Abstract
Utrophin gene is transcribed in a large mRNA of 13 kb that codes for a protein of 395 kDa. It shows amino acid identity with dystrophin of up to 73% and is widely expressed in muscle and non-muscle tissues. Up71 is a short utrophin product of the utrophin gene with the same cysteine-rich and C-terminal domains as full-length utrophin (Up395). Using RT-PCR, Western blots analysis, we demonstrated that Up71 is overexpressed in the mdx diaphragm, the most pathological muscle in dystrophin-deficient mdx mice, compared to wild-type C57BL/10 or other mdx skeletal muscles. Subsequently, we demonstrated that this isoform displayed an increased expression level up to 12 months, whereas full-length utrophin (Up395) decreased. In addition, β-dystroglycan, the transmembrane glycoprotein that anchors the cytoplasmic C-terminal domain of utrophin, showed similar increase expression in mdx diaphragm, as opposed to other components of the dystrophin-associated protein complex (DAPC) such as α-dystrobrevin1 and α-sarcoglycan. We demonstrated that Up71 and β-dystroglycan were progressively accumulated along the sarcolemma of regenerating clusters in mdx diaphragm. Our data provide novel functional insights into the pathological role of the Up71 isoform in dystrophinopathies.
Keywords: Aging; physiology; Animals; Desmin; metabolism; Diaphragm; cytology; metabolism; pathology; Dystroglycans; metabolism; Gene Expression Regulation; Lower Extremity; anatomy & histology; Mice; Mice, Inbred C57BL; Mice, Inbred mdx; Muscular Dystrophies; genetics; metabolism; pathology; Protein Isoforms; genetics; metabolism; Utrophin; genetics; metabolism
Keywords: Utrophin isoforms, beta-dystroglycan, Muscle, mdx, Sarcolemma, DMD
Duchenne muscular dystrophy (DMD) is the most prevalent inherited neuromuscular disorder, affecting ~1 in every 3,500 male births [1]. The disease results from mutations and/or deletions in the X-linked dystrophin gene, which prevents production of large cytoskeletal proteins of the spectrin superfamily [2, 3]. Although the exact function of dystrophin remains unclear, it has been shown to serve as a link between the internal actin cytoskeleton and the extracellular matrix via a complex of dystrophin-associated proteins (DAPC) [4, 5]. Through these interactions, dystrophin is thought to maintain the integrity of muscle cell membrane during the cycles of contraction and relaxation. Moreover, accumulating evidence suggests that some DAPC component has an important signaling role in muscle and is not merely a molecular scaffold serving the mechanical function [6].
In 1989, a gene that encodes a large cytoskeletal protein showing extensive sequence identity with dystrophin was identified on chromosome 6 [7, 8]. Interestingly, this protein, now referred to as utrophin, is more ubiquitously distributed than full-length dystrophin. In skeletal muscle fibers, utrophin also displays a clear difference in its pattern of distribution as compared with dystrophin. Whereas dystrophin is known to be expressed along the entire length of the sarcolemma in normal muscle fibers, utrophin preferentially accumulates at the neuromuscular junction in both normal and DMD muscles [4, 9, 10] where it may contribute to the full differentiation of the postsynaptic apparatus [11, 12]. Because of its high degree of identity with dystrophin [13], it has been suggested that upregulation of utrophin into extrasynaptic regions of DMD muscle fibers could serve as an adequate therapeutic strategy for this disease [10, 14, 15]. There has been considerable interest in understanding the mechanisms regulating the expression of utrophin in skeletal muscle. Several studies have shown that utrophin levels can be modulated according to the state of innervations, myogenic differentiation and the oxidative capacity of muscle cells [16–19]. In this context, regenerating muscle showed higher levels of utrophin but no increase in full-length utrophin mRNA was observed, leading to suggestions of post-transcriptional regulatory mechanisms or the presence of other utrophin transcription products [20]. Specific transcription factors and promoter elements were found to be involved in regulating the abundance and localization of utrophin transcripts within muscle fibers [21]. Recent studies have characterized C-terminal utrophin expression in normal and dystrophin-deficient mdx mouse tissues and have suggested a more complex array of utrophin gene products [22, 23]. There is still much to be learned about utrophin gene regulation, in particular regarding alternative isoforms and their role in muscle cell membrane. Several groups have observed small gene products using antibodies raised against the utrophin C-terminus. Up71 is a short transcript of utrophin with a unique first exon located in intron 62 and predicted to encode a 4.0 kb mRNA and a 71 kDa protein with the same cysteine-rich and C-terminal domains as full-length utrophin, commencing with exon 63 (dystrophin exon numbering) [24]. These domains are implicated in interactions with β-dystroglycan (β-DG), the transmenbrane glycoprotein that anchors utrophin and dystrophin in muscle cell membrane. [25, 26] Like full length utrophin, Up71 isoform has an ubiquitous expression and was detected by our team and others in the peripheral nervous system and small peripheral arteries [24, 27, 28]. To date there is little information about the expression of this short transcript in dystrophin deficient muscle and its implication in DMD pathology.
Several studies have shown that not all muscles that lack dystrophin are equally susceptible to muscle degeneration. The mdx mouse exhibits great variability in myofiber necrosis and contractile properties between different muscles [29]. These mice exhibit a mild form of muscular dystrophy, as seen by elevated serum level of creatine kinase and histological changes consistent with myofiber damage. The hind limb muscles undergo extensive myofiber degeneration and regeneration from 3 to 5 weeks of age [30]. Although hind limb muscles of mdx mice undergo successful regeneration, the mdx diaphragm exhibits high dystrophic pattern and responds to progressive muscle degeneration with transition to a slower phenotype associated with reduced power output andgreater endurance [31–33]. Interestingly, the diaphragm from mdx mice reproduces the degenerative pattern observed in DMD [34, 35] which suggest that the early dystrophic changes are most prominent in this muscle [33].
In this study, we demonstrated that the short utrophin isoform Up71 was overexpressed in mdx mouse diaphragm compared to wild-type C57BL/10 or other mdx skeletal muscles. We showed that Up71 and β-DG were accumulated in regenerative clusters and not exclusively in slow oxidative MHC (myosin heavy chain) type I fibers, as described previously for utrophin A in soleus muscle [19]. These results argue for a possible involvement of Up71 and β-DG in the regenerative process in mdx diaphragm and consequently may shed light on the pathological pattern of this muscle.
Materials and Methods
Laboratory Animals
Control (C57BL/10) and mdx mice were purchased from Jackson Laboratory (Bar Harbor, USA) and used at 2, 6, 8, 10, 12, 14 and 16 months of age. At each selected age, 5 animals were killed and tissues were analyzed separately. Diaphragms and TA muscles were dissected and divided into 2 parts. One part was rapidly frozen in 2-methylbutane, cooled in liquid nitrogen and stored at −80°C until use for immunohistochemical analysis, total protein preparation and RT-PCR analysis. The second part was immediately used for co-immunoprecipitation analysis. New Zealand white rabbits were used to produce polyclonal antibodies.
Antibodies
Polyclonal antibodies against β-DG (LG5), α-SG (Sarco3) and α-DB1 (D1) were produced and characterized as previously described [36]. Polyclonal antibody against the utrophin C- and N-terminal ends (respectively named K7 and B1) were produced as follows. Utrophin C-terminal end (residues 3423–3433) was used as a specific antigenic sequence and the K7 antibody was obtained by injecting the KLH-linked peptide as antigen, according to a previously described protocol [37]. The utrophin B1 antibody was produced as a recombinant protein. The corresponding DNA sequence (5302–5810; accession number NCBI X69086) coding for residues 1767–1810 of human utrophin was produced by RT-PCR using human total RNA from muscle cDNA, as previously described [38, 39]. All antibodies were tested in competition with corresponding synthetic peptides on cryostat sections or Western blots. No immunofluorescence labeling or protein bands were observed when peptides were applied. Antibodies against slow MHC I and fast MHC II isoforms were purchased from Sigma. MyoD antibody was purchased from BD Bioscience and desmin antibody from Chemicon International.
Histochemistry and MHC electrophoresis separation
ATPase myosin heavy chain (MHC) activity on a fresh serial cryostat section of muscles was used to determine fiber type in two conditions, an acid pH of 4.6 and a basic pH of 9.4, according to a previous protocol [40, 41]. MHC electrophoresis was performed according to a previously described protocols [29]. Myosin was isolated according to D’Albis et al. [42]. MHC isoforms were separated in 8% SDS-PAGE; gels were stained with a Biorad Silver Stain Plus Kit and scanned with the Fotolook/AGFA Duoscan system [43]. The percentages of the different MHC isoforms were quantified by the NIH Image program.
Immunofluorescence light microscopy
Cryostat sections (10 μm) of unfixed muscles were labeled with the antibodies described above. Immunoreactions were detected with Cy3- or FITC-conjugated sheep anti-rabbit IgG (Euromedex). Neuromuscular junctions were visualized with coupled bungarotoxin-fluorescein (FITC) (Sigma).
