Abstract
The X-linked Foxp3 is a member of the forkhead/winged helix transcription factor family. Germ-line mutations cause lethal autoimmune diseases in males. Serendipitously, we observed that Foxp3sf/+ heterozygous mice developed cancer at a high rate. The majority of the cancers were mammary carcinomas in which the wild-type Foxp3 allele was inactivated and ErbB2 was over-expressed. Foxp3 bound and repressed the ErbB2 promoter. Deletion, functionally significant somatic mutations and down-regulation of the FOXP3 gene were commonly found in human breast cancer samples and correlated significantly with HER-2 over-expression, regardless of the status of HER-2 amplification. In toto, the data demonstrate that FOXP3 is an X-linked breast cancer suppressor gene and an important regulator of the HER-2/ErbB2 oncogene.
Introduction
Identification of BRCA1 and BRCA2 marks a key advance in understanding the genetic defects responsible for breast cancer (Miki, 1994; Wooster, 1995). Several other genes, such as TP53, PIK3CA and PTEN, have also been implicated in familial and sporadic cancers (Samuels et al., 2004; Wooster, 2003). However, the genetic defects for breast cancer has yet to be fully elucidated. There is an important distinction between autosomal and X-linked genes, as many genes in the latter category are subject to X-inactivation, making it easier to fulfill Knudson’s two-hit theory (Knudson, 1971). As such, X-linked tumor suppressor genes can potentially be more important, as LOH or mutation of a single allele can in effect functionally silence the gene (Spatz, 2004). However, essentially all tumor suppressor genes are autosomal (Spatz, 2004), although tantalizing evidence concerning abnormalities in the X-chromosome, including LOH, skewed inactivation and selective loss, has been reported in breast cancer samples (Kristiansen et al., 2005; Piao and Malkhosyan, 2002; Richardson et al., 2006; Roncuzzi et al., 2002).
HER-2/Neu/ErbB2 is one of the first oncogenes to be identified (Schechter et al., 1984) and has been demonstrated to be expressed in a large proportion of cancer cells (Garcia de Palazzo et al., 1993). The level of HER-2/NEU is an important prognostic marker (Slamon et al., 1987). Anti-HER-2/NEU antibody Herceptin has emerged as an important therapeutic for patients with over-expressed HER-2/NEU on cancer tissues (Slamon et al., 2001). Given the clinical and therapeutic significance of Her-2/Neu/ErbB2 over-expression, it is important to identify the molecular mechanisms responsible for the over-expression. A well established mechanism responsible for HER-2 over-expression in human cancer is gene amplification (Slamon et al., 1987). However, it is unclear whether gene amplification alone is sufficient to cause HER-2 over-expression. Moreover, a significant proportion of human cancers with moderate over-expression of HER-2 do not show gene amplification (Bofin et al., 2004; Jimenez et al., 2000; Todorovic-Rakovic et al., 2005). It is therefore of great interest to identify regulators for HER-2 expression in breast cancer. In this context, Xing et al. (Xing, 2000) reported that DNA-binding protein PEA3 specifically targets a DNA sequence on the HER-2/neu promoter and down-regulates the promoter activity. It is less clear, however, whether genetic lesions of PEA3 can cause HER-2 over-expression.
Foxp3 was identified during position cloning of Scurfin, a gene responsible for X-linked autoimmune diseases in mice and humans (Immune dysregulation, polyendopathy, enterophathy, X-linked, IPEX) (Bennett, 2001; Brunkow, 2001; Chatila, 2000; Wildin, 2001). Serendipitously, we observed a high rate of spontaneous mammary cancer. Our systemic analyses reported herein demonstrate that the Foxp3 gene is a mammary tumor suppressor in mice and humans. Moreover, Foxp3 represses the transcription of the HER-2/ErbB2 gene via interaction with forkhead DNA binding motifs in the ErbB2 promoter.
Results
Spontaneous and carcinogen-induced mammary cancer in Foxp3sf/+ female mice
The mutant BALB/c mice we used for the initial study carried mutations in two closely linked X-chromosome genes, Foxp3sf and Otcspf. During the course of the study, a spontaneous segregation of Otcspf allowed us to obtain a BALB/c Otcspf/+ strain. Meanwhile, we obtained an independent line of Scurfy mice that had never been crossed to the Spf mutant mice and we backcrossed the Scurfy mutant allele (Foxp3sf) for more than 12 generations into the BALB/c background (Chang et al., 2005). Female mice with only one copy of the Foxp3 gene survived to adulthood and appeared normal within the first year of life (Godfrey et al., 1991) with normal T cell function (Fontenot et al., 2003; Fontenot et al., 2005; Godfrey et al., 1994). Our extended observations of the retired breeders for up to two years revealed that close to 90% of the Foxp3sf/+Otcspf/+ and Foxp3sf/+ mice spontaneously developed malignant tumors. Cancer incidences in the littermate controls and a line of congenic mice with a mutation in Otc but not Foxp3 were comparable with each other (Fig 1A, B). About 60% of the tumors were mammary carcinomas (Fig. 1A, C), although other tumors, such as lymphoma, hepatoma, and sarcoma were observed. Histological analyses revealed lung metastasis (Fig. 1A lower panels, based on expression of ER and/or PR, data not shown) in about 40% of the mice with mammary cancer. More than a third of the tumor-bearing mice had multiple lesions in the mammary glands. Most, although not all, mammary carcinomas expressed the estrogen receptor (ER+, 14/18) and progesterone receptor (PR+, 12/18).
Fig. 1.

Increased susceptibility to breast cancer in mice heterozygous for Foxp3sf. A. Representative breast cancers developed in female Foxp3sf/+Otcspf/+ mice. The top panel shows the gross anatomy while the lower panel shows the histology of local and metastatic lesions of a breast cancer. B. Cancer-free survival analysis of Foxp3sf/+, Foxp3sf/+Otcspf/+, Otcspf/+ and WT littermates. Mice were sacrificed when moribund to identify the tissue origins of cancers. Foxp3sf/+ vs. WT, P<0.0001; Foxp3sf/+ vs. Otcspf/+ , P=0.0003; Foxp3sf/+ vs. Foxp3sf/+Otcspf/+ , P=0 .9526; Foxp3sf/+Otcspf/+ vs. WT, P=0.0001; Foxp3sf/+Otcspf/+ vs. Otcspf/+, P=0.0001; Otcspf/+ vs. WT, P=0.4164. C. As in B, except that only incidences of mammary tumors were included. Foxp3sf/+ vs. Wt: P=0.00015, Foxp3sf/+Otcspf/+ vs. WT: P=0.00011. D. Increased susceptibility of Foxp3sf/+ mice to carcinogen DMBA and progesterone. The diagram on top depicts experimental protocol, while survival analysis is shown in the bottom panel. Foxp3sf/+ vs. WT, P<0.0001; Foxp3sf/+Otcspf/+ vs. Otcspf/+, P=0.0005; Otcspf/+ vs. WT, P=0.8157. In B & C, those mice that were observed for only part of the duration were incorporated as censored samples and were marked with a cross in the Kaplan-Meier survival curves. P values in B&C were derived from log-rank tests.
In order to focus on mammary cancer, we treated the mice with a carcinogen, 7,12-dimethylbenz [a] anthracene (DMBA), in conjunction with progesterone. Mice heterozygous for Foxp3sf, but not those heterozygous for Otcspf, showed substantially increased susceptibility to mammary cancer, as revealed by earlier onset, increased incidence (Fig. 1D) and multiplicity (data not shown) of the breast tumors. These data demonstrate that a mutation of Foxp3, but not Otc results in a major increase in susceptibility to mammary carcinoma.
