Abstract
Internal ribosome entry site (IRES) RNAs are necessary for successful infection of many pathogenic viruses, but the details of the RNA structure-based mechanism used to bind and manipulate the ribosome remain poorly understood. The IRES RNAs from the Dicistroviridae intergenic region (IGR) are an excellent model system to understand the fundamental tenets of IRES function, requiring no protein factors to manipulate the ribosome and initiate translation. Here, we explore the architecture of four members of the IGR IRESes, representative of the two divergent classes of these IRES RNAs. Using biochemical and structural probing methods, we show that despite sequence variability they contain a common three-dimensional fold. The three-dimensional architecture of the ribosome binding domain from these IRESes is organized around a core helical scaffold, around which the rest of the RNA molecule folds. However, subtle variation in the folds of these IRESes and the presence of an additional secondary structure element suggest differences in the details of their manipulation of the large ribosomal subunit. Overall, the results demonstrate how a conserved three-dimensional RNA fold governs ribosome binding and manipulation.
Keywords: Dicistroviridae, IRESes, translation initiation, RNA structure, TSV
Introduction
During viral infection, the host cell's translation initiation machinery is often inhibited by the host cell's antiviral response or through direct action of the virus. To initiate translation when the canonical cap-dependent mechanism is disabled, many viruses use internal ribosome entry sites (IRESes) to recruit the ribosome and drive protein synthesis. IRESes are cis-acting RNA sequences usually found in the untranslated region (UTR) of the viral RNA.1–3 Whereas the canonical pathway requires a modified nucleotide cap on the 5′ end to bind a specific initiation factor protein and begin the protein factor-dependent initiation process,4–6 IRES RNAs bypass the need for a cap and many of the initiation factors to recruit, position, and activate the host cell's protein-making apparatus by an alternate pathway.7 For medically and economically important viruses such as hepatitis C virus (HCV), foot-and-mouth disease virus (FMDV), poliovirus, and hepatitis A virus (HAV), production of all the viral proteins depends on the correct functioning of an IRES RNA. However, IRES RNAs are structurally and mechanistically diverse and thus deciphering fundamental rules governing IRES action is challenging.
A group of viruses that use two structurally and mechanistically distinct IRESes is the Dicistroviridae family (Figure 1(a)), which is single-stranded positive-sense RNA viruses whose genome is a single RNA molecule.8 The first IRES, located in the 5′ UTR, drives expression of the non-structural proteins. The second IRES, located in the intergenic region (IGR), drives expression of the structural proteins.9–11 This two-IRES strategy decouples synthesis of the two groups of proteins and allows the virus to respond to antiviral strategies of the cell, specifically the general shutdown of translation by eIF2 phosphorylation.12 Because they do not need initiation factors, the Dicistroviridae IGR IRESes serve as a powerful model for understanding fundamental tenets of RNA structure-driven manipulation of the ribosome and translation initiation. These IRES RNAs directly bind the ribosome near the mRNA binding groove, making specific contacts to both ribosomal subunits.13–16 Binding induces conformational changes in both the ribosome and the IRES RNA, and translation starts from the A-site of the ribosome, from a non-AUG start codon, and without initiation factors.11,17–23 Parallels exist between this simple mechanism and other more complex IRES mechanisms, namely HCV. These similarities include similar conformational changes in the ribosome induced by IRES binding and interactions with the same ribosomal proteins.24–26
Figure 1.
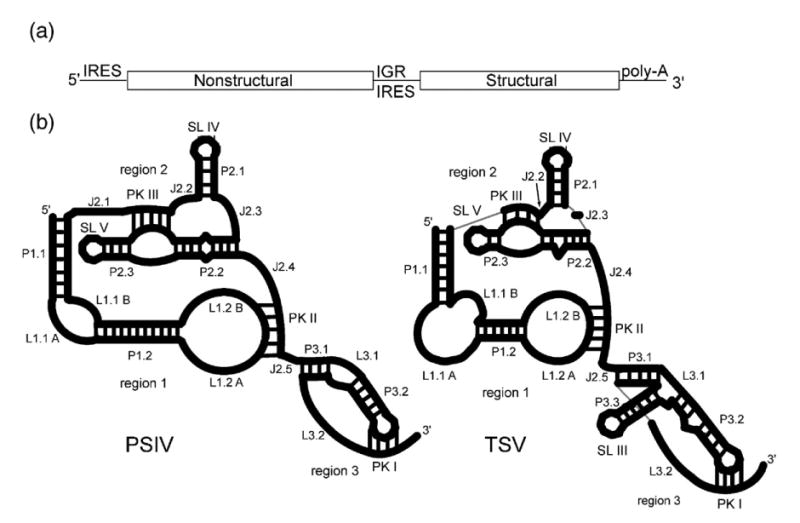
The Dicistroviridae virus genome and IGR IRES secondary structures. (a) The Dicistroviridae genome contains two cistrons on a single positive-sense RNA. The synthesis of the proteins from each cistron is governed by an IRES. The IRES, located between the two cistrons (intergenic region or IGR), is subdivided into two classes. (b) Cartoon representations of the two classes of IGR IRESes, drawn to scale relative to one another. The PSIV IGR IRES represents a typical class 1 IGR IRES while TSV represents a typical class 2 IGR IRES. Here, we use a detailed nomenclature to distinguish between various elements in the IRES structure. Nomenclature used to name the various structural elements follows standard RNA structure usage (J, junction; P, paired/helix; L, loop). SL III, IV, V, and the pseudoknots are also referred to with their traditional names. Furthermore, we reserve the term “domain” for folded into a higher-order structure, and collections of related secondary structures that may or may not adopt a higher-order fold are referred to as “regions.”
Genetic, biochemical, and structural studies coupled with mutagenesis have given considerable insight into the IGR IRES mechanism and have revealed that the three-dimensional folded architecture of the unbound RNA is seminal for function.9,19,20,27,28 The crystal structure of the ribosomal binding region of the Plautia stali intestine virus (PSIV), a member of the Dicistroviridae, IGR IRES reveals how the folded core structure positions recognition elements to interact with the ribosomal subunits,29 and these specific contacts have been explored biochemically.16 Both the crystal structure and cryo-electron microscopy reconstructions of the cricket paralysis virus IRES (CrPV) bound to the ribosome identified areas of stable and dynamic structure within the IRES RNA. These structures, coupled with the biochemical data, confirm that both the stable and dynamic region are vital for the IGR IRES mechanism.13,14,29
Despite evidence for the importance of a very specific RNA structure underlying the IGR IRES mechanism, diversity exists within the Dicistroviridae IGR IRESes in terms of their primary sequence and secondary structures (Figure 1(b)).9,28 Based on chemical and enzymatic probing, phylogeny, and mutagenesis, all are predicted to adopt a secondary structure that contains two highly conserved stem–loops that interact with the small ribosome subunit, multiple helical regions, and three pseudoknot tertiary interactions.9 However, differences in the specifics of the secondary structures are distinct enough to warrant division into two classes that are defined by differences in all three regions of the IRESes. (1) In region 1, loop L1.1 is quite different in sequence and length between the classes, but is highly conserved within each class. This loop makes direct contact to the 60 S subunit; loop L1.1 mutants interact with the 40 S subunit, but not the 80 S ribosome (Figure 1(b)).13,29 (2) In region 3, the class 2 IRESes have an additional stem–loop (SL III) that is absent in the class 1 (Figure 1(b)). Previous work shows this stem–loop is necessary for translation to occur, but does not elucidate its specific mechanistic contribution.30 In general, regions 1 and 2 contain fewer nucleotides and region 3 contains more nucleotides in the class 2 than in the class 1. Further more, examination of region 2 of the Taura Syndrome virus (TSV) IRES shows that some nucleotides revealed in the crystal structure to be involved in the PSIV IRES core fold are not present,29 and several structural elements (helices P1.2 and P2.2, J.2.2 and J2.3) contain fewer nucleotides.
