Abstract
Acanthamoeba class I myosins are unconventional, single-headed myosins that express actin-activated Mg2+-ATPase and in vitro motility activities only when a single serine or threonine in the heavy chain is phosphorylated by myosin I heavy chain kinase (MIHCK). Some other, but not most, class I myosins have the same consensus phosphorylation site sequence, and the two known class VI myosins have a phosphorylatable residue in the homologous position, where most myosins have an aspartate or glutamate residue. Recently, we found that the catalytic domain of Acanthamoeba MIHCK has extensive sequence similarity to the p21-activated kinase (PAK)/STE20 family of kinases from mammals and yeast, which are activated by small GTP-binding proteins. The physiological substrates of the PAK/STE20 kinases are not well characterized. In this paper we show that PAK1 has similar substrate specificity as MIHCK when assayed against synthetic substrates and that PAK1 phosphorylates the heavy chain (1 mol of Pi per mol) and activates Acanthamoeba myosin I as MIHCK does. These results, together with the known involvement of Acanthamoeba myosin I, yeast myosin I, STE20, PAK, and small GTP-binding proteins in membrane- and cytoskeleton-associated morphogenetic transformations and activities, suggest that myosins may be physiological substrates for the PAK/STE20 family and thus mediators of these events.
Conventional, two-headed, filamentous class II myosins and unconventional, single-headed, nonfilamentous class I myosins are the most widely distributed of the 12 classes of the myosin superfamily (1). Class I myosins have a single, relatively short (for myosins) heavy chain and one or more light chains (2, 3). As for all myosins, the N-terminal, globular domain of class I myosins has an ATP-sensitive actin-binding site and expresses actin-activated Mg2+-ATPase activity (4). The short C-terminal domain of the classic amoeba class I myosins contains an ATP-insensitive F-actin-binding site (5), an acidic phospholipid/membrane-binding site (6), and a Src homology 3 (SH3) region. Other class I myosins may lack the ATP-insensitive F-actin-binding site and SH3 region (7).
The actin-activated Mg2+-ATPase activity (2) and the in vitro motility activity (8, 9) of the three Acanthamoeba class I myosins depend on phosphorylation of a serine or threonine residue (depending on the isozyme) in the N-terminal domain of the heavy chain at a site (10) where almost every other myosin contains either an aspartate or glutamate residue (11). The known exceptions (11, 12) are class I myosins from Dictyostelium (five isozymes), Aspergillus, and Saccharomyces cerevisiae (two isozymes) and class VI myosins from Drosophila and pig, which also have either a serine or threonine residue at this position. However, thus far only Acanthamoeba (2, 8, 9) and Dictyostelium (13) myosins I have been shown to be activated by phosphorylation of their single heavy chains.
Acanthamoeba myosin I heavy chain kinase (MIHCK) is partially activated by association with phospholipids or membranes (14–16) and fully activated by autophosphorylation of multiple sites (14, 16), including at least one site in the C-terminal 35-kDa catalytic domain (35K; ref. 17). Studies with synthetic peptide substrates revealed an unusual requirement for a tyrosine residue two amino acids C-terminal to the serine or threonine that is phosphorylated, in addition to the common kinase requirement for basic amino acids on the N-terminal side of the phosphorylation site (18). This consensus MIHCK substrate site occurs in all but one of the myosins with a serine or threonine at the position of the phosphorylated residue in the eight known Acanthamoeba and Dictyostelium class I myosins (11, 12); in the exception, isoleucine replaces tyrosine as it does in the two class VI myosins (11, 12).
The recently determined sequences of MIHCK from Acanthamoeba (19) and Dictyostelium (20) show both to be members of the p21-activated kinase (PAK)/STE20 kinase family. PAK and STE20, which are generally thought to be involved in regulation of the stress-activated mitogen-activating protein kinases (21–23), are regulated by the small GTPases Rac and Cdc42 (24–26). These GTPases also induce membrane/cytoskeletal assembly, leading to the formation of distinct actin structures involved in cell motility and other cellular processes (27–29).