Foxp3 expression in normal and cancerous mammary tissues
Since expression of Foxp3 has not been reported in mammary tissue, we isolated normal and cancerous cells by laser-capture microdissection (Supplemental Fig. S1A) and compared expression of Foxp3 and Otc by real-time RT-PCR and histochemistry. The complete absence of the cd3 transcripts (Fig. S1B) indicated that the micro-dissected samples were devoid of T cells, the main cell types known to express Foxp3 (Fontenot et al., 2005). A representative profile and summarized data of Foxp3 expression in Foxp3sf/+Otcspf/+ mice and age-matched WT control mice are shown in Fig. 2A. Foxp3 mRNA was detected in normal mammary epithelium from both the WT and Foxp3sf/+Otcspf/+ mice, but not in mammary cancer cells from the same Foxp3sf/+Otcspf/+ mice. Immunohistochemical staining (Fig. 2B) confirmed the loss of expression of Foxp3 in the mammary carcinoma generated from the Foxp3sf/+Otcspf/+ mice.
Fig. 2.
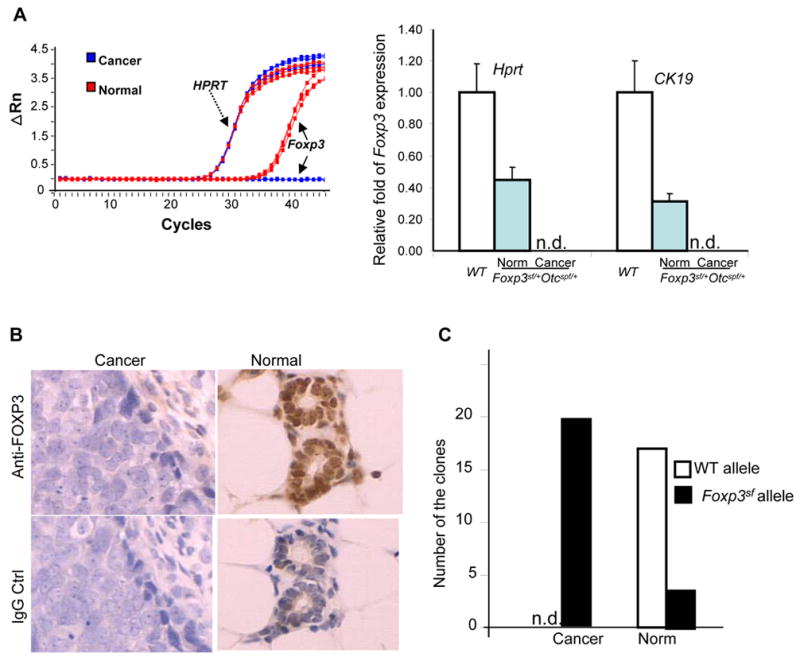
Inactivation of the WT Foxp3 allele in mammary cancer cells. A. Defective Foxp3 expression in breast cancer. RNA extracted from the cells isolated by LCM was subjected to quantitative real-time RT-PCR using primers specific for Foxp3, Hprt and CK19. In the left panel, fluorescence intensity (ΔRn) was plotted versus cycle number. Mean and S.D. from three individual mice per group are presented in the right panel (P<0.0001, one-way ANOVA test when either internal standard was used). B. Immunohistochemical staining of normal mammary glands and adenocarcinomas from a Foxp3sf/+Otcspf/+ mouse using rabbit anti-FOXP3 polyclonal antibody and normal rabbit IgG as the control. C. Specific silencing of the WT allele in breast cancer cells. Foxp3 transcripts were amplified from micro-dissected breast cancers or normal breast epithelium by two rounds of anchored PCR and were cloned into the TOPO vector and sequenced. The number of clones with sequences of WT or mutant alleles in the breast cancer and normal epithelium is presented. A total of 20 clones were sequenced from each group. Data shown are from pooled samples that lack CD3 transcripts. n.d., not detectable.
Foxp3 is an X-linked gene that is subject to X-chromosomal inactivation (Fontenot et al., 2005). We carried out an anchored RT-PCR and cloned the low levels of Foxp3 mRNA in the breast tissues. We sequenced the cDNA clones from pooled samples after ruling out potential T cell contamination (based on a lack of T-cell specific cd3 transcripts, Fig. S1B). As shown in Fig. 2C, 100% of the Foxp3 transcripts in the cancerous tissues were from the mutant alleles, which indicate that the wild-type allele was silenced in the tumor cells. In contrast, the transcripts from the mutant allele constituted 15% of the transcripts in the normal mammary samples from the same mice. Thus, the expression pattern of Foxp3 fulfills another criterion for a tumor suppressor gene.
FOXP3 is a repressor of ErbB2 transcription
Our characterization of the mammary tumors in the mutant mice revealed wide-spread up-regulation of ErbB2, in contrast to those rare tumors from WT mice, as shown in Figure 3A and supplemental Table S1. Using real-time RT-PCR, 8–12-fold more ErbB2 mRNA was found in the cancer cells than in normal epithelium (Fig. 3A). There was also more ErbB2 mRNA in the Foxp3sf/+spf/+ epithelium than in that of the WT female mice (Fig. 3A), which indicates a potential gene dosage effect of Foxp3 on the regulation of ErbB2 expression in vivo. Transfection of the TSA cell line with Foxp3 cDNA repressed ErbB2 levels on the TSA cell line (Fig. 3B).
Fig 3.

Foxp3 represses the expression of ErbB2. A. Over-expression of ErbB2 in mouse mammary cancers. The left panels show immunohistochemical staining of a non-cancerous mammary gland and an adjacent adenocarcinoma from a Foxp3sf/+Otcspf/+ mouse using anti-ErbB2 antibody. The right panel: Relative expression levels of ErbB2 in normal mammary epithelium of WT and Foxp3sf/+Otcspf/+ mice and of cancer tissues from Foxp3sf/+Otcspf/+ mice as revealed by real-time RT-PCR of LCM samples. Data shown are means and S.D. The expression of ErbB2 was normalized against either Hprt or CK19. Highly significant differences were observed between cancerous and normal tissue (P<0.001, ANOVA test when either internal standards were used). B. Transfection of Foxp3-V5 into TSA cells repressed expression of the ErbB2 locus. The left panel shows mRNA levels as measured by real-time PCR. Data shown are means and S.D. of triplicates. The right panel shows the protein levels by Western blot of the cell lysates using anti-ErbB2 antibody. The amount of actin β was used as loading control, while the amount of transfected Foxp3-V5 was measured by Western blot using anti-V5 antibodies. C. Binding of the Foxp3-V5 fusion protein to the promoter region of the ErbB2 gene. Top panel is a diagram of the 5′ region of the ErbB2 gene, including the promoter, exon 1, intron 1 and exon 3. The forkhead binding motifs are illustrated with small green bars, while the regions surveyed by real-time PCR are marked in red bars. The lower panel shows the amount of DNA precipitated by either control IgG or anti-V5 mAb expressed as % of the total input genomic DNA. D. Foxp3-mediated repression of the ErbB2 promotor requires forkhead binding motifs as evaluated by dual-luciferaseR reporter assay. The promoter regions differed in the number of forkhead binding motifs, as illustrated in the diagram on the left. Three different cell lines were transfected with either vector control or Foxp3 (1 μg/well) in conjunction with the luciferase reporter driven by different 5′ promoter regions of the ErbB2 gene (0.6 μg/well). pRL-TK was used as internal control.The luciferase activity from the cells transfected with the pGL2-basic vector was arbitrarily defined as 1.0. Data shown are means and S.D. of triplicates and have been repeated at least three times. E. Site-directed mutagenesis of one of two conserved regions with multiple forkhead binding motifs in the ErbB2/HER-2 promoter prevented repression of the ErbB2 promoter by Foxp3. The two binding sites, as illustrated in supplemental Fig. S2, were deleted individually (deleted DNA sequence, Mut A: AAATCTGGGATCATTTA; Mut B: TTTGAATTTCAGATAAA). Right panel shows that mutations of either site prevented FOXP3-mediated suppression. The promoter activity was measured and normalized as detailed in D, except that the amount of promoter DNA was 0.4 μg/sample. The promoter activities of the vector groups were artificially defined as 1.0. F. Foxp3-mediated binding to specific cis-elements in the ErbB2 promoter. Nuclear extracts from the Foxp3-V5- (F-NE) or vector-transfected control (C-NE) TSA cells were pre-incubated with 32P-labeled WT (*W) or mutant probes (*M) in the presence of an unlabeled WT (W) or mutant probe (M). The mixtures were analyzed by PAGE. NS, nonspecific; FP, free probe. The specific Foxp3-shifted band is marked by an arrow. Data shown have been repeated three times. G. Mutation of forkhead binding motifs (Mut C) abrogated FOXP3-mediated repression, but not basal promoter activity. As in E, except that WT, Mut B and Mut C (mutations that inactivate the Foxp3 binding as detailed in F) of the ErbB2 promoters were used. This experiment has been repeated twice with similar results. The differences were compared by student t-tests with P value provided.
