Abstract
In patients with chronic myeloid leukemia, the use of real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR) for measuring BCR-ABL1 transcripts has become standard methodology for the diagnosis and monitoring of minimal residual disease. In 2004 and 2005, 38 different laboratories from North America participated in three separate sample exchanges using real-time qRT-PCR to measure RNA transcript levels in unknown diluents of a BCR-ABL1-positive cell line, K562. In this study we compared results of quantitative testing for BCR-ABL1 from laboratories using different platforms, internal controls, reagents, and calculation methods. Our data showed that there can be considerable variability of results from laboratory to laboratory, with log reduction calculations varying from 1.6 to 3 log between laboratories at the same dilution. We found that none of the variables tested had a significant impact on the results reported, except for the use of ABL1 as the internal control (P < 0.001). Laboratories that used ABL1 consistently underreported their log reduction values. Regardless of the specific methodology and platform used for real-time qRT-PCR testing, it is important for laboratories to participate in proficiency testing to ensure consistent and acceptable test accuracy and sensitivity. Our study emphasizes the need for optimization of real-time qRT-PCR before offering clinical testing and the need for widely available universal standards that can be used for test calibration.
Chronic myeloid leukemia (CML) is a stem cell disease that is invariably associated with a reciprocal chromosomal translocation t(9;22)(q34;q11) called the Philadelphia chromosome (Ph).1 This translocation juxtaposes the upstream exons of the BCR (breakpoint cluster region) gene and the downstream exons of the ABL1 (v-abl Abelson murine leukemia viral oncogene homolog 1) gene, resulting in a novel fusion gene BCR-ABL1.2 This fusion gene is present in >99% of CML patients and 25 to 40% of adult patients with acute lymphoblastic leukemia.2,3 Three fusion transcripts, resulting from major (b3a2, b2a2) and minor (e1a2) breakpoints, can be distinguished according to the breakpoint within the BCR region. These transcripts encode the p210 (b3a2, b2a2) and p190 (e1a2) oncoproteins, both of which enhance tyrosine kinase activity and play a critical role in the pathogenesis of this leukemia.4,5 The chimeric BCR-ABL1 gene and its resulting transcripts provide specific markers for the diagnosis and monitoring of minimal residual disease (MRD).
Methods available to detect the BCR-ABL1 rearrangement include conventional cytogenetic analysis, fluorescence in situ hybridization and reverse transcription-polymerase chain reaction (RT-PCR).6 Real-time quantitative RT-PCR (qRT-PCR) provides not only detection of the fusion transcript but also accurate quantitation of the accumulated target PCR products. Real-time qRT-PCR is now widely used for the detection of MRD.7 The quantification of BCR-ABL1 transcripts is clinically useful for monitoring CML patient response to transplantation or to other treatments, including imatinib therapy. For example, early reduction of BCR-ABL1 transcripts can predict cytogenetic response.8,9 Recent data from the IRIS and TIDEL trials indicate that molecular response, as measured by real-time qRT-PCR, can also predict overall and progression-free survival after treatment with imatinib.10,11,12,13 Currently, many laboratories use log reduction as a measurement of MRD and response to treatment. Patients who achieve a 3-log reduction in BCR-ABL1 transcripts are considered to have achieved a major molecular response and to have a low incidence of disease progression.14,15 Conversely, rising levels of BCR-ABL1 transcripts are consistent with loss of response to treatment and can indicate relapse.