Recent insight into the RNA structure-based mechanism of these IRESes raises additional questions, including those pertaining to structural conservation within the folded core of the various diverse Dicistroviridae family members. Specifically, is the three-dimensional structure and RNA backbone packing strategy of the PSIV IRES RNA shared among all of the class 1 members despite sequence variation, and how does the class 2 IRES fold compare? Do the additional elements in the class 2 IRESes correlate with changes in the mechanism of action? While chemical probing, computational, and other analyses have established conserved secondary and tertiary elements, the presences of a conserved higher-order fold, with conserved RNA packing architectures, has not been established. Here, we use structural probing, ribosome binding, and biophysical analyses to address questions regarding conservation of folded structure in both classes of IGR IRESes. Our results show that although the sequences of these IRES RNAs vary, all adopt a similar RNA packing architecture, hence evolutionary pressure has maintained a conserved structural core. However, there is subtle diversity in the specific characteristics of each IRES structure. In addition, structural probing and footprinting of a representative class 2 family member, the IGR IRES of the TSV, suggest possible roles for the additional stem–loop found in this class.
Results
Analysis of the three-dimensional structural conservation in the class 1 IRESes
Although the secondary structure of the class 1 IGR IRESes is conserved, the sequences vary. An alignment of seven class 1 members shows 27% of the nucleotides are absolutely conserved, while 37% are conserved in six of the seven (Supplementary Data, Figure 1). Of these conserved bases, 58% are in non-Watson–Crick regions, consistent with their involvement in higher-order structure or direct interaction with the ribosome. If the specific three-dimensional fold revealed in the crystal structure of the PSIV IRES is necessary for IRES function, we expect it to be shared by all the members of the class 1 IGR IRESes (Figure 3).
Figure 3.
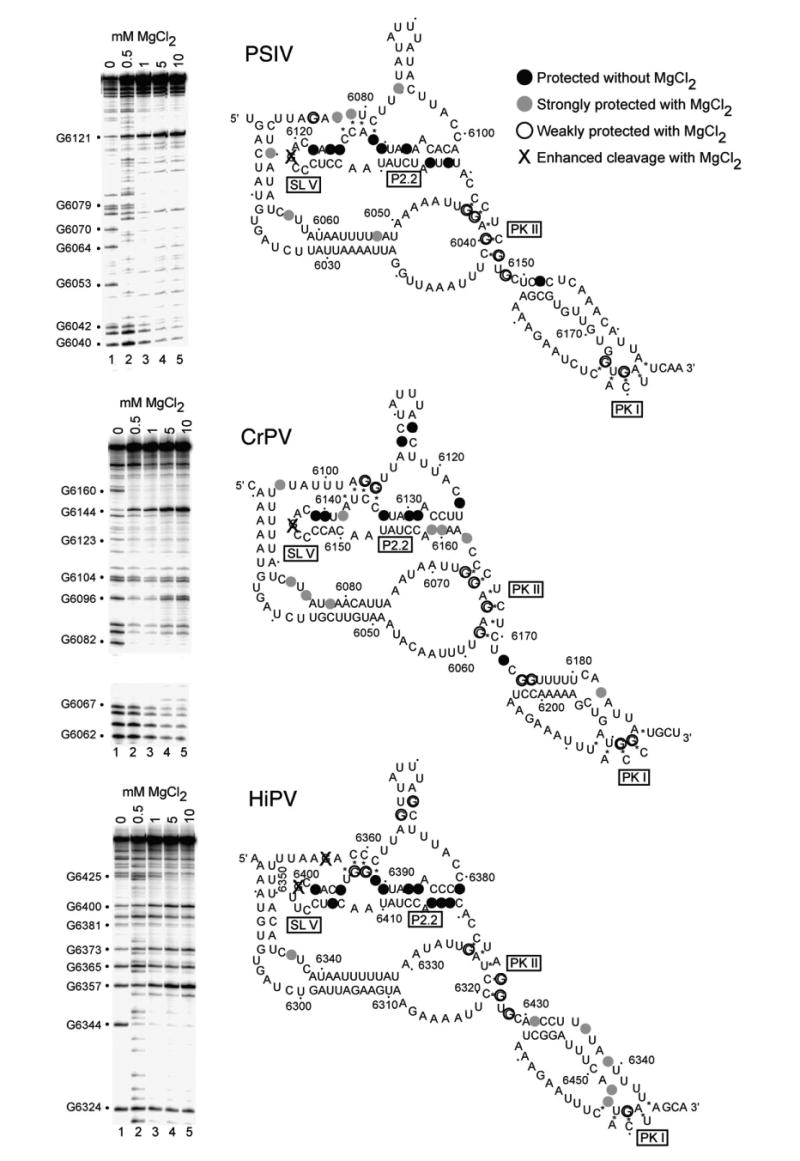
RNase T1 probing of the PSIV, CrPV, and HiPV IGR IRES RNAs. The gels shown on the left are examples of the data, in which the backbone was subjected to partial cleavage by RNase T1 (which cleaves after single-stranded G bases). Each gel contains a magnesium titration. At right are the results of this and other gels mapped onto the secondary structure of each IRES RNA. We did not obtain data for the entire backbone, but concentrated on the regions shown to be most structured in the crystal, especially the P2.2 helix and regions that interact with that helix. Bases that were protected from cleavage even in the absence of added magnesium are colored black. Those that were protected at low magnesium concentrations (0.5–1.0 mM) are indicated with filled gray circles. Bases that required elevated magnesium concentrations (5 mM–10 mM) are shown with open circles, and those that became hypersensitive to the enzyme when magnesium was added are shown with an X.
To explore the ability of the diverse class 1 IGR IRES sequences to fold and to bind the 40 S subunit, we chose three representative members for further experimentation: PSIV, cricket paralysis virus (CrPV), and Himetobi P virus (HiPV) IGR IRESes. These were chosen because their conservation relative to one another is 43%, reflecting the expected conservation of any three picked at random. As a first step in comparing these three IRES RNAs, we verified their ability to bind to the 40 S subunit. All three IRES RNAs bound to purified 40 S subunit with apparent Kd values between 2 nM and 10 nM (Figure 2(a)), matching our previously reported value for PSIV,27 and similar to the previously reported value for CrPV,19 confirming that all three RNAs are binding to the 40 S subunit as expected.
Figure 2.
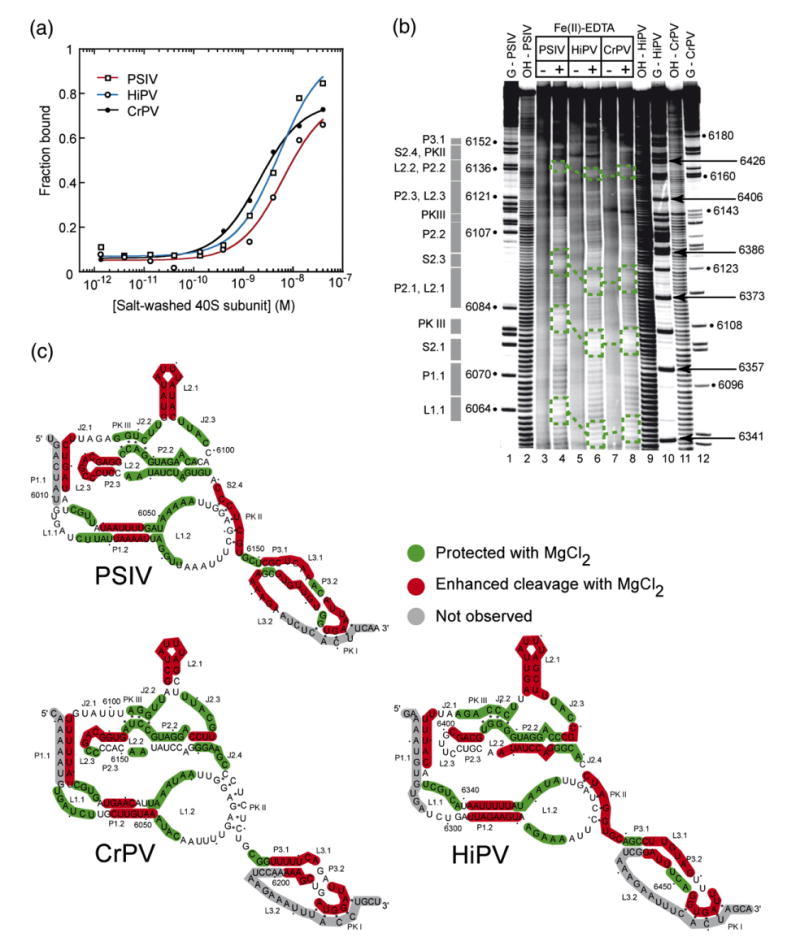
Small ribosomal subunit binding affinity comparison and structural comparison of class 1 IGR IRESes. (a) Binding isotherms of purified 40 S subunit to PSIV, CrPV, and HiPV IRES RNAs (a single experiment is shown) obtained from filter binding in a buffer containing 2.5 mM Mg2+ and 300 mM K+. These conditions minimize non-specific binding which occurs between purified ribosomes and RNA, and were chosen to match previous quantitative binding assays.27 (b) Example of a hydroxyl radical probing experiment of the PSIV (lanes 3 and 4), CrPV (lanes 7 and 8), and HiPV (lanes 5 and 6) IRES RNAs. A few examples of regions of the backbone protected in all three IRES RNAs are indicated with boxes. Repeating this experiment with different gel run times and quantitative analysis of the resulting gels led to a mapping of most of the RNA backbone. (c) Hydroxyl radical cleavage pattern for all three IRESes mapped onto their secondary structures.