Total protein extract and Western blotting
0.01 g of fresh muscle tissue was homogenized in 150 μl of 5% SDS buffer (50 mM Tris/HCl, pH 8.0, 10 mM EDTA, 5% SDS) supplemented with 1% trypsin inhibitor and 1% saponin. After centrifugation (10 min at 13000 g), protein concentration was estimated in the supernatant using the BCA protein assay kit (Pierce). 50 μl of SDS buffer containing 0.01% bromophenol blue, 10% glycerol and 5% beta-mercaptoethanol was added to the supernatant and each sample was denatured for 5 min at 100°C. Protein extracts were submitted in duplicate to SDS-PAGE (3–10% or 5–15%). One resulting gel was Coomassie blue stained and the other was transferred onto 0.2 μm nitrocellulose membrane. Each blot was blocked in Tris-buffered saline with 0.1% Tween 20 (TBST) containing 3% bovine serum albumin (w/v). All membranes were incubated with primary antibodies for 1 h at room temperature. After labeling, the membranes were washed in TBST and then incubated with a phosphatase-labeled second antibody (Jackson ImmunoResearch Laboratory, appropriate dilution). Antibody-bound proteins were detected with NBT/BCIP substrate.
Immunoprecipitation
Immunoprecipitaion was performed using monoclonal antibody 43DAG/8D5 as previously described [44, 45]. Crude muscle membrane fraction was prepared from fresh tissues homogenized in a Triton X100 lysis buffer (120 mM NaCl and 1% Triton X100) supplemented with 10 mM iodoacetamid, 1% saponin, 1% trypsin inhibitor, 20 mM PMSF and 1% leupeptin. Homogenates were centrifugated at 140000 g for 30 min at 4°C. The supernatant was incubated with 43DAG/8D5 antibody overnight at 4°C with gentile agitation. The protein-antibody complex was mixed with magnetic protein A micro-beads (MACS: Miltenyi Biotec). After extensive washing with a buffer containing 0.1% Triton X100, 50 mM Tris/HCl pH 8, 120 mM NaCl, 0.75 mM bezamedine and 0.1 mM PMSF, the magnetically labeled immune complex was purified over a micro-column placed in the magnetic field of the MACS separator. The bound fractions were eluted by a buffer containing 0.1% Triton X100, 50 mM Tris/HCl pH 7.4, 0.35 N-acetyl-D-glucosamine, 0.75 mM bezamedine and 0.1 mM PMSF. The experiments were performed in duplicate using 12-month-old mdx mice.
RT-PCR
Total RNA was isolated from cryoconserved muscle with SV total RNA isolation kit (Promega) following the manufacturer’s instructions. We used 0.1 microgram of total RNA to generate first strand cDNA sequences with random hexanucleotide primers and the M-MLV reverse transcriptase (invitrogen). Of the RT reaction, 12 μl were subsequently used for each PCR reaction in 25 μl using the Taq Polymerase from Qbiogene. Three PCR reactions were performed to amplify the mouse Up395 isoform using the primers (mUp-ex17/20-F and mUp-ex17/20-F) previously described [24], the mouse Up71 using a forward primer located in the unique first exon (FUP71, 5′-TTGAATACTGAGTAATAATTGAGT-ACTAG-3′), and a reverse primer located in exon 63 RUp71, 5′-AAGAGCT-CATTTTAGGATGAT-3′); and the mouse β-DG gene (Fdag1–5′-GGAGGCTGT-TCCCACCGTGGT-3′; Rdag1, 5′-CTCTGCATTCT-GTTCAACAGATCG-3′). The obtained PCR products were of 383-pb (mUp395), 220-pb (mUp71) and 474-pb (dag1), respectively. PCR conditions included 94°C for 5 min, 35 cycles of 94°C for 30s, 60°C for 30s, 72°C for 1 min, and a final extension of 72°C for 8 min. Amplification of the ubiquitously expressed phosphoglucomutase (PGM1) gene using the specific primers which amplify mouse PGM1 as previously described [24]. Negative controls in which total RNA was replaced with RNAse free water or reactions mixtures without RT (for DNA contamination) were performed. PCR products were visualized on 1.5% agarose gel. The 100-pb molecular mass markers (Promega) were used to estimate the molecular mass of the PCR products.
Morphometrical Analysis
Ten-μm transversally cryostat sections of mdx TA and diaphragm in each age were stained by hematoxylin and eosine (H&E), which allow to distinguish between healthy myofibers, showing peripheral nuclei, regenerating myofibers, and degenerating fibers, often present in areas where also small regenerating fibers and cell infiltrates were visible. Morphometrical analysis was performed on 10 cross-sections from each TA and diaphragm. The following parameters were evaluated and treated as previously described [46–48]: 1) percentage of centrally nucleated fibers referred to the total numer, 2) percentage of total nonmuscle area (fibrotic or adipose tissue) and 3) variance coefficient of the muscle fiber size determined with the minimal fret’s diameter method [47].
Scanning densitometry and statistical analysis
Using the NIH Image software package, the relative optical density of the protein band on Western blot membranes was calculated. Values are means ± SEM. Student’s test was used (P<0.05 was considered significant).
Results
Conserved β-DG level in mdx diaphragm
Immunofluorescence studies showed a decrease in α-DB1 and α-SG at the sarcolemma of both mdx diaphragm and TA compared with C57BL/10 muscles (not shown). Interestingly, β-DG seemed to be maintained in the mdx diaphragm membrane but not in the other mdx skeletal muscles such as TA (Fig. 1A). Western blot detection and protein quantification in each muscle confirmed this result and revealed a decreasing level of α-DB1 and α-SG in diaphragm and TA, whereas the β-DG level was maintained only in mdx diaphragm and significantly decreased in TA compared with wild-type C57BL/10 muscle (Fig. 1B).
Figure 1. β-DG, α-DB and α-SG in 2-month-old mdx and C57BL/10 mice.
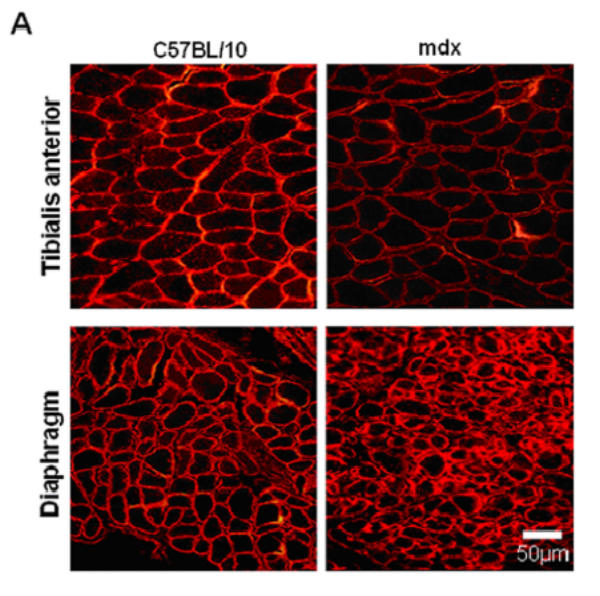
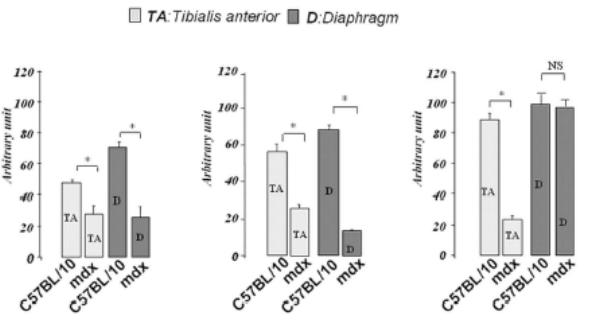
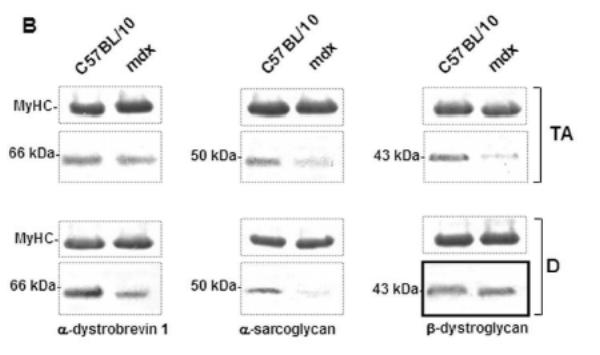
A: Immunofluorescence detection of β-DG on cryostat sections of diaphragm and tibialis anterior (TA) from mdx and wild-type C57BL/10 mice. As shown, high immunostaining of β-DG in mdx diaphragm was observed in comparison with TA muscle sections. B: Western blotting of α-DB1, α-SG and β-DG corresponds to total protein extracts of TA (tibialis anterior) and D (diaphragm) muscles. Quantification of protein bands by the NIH Image software package to determine the relative optical density of the protein band (arbitrary unit) is presented under the blots (histograms). The results are mean ± SEM of two independent experiments performed in triplicate. Asterisks denote significant difference (P ≤ 0.05) and NS indicates no significant result with the unpaired t test.