Data have shown that even when using a standardized protocol and common calibrator, there can be significant variation in results among different laboratories.16 Multilaboratory molecular testing using real-time qRT-PCR thus requires further standardization of techniques and robust quality control of results. A report from 26 laboratories in 10 European countries (the Europe against Cancer Network) has previously addressed the issue of standardization and quality control using real-time qRT-PCR for fusion gene quantification in leukemia and has made specific protocol recommendations.17 In North America, although many laboratories use real-time qRT-PCR to monitor patients with CML, no reports have compared results from laboratories doing the same test that may use different RNA extraction methods, different real-time qRT-PCR reagents or kits, and different instruments, standard curves, and internal controls. In 2004 and 2005, 38 different laboratories from North America (Table 1) participated in three separate sample exchanges using real-time qRT-PCR to measure RNA transcript levels in unknown diluents of a BCR-ABL1-positive cell line, K562. The two exchanges that involved only Canadian laboratories (six laboratories for GLEEM 2 and 13 laboratories for GLEEM 3) were supported by Novartis Canada (Montreal, QC, Canada), to ensure proficiency of laboratories participating in a trial (GLEEM trial) assessing the effects of 400 versus 800 mg of imatinib in patients that had not achieved an MMR within 12 months. The North American sample exchange involved 29 laboratories and was supported by the Association for Molecular Pathology as part of an initiative undertaken by the Hematopathology Subdivision of that organization. The overall purpose of these exchanges was to compare how similar or different results from participating laboratories were, without the use of a universally standardized protocol. We also aimed to identify variables that could potentially significantly affect test results to discern variables important in test standardization. In the present study we distributed unknown diluents of K562 and then collated and analyzed the data received back from participating laboratories, to assess the sensitivity and reproducibility of real-time qRT-PCR results from centers across North America.
Table 1.
List of 38 Laboratories in North America Participating in at Least One of the Three Sample Exchanges in 2004 and 2005
| Cancer Genetics Laboratory, BC Cancer Agency, Vancouver, BC, Canada | |
| Cancer Genetics Laboratory, Hamilton Health Sciences, Hamilton, ON, Canada | |
| Centre for Molecular Biology and Pathology, Laboratory Corporation of America, Research Triangle Park, NC | |
| Department of Clinical Pathology, The Cleveland Clinic Foundation, Cleveland, OH | |
| Department of Genetics, Credit Valley Hospital, Mississauga, ON, Canada | |
| Department of Genetics, St. Joseph’s Health Centre, Sudbury, ON, Canada | |
| Department of Hematology, Maisonneuve-Rosemont Hospital, Montreal, QC, Canada | |
| Department of Hematology, SMBD Jewish General Hospital, Montreal, QC, Canada | |
| Department of Hematopathology, University of Manitoba Health Science Centre, Winnipeg, MB, Canada | |
| Department of Laboratory Medicine and Pathology, University of Alberta, Edmonton, AB, Canada | |
| Department of Molecular Hematopathology and Genetics, ARUP Laboratories, Salt Lake City, UT | |
| Department of Pathology and Laboratory Medicine, Queen Elizabeth II Health Science Center, Halifax, NS, Canada | |
| Department of Pathology and Laboratory Medicine, Roswell Park Cancer Institute, Buffalo, NY | |
| Department of Pathology and Laboratory Medicine, Weill Medical College of Cornell University, New York, NY | |
| Department of Pathology, Carolinas Medical Centre, Charlotte, NC | |
| Department of Pathology, Rush University Medical Center, Chicago, IL | |
| Department of Pathology, The Methodist Hospital/Baylor College of Medicine, Houston, TX | |
| Department of Pathology, University of North Carolina, Chapel Hill, NC | |
| DNA Diagnostic Laboratory, Oregon Health and Sciences University, Portland, OR | |
| DNA Molecular Laboratory, John Hopkins University Hospital, Baltimore, MD | |
| Laboratory No. 2, Maisonneuve-Rosemont Hospital, Montreal, QC, Canada | |
| Laboratoire d’Hematologie , CHUM Hospital Notre Dame, Montreal, QC, Canada | |
| Molecular Diagnostics (Banting), Department of Pathology, University Health Network, Toronto, ON, Canada | |
| Molecular Diagnostics Laboratory, Brigham and Women’s Hospital, Boston, MA | |
| Molecular Diagnostics Laboratory, Yale New Haven Hospital, New Haven, CT | |
| Molecular Diagnostics Laboratory, Emory University Hospital, Atlanta, GA | |
| Molecular Diagnostics Laboratory, The Children’s Hospital, Denver, CO | |
| Molecular Diagnostics Laboratory, VCU Health System, Richmond, VA | |
| Molecular Diagnostics, London Health Sciences Centre, London, ON, Canada | |
| Molecular Diagnostics Section, University of Rochester Medical Center, Rochester, NY | |
| Molecular Diagnostics, The Blood Center of Southeastern Wisconsin, Milwaukee, WI | |
| Molecular Diagnostics, University of Pittsburgh Medical Centre, Pittsburgh, PA | |
| Molecular Genetics, Specialty Laboratories, Santa Monica, CA | |
| Molecular Hematopathology Laboratory, Mayo Clinic, Rochester, MN | |
| Molecular Pathology Laboratory Network, Maryville, TN | |
| Molecular Pathology Laboratory, Stanford University School of Medicine, Stanford, CA | |
| Stem Cell Transplant Laboratory, Royal Victoria Hospital, Montreal, QC, Canada | |
| St. Sacrement Hospital, Quebec City, QC, Canada |
This does not reflect the order of the laboratories in the results shown in Table 5.