Various members of the class 1 IRESes have been probed by chemicals or enzymes, identifying the common secondary structure and lending insight into some aspects of the tertiary structure. However, a detailed examination and side-by-side comparison of the RNA backbone packing strategies, and hence the folded RNA architecture at the core of these molecules, has not been done. This leaves open the question of whether specific RNA intramolecular interactions that pack RNA within the PSIV IRES (as revealed in the crystal structure) underlie all of the class 1 IRES RNAs. To address this, we conducted hydroxyl radical probing, a sensitive technique for solvent accessibility, and hence packing, of the RNA backbone, on the PSIV, CrPV, and HiPV IGR IRESes in the presence of physiological concentrations of magnesium (0.5–2 mM) (Figure 2(b) and (c)).
The hydroxyl radical cleavage of the folded PSIV, CrPV, and HiPV IGR IRES RNAs shows a pattern shared among all three (Figure 2(b) and (c)). Notably, pseudoknot III (PKIII), J2.2, J2.3, the 5′ part of P2.2, and parts of L1.2 are protected in all three IRESes. Likewise, L2.1 (SL IV) and L2.3 (SL V) show enhanced cleavage by the radicals in all three IRESes, as do parts of P1.2 and P1.1. It is important to note that similar secondary structures are not sufficient to give rise to these similar solvent accessibility patterns; similar three-dimensional structures and packing architectures are required. The pattern indicates the RNA backbone packs into a compact structure conserved among the three IRES RNAs although there are some differences in the cleavage patterns. Specifically, the 3′ half of P2.2 is protected in PSIV, less protected in CrPV, and somewhat enhanced in HiPV. These differences suggest that within the overall conserved fold, there are local variations in the structures of the three IRES RNAs. The crystal structure of the PSIV IRES ribosome-binding domain shows that the pattern of enhancement and protection of PSIV correlates with areas of the structure involved in intramolecular packing or extended into solution. Thus, the results shown here suggest the CrPV and HiPV IGR IRES ribosome binding domains fold like the PSIV IRES, with local variations that do not affect the ability of the RNA to interact with the 40 S subunit.
The class 1 IRESes have similar patterns of stability and flexibility
Regions of stable structure and dynamic structure have been identified in the IGR IRESes. To determine if a pattern is conserved among the PSIV, CrPV, and HiPV IGR IRESes, we conducted RNase T1 probing in a manner that allows the stability and flexibility of these three IRESes to be directly compared. Probing the IRES RNA structures in increasing concentrations of magnesium identified four classes of G bases.
The first class was not cleaved even in the absence of magnesium, identifying stable regions of structure that form even when the rest of the molecule is unstructured. In all three IRES RNAs, these protected bases were contained in the P2.2 helix of region 2. The second class of G bases become hypersensitive to RNase T1 cleavage when the RNA is folded. In all three IRESes a G of this type is located in the apical loop of SL V, which makes direct contact with the 40 S subunit.19,20,27,29 A third class of G bases became protected in the presence of magnesium, indicating that the fold places them into stable secondary or tertiary structures. The fourth class is only protected from the enzyme at higher magnesium concentrations (5 mM–10 mM), if at all. These likely reside in regions that are conformationally dynamic. For example, in all three IRESes, pseudoknot II (PKII) contains at least one G base that is cleaved even at elevated magnesium concentrations, indicating that PKII is likely unstable. This dynamic behavior is also true of PKI (which is outside of the ribosome affinity domain), where all three RNAs contain a G that is only weakly protected from enzyme upon folding. This is consistent with previous studies19 and suggests this is a common feature among these IRESes. Taken together, the location of these different classes of G bases is conserved among the three IRESes, suggesting common patterns of stability.
Although the overall pattern of cleavage by RNase T1 is conserved among the three IRES RNAs, there are differences. In general, the HiPV IGR IRES RNA has more G bases that are cleaved by RNase T1 in the folded state than do the other two. This may indicate that although the native conformations of all three IRESes are the same, HiPV is more prone to misfolding or less stable. This is consistent with the hydroxyl radical probing that shows HiPV is more accessible to the radicals, particularly in the P2.2 helix.
The TSV IGR IRES adopts a fold with an inner core
Having established a common three-dimensional RNA packing architecture among the class 1 IGR IRESes, we conducted hydroxyl radical probing of the TSV IGR IRES (a class 2) and compared the pattern to the class 1 (Figure 4(a)). Although analysis of the secondary structures and ribosome binding capabilities of the TSV IGR IRES have been published,30,31 its compact folded structure had not yet been analyzed. The differences in the TSV IGR IRES cleavage patterns between folded (10 mM Mg2+) and unfolded (0 Mg2+) are more subtle than the class 1 IRESes but the areas of protection and enhancements are similar (Figure 4(b) and (c)). Hence, regions 1 and 2 of the TSV IRES adopt a fold similar to the class 1 IRESes despite sequence and secondary structure deviation. Specifically, stem–loops SL IV, SL V and portions of PKII are solvent accessible, and a significant portion of the TSV IRES' region 2 is well protected. These areas of protection are in J2.2, J2.3, L2.1, P2.2 and PKIII (Figure 4(c)), corresponding to the packed core of the class 1. In region 1, loops L1.1A and L1.1B are protected as they are in the class 1 IGR IRESes despite the fact that their sequence deviates dramatically from the class 1. However, the TSV IRES cleavage pattern differs from the class 1 in several other parts of region 1 which may reflect a dynamic nature of this region of the IGR IRES and local differences in the fold.
Figure 4.