Utrophin accumulation in mdx diaphragm sarcolemma
Immunofluorescence using utrophin antibodies and α-bungarotoxin showed labeling of neuromuscular junctions in all C57BL/10 muscle sections (Fig. 2A). The labeling intensity between C-terminal and N-terminal antibodies was similar in mdx TA muscle (Fig. 2B, images c and d, respectively) but clearly different in diaphragm (Fig. 2B, images a and b). Interestingly, some specific areas and some cells appeared more intensively stained with C-terminal antibody but not with N-terminal antibody (Fig. 2B, dashed square).
Figure 2. Utrophin distribution in C57BL/10 and mdx diaphragm and Tibialis anterior membrane.
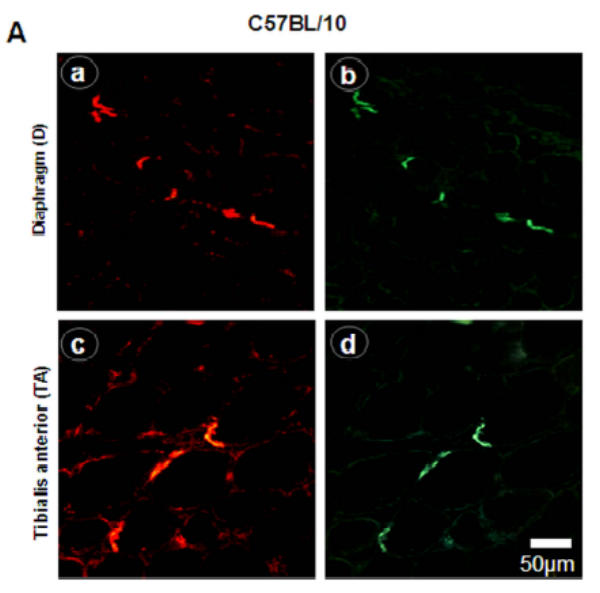
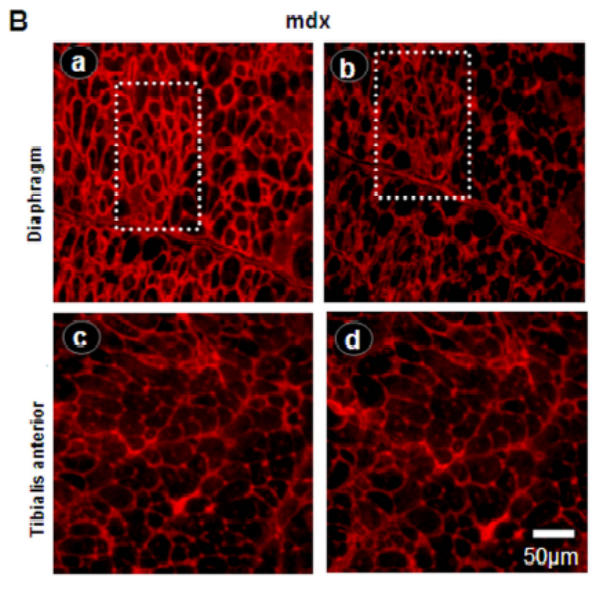
A: NMJ viewed by the C-terminal utrophin antibody (images a and c) and fluorescein α-bungarotoxin (images b and d) on serial cryostat sections C57BL/10 diaphragm and Tibialis anterior muscles C57BL/10. B: Utrophin stains on mdx diaphragm and Tibialis anterior muscles with C-terminal (images a and c) and N-terminal (images b and d) antibodies showing the utrophin distribution all along the sarcolemma. Importantly, only C-terminal antibody showed high fiber staining in mdx diaphragm (image a, dashed square).
Short utrophin isoform (Up71) in mdx diaphragm
A 70 kDa utrophin isoform was detected by the C-terminal antibody (K7) in mdx diaphragm but not in the mdx TA muscle extract (Fig. 3A). Up71 was detected in all analyzed mdx diaphragms, and it steadily increased between 6- and 12-months-old mice (Fig. 3A). Moreover, using the N-terminal antibody (B1) on the same extracts, the 70 kDa band was not detected in diaphragm or TA (Fig. 3B). Subsequently, β-DG Western blot detection on 6 to 16-month-old diaphragm extracts showed an increasing level of this protein until 12-month-old (Fig. 3C).
Figure 3. Up71, Up395 and β-DG expression in mdx diaphragm.
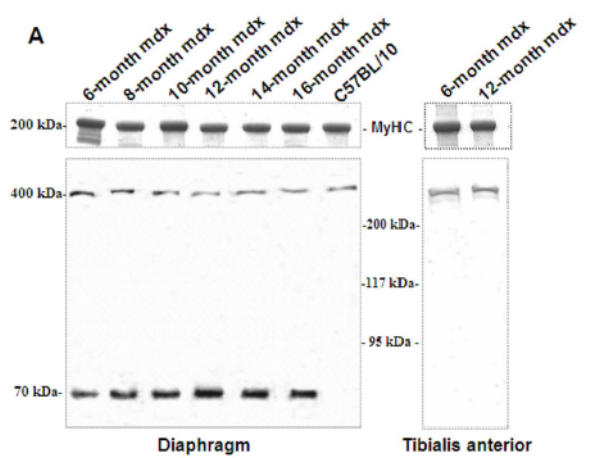
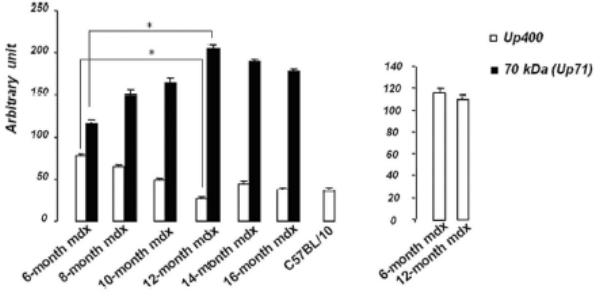
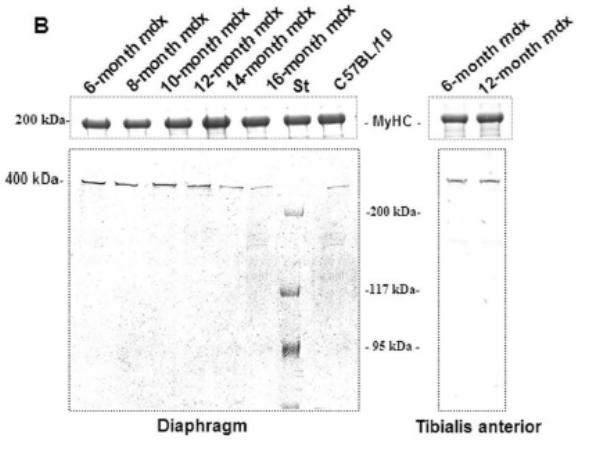
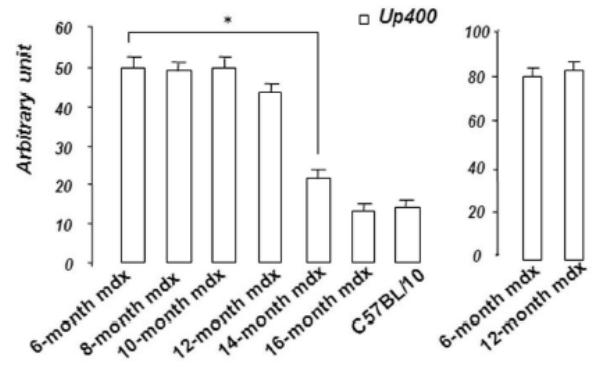
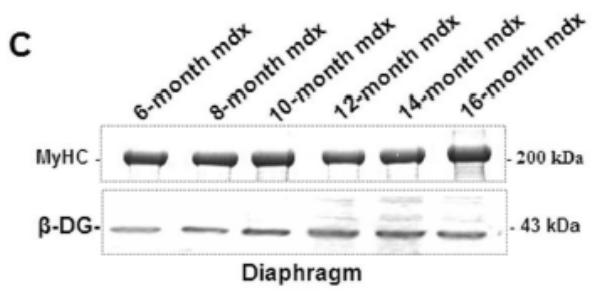
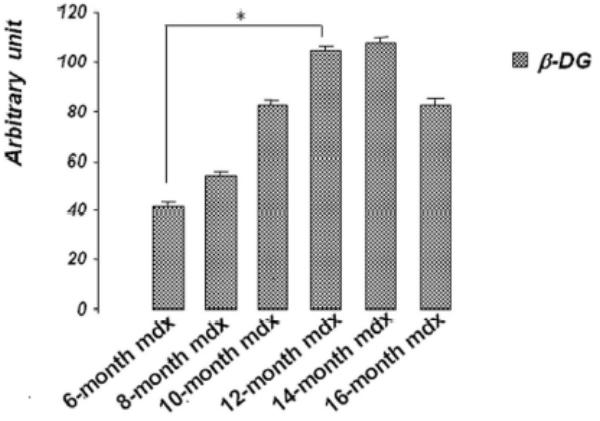
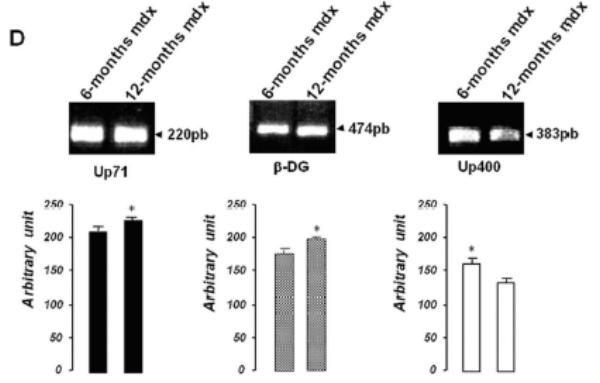
A: Western blot detection of utrophin isoforms by C-terminal utrophin antibody. Only the full-length Up395 isoform in mdx TA samples was detected, whereas mdx diaphragm (from 6 to 16 months) presented an additional protein band with Mr 70 kDa (Up71). In addition, a decline in full-length utrophin level was observed while the short isoform Up71 increase during the same time. B: Western blot detection with N-terminal utrophin antibody showed only full-length utrophin Up395 in both mdx diaphragm and TA samples. C: β-DG Western blots in mdx diaphragm extracts from 6- to 16-month-old mice revealed an increase in level up to 12 months. D: RT-PCR analysis of Up71, Up395 and β-DG transcripts. PCR products after cDNA amplification of Up71, Up395 and β-DG of 6- and 12-month-old mdx diaphragm samples revealed the abundance of Up71 and β-DG transcripts at these ages compared with full-length utrophin transcript. Increased expression of Up71 and β-DG was noted between 6- and 12-month-old mdx diaphragms, whereas Up395 was decreased. The 100-pb molecular mass markers (Promega) were used to estimate the molecular mass of the PCR products. Graphical representations show Western blot and PCR band quantification; the results are mean ± SEM of two independent experiments performed in triplicate. Asterisks denote significant difference (P ≤ 0.05).
