] [PubMed] [Google Scholar]
- Kargalioglu Y, Imlay JA. Importance of anaerobic superoxide dismutase synthesis in facilitating outgrowth of Escherichia coli upon entry into an aerobic habitat. J Bacteriol. 1994;176:7653–7658. doi: 10.1128/jb.176.24.7653-7658.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi K, Suzuki T, Suzuki F, Furuchi T, Kobayashi H, Igarashi K. Coexistence of the genes for putrescine transport protein and ornithine decarboxylase at 16 min on Escherichia coli chromosome. J Biol Chem. 1991;266:20922–20927. [PubMed] [Google Scholar]
- Kiley PJ, Beinert H. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol Rev. 1998;22:341–352. doi: 10.1111/j.1574-6976.1998.tb00375.x. [DOI] [PubMed] [Google Scholar]
- Klein JO. Role of nontypeable Haemophilus influenzae in pediatric respiratory tract infections. Pediatr Infect Dis J. 1997;16:S5–S8. doi: 10.1097/00006454-199702001-00002. [DOI] [PubMed] [Google Scholar]
- Levraut J, Ciebiera JP, Chave S, Rabary O, Jambou P, Carles M, Grimaud D. Mild hyperlactatemia in stable septic patients is due to impaired lactate clearance rather than overproduction. Am J Respir Crit Care Med. 1998;157:1021–1026. doi: 10.1164/ajrccm.157.4.9705037. [DOI] [PubMed] [Google Scholar]
- Liu X, De Wulf P. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J Biol Chem. 2004;279:12588–12597. doi: 10.1074/jbc.M313454200. [DOI] [PubMed] [Google Scholar]
- Loprasert S, Whangsuk W, Sallabhan R, Mongkolsuk S. DpsA protects the human pathogen Burkholderia pseudomallei against organic hydroperoxide. Arch Microbiol. 2004;182:96–101. doi: 10.1007/s00203-004-0694-0. [DOI] [PubMed] [Google Scholar]
- Lu S, Killoran PB, Fang FC, Riley LW. The Global Regulator ArcA controls resistance to reactive nitrogen and oxygen intermediates in Salmonella enterica serovar enteritidis. Infect Immun. 2002;70:451–461. doi: 10.1128/IAI.70.2.451-461.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch AS, Lin ECC. Responses to molecular oxygen. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, DC: ASM Press; 1996a. pp. 1526–1538. [Google Scholar]
- Lynch AS, Lin ECC. Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: characterization of DNA binding at target promoters. J Bacteriol. 1996b;178:6238–6249. doi: 10.1128/jb.178.21.6238-6249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver I, Hansen EJ. Lack of expression of the global regulator OxyR in Haemophilus influenzae has a profound effect on growth phenotype. Infect Immun. 1996;64:4618–4629. doi: 10.1128/iai.64.11.4618-4629.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuillen DP, Gulati S, Rice PA. Complement-mediated bacterial killing assays. Methods Enzymol. 1994;236:137–147. doi: 10.1016/0076-6879(94)36013-8. [DOI] [PubMed] [Google Scholar]
- Malpica R, Franco B, Rodriguez C, Kwon O, Georgellis D. Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc Natl Acad Sci USA. 2004;101:13318–13323. doi: 10.1073/pnas.0403064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manukhov IV, Bertsova YV, Trofimov DY, Bogachev AV, Skulachev VP. Analysis of HI0220 protein from Haemophilus influenzae, a novel structural and functional analog of ArcB protein from Escherichia coli. Biochemistry (Mosc) 2000;65:1321–1326. [PubMed] [Google Scholar]
- Martinez A, Kolter R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J Bacteriol. 1997;179:5188–5194. doi: 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita K, Ohnishi T, Kaback HR. NADH-ubiquinone oxidoreductases of the Escherichia coli aerobic respiratory chain. Biochemistry. 1987;26:7732–7737. doi: 10.1021/bi00398a029. [DOI] [PubMed] [Google Scholar]