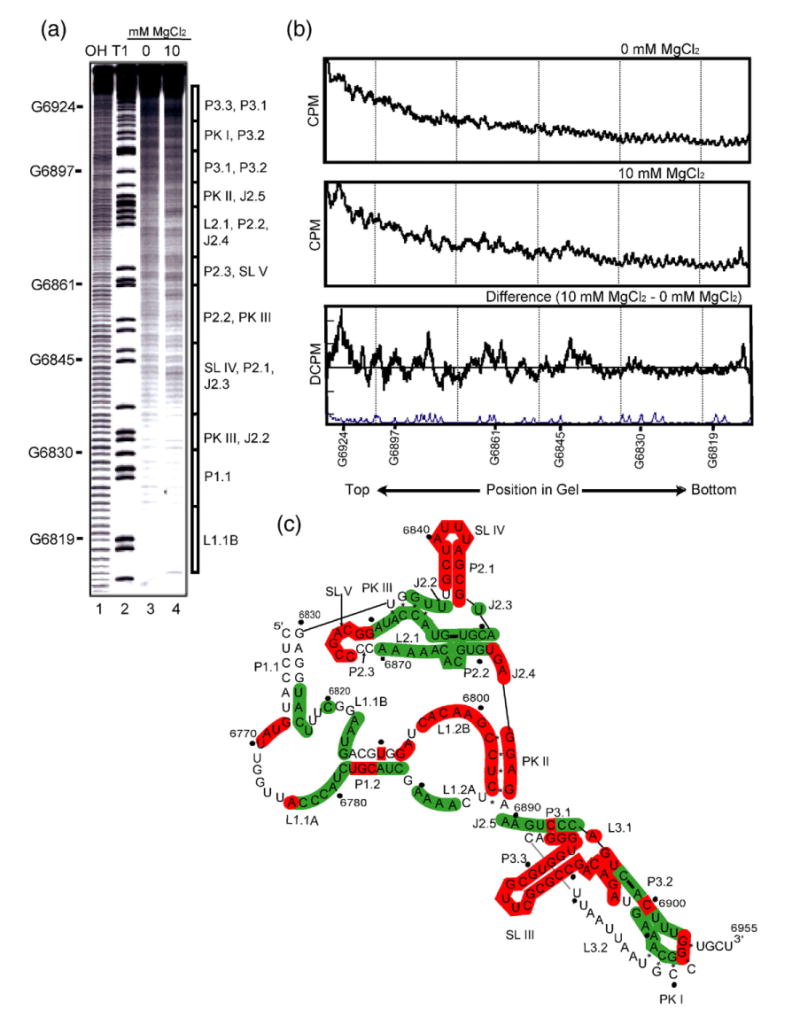
Hydroxyl radical probing of TSV full length IGR IRES. (a) An example of a hydroxyl radical probing experiment performed on 5′ end-labeled TSV full length IRES. Lanes 1 and 2 contain a hydrolysis ladder and an RNase T1 ladder, respectively. On the right of the gel, a schematic shows the location of various secondary structure elements. Lanes 3 and 4 show the TSV IRES hydroxyl radical cleavage pattern in the absence or presence of MgCl2, respectively. These lanes were analyzed as described.27 (b) Normalized quantified traces of lanes 3 and 4, and the difference trace shown with a trace of the RNase T1 lane. (c) Summary of all the hydroxyl radical probing data of the TSV IRES RNA. Solvent accessible or exposed regions of the backbone are colored red on the secondary structure, and protected or solvent inaccessible are colored green.
In region three of TSV, the extra stem–loop and its junction with region 3 (SL III) are solvent accessible. We also observed small areas of the backbone that are solvent accessible among areas near PKI where the backbone is protected. This is similar to what was observed in region three of the class 1 IRESes, suggesting that despite the presence of the extra stem–loop, the backbone packing interactions that underlie the structure are conserved.
Regions 1 and 2 of the TSV IRES fold independent of region 3
Regions 1 and 2 of the class 1 IGR IRESes adopt a compact fold crucial for the interaction with the small ribosomal subunit that is independent of region 3. To test if the class 2 IGR IRESes follow the same pattern, we constructed a mutant of TSV (nt 6761–6889) that lacks region 3. Hydroxyl radical probing shows that the removal of region 3 has little consequence on the overall protection and cleavage patterns. The folded core remains protected and the stem–loops IV and V are still positioned out into the solution (data not shown). Although there was subtle change in region 1, especially in L1.1 and P1.2, these differences were as not strong or reproducible enough for us to conclude this is due to a direct effect of region 3 changing the architecture of regions 1+2.
To further probe the fold, RNase T1 probing was performed on the truncated mutant and full length TSV. In general, the cleavage pattern resulting from the RNase T1 probing coincides with the predicted secondary structure, although many bases predicted to be in stable secondary structure regions are only weakly protected when the RNA folds, including some bases in P2.2 (Figure 5). As in the class 1 IRESes, a G at the end of SL V becomes hypersensitive to the enzyme. In addition, G bases within P1.1 are weakly protected, showing that like the class 1 IGR IRESes, P1.1 of the TSV IRES is not structured in the unbound form. Furthermore, the cleavage pattern around PKII reflects its dynamic nature. RNase T1 probing of the truncated mutant yielded a pattern similar to full-length (data not shown), providing additional supporting evidence that the architectural organization of the TSV IGR IRES regions 1+2 is not dependent on region 3.
Figure 5.
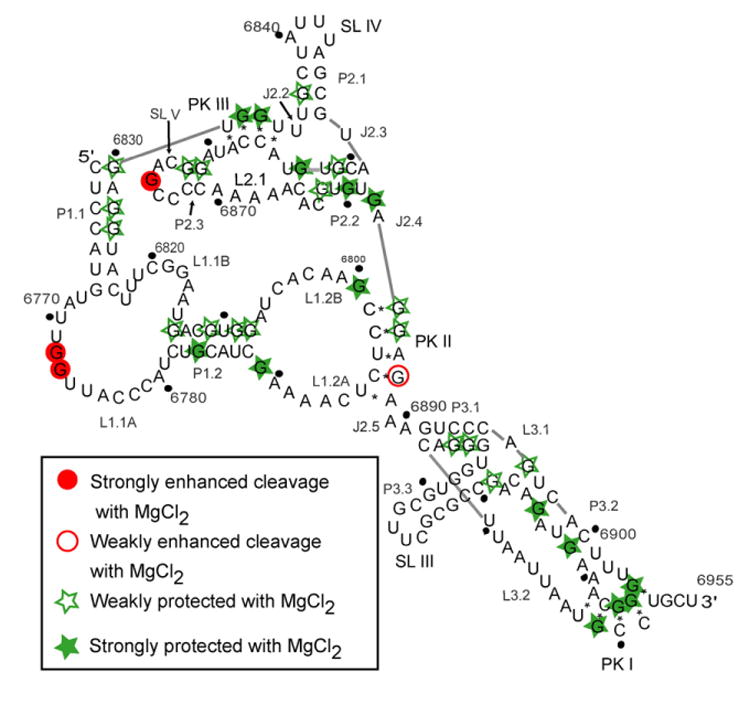
RNase T1 probing on the TSV IRES showing the cleavage pattern that resulted when the IRES was allowed to fold in the presence of MgCl2. Filled circles indicate that the G base was strongly protected from the RNase upon the addition of the cation. Open circles represent G bases that were weakly protected. Open stars are indicative of G bases that were weakly cleaved more in the presence of the cation. Filled stars depict G bases that were strongly cleaved in the presence of the Mg2+ cation.
[MgCl2] dependence of the TSV IGR IRES fold
To determine the magnesium ion dependence of the TSV IGR IRES' fold and to compare this to the class 1 IGR IRESes, we used hydroxyl radical probing with a magnesium titration. Several areas of the backbone were quantified at each magnesium concentration, converted to fraction folded, and plotted as a function of magnesium concentration (Figure 6(a)). These data show the RNA folds at 2.1(±0.6) mM MgCl2 with a Hill coefficient of 1.3±0.6. This is similar to the hydroxyl-radical monitored [MgCl2] dependence of the PSIV IRES (class 1),27 indicating that formation of the tightly packed core requires approximately the same amount of cation for both.
Figure 6.
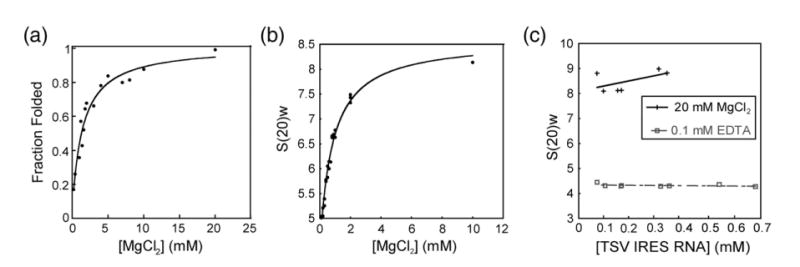
Folding of the TSV IRES as a function of MgCl2. (a) Graph of hydroxyl radical probing of full length TSV IRES RNA with varying concentrations of MgCl2. The plot shows fraction folded versus [MgCl2], fitted by a Langmuir isotherm. (b) Graph of sedimentation coefficient of the full-length TSV IRES RNA as a function of the concentration of MgCl2. (c) Graph of sedimentation coefficient of the full-length TSV IRES RNA at several [RNA], both in the absence and presence of MgCl2. These plots indicate no significant deviations from ideality that would affect our interpretation. The continuous line represents the 20 mM MgCl2 and the broken line represents 0.1 mM EDTA.