To further confirm our results, we performed RT-PCR analyses of Up71, β-DG and Up395 mRNA in mdx muscles using specific primers for each transcript. Up71 mRNA was highly detected in 6- and 12-month-old mdx diaphragms (Fig. 3D, left panel) but was weakly expressed in the other skeletal muscles (Tibialis anterior, Vastus lateralis) and C57BL/10 muscles (not shown). In contrast, Up395 expression was decreased from 6- to 12-month-old mdx diaphragms (Fig. 3D, right panel). The β-DG expression was very similar to that of Up71, with an increase up to 12 months (Fig. 3D, central panel). These results led us to assume that the intensive labeling in mdx diaphragm sarcolemma with C-terminal utrophin was due to the presence of the Up71 utrophin isoform, in accordance with previous results.
Immunoprecipitation assay
In an attempt to establish a relationship between Up71 and β-DG, we performed a co-immunoprecipitation assay using 43DAG/8D5 C-terminal β-DG antibody in diaphragm and TA muscle membrane preparations. Twelve-month-old mdx mice were selected according to the relative abundance of Up71 isoform. Our results showed that Up395 immunoprecipitated with β-DG antibody in the TA samples, whereas Up71 was largely detected in mdx diaphragm (Fig. 4A).
Figure 4. Co-immunoprecipitation of utrophin isoforms and β-DG.
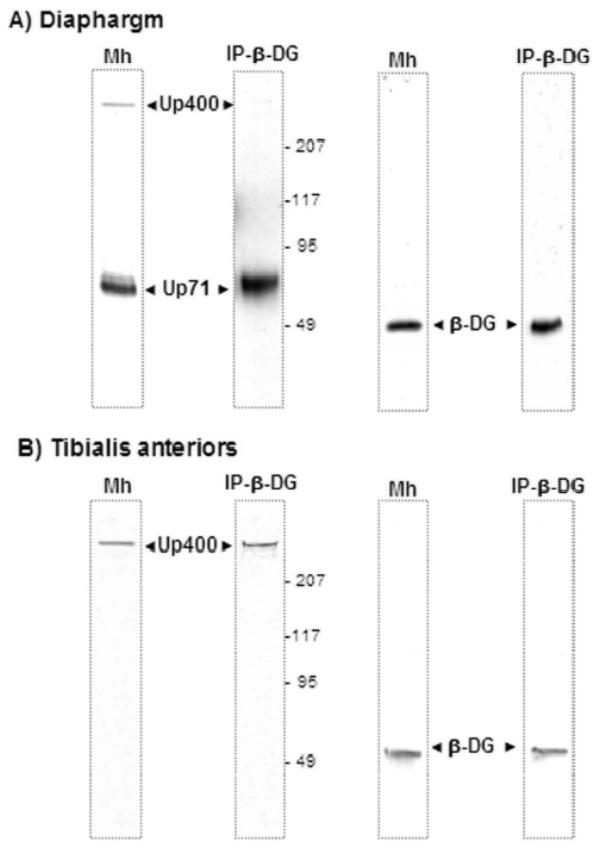
Western blots revealed by C-terminal utrophin and β-DG antibodies in membrane homogenate (Mh) and after co-immunoprecipitation of utrophin products with β-DG (IP-β-DG) respectively in mdx diaphragm (panel A) and Tibialis anterior (panel B) mdx muscles. Panel A showed that Up71 was present in both membrane homogenate (Mh) and eluted fraction of immunoprecipitation assay with β-DG antibody (IP-β-DG). In a second part Mh and eluted fraction were revealed by an antibody directed against β-DG. The same experiments were applied on Tibialis anterior muscle (panel B). No Up71 was detected in Mh and after IP-β-DG.
Up71 and β-DG are not preferentially accumulated in mdx diaphragm MHC I fibers
In order to establish a correlation between the time course expression of Up71 and MHC I fiber colonization of mdx diaphragm, we tested the level of MHC isoforms. In 6-month-old C57BL10 diaphragm, we observed a predominance of MHC IIa, MHC IIx and MHC IIb isoforms, whereas the MHC I phenotype was weakly represented (Fig. 5A). The 12-month-old mdx mice exhibited a number of important differences in MHC expression compared with the young mdx and control mice. The percentage of type I and type IIa MHC fibers was significantly higher in the 12-month-old mdx diaphragm; however, the IIb fiber population was even absent in the old mdx animal. In a second step of the experiment, serial sections of 12-month-old mdx diaphragm were labeled with MHC I (a), MHC II (b), C-terminal utrophin (c) and β-DG (d) antibodies (Fig. 5B). There was no evident relationship between utrophin and β-DG sarcolemmal accumulation with MHC type I fibers. Moreover, ATPase fiber activity on serial section showed that some MHC IIa and MHC IIb/IIx fibers have a high utrophin isoform and β-DG expression pattern in their sarcolemma (Fig. 5C).
Figure 5. Utrophin/β-DG accumulation and mdx diaphragm fiber types.