- Meng W, Green J, Guest JR. FNR-dependent repression of ndh gene expression requires two upstream FNR-binding sites. Microbiology. 1997;143:1521–1532. doi: 10.1099/00221287-143-5-1521. Part 5. [DOI] [PubMed] [Google Scholar]
- Moller LV, Regelink AG, Grasselier H, Dankert-Roelse JE, Dankert J, van Alphen L. Multiple Haemophilus influenzae strains and strain variants coexist in the respiratory tract of patients with cystic fibrosis. J Infect Dis. 1995;172:1388–1392. doi: 10.1093/infdis/172.5.1388. [DOI] [PubMed] [Google Scholar]
- Moxon ER, Murphy TF. Haemophilus influenzae. In: Mandell GL, Bennett JR, Dolin R, editors. Mandell, Douglas, and Bennett's Principles and Practices of Infectious Diseases. New York: Churchill Livingstone; 2000. pp. 2369–2378. [Google Scholar]
- Murphy TF, Sethi S. Chronic obstructive pulmonary disease: role of bacteria and guide to antibacterial selection in the older patient. Drugs Aging. 2002;19:761–775. doi: 10.2165/00002512-200219100-00005. [DOI] [PubMed] [Google Scholar]
- Murphy TF, Brauer AL, Schiffmacher AT, Sethi S. Persistent colonization by Haemophilus influenzae in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2004;170:266–272. doi: 10.1164/rccm.200403-354OC. [DOI] [PubMed] [Google Scholar]
- Nathan C, Shiloh MU. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc Natl Acad Sci USA. 2000;97:8841–8848. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizet V, Colina KF, Almquist JR, Rubens CE, Smith AL. A virulent nonencapsulated Haemophilus influenzae. J Infect Dis. 1996;173:180–186. doi: 10.1093/infdis/173.1.180. [DOI] [PubMed] [Google Scholar]
- Nunez MF, Kwon O, Wilson TH, Aguilar J, Baldoma L, Lin ECC. Transport of L-Lactate, D-Lactate, and glycolate by the LldP and GlcA membrane carriers of Escherichia coli. Biochem Biophys Res Commun. 2002;290:824–829. doi: 10.1006/bbrc.2001.6255. [DOI] [PubMed] [Google Scholar]
- Nystrom T, Larsson C, Gustafsson L. Bacterial defense against aging: role of the Escherichia coli ArcA regulator in gene expression, readjusted energy flux and survival during stasis. EMBO J. 1996;15:3219–3228. [PMC free article] [PubMed] [Google Scholar]
- O'Neill JM, St Geme JW, Cutter D, Adderson EE, Anyanwu J, Jacobs RF, Schutze GE. Invasive disease due to nontypeable Haemophilus influenzae among children in Arkansas. J Clin Microbiol. 2003;41:3064–3069. doi: 10.1128/JCM.41.7.3064-3069.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen KN, Larsen MH, Gahan CG, Kallipolitis B, Wolf XA, Rea R, et al. The Dps-like protein Fri of Listeria monocytogenes promotes stress tolerance and intracellular multiplication in macrophage-like cells. Microbiology. 2005;151:925–933. doi: 10.1099/mic.0.27552-0. [DOI] [PubMed] [Google Scholar]
- Oshima T, Aiba H, Masuda Y, Kanaya S, Sugiura M, Wanner BL, et al. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol Microbiol. 2002;46:281–291. doi: 10.1046/j.1365-2958.2002.03170.x. [DOI] [PubMed] [Google Scholar]
- Patschkowski T, Bates DM, Kiley PJ. Mechanisms for sensing and responding to oxygen deprivation. In: Storz G, Hengge-Aronis R, editors. Bacterial Stress Responses. Washington, DC: American Society for Microbiology Press; 2000. pp. 61–78. [Google Scholar]
- Pegg AE. Polyamine metabolism and its importance in neoplastic growth and a target for chemotherapy. Cancer Res. 1988;48:759–774. [PubMed] [Google Scholar]
- Pulliainen AT, Kauko A, Haataja S, Papageorgiou AC, Finne J. Dps/Dpr ferritin-like protein: insights into the mechanism of iron incorporation and evidence for a central role in cellular iron homeostasis in Streptococcus suis. Mol Microbiol. 2005;57:1086–1100. doi: 10.1111/j.1365-2958.2005.04756.x. [DOI] [PubMed] [Google Scholar]