The hydroxyl radical probing detects formation of the folded core, so to measure [MgCl2]–dependent changes in the size and shape of the TSV IGR IRES, we performed a titration experiment and monitored the changes of TSV's hydrodynamic properties using analytical ultracentrifugation (AUC). Figure 6(b) shows that the sedimentation coefficient increases with divalent cation concentration until it plateaus at the higher levels of MgCl2, indicating the folded IRES RNA is more compact than the unfolded RNA, with the transition occurring at 0.83(±0.11) mM MgCl2. The class 1 IGR IRESes show a similar change in the measured shape upon folding.27
The measured [MgCl2] dependence of the TSV IRES fold was different when monitored by hydroxyl radical probing or AUC. Similar behavior was observed with the class 1 IGR IRESes and with the group I intron,27,32 suggesting that like these RNAs, the TSV IGR IRES undergoes an initial collapse as secondary and some tertiary interactions form at [MgCl2]<1 mM, and adopts its tightly folded structure at [MgCl2]>2 mM. However, there is a difference between the TSV IRES and the PSIV IRES (class 1) in that the TSV IRES requires more magnesium to reach the first initial collapsed state, as indicated by the AUC data. Specifically, the TSV IRES requires 0.83(±0.11) mM MgCl2, whereas the PSIV IRES only needs ∼0.2 mM MgCl2. However, to assume the final tightly folded structure, the TSV IGR IRES requires essentially the same amount of MgCl2 as a class 1 IGR IRES, which is approximately 2 mM as indicated by the hydroxyl radical probing data. Therefore, we conclude that the conformational collapse of the TSV IRES is less stable than that of the class 1 IRESes, and this may correspond to the more subtle protections observed in the hydroxyl radical probing experiments. Furthermore, the fact that the TSV IRES RNA has fewer nucleotides in region 1 and 2 to form the stabilizing interactions observed in the PSIV IRES crystal structure may contribute to this reduced stability and requirement for more Mg2+.
The folded TSV IGR IRES is more compact than the class 1 IRESes
We used the AUC results to assess the overall dimensions of the TSV IRES and compare this class 2 IRES to the class 1 IRESes. Using the known molecular mass and the partial specific volume of 0.53 cm3/g, we calculated the Stokes radius and the ratio of experimental frictional coefficient to the theoretical frictional coefficient (f/fo). With these two values the ratios of length versus thickness for a prolate ellipsoid and oblate ellipsoid were calculated. For the TSV IRES RNA, we see that in the absence of MgCl2 the IRES has a Stokes radius of 57.1 Å and ratios of 32.7 for a prolate ellipsoid and 54.9 for an oblate ellipsoid. When the TSV IGR IRES is fully folded, the values decrease substantially to 30.8 Å, 3.8 and 6.9, respectively. Although both IRESes are approximately 180 nts in length, the TSV IRES' Stokes radius and the ratio of the ellipsoid dimensions are smaller than that of the representative class 1 IRES (Table 1), perhaps due in part to the shorter length of the TSV IRES' regions 1 and 2, which can presumably pack into a smaller particle.
Table 1.
Analytical ultracentrifugation results
| Measured values | Calculated values | ||||||
|---|---|---|---|---|---|---|---|
| IRES | [MgCl2](mM) | s20w | D20W | f/fo | RH (Å) | a/b prolate ellipsoid | a/b oblate ellipsoid |
| TSV | 0 | 4.39 | 3.29 | 2.45 | 57.1 | 32.7 | 54.9 |
| 10 | 8.14 | 7.02 | 1.31 | 30.8 | 3.8 | 6.9 | |
| PSIVa | 0 | 3.96 | 4.12 | 2.79 | 65.8 | 44.1 | 81.9 |
| 10 | 5.65 | 4.93 | 1.95 | 46.1 | 18.4 | 26.9 | |
Values from Costantino & Kieft.27
The extra stem–loop in region 3 of the TSV IGR IRES does not interact with the small ribosomal subunit
The class 1 IGR IRESes bind the 40 S subunit through SL IVand SLV, and region 3 is not needed to bind the subunit. However, since the class 2 IRESes have an extra stem–loop in region 3, we asked the question if this extra stem loop might provide additional affinity for the 40 S subunit. To address this, we measured the affinity of the 40 S subunit for the full-length TSV IRES RNA and an RNA lacking region 3 (Figure 7(a)). The full-length TSV IRES RNA bound to the subunit, but we were unable to measure a Kd due to high background on the nitrocellulose membrane and the inability to reach reproducibly binding saturation, which may be due to aggregation of the IRES RNA. The Kd value for this interactions has been reported as ∼56 nM, and those studies also showed saturation less than 100% fraction bound.31 However, when region 3 is removed, the high background effect disappeared and a more complete binding curve was obtained, although saturation was still well below 100%. Despite these technical complexities, Figure 7(a) indicates a similar binding affinity for these two RNAs, indicating that like the class 1 IRESes, the class 2 TSV IRES binds to the 40 S subunit through regions 1 and 2.
Figure 7.
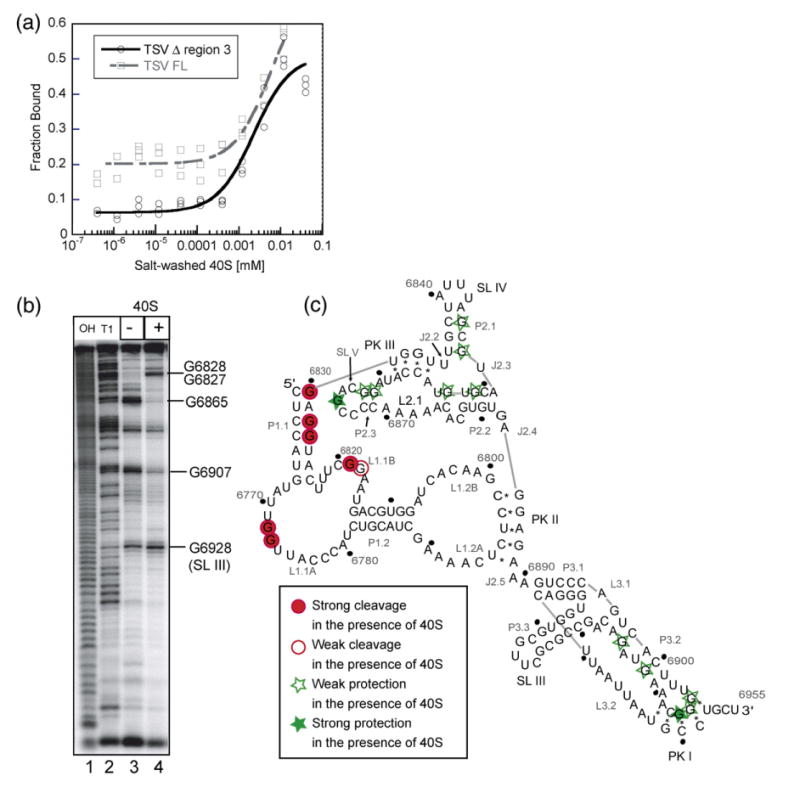
Binding of the small ribosomal subunit to the TSV IRES. (a) Fraction of TSV IRES bound plotted as a function of the small ribosomal subunit concentration. The broken line represents the wild-type and the solid line represents the mutant IRES. (b) A representative RNase T1 footprinting gel, in the absence (lane 3) and presence (lane 4) of 40 S ribosomal subunit. Lanes 1 and 2 are the hydrolysis ladder and RNase T1 ladder, respectively. Selected G residues are labeled to the right of the gel. (c) Footprinting results mapped on the secondary structure. Dark grey circles indicate that a G base was strongly cleaved when the small ribosome subunit was present. Light grey circles indicate that a G base was weakly cleaved when the small ribosome subunit was present. Light grey stars represent the weak protection offered by the 40 S subunit, while the dark grey stars represent G bases that were strongly protected.