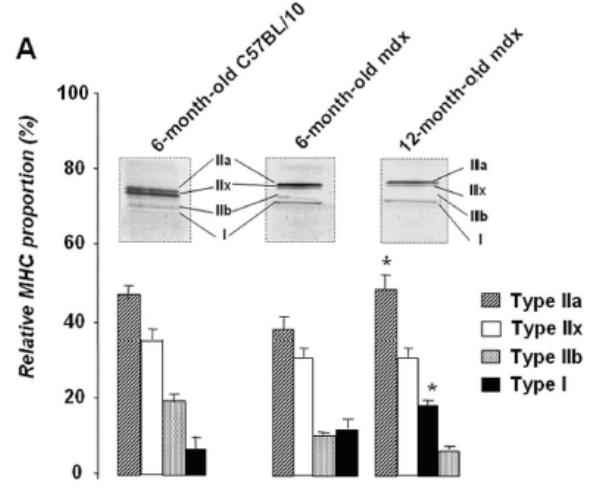
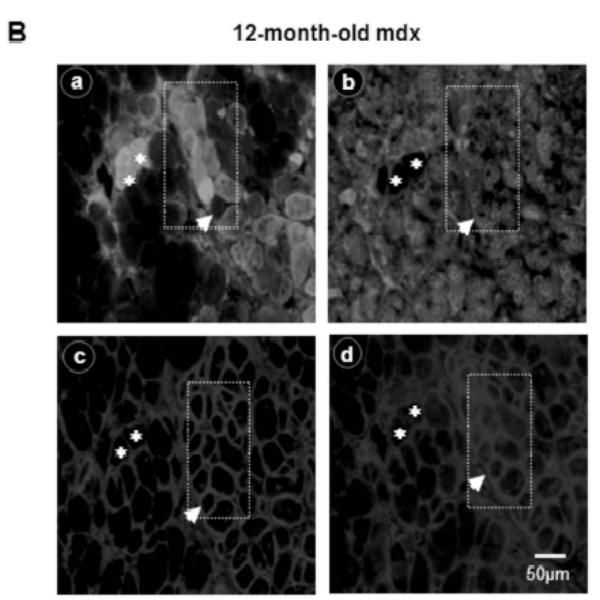
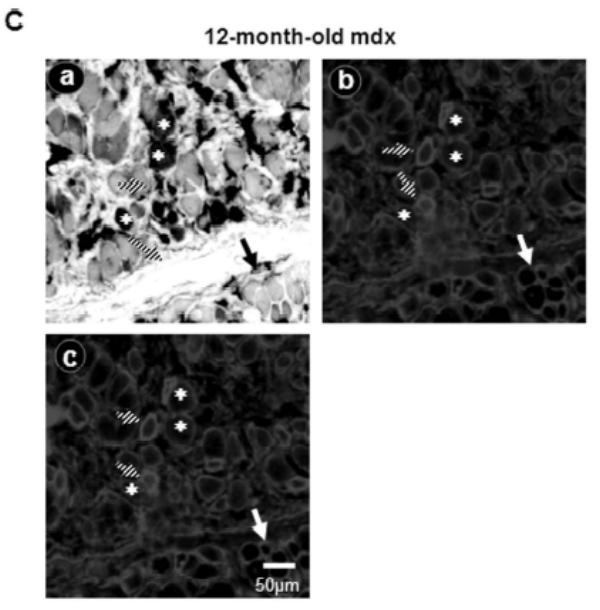
A: Relative proportion of diaphragm fiber types in 6- and 12-month-old mdx mice compared with C57BL/10. MHC isoforms were separated on 8% gel electrophoresis and the histogram shows the relative percentages of MHC isoforms of mdx and C57BL/10 diaphragms. B: Serial cross-sections of 12-month-old mdx stained with FITC-MHC I (a), FITC-MHC II (b), Cy3-C-terminal utrophin (c) and Cy3-β-DG antibodies (d). Asterisks show MHC I fibers without specifically high levels of utrophin and/or β-DG in their membranes. Dashed square and white arrowhead show fibers with high expression pattern of utrophin and β-DG in their membranes. C: Serial sections show ATPase activity (pH 4.6) of 12-month-old mdx diaphragm fibers (a), utrophin C-terminal (b) and β-DG antibodies (c). MHC IIa fibers have a milder ATPase activity in acid conditions (pH 4.6) and present an intermediate coloration, while MHC IIb fibers have white coloration. Asterisks show MHC I fibers (a) with a weak labelling of utrophin and β-DG in their membranes. Big arrows show some glycolytic MHC IIb/IIx fibers with a milder level of utrophin and β-DG expression pattern and hatched arrows show fibers with high utrophin and β-DG expression pattern.
Up71 is accumulated in regenerative clusters of mdx diaphragm
Serial sections labeled with C-terminal utrophin antibody and desmin antibody showed strong utrophin labeling in the multinucleated and central nucleated fibers observed in muscle regenerative processing (Fig. 6A) indicating a regenerative activating process in 12-month-old mdx diaphragm. These data demonstrate a possible implication of Up71, which is the major utrophin product, in association with β-DG in the regenerative process which takes place in mdx diaphragm. Western blot detection of MyoD (Fig. 6B) showed a significant increase in its level from 6-month to 12-month-old mdx diaphragm in comparison with the level in C57BL/10 diaphragm and TA muscle. Importantly the morphometrical study of mdx diaphragm compared with TA muscle showed that in 12-month-old mice this muscle showed an accentuated dystrophic pattern, elevated percentage of central nuclei fibers, invasion of connective tissue and high muscle fiber sizes heterogeneity (See Table).
Figure 6. Up71 and β-DG accumulation in regenerative/degenerative clusters of mdx diaphragm.
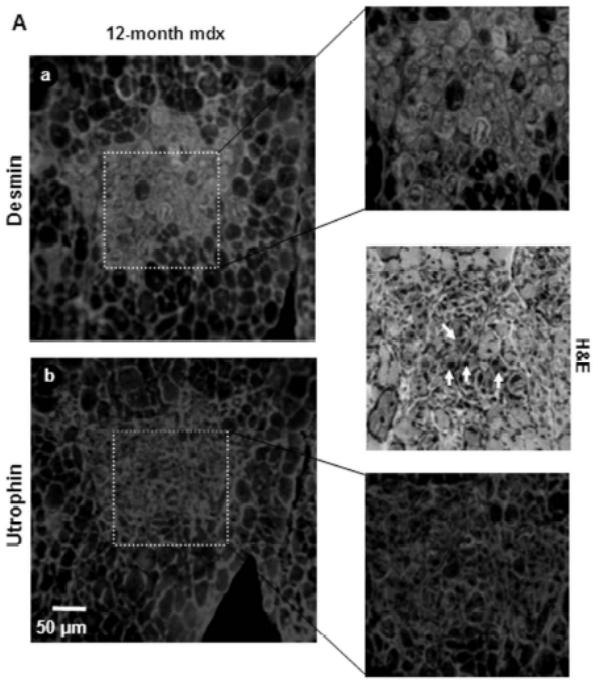
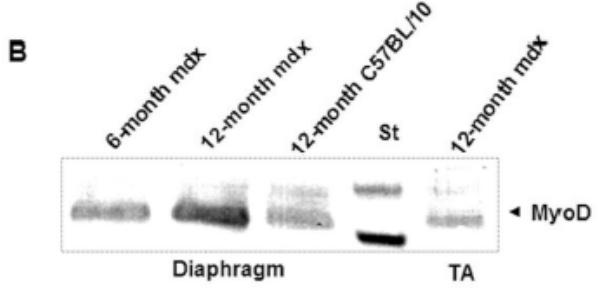
A: Serial sections of 12-month-old mdx diaphragm were labeled with desmin antibody revealed with FITC-conjugated antibody (a) and utrophin C-terminal antibody revealed with Cy3-conjugated antibody (b). Desmin labeling showed regenerative fibers (dashed square) with a high level of utrophin. When examined with H&E staining, these aggregate fibers were in majority centro-nucleated (white arrows). B: Western blot detection of MyoD in 6-month-old and 12-month-old mdx diaphragm compared with age-matched (12-month) mdx Tibialis anterior (TA) and C57BL/10 diaphragm.
Table - Morphometrical parameter of TA and diaphragm form 2 to 16-month-old mdx mice.
Variance coefficient of the muscle fiber sizes determined with the minimal ‘Fret’s diameter’ method in Tibialis anterior (TA) and diaphragm muscles from 2 to 16-month-old mdx mice. Percentage of centrally nucleated fibers and non muscle area (the space occupied by connective tissue over a standard section of 8 cem2) determined in hematoxylin-eosin stained sections (at least ten sections for each muscle and five animals per age) are also quantified to establish a comparative profile of the dystrophic pattern progression in these tow muscles. At 12-month-old mdx mice diaphragm showed a significant difference of the dystrophic pattern compared to age matched TA muscle.
| Mdx mice age (months) | Variance coefficient TA | Variance coefficient DIA | % Centrally nucleated fibers TA | % Centrally nucleated fibers DIA | Non-muscle area TA (cm2) | Non-muscle area DIA (cm2) |
|---|---|---|---|---|---|---|
| 2 | 375 ± 12 | 367 ± 14 | 42 ± 3 | 35 ± 2 | 0.633 ± 0.73 | 0.745 ± 0.064 |
| 6 | 395 ± 15 | 412 ± 17 | 34 ± 2 | 25 ± 4 | 0.007 ± 0.083 | 0.342 ± 0.078 |
| 8 | 425 ± 11 | 460 ± 21 | 45 ± 5 | 55 ± 6 | 0.0232 ± 0.0091 | 0.364 ± 0.082 |
| 10 | 456 ± 11 | 460 ± 21 | 55 ± 4 | 72 ± 7 | 0.442 ± 0.12 | 0.557 ± 0.094 |
| 12 | 485 ± 17 | 519 ± 26 | 70 ± 8 | 91 ± 4* | 0.625 ± 0.101 | 0.872 ± 0.105 |
| 14 | 490 ± 23 | 525 ± 32 | 75 ± 7 | 92 ± 11 | 0.675 ± 0.097 | 0.916 ± 0.103 |
| 16 | 500 ± 31 | 527 ± 33 | 85 ± 9 | 90 ± 12 | 0.713 ± 0.113 | 0.937 ± 0.110 |
Significantly different by impaired Student’s t-test for *P < 0.05.
Discussion
Although murine X-linked muscular dystrophy (mdx) and DMD muscle are both characterized by complete absence of dystrophin, the limb muscle weakness and progressive degeneration of adult mice is different from DMD muscles. The mdx diaphragm exhibits degenerative pattern similar to DMD skeletal muscle. Molecular mechanisms implied in this phenotypic difference are of major importance and may contribute to develop new therapeutic strategy to slow down or stop degenerative process in DMD muscles.