- Rocha ER, Owens G, Smith CJ. The redox-sensitive transcriptional activator OxyR regulates the peroxide response regulon in the obligate anaerobe Bacteroides fragilis. J Bacteriol. 2000;182:5059–5069. doi: 10.1128/jb.182.18.5059-5069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon KA, Hung SP, Steffen NR, Krupp R, Baldi P, Hatfield GW, Gunsalus RP. Global gene expression profiling in Escherichia coli K12: effects of oxygen availability and ArcA. J Biol Chem. 2005;280:15084–15096. doi: 10.1074/jbc.M414030200. [DOI] [PubMed] [Google Scholar]
- Sengupta N, Paul K, Chowdhury R. The global regulator ArcA modulates expression of virulence factors in Vibrio cholerae. Infect Immun. 2003;71:5583–5589. doi: 10.1128/IAI.71.10.5583-5589.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshu J, Boylan JA, Gherardini FC, Skare JT. Dissolved oxygen levels alter gene expression and antigen profiles in Borrelia burgdorferi. Infect Immun. 2004;72:1580–1586. doi: 10.1128/IAI.72.3.1580-1586.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi S, Murphy TF. Bacterial infection in chronic obstructive pulmonary disease in 2000: a state-of-the-art review. Clin Microbiol Rev. 2001;14:336–363. doi: 10.1128/CMR.14.2.336-363.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor CW, Tabor H. Polyamines in microorganisms. Microbiol Rev. 1985;49:81–99. doi: 10.1128/mr.49.1.81-99.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkachenko A, Nesterova L, Pshenichnov M. The role of the natural polyamine putrescine in defense against oxidative stress in Escherichia coli. Arch Microbiol. 2001;176:155–157. doi: 10.1007/s002030100301. [DOI] [PubMed] [Google Scholar]
- Ueshima J, Shoji M, Ratnayake DB, Abe K, Yoshida S, Yamamoto K, Nakayama K. Purification, gene cloning, gene expression, and mutants of Dps from the obligate anaerobe Porphyromonas gingivalis. Infect Immun. 2003;71:1170–1178. doi: 10.1128/IAI.71.3.1170-1178.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Boom TJ, Reed KE, Cronan JE., Jr Lipoic acid metabolism in Escherichia coli: isolation of null mutants defective in lipoic acid biosynthesis, molecular cloning and characterization of the E. coli lip locus, and identification of the lipoylated protein of the glycine cleavage system. J Bacteriol. 1991;173:6411–6420. doi: 10.1128/jb.173.20.6411-6420.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri GN, Eiteman MA, Altman E. Increased recombinant protein production in Escherichia coli strains with overexpressed water-forming NADH oxidase and a deleted ArcA regulatory protein. Biotechnol Bioeng. 2006;94:538–542. doi: 10.1002/bit.20853. [DOI] [PubMed] [Google Scholar]
- Vergauwen B, Pauwels F, Van Beeumen JJ. Glutathione and catalase provide overlapping defenses for protection against respiration-generated hydrogen peroxide in Haemophilus influenzae. J Bacteriol. 2003;185:5555–5562. doi: 10.1128/JB.185.18.5555-5562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidner U, Geier S, Ptock A, Friedrich T, Leif H, Weiss H. The gene locus of the proton-translocating NADH: ubiquinone oxidoreductase in Escherichia coli. Organization of the 14 genes and relationship between the derived proteins and subunits of mitochondrial complex I. J Mol Biol. 1993;233:109–122. doi: 10.1006/jmbi.1993.1488. [DOI] [PubMed] [Google Scholar]