Even though region 3 does not stabilize the interaction between the TSV IGR IRES and the 40 S subunit, it could still make contact with the subunit. To test this, we performed an RNase T1 footprinting experiment on end-labeled TSV IRES in the presence and absence of the small ribosomal subunit (Figure 7(b) and (c)). Areas that are protected from the RNase (such as nucleotides 6865 and 6907) correspond to predicted sites of interaction between the IRES and the 40 S subunit, which are regions within region 2 and PK1. We observed areas of enhanced cleavage in region 1 (such as nucleotides 6827 and 6828) upon the addition of the small ribosomal subunit, which could be due to local structure rearrangements upon binding. Interestingly, we did not observe any change in the digestion pattern of the RNase T1 for the region corresponding to the extra stem–loop in region 3, specifically G6928. Therefore, we conclude that the apical loop of SL III does not make contact with the 40 S subunit.
To understand the contribution of SL III in the translation initiation mechanism of TSV, we performed a ribosome assembly assay using a mutant that lacked the structural element (nts 6918–6934 deleted) (Figure 8(a)). In this assay, radiolabeled RNA was incubated in translationally competent rabbit reticulocyte lysate and the resultant preinitiation complexes were separated on a sucrose gradient. Full-length TSV IRES assembled both 40 S subunit and 80 S ribosome-containing complexes (Figure 8(b)). Although the efficiency of this process is low compared to other IRESes, it is expected given the low affinity the TSV IRES has for rabbit reticulocyte lysate-derived ribosomes.31 The SL III deletion mutant IRES forms complexes with a similar efficiency to the full-length, although there are reproducibly more 80 S-bound IRESes than 40 S-bound. Addition of cycloheximide to the reactions did not change this observation (data not shown). This result suggests that SL III may have a role in promoting 80 S formation or in subsequent downstream events.
Figure 8.
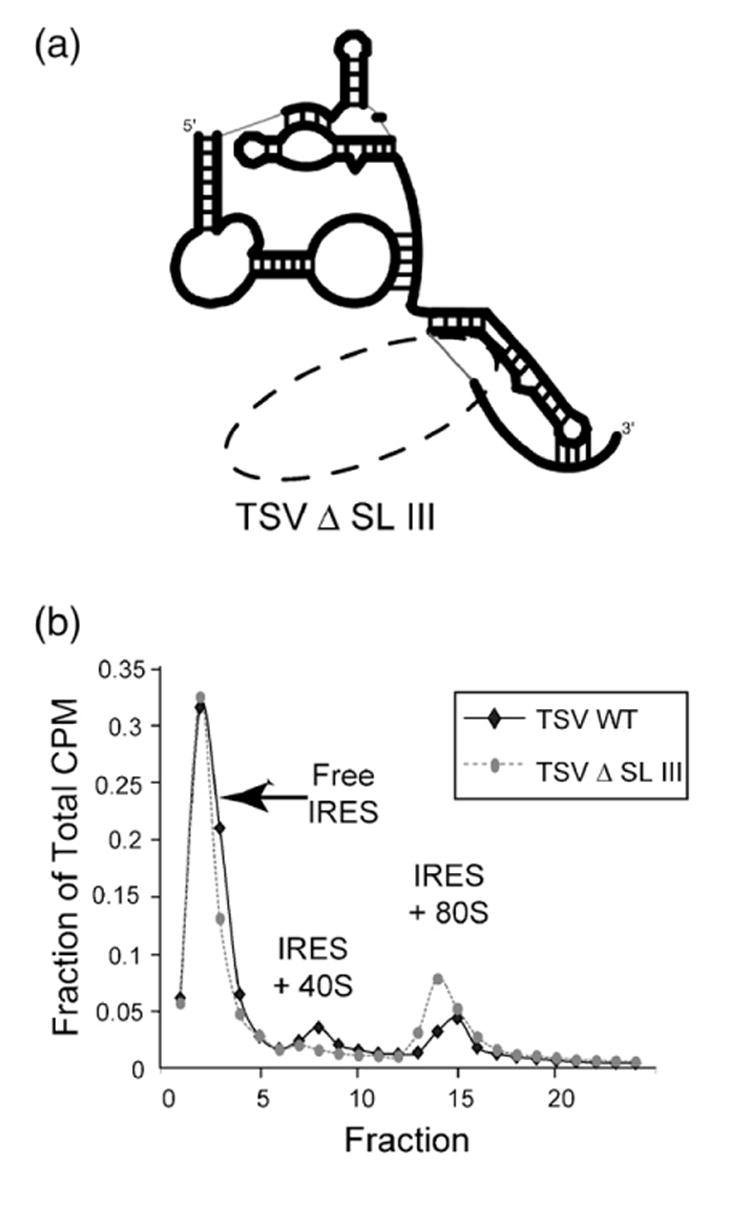
Ribosome assembly assay of TSV and a mutant lacking SL 3. (a) Schematic cartoon representation of the TSV IRES RNA mutant used in the assay; the broken oval shows where SL III has been removed. (b) Traces from the ribosome assembly assay with full-length TSV IRES RNA and the SL III deletion mutant. The profiles are plotted based on the percentage of the total counts per minute (CPM) in each fraction. Full-length is represented by a bold black line, while the mutant is represented by a broken line.
Discussion
IRES-driven translation initiation is critical for the infection of many viruses and for regulating the expression of many eukaryotic genes. The Dicistroviridae IGR IRESes are a useful model system to begin deciphering the structural basis of IRES function, because they are essentially all-RNA-based translation initiation apparatuses and their function requires a stable RNA fold. The recent crystal structure of the ribosome binding domain of one of these IRESes, coupled with a wealth of published biochemical data and data presented here, allow correlation of IRES sequence diversity, three-dimensional structure, and mechanism of ribosome interactions.
By probing the solvent accessibility of the RNA backbone, we show that despite significant sequence differences, three representative class 1 IRESes all fold into a similar three-dimensional structure. The similarity of folds among the three members of the class 1 IRESes is likely provided by a set of conserved nucleotides within regions 1 and 2 of the RNA (Supplementary Data, Figure 1). Many of these conserved nucleotides are not involved in direct interaction with the ribosome and therefore provide the stabilizing interaction within the common fold. It is evident that a subset of the nucleotides within the ribosome binding domain of these IRESes has been maintained in order to form the structure needed for initial interaction with the ribosome.
Within all of the Dicistroviridae IRESes that we tested, helix P2.2 is protected from solvent in the folded state, suggesting it lies at the heart of the folding of all these RNAs. In the crystal structure of the PSIV IRES ribosome binding domain, this helix is at the center of the stable prefolded RNA structure and forms many non-canonical tertiary interactions involving conserved nucleotides.29 Furthermore, nuclease probing of the class 1 IRESes shows that this helix can form even in the absence of fold-inducing magnesium ions, suggesting it is the core scaffold around which the rest of the molecule organizes. The fact that P2.2 is protected from solvent in the TSV IRES suggests that like the class 1 IGR IRESes, this class 2 IGR IRES' fold is organized by helix P2.2. This is noteworthy because some nucleotides observed in the PSIV IRES crystal structure to be involved in intramolecular interaction are missing or mutated in the P2.2 helix of the TSV IRES, again indicating the evolutionary pressure to maintain a folded architecture despite sequence change. Taken together, these data suggest that this helix has a conserved role in organizing the ribosome binding domain structure of all of the IGR IRES RNAs. Hence, recent structural information derived from crystallography and cryo-electron microscopy form the basis for understanding the function of all of the Dicistroviridae IGR IRESes, both class 1 and class 2.