Analysis of the 5′ sequence of the ErbB2 gene revealed multiple binding motifs for the forkhead domain (Fig. 3C). To test whether Foxp3 interacts with the ErbB2 promoter, we used anti-V5 antibody to precipitate sonicated chromatin from the TSA cells transfected with the Foxp3-V5 cDNA and used real-time PCR to quantitate the amounts of the specific ErbB2 promoter region precipitated by the anti-V5 antibodies in comparison to those that bound to mouse IgG control. As shown in Fig. 3C, the anti-V5 antibodies pulled down significantly higher amounts of ErbB2 promoter DNA than the IgG control, with the highest signal around 1.6 kb 5′ of the transcription starting site.
To test whether the binding correlated with the suppression by Foxp3, we produced luciferase reporter using the 1.8, 1.2 and 0.8 Kb upstream of the ErbB2 TSS and tested the ability of Foxp3 to repress ErbB2 promoter activity. In three separate cell lines, we observed that the region with the strongest ChIP signal was required for optimal repression by Foxp3 (Fig. 3D). Furthermore, we deleted two potential Foxp3-binding sites based on intensity of ChIP signal, abundance of consensus binding sites and conservation between mouse and human (Supplemental Fig. S2) by site-directed mutagenesis and measured the effect on Foxp3-mediated repression. As shown in Fig. 3E, deletion of either binding site substantially increased the ErbB2 promoter activity in the presence of Foxp3 and thus alleviated Foxp3-mediated repression.
Since the region deleted in mut B is 100% conserved between mouse and man and since this deletion completely wiped out repression, we used an electrophoretic mobility shift assay (EMSA) to determine whether the forkhead DNA-binding motifs in region B bound to Foxp3. As shown in Fig. 3F, the nuclear extracts from the Foxp3-expressing cells specifically retarded migration of the WT but not mutant 32P-labeled probes compared with control cells. While mutant cold probes did not affect Foxp3 binding activities, WT cold probes significantly diminished them, establishing that the binding of these complexes is specific to forkhead DNA-binding motifs. We therefore carried out site-directed mutagenesis to replace the 12 nucleotides (mut C) within the ErbB2 promoter and compared the promoter activity and Foxp3-repression by luciferase assays. While the wild-type promoter was repressed by Foxp3, no repression by Foxp3 was observed when the mutant promoter was used. Moreover, in contrast to the deletional Mut B, the mutations had no impact on the basal activity of the ErbB2 promoter (Fig. 3G). Taken together, our new data make a compelling case that Foxp3 represses the ErbB2 promoter via specific forkhead binding motifs.
FOXP3 defects in human breast cancer
We analyzed the levels and isoforms of the FOXP3 transcripts in a panel of normal human mammary epithelial cells (HMEC), an immortalized but non-malignant cell line (MCF-10A), and 10 malignant breast cancer cell lines differing in ER/PR and HER-2 status. Early passage of HMEC with no methylation in the CpG island of the P16 promoter (supplemental Fig. S3) was used to avoid effects associated with P16 inactivation in post-senescence HMEC cultures (Romanov et al., 2001). As shown in Fig. 4A, similar levels of FOXP3 transcripts were observed in two independent isolates of HMEC and in the immortalized cell line MCF-10A. Each of the 10 tumor cell lines had a different degree of reduction in FOXP3 mRNA levels in comparison to HMEC and MCF-10A. Among them, 2 were completely devoid of FOXP3 mRNA, while the others had a 1.5–20 fold reduction. We then used anchored primers spanning exons 1–12 to amplify the FOXP3 transcripts, and then we sequenced the PCR products. As shown in Fig. 4, none of the tumor cell lines expressed full-length FOXP3 transcripts. The HMEC expressed the same two isoforms as observed in the T cells, while MCF-10A expressed the exon 3-lacking. The same isoform was also found in 4 tumor cell lines at much lower levels. In addition, 3 tumor cell lines expressed an isoform lacking both exons 3 and 4. The alternative splicing resulted in a frame-shift beginning at codon 70 and an early termination at codon 172. Furthermore, 2 tumor cell lines expressed a FOXP3 isoform lacking exons 3 and/or 8. Exon 8 encodes the leucine-zipper domain that is frequently mutated in IPEX patients (Ziegler, 2006). Thus, FOXP3 is abnormal in breast cancer cell lines. Consistent with a role for FOXP3 in repressing HER-2 expression, the majority of the breast cancer cell lines had higher levels of HER-2 in comparison to normal HMEC (Fig. 4, lower panel). However, additional changes are also likely required for HER-2 over-expression, as three cell lines did not over express HER-2 even though the FOXP3 transcripts were greatly reduced.
Fig. 4.
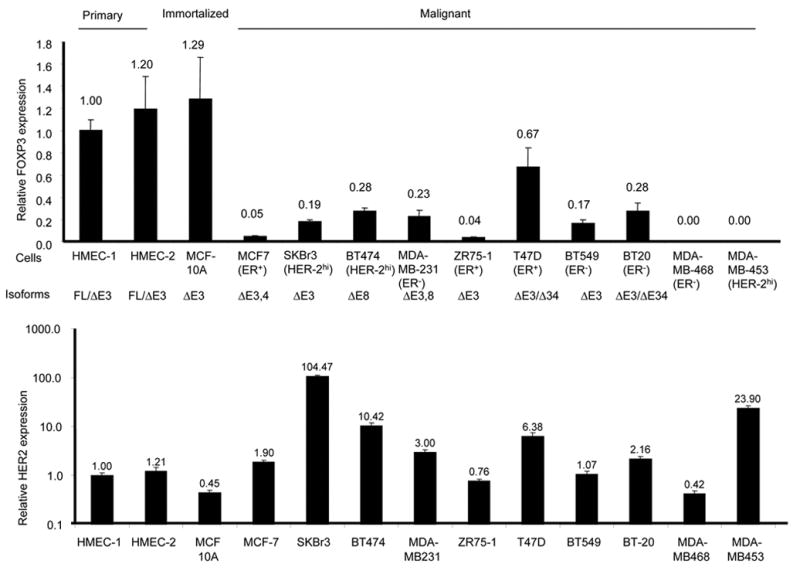
Characterization of FOXP3 transcripts in primary, immortalized and malignant mammary epithelial cells. Relative levels and isoforms of FOXP3 (the upper panel) and HER-2 (the lower panel) mRNA. After normalizing against endogenous GAPDH, the amounts of transcripts were compared to those found in HMEC-1, which was arbitrally defined as 1.0. The reported ER status and the isoforms of the FOXP3 transcripts detected are indicated. To characterize the isoforms, FOXP3 mRNA was amplified by two rounds of anchored PCR using primers annealing to exons 1 and 12. The bulk PCR products were sequenced only if one species was found in agarose electrophoresis. When more than one species was observed, the PCR products were cloned and multiple clones were sequenced until all of the species observed in the agarose gel were identified.