Materials and Methods
Cell Culture and Sample Preparation
U937 and K562 cells were obtained from American Type Culture Collection (Manassas, VA) and were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum. U937 is a BCR-ABL1-negative hematopoietic cell line, and K562 carries the t(9;22)(q34;q11) and expresses b3-a2 BCR-ABL1 transcripts.18 K562 cells were diluted with U937 cells to create diluents from 10−1 to 10−5, undiluted K562 cells were used as the neat sample, and undiluted U937 was used as the negative control. One million cells from each sample were washed once in phosphate-buffered saline (PBS) and suspended in RNAlater (Qiagen, Valencia, CA). RNAlater is an aqueous, nontoxic, tissue and cell storage reagent that stabilizes and protects cellular RNA from degradation by protecting against cell lysis.
The samples were shipped to each laboratory at room temperature, and laboratories were asked to store samples at 4°C until use. This was a blinded study, thus each laboratory received samples labeled only with a number. For the Association for Molecular Pathology sample exchange, 10 samples were sent, labeled as nos. 1 to 10. These included one aliquot of each dilution as well as one negative control and one undiluted aliquot of K562. Three cDNA samples were also sent, but these were subsequently excluded from the study because laboratories were confused as to what to do with them. For GLEEM 2 and GLEEM3, each laboratory received 10 samples labeled as nos. 1 to 10, including two negative controls, one aliquot of undiluted K562, one aliquot of K562 at 10−1, one aliquot of K562 at 10−2, one aliquot at 10−3, two aliquots at 10−4, and two aliquots of K562 at a 10−5 dilution. Each laboratory used their own standards and internal controls in their analysis.
RNA Isolation, Quantification, and Reverse Transcription
Total cellular RNA was isolated from the samples provided. Because of the use of RNAlater as a transport medium, we recommended that each laboratory first spin cells down for use in their RNA extraction protocol or spin cells and then wash in ice-cold PBS before extraction. Each laboratory used its own RNA extraction method according to their laboratory’s protocol, such as TRIzol reagent (Invitrogen, Carlsbad, CA), RNeasy kit (Qiagen), RNAwiz reagent (Ambion, Austin, TX), or PUREscript (Flowgen Bioscience, Nottingham, UK) (Table 2). Most laboratories quantified their total RNA using spectrophotometry. Only three laboratories evaluated their RNA quality by electrophoresis to determine the degree of degradation and the presence of contaminating DNA. Total RNA was reverse-transcribed using a variety of primers (gene-specific, oligo dT, random hexamer, or from the LightCycler kit). The amount of RNA and enzymes [Superscript, MMLV (both from Invitrogen), AMV (Fermentas, Glen Burnie, MD), Stratascript (Stratagene, La Jolla, CA), Omniscript (Qiagen)] used for reverse transcription varied in different laboratories (Table 2).
Table 2.