The result of the conserved fold organized around P2.2 is a prefolded structure in which each region within the ribosome-binding domain serves a particular function. Region 2 interacts with the 40 S subunit, while region 1 makes contact with the 60 S subunit.13,29 Interestingly, the ribosome-recruiting elements of region 2 (stem–loops IV and V) have remained conserved despite the subdivision of the family into two classes, while the ribosome binding elements of region 1 (L1.1) have changed dramatically. It may be that the differences seen in the sequence of L1.1 and region 3 of both classes are reflective of different mechanisms for interacting and manipulating the large ribosomal subunit.
A possible difference in the mechanism of action between the class 1 and class 2 IGR IRES is the presence of the additional stem–loop in region 3 of the class 2.30 Our experiments show that SL III is not necessary for recruiting either the large or small ribosomal subunit or for incorporation into the 80 S ribosome, and despite the likely position of region 3 in the P-site of the ribosome, the apical loop of SL III does not make contact to the 40 S subunit. SL III therefore may be functionally important in the context of the fully assembled ribosome. This would be in agreement with mutational studies of SL III that found it necessary for the TSV IRES to initiate translation.30 One possible role is that SL III could help release the 80 S complex from the IRES, explaining our observation of increased 80 S formation on a mutant with SL III deleted (Figure 8). Another explanation for our observation is that removal of SL III makes the TSV IRES more efficient at interacting with already assembled 80 S ribosomes, or that its removal enhances recruitment of 60 S subunit to 40 S-bound IRES. The possibilities imply the stem–loop may be a repressor of 80 S ribosome formation on, or binding to, the IRES. Clearly, further investigations are needed to resolve the role of this structural element.
In addition to the mechanistic differences, variations with the folding behavior between the two classes were also observed. From our ultracentrifugation experiments, it is evident that the TSV IRES RNA (class 2) forms a smaller structure than the PSIV IRES RNA (class 1). This is not unexpected because in the secondary structures of the two classes, the TSV IRES lacks certain junction regions (e.g. J2.3) and has shorter helices (e.g. P2.2, P2.3, P1.2). To stabilize this compact structure, TSV requires more magnesium than the class 1 IGR IRESes, perhaps due to the loss of key intramolecular contacts present in the larger cores of the class 1. In addition, the results presented here suggest that the fold of the TSV IRES, while similar to the class 1 IGR IRESes, is not as stable as the class 1 IRESes that we tested. We base this on the weaker protection from hydroxyl radicals, the lower efficiency of preinitiation complex formation, the requirement for more Mg2+ for initial folding, and the lower affinity for 40 S subunits. It could be that the TSV IRES requires a more specialized set of conditions for optimal folding and function.
From this work, it is evident that the overall architecture of the Dicistroviridae IGR IRES ribosome binding domain is conserved. The presence of similarities both in structure and likely mode of binding to the small ribosomal subunit illustrate that the two classes use a fundamental approach that is structure-driven for interacting with the translation machinery. However, distinct structural differences between the two classes may reflect differences in the approach each uses to manipulate the ribosome, especially at the steps involving interaction with the large subunit. More detailed analysis of the mechanism of both classes will provide a deeper understanding of how structural differences alter the ways IRESes initiate translation. This knowledge will augment our understanding of how other IRESes, like the HCV IRES, interact with the same ribosomal proteins as the IGR IRESes while using a different structure.24,26,33 Hence, a detailed understanding of the RNA structure-based mechanism of the Dicistroviridae IRESes lays the foundation for understanding more complex and more divergent IRES structures and mechanisms.
Materials and Methods
Cloning and plasmid production
Plasmids pT7CAT-5375 (encoding the PSIV IRES) and pT7CAT-HiPV6201-7259 (encoding the HiPV IRES) were a kind gift from Nobuhiko Nakashima. Plasmid pCrPV1-1 (encoding the cricket paralysis virus IRES, CrPV IRES) and pRC1 (encoding the TSV IRES) was a kind gift of Eric Jan and Peter Sarnow. These plasmids were used as templates for PCR reactions to generate inserts that contain the T7 polymerase promoter and the IRES sequence flanked by cis-acting ribozymes. Inserts were ligated into the EcoRI/BamHI site of pUC-19, amplified in DH5α cells and sequenced. Full-length IRES sequences included: nt 6002–6195 of PSIV, nt 6029–6219 of CrPV, and nt 6087–6275 of HiPV, and nt 6761–6958 of TSV. Mutants were generated using the QuickChange mutagenesis kit (Stratagene) or by standard cloning techniques. The clones were sequenced to verify the sequence.
RNA transcription, purification, and end-labeling
RNA was generated from in vitro transcriptions as described.34 RNA was purified on denaturing polyacrylamide gels, passively eluted from the gel, concentrated in ultrafiltration devices (Amicon) and stored in DEPC-treated water at −20 °C. RNA was 5′ or 3′ end-labeled as described.34,35
Chemical and enzymatic probing
Hydroxyl radical probing was performed as described.34 Buffer conditions were 30 mM Hepes-KOH (pH 7.4), 0-10 mM MgCl2, 0.1 mg/ml tRNA (0.3 mg/ml tRNA for TSV). Reactions were conducted at 37 °C for 2 min and analyzed on 10% (w/v) sequencing denaturing polyacrylamide gels. Gels were dried and imaged on a phosphorimager and analyzed quantitatively as described.27 Sequencing ladders using RNase T1 and hydrolysis ladders were generated as described.34
RNase T1 probing was conducted as described.34 Buffer conditions were identical to those used in hydroxyl radical probing. To initiate cleavage, 1 μl of RNase T1 at 0.1unit/μl was added to the 10 μl (total) reaction and incubated at 37 °C for 5 min, then quenched with 10 μl of 7M urea in 1× TBE. Reactions were resolved on 10% sequencing denaturing polyacrylamide gels which were dried and analyzed on a phosphorimager.
Analytical ultracentrifugation
To anneal and fold the RNA, it was suspended in 10 mM sodium cacodylate buffer (pH 7.5), heated to 80 °C for 1 min, cooled on the bench to room temperature, and MgCl2 was added to the desired concentration. Analytical ultracentrifugation was performed in a Beckman XL-A centrifuge with an An-60Ti rotor at 40,000 RPM at 12 °C. The sample was scanned every 5 min at 260 nm. For each experiment, at least 30 boundary traces were collected; in most cases, 60 were collected.
The traces were analyzed as dc/R using the program SVEDBERG V6.39.36 In each case, at least 15 traces were used simultaneously. The fit directly yielded the parameters S and D. We adjusted these values to standard conditions using the program SEDNTRP V1.08. A partial specific volume of 0.53 cm3/g and the calculated molecular mass was used to obtain the parameters RH, f/fo, and a/b. This analysis is similar to that employed in the analysis of other folded RNAs.32,37
40 S ribosomal subunit binding assays
The 40 S ribosomal subunit was purified from bulk rabbit reticulocyte lysate (Green Hectares, WI) as described.27,35 These subunits were used in filter binding assays.27,35 The buffer was 20 mM Tris–HCl (pH 7.0), 2.5 mM MgCl2, 100 mM potassium acetate, 200 mM KCl, 1 mM DTT.
40 S ribosomal subunit footprinting
RNase T1 probing was conducted as described.34 Buffer conditions were identical to those used in hydroxyl radical probing. After the TSV RNA was denatured and allowed to anneal, it was incubated at 37 °C for 10 min. Purified 40 S from rabbit reticulocyte lysate was added at a level to achieve maximal interaction with the IRES and allowed to incubate at 37 °C for 10 to 15 min. To initiate cleavage, 1 μl of RNase T1 at 0.1 unit/μl was added to the 10 μl (total) reaction and incubated at 37 °C for 5 min, then quenched with 10 μl of 7M urea in 1× TBE. Reactions were resolved on 10% sequencing denaturing polyacrylamide gels which were dried, analyzed on a phosphorimager, and quantitated using Imagequant.