Little is known about the biological substrates of PAK and related kinases in general, although PAK has been shown to phosphorylate the p47phox component of the NADPH oxidase in phagocytic leukocytes (25, 30). PAK1 has been localized to membranes where active membrane/cytoskeletal changes are occurring in mammalian fibroblasts (S. Dharmawawardhane, R. H. Daniels, and G.M.B., unpublished data), and PAK1 is capable of inducing a polarized rearrangement of the actin cytoskeleton (M. A. Sells, U.G.K., S. Bagrodia, D. Ambrose, G.M.B., and J. Chernoff, unpublished data). Acanthamoeba and Dictyostelium myosin I are known to be involved in membrane-associated activities (31–35), and myosin I in S. cerevisiae is required for receptor-mediated endocytosis (36) and cytoskeleton polarization (37). Based on the identification of MIHCK as a member of the PAK family, it is intriguing to speculate that PAK/STE20 might regulate morphological processes in yeast and mammalian cells by phosphorylating and activating myosins.
The present study shows that PAK shares the unusual peptide substrate specificity of Acanthamoeba MIHCK, phosphorylates Acanthamoeba myosin I heavy chain, and activates its actin-activated Mg2+-ATPase activity in vitro. These results support the hypothesis that the PAK/STE20 family regulates cell morphology and function, at least in part, through phosphorylation and activation of myosins.
MATERIALS AND METHODS
Proteins and Peptides.
MIHCK was purified from Acanthamoeba as previously described (38). MIHCK catalytic domain (35K) (18) and myosin IC (39) were expressed in insect cells and purified as described elsewhere. Recombinant human PAK1 was expressed in Escherichia coli as a poly(His) N-terminal fusion protein and affinity-purified on Ni2+-NTA agarose beads. The peptide substrates were synthesized and purified as previously described (18).
Enzyme Assays.
Kinase activity was assayed as described (18) by incubation in 50 mM imidazole buffer (pH 7.0), containing 2.5 mM [γ-32P]ATP (30 cpm/pmol), 3.5 mM MgCl2, 1 mM EDTA, and 0.2 mg/ml of bovine serum albumin for 2 min at 30°C. The concentration of each of the peptide substrates was 0.4 mM and the kinase concentrations were as follows: MIHCK, 3.36 μg/ml; 35K, 5.36 μg/ml; and PAK, 2.36 μg/ml. MIHCK was fully activated by autophosphorylation (14) before the assay. The Mg2+-ATPase activity of myosin I was determined by incubating myosin IC (11 μg/ml) in 20 mM imidazole (pH 7.5), containing 3 mM [γ-32P]ATP (2 cpm/pmol), 4 mM MgCl2, 0.4 mM EGTA, and 0.4 mg/ml of bovine serum albumin without and with 90 μM rabbit skeletal F-actin for 2 min at 30°C.
Phosphorylation of Myosin IC.
Myosin IC (110 μg/ml) was phosphorylated by incubation with either 35K or PAK at 30°C in 10 mM imidazole (pH 7.5), containing 1 mM ATP, 5 mM MgCl2, and 1 mM EGTA. When maximum incorporation of phosphate was determined, [γ-32P]ATP (300 cpm/pmol) was used and the concentrations of 35K and PAK were 27 μg/ml and 170 μg/ml, respectively. Samples were separated by SDS/PAGE and imaged by autoradiography, and the bands corresponding to the heavy chain of myosin IC were excised, solubilized, and counted by liquid scintillation counting as described (40). When myosin IC was phosphorylated for determination of its Mg2+-ATPase activity, nonradioactive ATP was used and the concentrations of 35K and PAK were 32 μg/ml and 25 μg/ml, respectively.
RESULTS
Substrate Specificity.