Summary of Reagents, Platforms, Internal Controls, and Type of Standard Curve Used
| Number of laboratories | |
|---|---|
| Extraction method | |
| Qiagen | 23 |
| TRIzol | 13 |
| Ambion | 4 |
| Others | 3 |
| RT primer | |
| Random hexamer | 36 |
| Gene specific | 3 |
| Others | 2 |
| RT enzyme | |
| MMLV | 15 |
| Superscript | 9 |
| AMV | 14 |
| Others | 5 |
| Internal control | |
| ABL1 | 16 |
| GAPDH | 7 |
| BCR | 4 |
| G6PD | 13 |
| GUSB/B2M | 3 |
| PCR kit | |
| Homebrew | 16 |
| Roche | 15 |
| Ipsogen | 3 |
| ABI | 9 |
| Instrument | |
| ABI Prism 7700 | 6 |
| ABI Prism 7900 | 6 |
| ABI Prism 7000/7500 | 8 |
| Roche LightCycler | 22 |
| BioRad iCycler | 2 |
| Standard curve | |
| Diluted RNA | 8 |
| Diluted cDNA | 4 |
| Diluted plasmid | 13 |
| Diluted cell line | 16 |
| NA | 2 |
NA, not available.
Real-Time qRT-PCR
Real-time qRT-PCR was performed using different platforms, such as the ABI7700, ABI7900, or ABI7000, the Lightcycler (Roche, Indianapolis, IN), or the iCycler (Bio-Rad, Hercules, CA) (Table 2). Reaction conditions and primer/probe selections for optimal amplification of both BCR-ABL1 and internal controls were determined by each individual laboratory according to their own protocol (Table 2). The size of BCR-ABL1 PCR products from different laboratories ranged from 66 to 596 bp with most laboratories generating amplicons less than 200 bp. Probes were located either at the 5′ end, 3′ end, or middle of BCR-ABL1. Because internal control genes can reflect the degree of degradation of the samples being tested, amplification of internal controls was performed to normalize the amount of sample RNA present in a reaction. Various internal control genes were used by laboratories, including GAPDH (glyceraldehyde-3-phosphate dehydrogenase), ABL1, BCR, G6PD (glucose-6-phosphate dehydrogenase), B2M (β-2-microglobulin), and GUSB (glucuronidase, β) (Table 2). The expression levels of the target gene and of the internal control were quantified and analyzed automatically using instrument-specific software, such as Sequence Detection Systems 2.1 (Applied Biosystems, Foster City, CA).
Standard Curve and Comparative CT
For quantitation of BCR-ABL1 normalized to an internal control, standard curves were constructed for both BCR-ABL1 and the internal control. Laboratories analyzed their data using the standard curve method or the comparative (ΔΔCT) method. Five to six serial samples of diluted RNA, cDNA, plasmid DNA, or a cell line were used by laboratories to generate standard curves (Table 2). Using this methodology, samples are generally rejected in the analysis and are repeated if the correlation coefficient (R2) is less than 0.98.19 The observed CT measurements of the BCR-ABL1 fusion transcripts or the internal controls in the tested samples are then calculated against the standard curve plots. The normalized measurements of the BCR-ABL1 fusion transcripts are determined according to the following formula: ratio = BCR-ABL1 levels/internal control levels. The rationale for the need of such normalization is that PCR amplification efficiency can vary between runs, and RNA quality can vary from sample to sample within a run.
Some laboratories used the comparative CT method, which is similar to the standard curve method, except that it uses an arithmetic formula to achieve the same result as relative quantitation. The amount of BCR-ABL1 normalized to an internal control and relative to a calibrator is given by 2−ΔΔCT, where ΔCT = CT (BCR-ABL1) − CT (internal control) and ΔΔCT = ΔCT (patient sample) − ΔCT (reference sample). Before using this CT method, however, it is recommended that a validation experiment is performed to demonstrate that the amplification efficiencies of BCR-ABL1 and the selected internal control are approximately equal.
Log Reduction
In all three exchanges, to compare real-time qRT-PCR results obtained by different laboratories, laboratories were asked to send their results as a ratio and to also convert their results to a log reduction. To do this, the ratio of the BCR-ABL1 value versus internal control value was converted to a logarithmic (base 10) scale. This log reduction was calculated in each laboratory by using the sample with the highest ratio as the baseline or diagnostic level. The reduction in BCR-ABL1 levels from this baseline value was then calculated for each subsequent sample and reported as a log reduction.