We took three approaches to determine whether the findings in the mutant mice and human breast cancer cell lines are relevant to the pathogenesis of human breast cancer. First, we used immunohistochemistry to determine expression of FOXP3 in normal vs. cancerous tissue. As shown in Fig. 5A, while more than 80% of the normal breast samples expressed FOXP3 in the nuclei of the epithelial cells, less than 20% of the cancerous tissue showed nuclear staining. Second, we used fluorescence in situ hybridization (FISH) to determine whether the FOXP3 gene was deleted in the breast cancer samples. The minimal common region of deletion was identified using flanking p-telomeric and centromeric clones. Out of 223 informative samples, we observed 28 cases (12.6%) with deletions in any of the three loci. Interestingly, deletion of the FOXP3 locus was found in all of the 28 cases (Fig. 5B and supplemental Table S2). These data suggest that FOXP3 is likely within the minimal region of deletion in the Xp11 region studied. Although all deletions were heterozygous, the FOXP3 protein was undetectable in 26/28 cases. Thus, it appears that for the majority of the breast cancer samples, LOH alone was sufficient to inactivate the locus, perhaps due to X-chromosomal inactivation. The two cases with both deletion and FOXP3 expression had X-polysomy with 3 and 4 X-chromosomes respectively (Table S2). Thirdly, we isolated DNA from matched normal and cancerous tissues (50 cases with formalin fixed samples and 15 cases of frozen samples) from patients with invasive ductal carcinoma and amplified all 11 coding exons and intron-exon boundary regions by PCR. Two independent PCR products were sequenced in order to confirm the mutations. Unless the bulk sequencing data were unambiguous (Supplemental Fig S4a & c), the PCR products were cloned and 5–10 independent clones from each reaction were sequenced (Supplemental Fig. S4b). Among the formalin fixed samples, we only used the cases in which the normal tissue samples gave unambiguous sequencing data that matched the wild-type FOXP3 sequence. When the cancerous tissues were compared with normal tissues from the same patient, 36% (18/50 formalin-fixed samples and 5/15 frozen samples) showed somatic mutations (Table S3). Loss of the wild-type allele was found in 6/23 cases (38%) of cancer samples with somatic FOXP3 mutations (See supplemental Fig. 4c for an example). The other cases had heterozygous mutations (Supplemental Fig. S4a). Eighteen mutations resulted in the replacement of amino acids. Most are likely to be critical for FOXP3 function, as judged from the pattern of mutation in IPEX patients (Ziegler, 2006) or in the conserved zinc finger domain that has so far not been implicated (Fig. 5C).
Fig. 5.
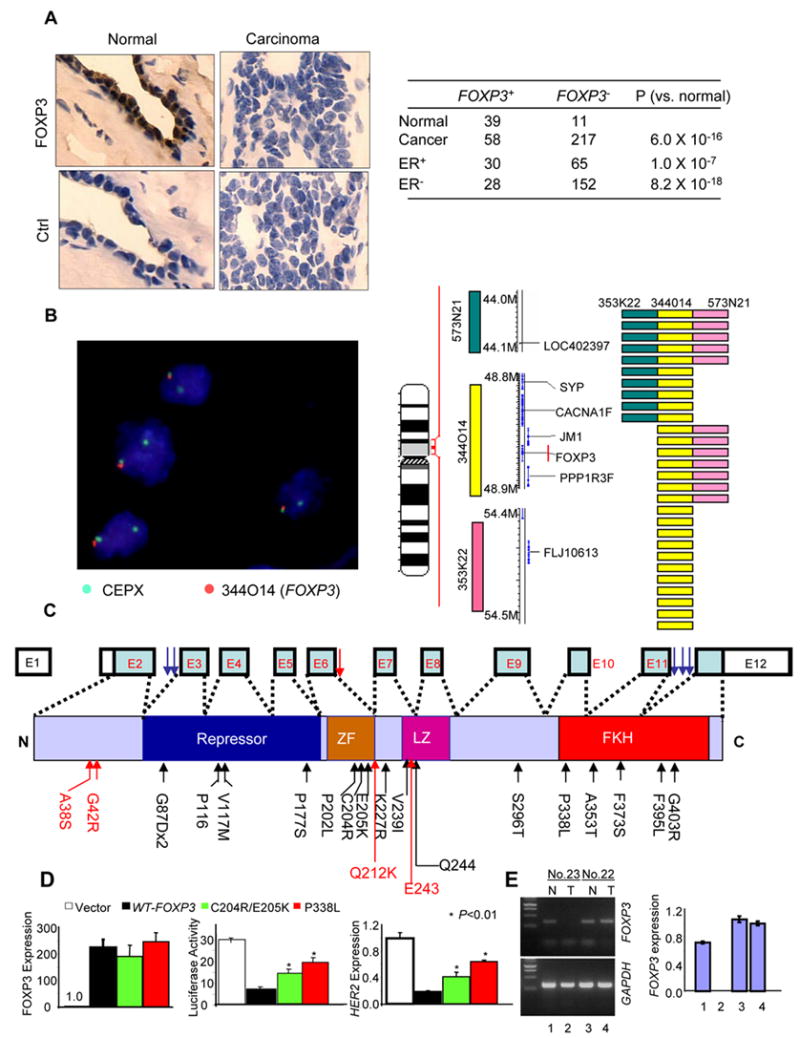
FOXP3 defects in human breast cancer. A. Down-regulation of the FOXP3 protein among human breast cancer cells. Photographs in the top panels show anti-FOXP3 staining of normal and carcinoma tissues from the same patient, with specificity control shown at the bottom. The number and percentage of FOXP3 positive tissues are shown in the lower panel. Samples with nuclear staining by the anti-Foxp3 antibody were scored as positive. B. Deletion of the FOXP3 locus in breast cancer cells. Breast cancer tissue microarray samples were analyzed by FISH using 3 BAC clone probes surrounding a 10MB region in Xp11.2. A typical FISH for the CEPX (green) and FOXP3 (orange) probes is shown on the left, while the genomic structure of the X-chromosome and probe positions are illustrated in the middle panel. A total of 238 samples were analyzed for all probes, with 223 samples providing definitive FISH data. 28/223 samples showed deletions as detected by at least one of the 3 probes. The positions of the deletions in the 28 samples are summarized in the right panel. C. Somatic mutation of the FOXP3 gene in breast cancer samples: summary of sequencing data from 65 cases, including 50 formalin-fixed samples and 15 frozen samples. Genomic DNA was isolated from matched normal and cancerous tissues from the same patients and amplified with primers for individual exons and intron-exon boundary regions. Somatic mutations were identified by comparing sequences from normal and cancerous samples from the same patients. The data are from either bulk sequencing of PCR products or from the sequencing of 5–10 clones from PCR products. Only those mutations that were observed in multiple clones were scored. Mutations identified from 50 cases of formalin fixed samples are marked in black, while those identified from 15 cases of frozen tissue samples are marked in red. D. FOXP3 mutations reduced its repressor activity for the HER-2 promoter in the SKBr-3 cell line. The left panel shows expression of mutant cDNA. The middle panel shows luciferase activity, while the right panel shows the levels of HER-2 transcripts. The difference between WT and 318 P> L and that between WT and 204C>R205E>K are highly significant (P<0.01). Data shown are representative of at least 2–3 independent experiments. E. A breast cancer sample with a somatic mutation in intron 6 (case 23) had an inactivated FOXP3 locus. Normal mammary epithelial (N) and tumor (T) cells were isolated by LCM. The FOXP3 transcripts were determined either by PCR using primers spanning exons 5–8 to detect alternatively spliced products or by real-time PCR using primers spanning exons 10–12. The upper and middle panels show photographs of PCR products of FOXP3 or GAPDH loci, while the right panel shows the relative level of FOXP3 transcripts as determined by real time PCR. Neither assay detected any FOXP3 transcripts in the tumor of case No. 23. Substantial amounts of FOXP3 transcripts were detected in normal samples and tumors in case 22 (with a synonymous mutation in exon 7), which was artificially defined as 1.0.