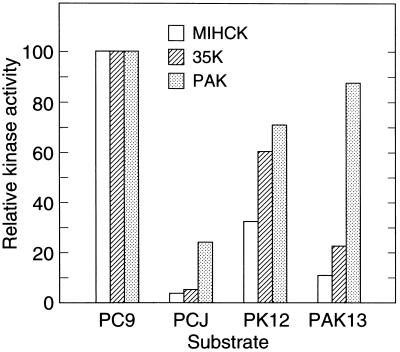
The synthetic peptides used as substrates for PAK1 and MIHCK are shown in Table 1. PC9 corresponds to the established phosphorylation site of Acanthamoeba myosin IC (10, 18). PCJ is the same peptide, but with the essential tyrosine residue replaced by leucine, and is not a substrate for MIHCK (18). PK12 corresponds in sequence to the phosphorylation site within the catalytic domain of Acanthamoeba MIHCK (19) that must be autophosphorylated for kinase activity (14, 41). PAK13 is the corresponding site in PAK1, which has been shown to be autophosphorylated in active kinase (42). The strong sequence similarity between PK12 and PAK13 is indicative of the highly conserved nature of this region of the catalytic domain of the MIHCK/PAK/STE20 family.
Table 1.
Synthetic peptides used as kinase substrates
| Name | Sequence | Identification |
|---|---|---|
| PC9 | GRGRSS*VYS | Phosphorylation site of Acanthamoeba myosin IC |
| PCJ | GRGRSS*VLS | As above, with leucine replacing tyrosine |
| PK12 | KRASVVGT*TYWM | Phosphorylation site of MIHCK |
| PAK13 | SKRSTMVGT*PYWM | Phosphorylation site of PAK |
See text for references to peptide sequences. The asterisks identify the serine that has been shown to be phosphorylated in PC9 by MIHCK (MICHK phosphorylates the same residue in myosin IC) and the serine and threonines that are in the corresponding locations in PCJ and PK12 and PAK13, respectively. The tyrosine that is essential for phosphorylation of PC9 by MIHCK is underlined, as are the leucine and tyrosines in the corresponding locations in PCJ and PK12 and PAK13, respectively. See text for details.
The data in Fig. 1 show that PAK1, like MIHCK and 35K, strongly prefers a substrate with a tyrosine residue two positions C-terminal to the phosphorylatable serine (PC9 vs. PCJ) and that PAK1 has a somewhat broader peptide specificity than MIHCK and 35K, in that it phosphorylates both its own and the MIHCK putative phosphorylation sites almost as well as PC9, while MIHCK and 35K show less activity for PK12 than for PC9 and even lower activity with PAK13. It is likely that PAK1 phosphorylates the same residue in PC9 as MIHCK (see Table 1). We do not know which of the serine/threonine residues were phosphorylated in PK12 and PAK13. In previous studies with synthetic peptides (18), phosphorylation with MIHCK always occurred at the hydroxyamino acid two positions N-terminal to Tyr and was unaffected by the exact position of the basic amino acids. The principal difference between PC9 and PK12 and PAK13 may be the greater distance between the basic amino acids and Tyr in PK12 and PAK13, and the principal difference between PAK13 and PK12 may be the presence in PAK13 of a proline between the phosphorylated threonine and tyrosine; similar conclusions were reached in a separate study (U.G.K., unpublished observations). These differences might shift the phosphorylation site to a more N-terminal position. The presence of threonine or serine in the phosphorylation site seems unlikely to be of significance as MIHCK phosphorylates Acanthamoeba myosin IA (which has a threonine in that position) as well as it phosphorylates Acanthamoeba myosin IB and IC (which have serines).
Figure 1.
Substrate specificities of Acanthamoeba MIHCK, 35K, and PAK1. Kinase activities were determined as described with the synthetic peptides described in Table 1 as substrates. MIHCK was fully activated by autophosphorylation (11) before its assay; expressed 35K is fully active without autophosphorylation (17). His-PAK1 is constitutively active (U.G.K., unpublished observations). To compare their relative substrate specificities, the activities were normalized to their activities with PC9 as substrate as 100; the actual specific activities with PC9 were: MIHCK, 26 μmol·min−1·mg−1; 35K, 27 μmol·min−1·mg−1; and His-PAK1, 0.37 μmol·min−1·mg−1.