Results
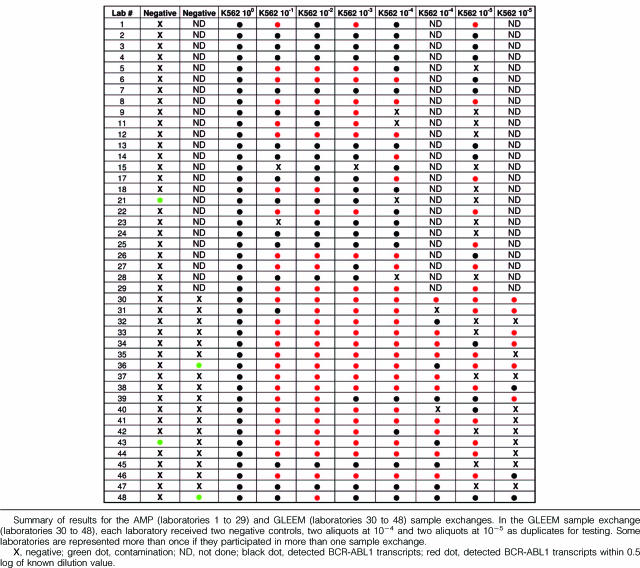
A total of 42 laboratories (29 laboratories participated in the Association for Molecular Pathology exchange, six in GLEEM 2, and 13 in GLEEM 3; some laboratories participated in more than one sample exchange) were provided with samples. Two laboratories were unable to provide results because of lack of time and resources, and another two laboratories had technical difficulties; therefore results were obtained from 38 laboratories. Twenty-nine laboratories had quantitative results starting from the 10−5 dilution, 40 laboratories had results including the 10−4 dilution, 43 laboratories amplified transcripts at the 10−3 dilution, 43 laboratories amplified at the 10−2 dilution, and 42 laboratories amplified transcripts at the 10−1 dilution (Table 3). The mean, median, and SD of log reduction values (accuracy) from each laboratory are pooled and summarized in Table 4. The mean and median results obtained for all dilutions were within 0.5 log of the known dilution value (expected value) except for at 10−5, where it was 0.55 log, and the SD was 0.6 log at all dilutions except for 10−1, where it was 0.4 log. When the data from laboratories using ABL1 as an internal control were analyzed separately (Table 4), the mean and median log reduction values were ∼1 log less than the known dilution value with the exception of the 10−1 dilution in which the value was within 0.6 log of the expected value. Laboratories using other internal controls (Table 4) had mean and median values within 0.2 to 0.4 log of the known dilution values. Four laboratories detected BCR-ABL1 in a U937 sample that should have been negative (Table 5). This false-positive finding could have been a result of contamination or sample mix up. At this time, based on our experience and that of others, reproducible results are those that are different by less than 0.5 log in duplicate samples at dilutions as high as 10−4 and 10−5. Duplicate samples at lower dilutions (diagnostic and 10−1 to 10−3) should have nearly identical values. When laboratories were given known dilutions in duplicate, 10 of 19 did not have reproducible results (±0.5 log) at the 10−4 level and 16 of 19 did not have reproducible results at the 10−5 level (Table 5). The effect of different variables on the reported log reduction at each dilution was assessed using one-way analysis of variance (Table 6). None of the tested variables consistently and significantly affected the mean log reduction reported, except for the internal control.
Table 3.
Summary of Test Sensitivity at Different Dilutions
| Known dilution (K562 in U937) | Number of laboratories with results/total laboratories (%) |
|---|---|
| 10−5 | 29/44 (65.9%) |
| 10−4 | 40/44 (90.9%) |
| 10−3 | 43/44 (97.7%) |
| 10−2 | 43/44 (97.7%) |
| 10−1 | 42/44 (95.5%) |
Table 4.