Although most samples had a single mutation of the FOXP3 gene, we did observe two cases with multiple mutations. In the first sample (supplemental Fig S4b, case 3 in Table S3), the two mutations occurred in consecutive codons, resulting in two nonconservative replacements of amino acid residues. Clonal analysis revealed that both mutations occurred in the same clone (Fig. S4b). In the second sample (Table S3, case 16), three mutations occurred in intron 11. Since this case lacked a WT allele (Supplemental Fig. S4d), it is likely that all of the mutations occurred in the same allele. The possibility of a mismatch in the cancer and normal samples was ruled out by comparing the normal and cancer samples for polymorphism of two unrelated genes (data not shown).
To directly test whether FOXP3 mutations affect the repressor activity for the HER-2 gene, we chose two representative somatic FOXP3 mutants isolated in the cancer cells and tested their repressor activity for the HER-2 promoter. One mutation (338P>L) resided in the signature forkhead domain which is often mutated in the IPEX patient, while the other double mutation (204C>R205E>K) was from the zinc finger domain that has not been implicated in IPEX patients. As shown in Fig. 5E, both mutations significantly reduced the repressor activity of FOXP3. The reduced repression of the HER-2 promoter correlates with a significantly reduced inhibition of HER-2 mRNA (Fig. 5D).
Four cases had mutations in introns that may potentially affect RNA splicing. We used laser-guided micro-dissection to isolate normal and cancerous epithelial cells from one case with a mutation in intron 6 (case 23, Supplemental Table S3). RNA was isolated and tested for the potential effects of the mutation on RNA splicing (using primers on exons 5 and 8) and total FOXP3 transcript, as quantitated by real time PCR using primers spanning exons 10–12. Tissues from another patient with a mutation in exon 7 were used as control. As show in Fig. 5E left panel, primers spanning exons 5 and 8 failed to detect FOXP3 mRNA from the cancerous tissue of case No. 23. Furthermore, primers spanning exons 10–12 also failed to detect any FOXP3 transcripts. Substantial levels were detected in the normal epithelial cells of the same patients as well as in normal and cancerous tissues from case No. 22. Since the wild-type allele had been lost in the cancer cells of case No. 23, it is likely that the mutation in intron 6 inactivated FOXP3. With an intron of 944 nucleotides, a mutation that prevented splicing of intron 6 would cause premature-termination codon-mediated RNA decay, which is operative in the FOXP3 gene (Chatila et al., 2000).
FOXP3 defects and HER-2 over-expression
To demonstrate a role for FOXP3 defect in HER-2 overexpression, we first silenced the FOXP3 gene in early passage of primary HMEC (Supplemental Fig. S3) using a lentiviral vector expressing FOXP3 siRNA. As shown in Fig. 6A, the FOXP3 siRNA reduced FOXP3 expression by more than 100-fold while increasing HER-2 mRNA by 7-fold. A corresponding increase in cell surface HER-2 was also observed (Fig. 6B). These results implicate FOXP3 as a repressor of HER-2 in human breast epithelial cells.
Fig. 6.
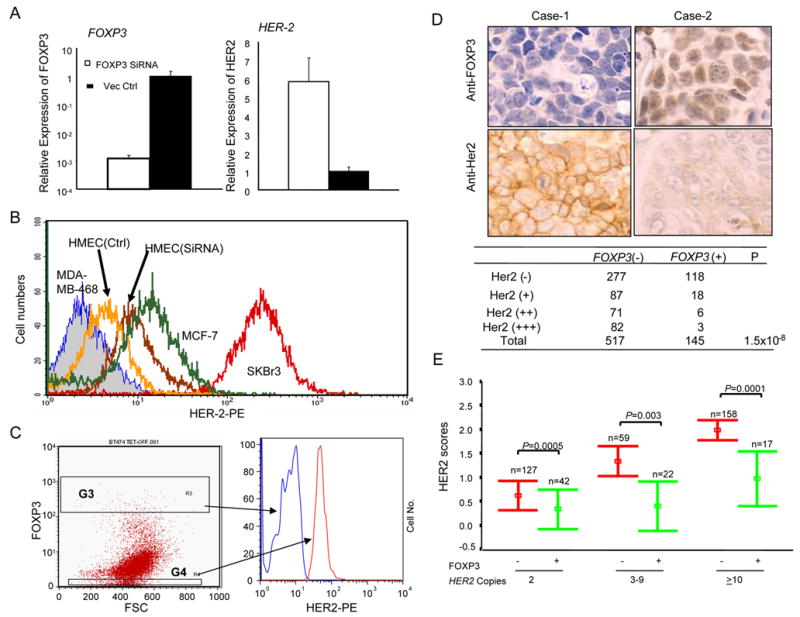
FOXP3 is an important HER-2 repressor. A. Silencing of FOXP3 resulted in the up-regulation of HER-2 in primary human mammary epithelial cells (HMEC). Early passage of HMEC were transduced with lentiviral vector for either control sequence or FOXP3 siRNA. The untransfected cells were removed by selection with blasticidin. At one week after transduction, the levels of the FOXP3 and HER-2 transcripts were quantitated by real-time PCR. Data shown are mean and SEM of relative levels of transcripts (with that in the vector-transduced cells defined as 1.0) and represent those of three independent experiments. B. Flow cytometry data showing the effect of FOXP3 silencing on HMEC surface HER-2 levels. HER-2-negative MDA-MB468, HER-2lo MCF-7 and HER-2hi SKBr3 cell lines were included for comparison. C. In the Tet-off inducible FOXP3-expressing BT474, FOXP3 repressed HER-2. BT474 cells were first transfected with pTet-Off vector. The transfectants were selected by both blasticidin and G418 in doxycycline-containing medium. The drug-resistant cells were cultured in the absence of doxycycline for 5 days to induce FOXP3. The cells were stained for FOXP3 and HER-2 proteins by flow cytometry. Data shown are histograms depicting HER-2 levels among the gated FOXP3hi and FOXP3− cells based on reactivity to the anti-Foxp3 antibody and are representative of those from two independent experiments. D. Inverse correlation between FOXP3 expression (the top panel) and that of the HER-2 (middle panel) among the human breast cancer samples. Tissue microassay samples were stained with either anti-FOXP3 antibodies or anti-HER-2 antibodies and were scored by two different pathologists in a double blind fashion. FOXP3 staining was scored by nuclear staining with affinity-purified anti-FOXP3 antibodies. A summary of 517 FOXP3+ and 145 FOXP3− samples is shown in the bottom panel. E. Inverse correlations between FOXP3 expression and HER-2 scores in cells with or without HER-2 amplification. The HER-2 gene copy number was determined by FISH, while nuclear expression of FOXP3 was determined by immunohistochemistry. Data shown are mean and S.D. of HER-2 scores of 425 cases of breast cancers grouped by HER-2 copy number. P values were generated by the Mann-Whitney test.