Phosphorylation and Activation of Myosin I.
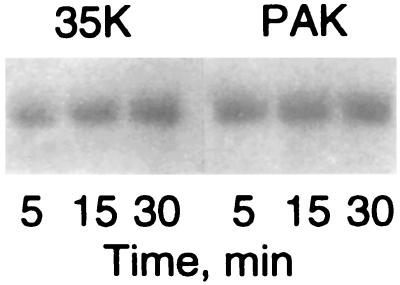
35K catalyzed the incorporation of 1 mol of Pi per mol of baculovirus-expressed Acanthamoeba myosin IC, as was previously shown for native MIHCK and native myosin IC (43). Under identical conditions, PAK1 phosphorylated the heavy chain of myosin IC to the same extent as MIHCK (Fig. 2). In this experiment, phosphate incorporation by PAK1 was complete in 5 min and never exceeded 1 mol of Pi per mol, despite the excess of kinase and the length of the incubation. These results indicate that PAK1, like MIHCK, phosphorylates only one specific site in the myosin IC heavy chain. Moreover, the actin-dependent Mg2+-ATPase activity of myosin IC was stimulated ≈15-fold after phosphorylation by either PAK1 or 35K (Table 2), which, together with the data for the synthetic peptides, strongly indicates that PAK1 phosphorylates the same residue in the myosin IC heavy chain as MIHCK.
Figure 2.
Autoradiography of the heavy chain of Acanthamoeba myosin IC after phosphorylation with expressed 35K and PAK1. Myosin IC was incubated with [γ-32P]ATP and either 35K (Left) or His-PAK1 (Right) as described for the indicated times. The reactions were stopped by addition of SDS sample buffer and the samples separated by PAGE. The autoradiogram of the myosin IC heavy chains is shown. The level of phosphorylation was ≈1 mol of Pi per mol in all samples except the 5-min 35K sample, which was ≈0.8 mol of Pi per mol.
Table 2.
Activation of Acanthamoeba myosin IC by phosphorylation of its heavy chain by PAK1
| Kinase | Mg2+-ATPase activity, s−1
|
|
|---|---|---|
| − actin | + actin | |
| None | 0.5 | 1.3 |
| PAK1 | 0.5 | 7.5 |
| 35K | 0.5 | 8.5 |
Myosin IC was incubated in the presence of ATP either alone or with PAK1 or 35K for 30 min at 30°C. The Mg2+-ATPase activity of the myosin was then measured in the absence or presence of 90 μM F-actin, as described in more detail in text.
DISCUSSION
The data in this paper indicate that, in addition to their sequence similarity, PAK1 and MIHCK have similar substrate specificities and that PAK1 phosphorylates the heavy chain of myosin I and stimulates its actin-dependent Mg2+-ATPase activity. Thus PAK1 and other members of the PAK/STE20 family are likely to function in vivo to phosphorylate and activate myosins in addition to their roles in regulating the mitogen-activating protein kinase cascade. The myosins so regulated might extend beyond those class I and class VI myosins that, as mentioned above, have an hydroxyamino acid at the same position in the heavy chain as Acanthamoeba myosin IC, because Acanthamoeba MIHCK has long been known (44) and PAK has recently been shown (G.M.B., unpublished observations; refs. 45–47) to phosphorylate the regulatory light chain and enhance the actin-activated Mg2+-ATPase activity of class II myosins.
That the regulatory light chain of myosin II is phosphorylated by MIHCK and PAK, even though it lacks the exact consensus MIHCK/PAK phosphorylation site, as defined by studies with synthetic peptides¶, illustrates one of the limitations of determining substrate specificity based on synthetic peptides. While it is likely that protein sites with the same sequence as the peptide substrates will also be substrates (if accessible to the enzyme), the peptide studies do not exclude other sites that either were not tested as synthetic peptides or that were made to resemble the synthetic substrate as a consequence of the tertiary structure of the protein.