Summary of Test Accuracy at Different Dilutions (Based on Log Reduction), Calculated Based on All Internal Controls Used or ABL1 as the Internal Control, or GAPDH, BCR, G6PD, or B2Mas the Internal Control
| 10−5 | 10−4 | 10−3 | 10−2 | 10−1 | |
|---|---|---|---|---|---|
| All internal controls | |||||
| Mean | 4.4555 | 3.5233 | 2.5840 | 1.5367 | 0.6674 |
| SD | 0.60949 | 0.57820 | 0.57493 | 0.58402 | 0.39451 |
| Median | 4.5200 | 3.5650 | 2.6300 | 1.6000 | 0.6050 |
| Minimum | 3.26 | 2.18 | 1.03 | 0.26 | 0.14 |
| Maximum | 6.30 | 4.71 | 3.70 | 3.00 | 1.70 |
| Range | 3.04 | 2.53 | 2.67 | 2.74 | 1.56 |
| n | 29 | 40 | 43 | 43 | 42 |
| ABL1 as the internal control | |||||
| Mean | 4.1490 | 3.0607 | 2.0913 | 1.1225 | 0.3773 |
| SD | 0.4860 | 0.3854 | 0.5464 | 0.4464 | 0.3404 |
| Median | 4.100 | 3.0800 | 2.1450 | 1.0100 | 0.3000 |
| Minimum | 3.26 | 2.34 | 1.03 | 0.50 | 0.14 |
| Maximum | 4.80 | 3.85 | 3.20 | 2.20 | 1.50 |
| Range | 1.54 | 1.51 | 2.17 | 1.70 | 1.36 |
| n | 10 | 14 | 16 | 16 | 15 |
| GAPDH, BCR, G6PD, and B2Mas the internal control | |||||
| Mean | 4.6168 | 3.7723 | 2.8759 | 1.7823 | 0.8285 |
| SD | 0.6165 | 0.4012 | 0.3518 | 0.4270 | 0.3279 |
| Median | 4.5800 | 3.7800 | 2.8000 | 1.7550 | 0.7100 |
| Minimum | 3.52 | 2.18 | 2.30 | 0.26 | 0.38 |
| Maximum | 6.30 | 4.71 | 3.70 | 3.00 | 1.70 |
| Range | 2.78 | 2.53 | 1.40 | 2.74 | 1.32 |
| n | 19 | 26 | 27 | 27 | 27 |
Table 5.
Summary of Results for the AMP (Laboratories 1 to 29) and GLEEM (Laboratories 30 to 48) Sample Exchanges
Table 6.
Effect of Different Variables on Reported Log Reduction at Different Dilutions
| Dilution | Variables
|
||||||
|---|---|---|---|---|---|---|---|
| Extraction method | RT primer | RT enzyme | PCR kit | Instrument | Standard curve | Internal control | |
| 10−5 | 0.89 | 0.41 | 0.90 | 0.36 | 0.66 | 0.16 | 0.16 |
| 10−4 | 0.84 | 0.52 | 0.40 | 0.21 | 0.75 | 0.11 | 0.001 |
| 10−3 | 0.78 | 0.60 | 0.05 | 0.09 | 0.61 | 0.01 | <0.001 |
| 10−2 | 0.39 | 0.42 | 0.08 | 0.07 | 0.48 | 0.05 | 0.001 |
| 10−1 | 0.16 | 0.32 | 0.75 | 0.17 | 0.02 | 0.06 | <0.001 |
One-way ANOVA was performed to compare the mean log reduction reported at each dilution from different laboratories using the same extraction methods, reagents, kits, platforms, standard curves, and internal controls. P values are listed in each column.
RNA Quality and cDNA Synthesis
Laboratories measured their RNA quality and quantity by spectrophotometry and/or gel electrophoresis. In some cases, the yields were low, but this did not seem to affect results (data not shown). Storage time did not affect the sensitivity or accuracy of results because laboratories that achieved log reduction values closest to expected values had storage times that ranged from 1 to 25 days (data not shown). For the cDNA synthesis, reverse transcription was done with gene-specific primers, random hexamers, oligo dTs or kits, and the reverse transcription enzymes used were either Superscript, MMLV, AMV, Stratascript, or Omniscript (Table 2). Overall, the primers and enzymes used did not affect result sensitivity or accuracy (Table 6).