In the present study, we demonstrated that in mdx mice, β-DG expression was dramatically reduced in skeletal muscles such TA and VL but not in diaphragm. The short utrophin isoform Up71 is detected in mdx diaphragm but not in other mdx nor in C57BL/10 muscle. Wilson et al have detected the mRNA of Up71 and the Up140 (another short utrophin transcript) transcripts in adult and fetal human and mouse tissue. In that study no western blot detection of these isoforms was shown and no precision about the analyzed muscles was cited. In another hand the authors showed that the Up71 mRNA was weakly expressed in muscle tissue. In our study we have analyzed three skeletal muscles, TA, VL (Not shown) and diaphragm in normal and mdx mice (from 2 to 12-month old). Despite the mdx diaphragm no Up71 was detected in western blot in the other tissue. In accordance with Wilson et al the RT-PCR analysis showed a very weak expression of this isoform in all normal muscles, but in mdx mouse diaphragm, Up71 was largely expressed. This difference suggest that posttranscriptional and posttranslational events could take place in the regulation of Up71 in diaphragm and the other skeletal muscles. To date, this is the first demonstration that Up71 is specifically expressed in mdx diaphragm sarcolemma and its expression correlates with evolution of pathological pattern of this muscle according to the morphometrical study (see Table). Subsequently, full-length utrophin (Up395) showed a decreased expression in mdx diaphragm while it remains stable in age-matched mdx TA muscles. The short transcript Up71 is predicted to encode protein that has lost the N-terminal actin-binding domain and part of or the entire long spectrin-like rod region. Up71 retains sequences that encode the C-terminal domain that mediates binding to membrane proteins and it seems probable that their cellular function involves interactions with cell membrane, especially with β-DG. As in Up71, the Dp71 (dystrophin product with a Mr 71 kDa) did not contain the N-terminal part of the parental molecule (dystrophin full length), and fails to link F-actin. Then even with partial restoration of the DAPC such Dp71 transgenes fails to prevent muscle degeneration [49–51]. It is possible that the pathological pattern of the mdx diaphragm is due to the concurrent presence of full-length utrophin and the short isoform Up71. By co-immunoprecipitation analysis, our results showed that Up71 may compete with Up395 in β-DG site attachment in mdx diaphragm sarcolemma. In the other analyzed muscles immpunoprecipitation with the β-DG antibody showed exclusively the Up395 in the crude membrane fraction. Likeness in the dystrophic process between mdx diaphragm and DMD muscle led suggested the same competition between full and short length utrophin for β-DG anchoring at the cell membrane of DMD muscle.
In Our experiment we demonstrated that Up71 and β-DG accumulation does not specifically concern slow oxidative MHC I fibers, as previously described with utrophin A expression in soleus muscle [19]. The utrophin A promoter contains an NFAT-binding site, and calcineurin and NFATc1 can increase the transcriptional activity of this promoter in cultured myogenic cells, as well as in oxidative slow MHC I fibers of soleus muscle [19, 52]. In our experiment, no specific activation of the calcineurin/NFATc1 pathway was observed between 6- and 12-month-old mdx diaphragms (Not shown) which demonstrate that utrophin A and the short isoform Up71 are differently regulated in muscle cells and possibly also during muscle development. Previous reports have shown in regenerating fibers that increase in full-length utrophin mRNA is transient and relatively rapid and that one week after muscle damage, utrophin returns to a low level [20]. This kinetic induction correlates with satellite cell activation. Thus, utrophin transcription is already activated in satellite cells before or at the time of fusion into fibers, suggesting that most of the utrophin present along the newly formed fibers is already synthesized at the time of myoblast fusion to myotubes [20]. However, further investigations about the role of the short utrophin isoforms in satellite cell differentiation and myotubes formation should clarify the implication of these isoforms in myogenisis, as recently described for short dystrophin isoform, Dp71 [53].
The mdx diaphragm reproduces the degenerative pattern observed in DMD muscles and showed a high regenerative/degenerative pattern. We tried to correlate Up71 and β-DG overexpression and the expression pattern of MyoD and desmin. Our results showed a high Up71 staining in regenerating fibers of mdx mice diaphragm according to desmin labeling and H&E staining in 12-month-old mdx diaphragm sections. These observations lead us to propose that β-DG may specifically anchor the short utrophin isoform Up71 in regenerative clusters of mdx diaphragm sarcolemma. Since diaphragm is one of the organs with higher presence of Up71, among striated muscle studied, we decided to start a length-term study focusing on the identification of any potential function of Up71 in diaphragm. Interestingly the heregulin and the phosphatase inhibitor, the okadoic acid and the GTPase Rho, all these signaling molecules have recently been described as activators of utrophin promoter A [55–58]. GTPase Rho is also found crucial for myogenesis induction such as the muscle-determining Factor MyoD, as well as that of few other muscle-specific genes [59]. Then the plasma membrane association and the stability of utrophin is significatively increased upon effect of an activate form of RhoA GTPase [58]. Even cytochalasin B treatment, which disrupt the F-actin cytoskeleton, appears to translocate utrophin from the plasma membrane, indicating that utrophin localization is dependent one the assembly of the F-actin submembranous distribution [58]. However, the Up71 found in abundancy in diaphragm was not containing the actin binding site present only in full length utrophin. So, the competitive binding to beta-dystroglycan between utrophin and Up71 to be anchor at the muscle cell membrane may involve lost in the F-actin network stability when Up71 win. The observation that significant amount of Up71 along the plasma membrane places this protein on the list of potential competitor toward utrophin leading to maintain membrane instability due to dystrophin deficiency. Then in diaphragm, the compensation of dystrophin loss by replacement with utrophin cannot be reached because of Up71 presence. This is mainly due to the particularity of this muscle. In fact it is now established that mechanical stress applied axially and transversally to skeletal muscle fibers involves distinct signaling pathway in response [60]. In particular, diaphragm, that is have axial muscle that answer to both axial and transverse mechanical forces reveals also even more specific signal transduction pathways in response to various environmental stimuli and this feature was clearly demonstrate for example when integrin alpha7 is absent [61]. In addition diaphragm is the most damaged muscle among all other dystrophin-deficient muscles in mdx mouse. There is more regeneration cell clusters in this muscle in attempt to maintain performance in muscle contraction. These regenerating cells are differently regulated than mature muscle cells and there is certainly a regulatory molecular mechanism that could favors Up71 expression in an early stage of myogenisis. Preliminary results showed that expression level of Up71 in primary myoblastes of mdx diaphragm was higher than that observed when cells fused in myotubes (No published data).
The presence of Up71 protein appears particular and its function needs to be identified by further studies in diaphragm. We need also to define more precisely the regulatory molecular mechanism of its expression, that could corresponds to novel alternate mechanism, to hope mdx diaphragm and powerful all DMD muscles with complete utrophin compensation as this is reached in other mdx skeletal muscles where Up71 is absent.
Acknowledgments
We thank Dr. Guillaume Py (UFR STAPS, Montpellier, FRANCE) for his technical help. This work was supported by the “Association Française contre les Myopathies” (AFM, Fellowship N° 10529), INSERM and CNRS.