The current findings suggest that at least part of the effects of Rac and/or Cdc42 on cell morphology and motility results from their ability to regulate myosin phosphorylation via kinases of the PAK family. In light of the recent observations that myosin II is also phosphorylated and activated by Rho-associated kinase (48, 49), this may be a common feature of cytoskeletal regulation by Rho GTPases. These results also support a role for PAKs as mediators of actin cytoskeleton regulation (S. Dharmawawardhane, R. H. Daniels, and G.M.B., unpublished data; M. A. Sells, U.G.K., S. Bagrodia, D. Ambrose, G.M.B., and J. Chernoff, unpublished data). Interestingly, modulation of the cytoskeleton by PAK may require both the catalytic serine/threonine kinase activity, which resides in the C terminus, and interactions of PAK with SH3-containing regulatory proteins via N-terminal, proline-rich, SH3-binding motifs (50, 51). These data and the aforementioned involvement of STE20 kinases and/or class I myosins in endocytic events and other morphological transformations in S. cerevisiae, Acanthamoeba, and Dictyostelium, indicate that the underlying molecular basis of these processes has been highly conserved in eukaryotes from fungi to mammals.
Acknowledgments
We thank the authors of ref. 52 for sharing their results with us before publication, Ms. A. Corigliano-Murphy for peptide syntheses, and Dr. M. J. Redowicz for skeletal muscle actin. This research was supported in part by grants from the National Institutes of Health (GM39434 to G.M.B. and A135947 to U.G.K.). This paper is The Scripps Research Institute (TSRI) manuscript no. 10504-IMM.
Note.
After this research was completed, we learned that Wu et al. (52) had reached conclusions similar to those reported in this paper after finding that Dictyostelium myosin ID was phosphorylated and activated by several members of the PAK/STE family.
Footnotes
Abbreviations: MIHCK, myosin I heavy chain kinase; 35K, the 35-kDa catalytic domain of MIHCK; SH3, Src homology 3; PAK, p21-activated kinase.
Thr-18, the site in the myosin II light chain phosphorylated by MIHCK (H. Brzeska, J. R. Sellers, and E. D. Korn, unpublished observations), has basic residues N-terminal to it, like the consensus MIHCK phosphorylation site, but Phe (not Tyr) four (not two) positions on its C-terminal side; substitution of Phe for Tyr in PC9 drastically reduces its phosphorylation by MIHCK (18).
References
- 1.Sellers J R, Goodson H V, Wang F. J Muscle Res Cell Motil. 1996;17:67–75. doi: 10.1007/BF00140320. [DOI] [PubMed] [Google Scholar]
- 2.Korn E D. Curr Top Membr Transp. 1991;38:13–30. [Google Scholar]
- 3.Wolenski J A. Trends Cell Biol. 1995;5:310–317. doi: 10.1016/s0962-8924(00)89053-4. [DOI] [PubMed] [Google Scholar]
- 4.Brzeska H, Lynch T J, Korn E D. J Biol Chem. 1988;263:427–435. [PubMed] [Google Scholar]
- 5.Jung J, Hammer J A., III FEBS Lett. 1994;342:197–202. doi: 10.1016/0014-5793(94)80500-8. [DOI] [PubMed] [Google Scholar]