40 S/80 S pre-initiation complex assembly assays
Approximately 1000 CPM of 32P-labeled IRES RNA in 30 mM Hepes (pH 7.5) buffer was heat denatured at 85 °C for about 50 s. MgCl2 was added to a final concentration of 2 mM in a 10 μl volume. To assemble pre-initiation complexes in cell-free lysate, we added the prefolded 32P-labeled IRES RNA to 33 μl of rabbit reticulocyte lysate and RNase-free water to a final volume of 50 μl. The reactions were incubated at 30 °C, then quenched with 250 μl of buffer (50 mM Tris–HCl pH 7.5, 50 mM NaCl, 5 mM MgCl2, 1 mM DTT) and placed on ice. Reactions were immediately loaded onto sucrose gradients prepared as described below and analyzed by ultracentrifugation. Assays also were done in the presence of 100 μg/ml cycloheximide.
Assembly reactions were analyzed on 15%–30% (w/v) sucrose gradients in 50 mM Tris–HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2, 1 mM DTT. Gradients were prepared in SW41 tubes using the automated mixing procedure of a BioComp Gradient Master apparatus and used immediately after preparation. Reactions were layered on the gradients and spun at 4 °C for 3 h at 36,000 RPM using a SW41 rotor. After spinning, the gradients were fractionated using the BioComp Gradient Master automated procedure. Fractions were analyzed by blotting onto a sandwich of nitrocellulose and nylon membranes and quantitation of the radioactivity on each membrane using a phosphorimager. The 80 S ribosomes and 40 S complex peaks were assigned based on location in the gradient and previous studies. Reactions were incubated for 15 min. For the assays that involved the use of cyclohexamide, the compound was included in the gradients.
Supplementary Material
Supplementary Data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2007.04.076
Acknowledgments
The authors thank P. Sarnow, E. Jan, and N. Nakashima for sharing plasmids, and R. Batey, D. Bentley, and E. Eisenmesser for a critical reading of the manuscript.
Abbreviations used
- IRES
internal ribosome entry site
- IGR
intergenic region
- TSV
Taura Syndrome virus
- UTR
untranslated region
- HCV
hepatitis C virus
- PSIV
Plautia stali intestine virus
- CrPV
cricket paralysis virus
- HiPV
Himetobi P virus
- PK
pseudoknot
- AUC
analytical ultracentrifugation
- SL
stem–loop
References
- 1.Jang SK. Internal initiation: IRES elements of picornaviruses and hepatitis c virus. Virus Res. 2006;119:2–15. doi: 10.1016/j.virusres.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 3.Stoneley M, Willis AE. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- 4.Hershey JWB, Merrick WC. Pathway and mechanism of initiation of protein synthesis. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 33–88. [Google Scholar]
- 5.Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 6.Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CU. Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci USA. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bushell M, Sarnow P. Hijacking the translation apparatus by RNA viruses. J Cell Biol. 2002;158:395–399. doi: 10.1083/jcb.200205044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christian P, Castens E, Domier L, Johnson K, Nakashima N, Scotti P, van der Wilk F. In: Virus Taxonomy. Regenmortel MHV, Fauquet CM, Bishop DHL, Carstens EB, Estes MK, Lemon SL, Maniloff J, Mayo MA, McGeoch DJ, Pringle CR, Wickner RB, editors. Academic Press; New York, NY: 2000. [Google Scholar]
- 9.Jan E. Divergent IRES elements in invertebrates. Virus Res. 2006;119:16–28. doi: 10.1016/j.virusres.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 10.Wilson JE, Powell MJ, Hoover SE, Sarnow P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol Cell Biol. 2000;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasaki J, Nakashima N. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc Natl Acad Sci USA. 2000;97:1512–1515. doi: 10.1073/pnas.010426997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jan E, Thompson SR, Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiator Met-tRNA-independent translation mediated by an internal ribosome entry site element in cricket paralysis virus-like insect viruses. Cold Spring Harbor Symp Quant Biol. 2001;66:285–292. doi: 10.1101/sqb.2001.66.285. [DOI] [PubMed] [Google Scholar]
- 13.Schuler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, et al. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nature Struct Mol Biol. 2006;13:1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- 14.Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes; the IRES functions as an RNA-based translation factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Pestova TV, Lomakin IB, Hellen CU. Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep. 2004;5:906–913. doi: 10.1038/sj.embor.7400240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishiyama T, Yamamoto H, Uchiumi T, Nakashima N. Eukaryotic ribosomal protein RPS25 interacts with the conserved loop region in a dicistroviral intergenic internal ribosome entry site. Nucl Acids Res. 2007 doi: 10.1093/nar/gkl1121. In the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson SR, Gulyas KD, Sarnow P. Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element. Proc Natl Acad Sci USA. 2001;98:12972–12977. doi: 10.1073/pnas.241286698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pestova TV, Hellen CU. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jan E, Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol. 2002;324:889–902. doi: 10.1016/s0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- 20.Nishiyama T, Yamamoto H, Shibuya N, Hatakeyama Y, Hachimori A, Uchiumi T, Nakashima N. Structural elements in the internal ribosome entry site of Plautia stali intestine virus responsible for binding with ribosomes. Nucl Acids Res. 2003;31:2434–2442. doi: 10.1093/nar/gkg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki J, Nakashima N. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J Virol. 1999;73:1219–1226. doi: 10.1128/jvi.73.2.1219-1226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto H, Nakashima N, Ikeda Y, Uchiumi T. Binding mode of the first aminoacyl-tRNA in translation initiation mediated by Plautia stali intestine virus IRES. J Biol Chem. 2007 doi: 10.1074/jbc.M610887200. In the press. [DOI] [PubMed] [Google Scholar]
- 24.Fukushi S, Okada M, Stahl J, Kageyama T, Hoshino FB, Katayama K. Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J Biol Chem. 2001;276:20824–20826. doi: 10.1074/jbc.C100206200. [DOI] [PubMed] [Google Scholar]
- 25.Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 26.Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H. Structure of the hepatitis C Virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure (Camb) 2005;13:1695–1706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 27.Costantino D, Kieft JS. A preformed compact ribosome-binding domain in the cricket paralysis-like virus IRES RNAs. RNA. 2005;11:332–343. doi: 10.1261/rna.7184705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanamori Y, Nakashima N. A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA. 2001;7:266–274. doi: 10.1017/s1355838201001741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfingsten JS, Costantino DA, Kieft JS. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science. 2006;314:1450–1454. doi: 10.1126/science.1133281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatakeyama Y, Shibuya N, Nishiyama T, Nakashima N. Structural variant of the intergenic internal ribosome entry site elements in dicistroviruses and computational search for their counterparts. RNA. 2004;10:779–786. doi: 10.1261/rna.5208104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cevallos RC, Sarnow P. Factor-independent assembly of elongation-competent ribosomes by an internal ribosome entry site located in an RNA virus that infects penaeid shrimp. J Virol. 2005;79:677–683. doi: 10.1128/JVI.79.2.677-683.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takamoto K, He Q, Morris S, Chance MR, Brenowitz M. Monovalent cations mediate formation of native tertiary structure of the Tetrahymena thermophila ribozyme. Nature Struct Biol. 2002;9:928–933. doi: 10.1038/nsb871. [DOI] [PubMed] [Google Scholar]
- 33.Otto GA, Lukavsky PJ, Lancaster AM, Sarnow P, Puglisi JD. Ribosomal proteins mediate the hepatitis C virus IRES-HeLa 40S interaction. RNA. 2002;8:913–923. doi: 10.1017/s1355838202022057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J Mol Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 35.Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Philo JS. An improved function for fitting sedimentation velocity data for low- molecular-weight solutes. Biophys J. 1997;72:435–444. doi: 10.1016/S0006-3495(97)78684-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deras ML, Brenowitz M, Ralston CY, Chance MR, Woodson SA. Folding mechanism of the Tetrahymena ribozyme P4-P6 domain. Biochemistry. 2000;39:10975–10985. doi: 10.1021/bi0010118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2007.04.076