The abbreviations used are
- mdx
X-chromosome-linked muscular dystrophy
- Up
utrophin product
- NMJ
neuromuscular junction
- DMD
Duchenne muscular dystrophy
- DAPC
dystrophin-associated protein complex
- TA
Tibialis anteriors
- MHC
myosin heavy chain
- β-DG
beta-dystroglycan
- α-SG
alpha-sarcoglycan
- NBT
p-nitroblue tetrazolium
- BCIP
5-bromo-4-chloro-3-indoylphosphate substrate
- NFATc1
nuclear factor-activated T cells 1
- MyoD
myogenic differentiation protein
References
- 1.Chakkalakal JV, Thompson J, Parks RJ, Jasmin BJ. Molecular, cellular, and pharmacological therapies for duchenne/becker muscular dystrophies. Faseb J. 2005;19:880–91. doi: 10.1096/fj.04-1956rev. [DOI] [PubMed] [Google Scholar]
- 2.Ahn AH, Kunkel LM. The structural and functional diversity of dystrophin. Nat Genet. 1993;3:283–91. doi: 10.1038/ng0493-283. [DOI] [PubMed] [Google Scholar]
- 3.De Bleecker JL, Engel AG. Expression of cell adhesion molecules in inflammatory myopathies and duchenne dystrophy. J Neuropathol Exp Neurol. 1994;53:369–76. doi: 10.1097/00005072-199407000-00008. [DOI] [PubMed] [Google Scholar]
- 4.Matsumura K, Campbell KP. Dystrophin-glycoprotein complex: Its role in the molecular pathogenesis of muscular dystrophies. Muscle Nerve. 1994;17:2–15. doi: 10.1002/mus.880170103. [DOI] [PubMed] [Google Scholar]
- 5.Higuchi I, Niiyama T, Fukunaga H, Nakamura K, Nakagawa M, Osame M. Dystrophin-related protein in becker muscular dystrophy. Intern Med. 1994;33:334–6. doi: 10.2169/internalmedicine.33.334. [DOI] [PubMed] [Google Scholar]
- 6.Blake DJ, Weir A, Newey SE, Davies KE. Function and genetics of dystrophin and dystrophin-related proteins in muscle. Physiol Rev. 2002;82:291–329. doi: 10.1152/physrev.00028.2001. [DOI] [PubMed] [Google Scholar]
- 7.Love DR, Hill DF, Dickson G, Spurr NK, Byth BC, Marsden RF, Walsh FS, Edwards YH, Davies KE. An autosomal transcript in skeletal muscle with homology to dystrophin. Nature. 1989;339:55–8. doi: 10.1038/339055a0. [DOI] [PubMed] [Google Scholar]
- 8.Tinsley JM, Blake DJ, Roche A, Fairbrother U, Riss J, Byth BC, Knight AE, Kendrick-Jones J, Suthers GK, Love DR, et al. Primary structure of dystrophin-related protein. Nature. 1992;360:591–3. doi: 10.1038/360591a0. [DOI] [PubMed] [Google Scholar]
- 9.Blake DJ, Nawrotzki R, Peters MF, Froehner SC, Davies KE. Isoform diversity of dystrobrevin, the murine 87-kda postsynaptic protein. J Biol Chem. 1996;271:7802–10. doi: 10.1074/jbc.271.13.7802. [DOI] [PubMed] [Google Scholar]
- 10.Gramolini AO, Wu J, Jasmin BJ. Regulation and functional significance of utrophin expression at the mammalian neuromuscular synapse. Microsc Res Tech. 2000;49:90–100. doi: 10.1002/(SICI)1097-0029(20000401)49:1<90::AID-JEMT10>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 11.Deconinck AE, Potter AC, Tinsley JM, Wood SJ, Vater R, Young C, Metzinger L, Vincent A, Slater CR, Davies KE. Postsynaptic abnormalities at the neuromuscular junctions of utrophin-deficient mice. J Cell Biol. 1997;136:883–94. doi: 10.1083/jcb.136.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grady RM, Merlie JP, Sanes JR. Subtle neuromuscular defects in utrophin-deficient mice. J Cell Biol. 1997;136:871–82. doi: 10.1083/jcb.136.4.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsumura K, Ervasti JM, Ohlendieck K, Kahl SD, Campbell KP. Association of dystrophin-related protein with dystrophin-associated proteins in mdx mouse muscle. Nature. 1992;360:588–91. doi: 10.1038/360588a0. [DOI] [PubMed] [Google Scholar]
- 14.Blake DJ, Tinsley JM, Davies KE. Utrophin: A structural and functional comparison to dystrophin. Brain Pathol. 1996;6:37–47. doi: 10.1111/j.1750-3639.1996.tb00781.x. [DOI] [PubMed] [Google Scholar]
- 15.Jasmin BJ, Angus LM, Belanger G, Chakkalakal JV, Gramolini AO, Lunde JA, Stocksley MA, Thompson J. Multiple regulatory events controlling the expression and localization of utrophin in skeletal muscle fibers: Insights into a therapeutic strategy for duchenne muscular dystrophy. J Physiol Paris. 2002;96:31–42. doi: 10.1016/s0928-4257(01)00078-x. [DOI] [PubMed] [Google Scholar]
- 16.Jasmin BJ, Alameddine H, Lunde JA, Stetzkowski-Marden F, Collin H, Tinsley JM, Davies KE, Tome FM, Parry DJ, Cartaud J. Expression of utrophin and its mrna in denervated mdx mouse muscle. FEBS Lett. 1995;374:393–8. doi: 10.1016/0014-5793(95)01131-w. [DOI] [PubMed] [Google Scholar]
- 17.Gramolini AO, Jasmin BJ. Expression of the utrophin gene during myogenic differentiation. Nucleic Acids Res. 1999;27:3603–9. doi: 10.1093/nar/27.17.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson LA, Cooper BJ, Dux L, Dubowitz V, Sewry CA. Expression of utrophin (dystrophin-related protein) during regeneration and maturation of skeletal muscle in canine x-linked muscular dystrophy. Neuropathol Appl Neurobiol. 1994;20:359–67. doi: 10.1111/j.1365-2990.1994.tb00981.x. [DOI] [PubMed] [Google Scholar]
- 19.Chakkalakal JV, Stocksley MA, Harrison MA, Angus LM, Deschenes-Furry J, St-Pierre S, Megeney LA, Chin ER, Michel RN, Jasmin BJ. Expression of utrophin a mrna correlates with the oxidative capacity of skeletal muscle fiber types and is regulated by calcineurin/nfat signaling. Proc Natl Acad Sci U S A. 2003;100:7791–6. doi: 10.1073/pnas.0932671100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galvagni F, Cantini M, Oliviero S. The utrophin gene is transcriptionally up-regulated in regenerating muscle. J Biol Chem. 2002;277:19106–13. doi: 10.1074/jbc.M109642200. [DOI] [PubMed] [Google Scholar]
- 21.Gramolini AO, Jasmin BJ. Molecular mechanisms and putative signalling events controlling utrophin expression in mammalian skeletal muscle fibres. Neuromuscul Disord. 1998;8:351–61. doi: 10.1016/s0960-8966(98)00052-2. [DOI] [PubMed] [Google Scholar]
- 22.Weir AP, Burton EA, Harrod G, Davies KE. A- and b-utrophin have different expression patterns and are differentially up-regulated in mdx muscle. J Biol Chem. 2002;277:45285–90. doi: 10.1074/jbc.M205177200. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi J, Itoh Y, Fujimori K, Imamura M, Wakayama Y, Miyagoe-Suzuki Y, Takeda S. The utrophin promoter a drives high expression of the transgenic lacz gene in liver, testis, colon, submandibular gland, and small intestine. J Gene Med. 2005;7:237–48. doi: 10.1002/jgm.651. [DOI] [PubMed] [Google Scholar]
- 24.Wilson J, Putt W, Jimenez C, Edwards YH. Up71 and up140, two novel transcripts of utrophin that are homologues of short forms of dystrophin. Hum Mol Genet. 1999;8:1271–8. doi: 10.1093/hmg/8.7.1271. [DOI] [PubMed] [Google Scholar]
- 25.Ibraghimov-Beskrovnaya O, Ervasti JM, Leveille CJ, Slaughter CA, Sernett SW, Campbell KP. Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature. 1992;355:696–702. doi: 10.1038/355696a0. [DOI] [PubMed] [Google Scholar]
- 26.Hoyte K, Jayasinha V, Xia B, Martin PT. Transgenic overexpression of dystroglycan does not inhibit muscular dystrophy in mdx mice. Am J Pathol. 2004;164:711–8. doi: 10.1016/S0002-9440(10)63158-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fabbrizio E, Latouche J, Rivier F, Hugon G, Mornet D. Re-evaluation of the distributions of dystrophin and utrophin in sciatic nerve. Biochem J. 1995;312(Pt 1):309–14. doi: 10.1042/bj3120309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rivier F, Robert A, Latouche J, Hugon G, Mornet D. Presence of long and short dystrophin and/or utrophin products in torpedo marmorata peripheral nerves. FEBS Lett. 1996;378:272–6. doi: 10.1016/0014-5793(95)01476-4. [DOI] [PubMed] [Google Scholar]
- 29.Muller J, Vayssiere N, Royuela M, Leger ME, Muller A, Bacou F, Pons F, Hugon G, Mornet D. Comparative evolution of muscular dystrophy in diaphragm, gastrocnemius and masseter muscles from old male mdx mice. J Muscle Res Cell Motil. 2001;22:133–9. doi: 10.1023/a:1010305801236. [DOI] [PubMed] [Google Scholar]
- 30.Dupont-Versteegden EE, McCarter RJ. Differential expression of muscular dystrophy in diaphragm versus hindlimb muscles of mdx mice. Muscle Nerve. 1992;15:1105–10. doi: 10.1002/mus.880151008. [DOI] [PubMed] [Google Scholar]