Second, since a major mechanism for HER-2 up-regulation in breast cancer is gene amplification (Kallioniemi et al., 1992), an intriguing issue is whether FOXP3 is capable of repressing HER-2 in cancer cells with an amplified HER-2 gene. We produced a Tet-off line of BT474, a breast cancer cell line known to have HER-2 gene amplification (Kallioniemi et al., 1992), and transiently transfected it with a pBI-EGFP-FOXP3- vector. After drug selection, the cells were cultured either in the presence or absence of doxycycline. While the cells cultured with doxycycline did not express FOXP3 (data not shown), removal of doxycycline resulted in induction of FOXP3 in a significant fraction of the cancer cells, which allowed us to compare HER-2 levels in the FOXP3+ and FOXP3− cells in the same culture by flow cytometry. As shown in Fig. 6C, FOXP3−cells had about a 5–10-fold higher level of the HER-2 protein on the cell surface in comparison to the FOXP3+ cells.
Thirdly, we compared the expression of FOXP3 with HER-2 expression in breast cancer tissues. As shown in Fig. 6D, down-regulation of FOXP3 was strongly associated with the over-expression of HER-2, which supports a role for FOXP3 inactivation in HER-2 over-expression in breast cancer. Nevertheless, since many of the FOXP3− cells remained HER-2−, it is likely that dis-regulation of FOXP3 is insufficient for HER-2 up-regulation. On the other hand, since only 3/82 FOXP3+ cancer cells expressed high levels of HER-2, FOXP3 inactivation is likely important for HER-2 up-regulation under most circumstances.
Fourth, we divided breast cancer samples based on their HER-2 gene copy numbers and compared the FOXP3+ and FOXP3− cancer samples for the relative amounts of cell surface HER-2 expression. As shown in Fig. 6E, in each of the gene dose categories, FOXP3+ samples had reduced HER-2 scores in comparison to the FOXP3− samples. These results strongly suggest a critical role for FOXP3 in repressing HER-2 expression even in the cases of HER-2 gene amplification.
Fifth, of the 223 informative samples among the 238 that we screened for Xp11.2 deletions, those with deletions encompassing the FOXP3 locus had significantly higher HER-2 scores compared to those without deletions (P=0.03) (Supplemental Table S4). Likewise, we compared the relative HER-2 scores among the 50 samples in which we had sequenced all FOXP3 exons. As shown in Supplemental Table S5, the mutations in the FOXP3 gene correlated with higher levels of HER-2 (P=0.0083).
Foxp3/FOXP3 inhibits tumorigenicity of breast cancer cells
To test whether the Foxp3 gene can suppress the growth of breast cancer cells, we transfected the empty vector or the vectors carrying either Foxp3 (mouse or human origin) or Otc cDNA into three breast cancer cell lines, including mouse mammary tumor cell line TSA or human breast cancer cell lines MCF7 (ER+HER-2low, no HER-2 amplification) and SKBr3 (ER-HER-2high with HER-2 amplification). The untransfected cells were removed by a selection with G418. While the vector-transfected cells grew rapidly, the Foxp3-transfected cell lines seldom grew into large colonies. The Foxp3-transfected culture had a drastic reduction in both the size and the number of the drug-resistant colonies. No effect was observed when the Otc cDNA was used (Fig. 7A&B).
Fig. 7.
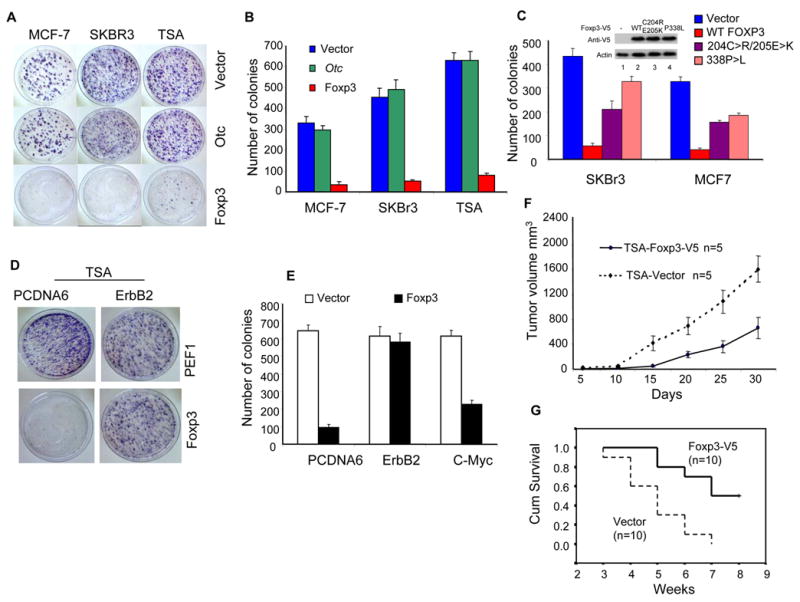
Foxp3 inhibits the growth and tumorigenicity of multiple breast cancer cell lines. A. Breast cancer cell lines MCF-7, SKBr3 and TSA were transfected with equal concentrations of either vector alone (Vector), Foxp3 or Otc cDNA. After 3 weeks of G-418 selection, the drug resistant clones were visualized by crystal violet dye. B. Summary of the colony numbers in three independent experiments as described in A. Data shown are means and S.D. C. Somatic mutations identified from breast cancer samples attenuated the growth suppression of the FOXP3. As in A & B, except that two somatic mutants were compared with WT FOXP3 cDNA using the two human breast cancer cell lines. Expression of WT and mutant proteins at 1 week after transfection is presented in the insert. D&E. Ectopic expression of the ErbB2 but not the c-Myc cDNA abrogated Foxp3-mediated repression. TSA cells were transfected with either pcDNA6-blasticidin vector or ErbB2 cDNA and selected with blasticidin for two weeks. Pools of blasticidin-resistant cells were super-transfected with the pEF1-G418 vector or Foxp3 cDNA. The cells were then plated and selected with blasticidin and G418 for three weeks. The viable colonies were visualized after staining with the crystal violet dye. D. Photographs of a representative plate showing abrogation of Foxp3-mediated suppression by ErbB2. E. The mean and S.D. of the colony numbers. This experiment has been repeated twice with essentially identical results. F. Expression of Foxp3 reduced growth rate of tumors. Syngeneic BALB/c mice were injected with 5x105/mouse Foxp3 or vector-transfected TSA cells in the flank and the sizes of the local tumor mass were measured using a caliper. Data shown are means and S.D. and have been repeated once. G. The survival of tumor-bearing mice was monitored over a 7-weeks period (P=0.0015, log-rank test). As in F, except that 106 tumor cells /mouse were injected and the mice were euthanized when they became moribund.
To test whether the somatic mutations uncovered from cancerous tissues ablated their growth inhibition, we transfected WT and two mutant Foxp3 cDNA into SKBr3 and MCF7 cell lines. As shown in Fig. 7C, in both cell lines, the mutants had a greatly reduced ability to suppress tumor growth.