- 6.Doberstein S K, Pollard T D. J Cell Biol. 1992;117:1241–1249. doi: 10.1083/jcb.117.6.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mooseker M S, Cheney R E. Annu Rev Cell Dev Biol. 1995;4:633–675. doi: 10.1146/annurev.cb.11.110195.003221. [DOI] [PubMed] [Google Scholar]
- 8.Albanesi J P, Fujisaki H, Hammer J A, III, Korn E D, Jones R, Sheetz M. J Biol Chem. 1985;260:8649–8652. [PubMed] [Google Scholar]
- 9.Zot H G, Doberstein S K, Pollard T D. J Cell Biol. 1992;116:367–376. doi: 10.1083/jcb.116.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brzeska H, Lynch T J, Martin B, Korn E D. J Biol Chem. 1989;264:19340–19348. [PubMed] [Google Scholar]
- 11.Bement W H, Mooseker M S. Cell Motil Cytoskeleton. 1995;31:87–92. doi: 10.1002/cm.970310202. [DOI] [PubMed] [Google Scholar]
- 12.Brzeska H, Korn E D. J Biol Chem. 1996;271:16983–16986. doi: 10.1074/jbc.271.29.16983. [DOI] [PubMed] [Google Scholar]
- 13.Lee S F, Côté G P. J Biol Chem. 1995;270:11776–11782. doi: 10.1074/jbc.270.20.11776. [DOI] [PubMed] [Google Scholar]
- 14.Brzeska H, Lynch T J, Korn E D. J Biol Chem. 1990;265:3591–3594. [PubMed] [Google Scholar]
- 15.Kulesza-Lipka D, Brzeska H, Baines I C, Korn E D. J Biol Chem. 1993;268:17995–18001. [PubMed] [Google Scholar]
- 16.Wang Z-Y, Brzeska H, Baines I C, Korn E D. J Biol Chem. 1995;270:27969–27976. doi: 10.1074/jbc.270.46.27969. [DOI] [PubMed] [Google Scholar]
- 17.Brzeska H, Martin B, Korn E D. J Biol Chem. 1996;271:27049–27055. doi: 10.1074/jbc.271.43.27049. [DOI] [PubMed] [Google Scholar]
- 18.Brzeska H, Lynch T J, Martin B, Corigliano-Murphy A, Korn E D. J Biol Chem. 1990;265:16138–16144. [PubMed] [Google Scholar]
- 19.Brzeska H, Szczepanowska J, Hoey J, Korn E D. J Biol Chem. 1996;271:27056–27062. doi: 10.1074/jbc.271.43.27056. [DOI] [PubMed] [Google Scholar]
- 20.Lee S-F, Egelhoff T T, Mahasneh A, Côté G P. J Biol Chem. 1996;271:27044–27048. doi: 10.1074/jbc.271.43.27044. [DOI] [PubMed] [Google Scholar]
- 21.Zhang S, Han J, Sells M A, Knaus U G, Ulevitch R, Bokoch G M. J Biol Chem. 1995;270:23934–23936. doi: 10.1074/jbc.270.41.23934. [DOI] [PubMed] [Google Scholar]
- 22.Bagrodia S, Dérijard B, Davis R J, Cerione R A. J Biol Chem. 1995;270:27995–27998. doi: 10.1074/jbc.270.47.27995. [DOI] [PubMed] [Google Scholar]
- 23.Frost J A, Xu S, Hutchinson M R, Marcus S, Cobb M H. Mol Cell Biol. 1996;16:3707–3713. doi: 10.1128/mcb.16.7.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manser E, Leung T, Sallihudin H, Zhao Z-S, Lim L. Nature (London) 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 25.Knaus U G, Morris S, Dong H-J, Chernoff J, Bokoch G M. Science. 1995;269:221–223. doi: 10.1126/science.7618083. [DOI] [PubMed] [Google Scholar]
- 26.Martin G A, Bollag G, McCormick F, Abo A. EMBO J. 1995;14:1970–1978. doi: 10.1002/j.1460-2075.1995.tb07189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall A. Annu Rev Cell Biol. 1994;10:31–54. doi: 10.1146/annurev.cb.10.110194.000335. [DOI] [PubMed] [Google Scholar]
- 28.Ridley A J. BioEssays. 1994;16:321–327. doi: 10.1002/bies.950160506. [DOI] [PubMed] [Google Scholar]