- 31.Coirault C, Lambert F, Marchand-Adam S, Attal P, Chemla D, Lecarpentier Y. Myosin molecular motor dysfunction in dystrophic mouse diaphragm. Am J Physiol. 1999;277:C1170–6. doi: 10.1152/ajpcell.1999.277.6.C1170. [DOI] [PubMed] [Google Scholar]
- 32.Petrof BJ, Stedman HH, Shrager JB, Eby J, Sweeney HL, Kelly AM. Adaptations in myosin heavy chain expression and contractile function in dystrophic mouse diaphragm. Am J Physiol. 1993;265:C834–41. doi: 10.1152/ajpcell.1993.265.3.C834. [DOI] [PubMed] [Google Scholar]
- 33.Stedman HH, Sweeney HL, Shrager JB, Maguire HC, Panettieri RA, Petrof B, Narusawa M, Leferovich JM, Sladky JT, Kelly AM. The mdx mouse diaphragm reproduces the degenerative changes of duchenne muscular dystrophy. Nature. 1991;352:536–9. doi: 10.1038/352536a0. [DOI] [PubMed] [Google Scholar]
- 34.Reid WD, MacGowan NA. Respiratory muscle injury in animal models and humans. Mol Cell Biochem. 1998;179:63–80. doi: 10.1023/a:1006803703128. [DOI] [PubMed] [Google Scholar]
- 35.Ragusa RJ, Chow CK, St Clair DK, Porter JD. Extraocular, limb and diaphragm muscle group-specific antioxidant enzyme activity patterns in control and mdx mice. J Neurol Sci. 1996;139:180–6. [PubMed] [Google Scholar]
- 36.Rivier F, Robert A, Royuela M, Hugon G, Bonet-Kerrache A, Mornet D. Utrophin and dystrophin-associated glycoproteins in normal and dystrophin deficient cardiac muscle. J Muscle Res Cell Motil. 1999;20:305–14. doi: 10.1023/a:1005426920070. [DOI] [PubMed] [Google Scholar]
- 37.Rivier F, Robert A, Hugon G, Bonet-Kerrache A, Nigro V, Fehrentz JA, Martinez J, Mornet D. Dystrophin and utrophin complexed with different associated proteins in cardiac purkinje fibres. Histochem J. 1999;31:425–32. doi: 10.1023/a:1003805905456. [DOI] [PubMed] [Google Scholar]
- 38.Fabbrizio E, Leger J, Anoal M, Leger JJ, Mornet D. Monoclonal antibodies targeted against the c-terminal domain of dystrophin or utrophin. FEBS Lett. 1993;322:10–4. doi: 10.1016/0014-5793(93)81100-e. [DOI] [PubMed] [Google Scholar]
- 39.Pons F, Nicholson LV, Robert A, Voit T, Leger JJ. Dystrophin and dystrophin-related protein (utrophin) distribution in normal and dystrophin-deficient skeletal muscles. Neuromuscul Disord. 1993;3:507–14. doi: 10.1016/0960-8966(93)90106-t. [DOI] [PubMed] [Google Scholar]
- 40.Sher J, Cardasis C. Skeletal muscle fiber types in the adult mouse. Acta Neurol Scand. 1976;54:45–56. doi: 10.1111/j.1600-0404.1976.tb07619.x. [DOI] [PubMed] [Google Scholar]
- 41.Florini JR, Ewton DZ. Skeletal muscle fiber types and myosin atpase activity do not change with age or growth hormone administration. J Gerontol. 1989;44:B110–7. doi: 10.1093/geronj/44.5.b110. [DOI] [PubMed] [Google Scholar]
- 42.d’Albis A, Couteaux R, Janmot C, Mira JC. Myosin isoform transitions in regeneration of fast and slow muscles during postnatal development of the rat. Dev Biol. 1989;135:320–5. doi: 10.1016/0012-1606(89)90182-6. [DOI] [PubMed] [Google Scholar]
- 43.Gondret F, Lefaucheur L, D’Albis A, Bonneau M. Myosin isoform transitions in four rabbit muscles during postnatal growth. J Muscle Res Cell Motil. 1996;17:657–67. doi: 10.1007/BF00154060. [DOI] [PubMed] [Google Scholar]
- 44.Saito F, Masaki T, Kamakura K, Anderson LV, Fujita S, Fukuta-Ohi H, Sunada Y, Shimizu T, Matsumura K. Characterization of the transmembrane molecular architecture of the dystroglycan complex in schwann cells. J Biol Chem. 1999;274:8240–6. doi: 10.1074/jbc.274.12.8240. [DOI] [PubMed] [Google Scholar]
- 45.Yamada H, Saito F, Fukuta-Ohi H, Zhong D, Hase A, Arai K, Okuyama A, Maekawa R, Shimizu T, Matsumura K. Processing of beta-dystroglycan by matrix metalloproteinase disrupts the link between the extracellular matrix and cell membrane via the dystroglycan complex. Hum Mol Genet. 2001;10:1563–9. doi: 10.1093/hmg/10.15.1563. [DOI] [PubMed] [Google Scholar]
- 46.De Luca A, Nico B, Liantonio A, Didonna MP, Fraysse B, Pierno S, Burdi R, Mangieri D, Rolland JF, Camerino C, Zallone A, Confalonieri P, Andreetta F, Arnoldi E, Courdier-Fruh I, Magyar JP, Frigeri A, Pisoni M, Svelto M, Conte Camerino D. A multidisciplinary evaluation of the effectiveness of cyclosporine a in dystrophic mdx mice. Am J Pathol. 2005;166:477–89. doi: 10.1016/S0002-9440(10)62270-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Briguet A, Courdier-Fruh I, Foster M, Meier T, Magyar JP. Histological parameters for the quantitative assessment of muscular dystrophy in the mdx-mouse. Neuromuscul Disord. 2004;14:675–82. doi: 10.1016/j.nmd.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 48.Chazalette D, Hnia K, Rivier F, Hugon G, Mornet D. Alpha7b integrin changes in mdx mouse muscles after l-arginine administration. FEBS Lett. 2005;579:1079–84. doi: 10.1016/j.febslet.2004.12.081. [DOI] [PubMed] [Google Scholar]
- 49.Schulz RA, Yutzey KE. Calcineurin signaling and nfat activation in cardiovascular and skeletal muscle development. Dev Biol. 2004;266:1–16. doi: 10.1016/j.ydbio.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 50.Parsons SA, Wilkins BJ, Bueno OF, Molkentin JD. Altered skeletal muscle phenotypes in calcineurin aalpha and abeta gene-targeted mice. Mol Cell Biol. 2003;23:4331–43. doi: 10.1128/MCB.23.12.4331-4343.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Allen DL, Sartorius CA, Sycuro LK, Leinwand LA. Different pathways regulate expression of the skeletal myosin heavy chain genes. J Biol Chem. 2001;276:43524–33. doi: 10.1074/jbc.M108017200. [DOI] [PubMed] [Google Scholar]
- 52.Kegley KM, Gephart J, Warren GL, Pavlath GK. Altered primary myogenesis in nfatc3(−/−) mice leads to decreased muscle size in the adult. Dev Biol. 2001;232:115–26. doi: 10.1006/dbio.2001.0179. [DOI] [PubMed] [Google Scholar]
- 53.Swoap SJ, Hunter RB, Stevenson EJ, Felton HM, Kansagra NV, Lang JM, Esser KA, Kandarian SC. The calcineurin-nfat pathway and muscle fiber-type gene expression. Am J Physiol Cell Physiol. 2000;279:C915–24. doi: 10.1152/ajpcell.2000.279.4.C915. [DOI] [PubMed] [Google Scholar]
- 54.de Leon MB, Montanez C, Gomez P, Morales-Lazaro SL, Tapia-Ramirez V, Valadez-Graham V, Recillas-Targa F, Yaffe D, Nudel U, Cisneros B. Dystrophin dp71 expression is down-regulated during myogenesis: Role of sp1 and sp3 on the dp71 promoter activity. J Biol Chem. 2005;280:5290–9. doi: 10.1074/jbc.M411571200. [DOI] [PubMed] [Google Scholar]
- 55.Courdier-Fruh I, Barman L, Briguet A, Meier T. Glucocorticoid-mediated regulation of utrophin levels in human muscle fibers. Neuromuscul Disord. 2002;12(Suppl 1):S95–104. doi: 10.1016/s0960-8966(02)00089-5. [DOI] [PubMed] [Google Scholar]
- 56.Khurana TS, Rosmarin AG, Shang J, Krag TO, Das S, Gammeltoft S. Activation of utrophin promoter by heregulin via the ets-related transcription factor complex ga-binding protein alpha/beta. Mol Biol Cell. 1999;10:2075–86. doi: 10.1091/mbc.10.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodova M, Brownback K, Werle MJ. Okadaic acid augments utrophin in myogenic cells. Neurosci Lett. 2004;363:163–7. doi: 10.1016/j.neulet.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 58.Bonet-Kerrache A, Fortier M, Comunale F, Gauthier-Rouviere C. The gtpase rhoa increases utrophin expression and stability, as well as its localization at the plasma membrane. Biochem J. 2005;391:261–8. doi: 10.1042/BJ20050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carnac G, Primig M, Kitzmann M, Chafey P, Tuil D, Lamb N, Fernandez A. Rhoa gtpase and serum response factor control selectively the expression of myod without affecting myf5 in mouse myoblasts. Mol Biol Cell. 1998;9:1891–902. doi: 10.1091/mbc.9.7.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakamura A, Yoshida K, Ueda H, Takeda S, Ikeda S. Up-regulation of mitogen activated protein kinases in mdx skeletal muscle following chronic treadmill exercise. Biochim Biophys Acta. 2005;1740:326–31. doi: 10.1016/j.bbadis.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 61.Lopez MA, Mayer U, Hwang W, Taylor T, Hashmi MA, Jannapureddy SR, Boriek AM. Force transmission, compliance, and viscoelasticity are altered in the alpha7-integrin-null mouse diaphragm. Am J Physiol Cell Physiol. 2005;288:C282–9. doi: 10.1152/ajpcell.00362.2003. [DOI] [PubMed] [Google Scholar]