To test whether repression of ErbB2 explains the tumor suppressor activity of the Foxp3 gene in the ErbB2+ cancer cell line, we transfected TSA cells with mouse CMV promoter-driven ErbB2 cDNA cloned into the pcDNA6 vector and evaluated their susceptibility to Foxp3-mediated growth suppression. In this setting, the expression of ErbB2 was resistant to Foxp3-mediated repression (data not shown). If repression of endogenous ErbB2 is critical for Foxp3-mediated tumor suppression, ectopic expression of ErbB2 should alleviate the growth inhibition by Foxp3. As shown in Fig. 7D & E, while the pcDNA6-vector-transfected TSA cells remained susceptible to Foxp3-mediated repression, the ErbB2-transfected TSA cells were completely resistant. In contrast, transfection of c-Myc barely alleviated the growth inhibition by FOXP3 (Fig. 7E). These results suggest that Foxp3 suppresses TSA growth by repressing transcription of ErbB2.
We transfected TSA cells with either empty vector or V5-tagged Foxp3 cDNA. The stable transfectant cell lines were selected by G-418. The vector and Foxp3-V5-transfected cell lines were injected into syngeneic BALB/c mice, which were then observed for tumor growth and mouse survival. As shown in Fig. 7F, Foxp3-transfectants showed reduced growth in vivo. The mice that received TSA-vector cells became moribund earlier with higher incidence, while about 50% of the mice that received the Foxp3-V5-transfected cells survived more than 7 weeks (Fig. 7G). Similarly, Foxp3-transfected 4T1, a mouse mammary cancer cell line with ErbB2 over-expression, also showed reduced tumorigenicity in vivo (data not shown).
Discussion
Foxp3 is an X-linked mammary tumor suppressor gene
Serendipitously, we observed that mice heterozygous for the Foxp3 mutation spontaneously developed mammary cancer at a high rate. Since two independently maintained lines sharing the Foxp3 mutation have a comparably higher incidence of mammary cancer, the Foxp3 mutation is likely responsible for the increased rate of breast cancer. Unlike essentially all cancer suppressor genes identified to date, Foxp3 is X-linked and inactive in cells in which the WT allele was silenced by X-inactivation. This is indeed the case as the low levels of Foxp3 transcripts in the cancer cells were derived exclusively from the mutant alleles.
Our analysis of human breast cancer samples provides strong support for an important role for the FOXP3 gene in the development of breast cancer. First, we searched X chromosomal deletion using three markers encompassing more than 10MB of Xp11 and found that FOXP3 is likely the minimal region of deletion. Second, we revealed a high proportion of somatic mutations in the FOXP3 gene (23/65 cases over about 2,000 bp exon and intron sequence scanned). The significance of our finding can be discerned indirectly based on the fact that the mutations tended to cluster around important domains, such as the FKH and the zinc finger domains. In addition, most of the mutations resulted in the nonconservative replacement of amino acids, and cancers with mutations identified had higher levels of HER-2 than those without mutations. The rate of missense to synonymous mutation was 18/3, which greatly exceeds what would be predicted if the mutations were not relevant to tumor development. More importantly, we demonstrated that two tested mutations in the FKH and zing finger domains inactivated the repressor activity and tumor growth inhibition, and that cancer tissues bearing an intronic mutation had an inactive FOXP3 locus. Moreover, mutations and deletions of the FOXP3 locus corresponded to increased HER-2 levels. Third, we have documented extensive down-regulation of FOXP3 among more than 600 cases of breast cancer tissues.
Foxp3 is a major transcriptional repressor for ErbB2
The molecular lesions leading to HER-2 over-expression remain poorly understood. Here we showed that the Foxp3 mutation resulted in over-expression of ErbB2, the murine homologue of HER-2. In addition, transfection of Foxp3 repressed ErbB2 transcription. More importantly, chromatin immunoprecipitation and EMSA analyses revealed that Foxp3 binds specifically to its consensus sequence in the 5′ of the ErbB2 gene. Since specific mutations in the promoter abrogate its susceptibility to repression by Foxp3, such binding is likely responsible for it.
Importantly, we have demonstrated that, for TSA cell line which has ErbB2 over-expression, repressing the ErbB2 locus is responsible for Foxp3′s tumor suppressor activity. The requirement for continuous expression of ErbB2 is best explained by the concept of oncogene addiction (Weinstein, 2002). However, FOXP3 can also suppress the growth of tumor cell lines that do not grossly over-express HER-2/ErbB2, such as MCF-7. In an effort to identify other potential FOXP3 targets, we have produced a FOXP3-Tet-off MCF-7 cell line that expresses FOXP3 upon removal of tetracycline (Fig. S5a). Using the most current version of Entrez Gene-based CDFs for a more accurate GeneChip analysis (Dai et al., 2005), we uncovered wide-spread changes in the expression of genes that are involved in several pathways critical for cancer cell growth (Fig. S5b). Interestingly, 10 genes involved in ErbB2 signaling pathway were repressed by FOXP3 (Fig. S5c). Thus, multiple oncogenes can potentially be up-regulated by FOXP3 inactivation. Taken together, we have demonstrated that FOXP3 is the first X-linked breast cancer suppressor that represses the HER-2/ErbB2 oncogene. Given the significant role of HER-2 in the pathogenesis of human breast cancer and the wide-spread defects of the FOXP3 locus, it is likely that FOXP3 is an important suppressor for human breast cancer.
Experimental Procedures
Quantitative real-time PCR
Relative quantities of mRNA expression were analyzed using real-time PCR (Applied Biosystems ABI Prism 7700 Sequence Detection System, Applied Biosystems). The SYBR (Qiagen) green fluorescence dye was used in this study. The primer sequences (5′–3′) are listed in Table S6.
Chromatin immunoprecipitation was carried out according to published procedure (Im et al., 2004). Briefly, the Foxp3-V5-transfected TSA cells were sonicated and fixed with 1% paraformaldehyde. The anti-V5 antibodies or control mouse IgG were used to pull down chromatin associated with Foxp3-V5. The amounts of the specific DNA fragment were quantitated by real-time PCR and normalized against the genomic DNA preparation from the same cells.
FOXP3-silencing lentiviral vector
The lentivirus-based siRNA expressing vectors were created by introducing the murine U6 RNA polymerase III promoter and a murine phosphoglycerate kinase promoter (pGK)-driven EGFP expression cassette into a vector of pLenti6/V5-D-TOPO back bone without CMV promoter. A hairpin siRNA sequence of FOXP3 (target sequence at the region of 1256 to 1274 nucleotides; 5′-GCAGCGGACACTCAATGAG-3′) was cloned into the lentiviral siRNA expressing vectors by restriction sites of ApaI and EcoRI.
Immunohistochemistry and Fluorescence In-situ Hybridization (FISH)
HER-2 expression was performed using PathwayTM HER-2 (Clone CB11) (Ventana Medical Systems, Inc., Tucson, AZ) on the BenchMark® XT automated system per the manufacturer’s recommended protocol. The HER-2 levels were scored by commonly used criteria (Yaziji et al., 2004).
FISH for FOXP3 deletion was done using BAC clone RP11-344O14 (ntLocus X: 48,817,975- 48,968,223), which was verified by PCR to contain the FOXP3 gene. The minimal common region of deletion was done using flanking p-telomeric and centromeric clones, RP11-573N21 (ntLocus X: 43,910,391-44,078,600) and RP11-353K22 (ntLocus X: 54,416,890- 54,545,788), respectively.
EMSA
Nuclear extracts were prepared as described previously (Wang et al., 1999). The sequence for the WT probe (W) was AGTTCAATTTGAATTTCAGATAAACG. Mutant probe (M) (AGTTCAGCGCGAGCGCCAGAGCGCCG) with mutations of all three potential forkhead binding sites was used as specificity control.