Reagents for Quantitative PCR
Different laboratories performed real-time quantitative PCR using their own protocols. Some laboratories used the Applied Biosystems kit and Applied Biosystems instruments, some used the Roche t(9;22) quantification kit and LightCycler, others used either the Ipsogen (Windsor, CT) FusionQuant kit or homebrew buffers on various instruments (Table 2). Overall, the different PCR kits and reagents used did not affect reported log reduction results (Table 6).
Platforms
Samples prepared from serial dilutions of K562 cells were analyzed on ABI Prism 7000, ABI Prism 7700, ABI Prism 7900, Roche LightCycler, or Bio-Rad iCycler instruments. Quantification was performed using either a standard curve or comparative CT method. Ninety-one percent of laboratories were able to amplify transcripts from samples diluted at 10−4, and 66% achieved amplification of samples diluted at 10−5 (Table 3), regardless of the platform or reagents that were used (Table 6).
Calculation and Use of Standard Curve
Various templates and calculation methods were used to generate standard curves. For the laboratories that included the slope and R2 value in the data they sent to us, these ranged from −2.96 to −4.96 for the slope and 0.99 to 1.00 for the R2 value. All but two laboratories generated their own standard curves. Two laboratories used the comparative CT method. One laboratory used a freshly diluted cell line each time to generate the standard curve and although this method may appear optimal (their results were within 0.2 log at all dilutions), it is however, quite labor intensive. Laboratories that used a previously diluted and aliquoted cell line to generate their standard curve had mean values that were close to the known dilution value (Table 7). Because this was not statistically significant at all dilutions (Table 6), however, we must conclude that overall it does not seem to make a difference whether a laboratory uses diluted RNA, cDNA, plasmid DNA, or cell lines for generation of the standard curve (Table 7).
Table 7.
Comparison of Log Reduction Results Using Standard Curves from Different Sources
| Standard curve derived from | 10−5 | 10−4 | 10−3 | 10−2 | 10−1 |
|---|---|---|---|---|---|
| Diluted RNA | |||||
| Mean | 4.2500 | 3.2788 | 2.4550 | 1.0975 | 0.8057 |
| n | 6 | 8 | 8 | 8 | 7 |
| SD | 0.79405 | 0.86717 | 0.72012 | 0.58137 | 0.54427 |
| Diluted cDNA | |||||
| Mean | 4.0967 | 3.3875 | 2.5400 | 1.5450 | 0.6100 |
| n | 3 | 4 | 4 | 4 | 4 |
| SD | 0.53257 | 0.11442 | 0.21909 | 0.19330 | 0.15811 |
| Diluted plasmid | |||||
| Mean | 4.3033 | 3.3460 | 2.2908 | 1.4623 | 0.4623 |
| n | 6 | 10 | 13 | 13 | 13 |
| SD | 0.51282 | 0.56708 | 0.57419 | 0.72209 | 0.33164 |
| Diluted cell line | |||||
| Mean | 4.7717 | 3.8231 | 2.9544 | 1.8650 | 0.8300 |
| n | 12 | 16 | 16 | 16 | 16 |
| SD | 0.52719 | 0.38922 | 0.38550 | 0.34892 | 0.35208 |
| ΔΔCT | |||||
| Mean | 4.1700 | 3.2600 | 2.1300 | 1.1350 | 0.3300 |
| n | 2 | 2 | 2 | 2 | 2 |
| SD | 0.41012 | 0.35355 | 0.24042 | 0.31820 | 0.07071 |
| Total | |||||
| Mean | 4.4555 | 3.5233 | 2.5840 | 1.5367 | 0.6674 |
| n | 29 | 40 | 43 | 43 | 42 |
| SD | 0.60949 | 0.57820 | 0.57493 | 0.58402 | 0.39451 |
Results are reported as a log reduction from the sample with the highest transcript levels (diagnostic sample). n = number of laboratories reporting a result for that dilution.