- 29.Nobes C D, Hall A. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 30.Ding J, Knaus U G, Lian J P, Bokoch G M, Badwey J A. J Biol Chem. 1996;271:24869–24873. doi: 10.1074/jbc.271.40.24869. [DOI] [PubMed] [Google Scholar]
- 31.Baines I C, Corigliano-Murphy A, Korn E D. J Cell Biol. 1995;130:591–603. doi: 10.1083/jcb.130.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doberstein S K, Baines I C, Wiegand C, Korn E D, Pollard T D. Nature (London) 1995;365:841–843. doi: 10.1038/365841a0. [DOI] [PubMed] [Google Scholar]
- 33.Fukui Y, Lynch T J, Brzeska H, Korn E D. Nature (London) 1989;341:328–321. doi: 10.1038/341328a0. [DOI] [PubMed] [Google Scholar]
- 34.Jung G, Wu X, Hammer J A., III J Cell Biol. 1996;122:305–323. doi: 10.1083/jcb.133.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novak K D, Peterson M D, Reedy M C, Titus M A. J Cell Biol. 1996;131:1205–1221. doi: 10.1083/jcb.131.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geli M I, Riesman H. Science. 1996;272:533–535. doi: 10.1126/science.272.5261.533. [DOI] [PubMed] [Google Scholar]
- 37.Goodson H V, Anderson B L, Warrick H M, Pon L A, Spudich J A. J Cell Biol. 1996;133:1277–1291. doi: 10.1083/jcb.133.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lynch T J, Brzeska H, Baines I C, Korn E D. Methods Enzymol. 1990;196:12–23. doi: 10.1016/0076-6879(91)96004-b. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z-Y, Wang F, Korn E D, Hammer J A., III Mol Biol Cell. 1996;7:374a. (abstr.). [Google Scholar]
- 40.Brzeska H, Martin B, Kulesza-Lipka D, Baines I C, Korn E D. J Biol Chem. 1992;267:4949–4956. [PubMed] [Google Scholar]
- 41.Szczepanowska J, Qin J, Martin B, Korn E D, Brzeska H. Mol Biol Cell. 1996;7:373a. (abstr.). [Google Scholar]
- 42.Benner G E, Dennis P B, Masaracchia R A. J Biol Chem. 1995;270:21121–21128. doi: 10.1074/jbc.270.36.21121. [DOI] [PubMed] [Google Scholar]
- 43.Lynch T J, Brzeska H, Miyata H, Korn E D. J Biol Chem. 1988;264:19333–19339. [PubMed] [Google Scholar]
- 44.Hammer J A, III, Sellers J R, Korn E D. J Biol Chem. 1984;259:21121–21128. [PubMed] [Google Scholar]
- 45.Tuazon P T, Traugh J A. J Biol Chem. 1984;259:541–546. [PubMed] [Google Scholar]
- 46.Jakobi R, Chen C-J, Tuazon P T, Traugh J A. J Biol Chem. 1996;271:6206–6211. doi: 10.1074/jbc.271.11.6206. [DOI] [PubMed] [Google Scholar]
- 47.Masaracchia R, Ramos E. FASEB J. 1996;10:1538. (abstr.). [Google Scholar]
- 48.Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- 49.Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K. J Biol Chem. 1996;271:20246–20249. doi: 10.1074/jbc.271.34.20246. [DOI] [PubMed] [Google Scholar]
- 50.Galisteo M L, Chernoff J, Su Y-C, Skolnik E Y, Schlessinger J. J Biol Chem. 1996;271:20997–21000. doi: 10.1074/jbc.271.35.20997. [DOI] [PubMed] [Google Scholar]
- 51.Bokoch G M, Wang Y, Bohl B P, Sells M A, Quilliam L A, Knaus A G. J Biol Chem. 1996;271:25746–25749. doi: 10.1074/jbc.271.42.25746. [DOI] [PubMed] [Google Scholar]
- 52.Wu C, Lee S-F, Furmaniak-Kazmierczak E, Côté G P. J Biol Chem. 1996;271:31787–31790. doi: 10.1074/jbc.271.50.31787. [DOI] [PubMed] [Google Scholar]