Supplementary Material
Acknowledgments
We thank Dr. Fan Meng for assistance in bioinformatics, Drs. Eric Fearon, Albert de la Chapelle, Zhaohui Qin, Michael Caligiuri, and Charis Eng for their valuable discussions and/or critical reading of the manuscript and Lynde Shaw for secretarial assistance. This study is supported by grants from the National Institutes of Health and the Department of Defense.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, Kelly TE, Saulsbury FT, Chance PF, Ochs HD. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- Bofin AM, Ytterhus B, Martin C, O'Leary JJ, Hagmar BM. Detection and quantitation of HER-2 gene amplification and protein expression in breast carcinoma. Am J Clin Pathol. 2004;122:110–119. doi: 10.1309/8A2D-JFT0-7NE6-EWHE. [DOI] [PubMed] [Google Scholar]
- Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF, Ramsdell F. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- Chang X, Gao JX, Jiang Q, Wen J, Seifers N, Su L, Godfrey VL, Zuo T, Zheng P, Liu Y. The Scurfy mutation of FoxP3 in the thymus stroma leads to defective thymopoiesis. J Exp Med. 2005;202:1141–1151. doi: 10.1084/jem.20050157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatila TA, Blaeser F, Ho N, Lederman HM, Voulgaropoulos C, Helms C, Bowcock AM. JM2, encoding a fork head-related protein, is mutated in X-linked autoimmunity-allergic disregulation syndrome. J Clin Invest. 2000;106:R75–81. doi: 10.1172/JCI11679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatila TA, Blaeser F, Ho N, Lederman HM, Voulgaropoulos C, Helms C, Bowcock AM. JM2, encoding a fork head-related protein, is mutated in X-linked autoimmunity-allergic disregulation syndrome. J Clin Invest. 2000;106:R75–81. doi: 10.1172/JCI11679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Garcia de Palazzo I, Klein-Szanto A, Weiner LM. Immunohistochemical detection of c-erbB-2 expression by neoplastic human tissue using monospecific and bispecific monoclonal antibodies. Int J Biol Markers. 1993;8:233–239. doi: 10.1177/172460089300800406. [DOI] [PubMed] [Google Scholar]
- Godfrey VL, Rouse BT, Wilkinson JE. Transplantation of T cell-mediated, lymphoreticular disease from the scurfy (sf) mouse. Am J Pathol. 1994;145:281–286. [PMC free article] [PubMed] [Google Scholar]
- Godfrey VL, Wilkinson JE, Rinchik EM, Russell LB. Fatal lymphoreticular disease in the scurfy (sf) mouse requires T cells that mature in a sf thymic environment: potential model for thymic education. Proc Natl Acad Sci U S A. 1991;88:5528–5532. doi: 10.1073/pnas.88.13.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im H, Grass JA, Johnson KD, Boyer ME, Wu J, Bresnick EH. Measurement of protein-DNA interactions in vivo by chromatin immunoprecipitation. Methods Mol Biol. 2004;284:129–146. doi: 10.1385/1-59259-816-1:129. [DOI] [PubMed] [Google Scholar]
- Jimenez RE, Wallis T, Tabasczka P, Visscher DW. Determination of Her-2/Neu status in breast carcinoma: comparative analysis of immunohistochemistry and fluorescent in situ hybridization. Mod Pathol. 2000;13:37–45. doi: 10.1038/modpathol.3880007. [DOI] [PubMed] [Google Scholar]
- Kallioniemi OP, Kallioniemi A, Kurisu W, Thor A, Chen LC, Smith HS, Waldman FM, Pinkel D, Gray JW. ERBB2 amplification in breast cancer analyzed by fluorescence in situ hybridization. Proc Natl Acad Sci U S A. 1992;89:5321–5325. doi: 10.1073/pnas.89.12.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen M, Knudsen GP, Maguire P, Margolin S, Pedersen J, Lindblom A, Orstavik KH. High incidence of skewed X chromosome inactivation in young patients with familial non-BRCA1/BRCA2 breast cancer. J Med Genet. 2005;42:877–880. doi: 10.1136/jmg.2005.032433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- Piao Z, Malkhosyan SR. Frequent loss Xq25 on the inactive X chromosome in primary breast carcinomas is associated with tumor grade and axillary lymph node metastasis. Genes Chromosomes Cancer. 2002;33:262–269. doi: 10.1002/gcc.10024. [DOI] [PubMed] [Google Scholar]
- Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Romanov SR, Kozakiewicz BK, Holst CR, Stampfer MR, Haupt LM, Tlsty TD. Normal human mammary epithelial cells spontaneously escape senescence and acquire genomic changes. Nature. 2001;409:633–637. doi: 10.1038/35054579. [DOI] [PubMed] [Google Scholar]
- Roncuzzi L, Brognara I, Cocchi S, Zoli W, Gasperi-Campani A. Loss of heterozygosity at pseudoautosomal regions in human breast cancer and association with negative hormonal phenotype. Cancer Genet Cytogenet. 2002;135:173–176. doi: 10.1016/s0165-4608(01)00652-5. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Schechter AL, Stern DF, Vaidyanathan L, Decker SJ, Drebin JA, Greene MI, Weinberg RA. The neu oncogene: an erb-B-related gene encoding a 185,000-Mr tumour antigen. Nature. 1984;312:513–516. doi: 10.1038/312513a0. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- Spatz A, Borg C, Feunteun J. X-Chromosome genetics and human cancer. Nat Rev Cancer. 2004;4:617–629. doi: 10.1038/nrc1413. [DOI] [PubMed] [Google Scholar]
- Todorovic-Rakovic N, Jovanovic D, Neskovic-Konstantinovic Z, Nikolic-Vukosavljevic D. Comparison between immunohistochemistry and chromogenic in situ hybridization in assessing HER-2 status in breast cancer. Pathol Int. 2005;55:318–323. doi: 10.1111/j.1440-1827.2005.01831.x. [DOI] [PubMed] [Google Scholar]
- Wang CY, Cusack JC, Jr, Liu R, Baldwin AS., Jr Control of inducible chemoresistance: enhanced anti-tumor therapy through increased apoptosis by inhibition of NF-kappaB. Nat Med. 1999;5:412–417. doi: 10.1038/7410. [DOI] [PubMed] [Google Scholar]
- Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- Wildin RS, Ramsdell F, Peake J, Faravelli F, Casanova JL, Buist N, Levy-Lahad E, Mazzella M, Goulet O, Perroni L, Bricarelli FD, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet. 2001;27:18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- Wooster RaWBL. Breast and Ovarian Cancer. The New England Journal of Medicine. 2003;348:2339–2347. doi: 10.1056/NEJMra012284. [DOI] [PubMed] [Google Scholar]
- Xing X, Wang SC, Xia W, Zou Y, Shao R, Kwong KY, Yu Z, Zhang S, Miller S, Huang L, Hung MC. The ets protein PEA3 suppresses HER-2/neu overexpression and inhibits tumorigenesis. Nat Med. 2000;6:189–195. doi: 10.1038/72294. [DOI] [PubMed] [Google Scholar]
- Yaziji H, Goldstein LC, Barry TS, Werling R, Hwang H, Ellis GK, Gralow JR, Livingston RB, Gown AM. HER-2 testing in breast cancer using parallel tissue-based methods. JAMA. 2004;291:1972–1977. doi: 10.1001/jama.291.16.1972. [DOI] [PubMed] [Google Scholar]
- Ziegler SF. FOXP3: Of Mice and Men. Annu Rev Immunol. 2006;24:209–226. doi: 10.1146/annurev.immunol.24.021605.090547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