- Weiser JN, Chong ST, Greenberg D, Fong W. Identification and characterization of a cell envelope protein of Haemophilus influenzae contributing to phase variation in colony opacity and nasopharyngeal colonization. Mol Microbiol. 1995;17:555–564. doi: 10.1111/j.1365-2958.1995.mmi_17030555.x. [see comments]. [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, Mosolf J, Willits D, Yeager M, Dryden KA, Young M, Douglas T. An archaeal antioxidant: characterization of a Dps-like protein from Sulfolobus solfataricus. Proc Natl Acad Sci USA. 2005;102:10551–10556. doi: 10.1073/pnas.0501497102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SM, Akerley BJ. Inducible expression system and marker-linked mutagenesis approach for functional genomics of Haemophilus influenzae. Gene. 2003;316:177–186. doi: 10.1016/s0378-1119(03)00762-5. [DOI] [PubMed] [Google Scholar]
- Wong SM, Akerley BJ. Environmental and genetic regulation of the phosphorylcholine epitope of Haemophilus influenzae lipooligosaccharide. Mol Microbiol. 2005;55:724–738. doi: 10.1111/j.1365-2958.2004.04439.x. [DOI] [PubMed] [Google Scholar]
- Zhao G, Ceci P, Ilari A, Giangiacomo L, Laue TM, Chiancone E, Chasteen ND. Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A ferritin-like DNA-binding protein of Escherichia coli. J Biol Chem. 2002;277:27689–27696. doi: 10.1074/jbc.M202094200. [DOI] [PubMed] [Google Scholar]
- Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol. 2001;183:4562–4570. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Microarray data of parent vs. ΔarcA mutant in H. influenzae Rd. Microarray data was analyzed using the Cyber T statistics program. Input for the Cyber T program is the corrected signal intensity values from 1697 genes representing the genome of H. influenzae and control probes (see Col. 1 definitions in table) hybridized with cDNA from quadruplicate cultures of the RdV parental strain, ΔarcA mutant, RAA6V, and complemented strain, RAA6C grown anaerobically. In this table, the Cyber T analysis represents the comparison between the parent strain and ΔarcA mutant. The column heading definitions are listed at the end of the data file.
Microarray data of arcA complemented strain vs. ΔarcA mutant in H. influenzae Rd. Microarray data was analyzed using the Cyber T statistics program. Input for the Cyber T program is the corrected signal intensity values from 1697 genes representing the genome of H. influenzae and control probes (see Col. 1 definitions in table) hybridized with cDNA from quadruplicate cultures of the RdV parental strain, ΔarcA mutant, RAA6V, and complemented strain, RAA6C grown anaerobically. In this table, the Cyber T analysis represents the comparison between the complemented strain and ΔarcA mutant. The column heading definitions are listed at the end of the data file.
Comparsion of arcA microarray expression data between H. influenzae and E. coli. Similarities and differences across species in putative ArcA regulons. H. influenzae arcA microarray expression data (from Tables 1 and 2, columns RAA6V/RdV) was compared to E. coli arcA microarray expression data of Salmon et al. (Salmon, K.A., et al. 2005. J. Biol. Chem. 280: 15084–15096) and Liu and DeWulf (Liu, X. and P. De Wulf. 2004. J. Biol. Chem. 279: 12588–12597). Putative E. coli homologs in bold have expression data from either microarray sets of Salmon et al. or Liu and De Wulf that show agreement with the H. influenzae array data in anaerobic expression ratios in the ΔarcA mutant, RAA6V vs. the parent strain, RdV. Fold change values represent an increase (positive) or decrease (negative) in gene expression in the ΔarcA mutant vs. parent strain, RdV. E. coli homologs with blank entries are genes whose expression levels did not meet the authors′ criteria for minimum signal detection on the array.