Internal Controls
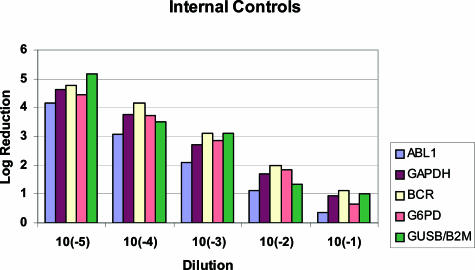
Optimally, internal control genes should be expressed at similar levels as the BCR-ABL1 target gene. In our study, participating laboratories used a variety of genes as internal controls, including GUSB, ABL1, GAPDH, BCR, G6PD, and B2M (Table 2 and Figure 1). The most frequently used internal controls were G6PD and ABL1. The four laboratories that used BCR as their internal control seemed to achieve the most accurate and sensitive results (Figure 1). Several laboratories were using ABL1, which results in log reductions that are consistently lower than expected values because of the undercalling of the ratio of the diagnostic sample. This overestimation of ABL1 is attributable to the ABL1-specific primer binding to both BCR-ABL1 and the endogenous normal copy of the ABL1 gene. Laboratories using ABL1 as the internal control showed log reduction values that were significantly different from those that used other internal control genes, at four of five dilutions tested (Table 6).
Figure 1.
Mean of the log reduction variation using different internal controls. The mean log reduction for all laboratories using the same internal control was calculated for each dilution. Six internal controls (ABL1, GAPDH, BCR, G6PD, GUSB, and B2M) were used from different laboratories for this analysis. Note that the log reduction calculated when using ABL1 as an internal control is consistently lower than the known dilution value.
Summary of Results
We compared many variables (RNA quality, cDNA synthesis enzymes, and primers, reagents, or kits used for PCR, platforms, standard curves, and internal controls) that could affect the accuracy and sensitivity of real-time qRT-PCR testing in CML. Overall, the use of specific platforms or reagents did not seem to significantly affect results. However, the limited number of laboratories and the number of variables that were considered made statistical analyses difficult. The only variable that consistently made a difference was the type of internal control used, with ABL1 consistently undercalling the reported log reduction value (Tables 4and 6). Laboratories that used a cell line to generate their standard curve had mean values that were close to the known dilution value although this was not statistically significant at all dilutions tested (Tables 6and 7). GLEEM study results were more consistent than the Association for Molecular Pathology study results, probably attributable to the smaller number of laboratories involved and thus less variation in techniques and platforms used (Table 5).
Discussion
Currently, real-time qRT-PCR is considered the standard of care for the monitoring of MRD in patients with CML. Treatment decisions are made based on this technology, yet standard operating procedures can vary from laboratory to laboratory because of the use of different reagents, controls, and equipment. Intra- and interlaboratory standardization of test procedures have become important because results from different laboratories should be consistent from center to center, thus allowing patients a more universal standard of care. The accuracy, reproducibility, and reliability of real-time qRT-PCR to quantitate BCR-ABL1 transcript levels are dependent on many factors, including stringent quality control measures and robust test validation. In 2004 and 2005, we organized three sample exchanges in which 38 different laboratories were required to use real-time qRT-PCR to measure transcript levels in a series of diluents derived from a BCR-ABL1-positive cell line, K562. The purpose of these exchanges was to compare test results from different centers and to determine how similar they were in the absence of a universally standardized protocol.
The introduction of real-time qRT-PCR platforms to the general marketplace now makes the development of real-time qRT-PCR assays more widely available. In this study, the two most commonly used platforms for real-time qRT-PCR assays were the Applied Biosystems platform and the Roche LightCycler platform. The Applied Biosystems platform uses a 5′ nuclease assay with a TaqMan probe,7,20 whereas Roche uses the LightCycler FastStart DNA MasterPLUS HybProbe technology.21,22 These instruments differ in several respects, including the light source and the approach to acquisition of fluorescence data. Published data have confirmed the ability of the LightCycler to use TaqMan technology as an alternative, if desired.23 Silvy and colleagues24 demonstrated that the Europe against Cancer standardized protocol could be transferred to seven different real-time qRT-PCR instruments without any modifications. The results from our studies agree with these data, as our results show that the ABI platform, the Roche LightCycler, and the Bio-Rad iCycler all performed similarly as long as the conditions and reagents were optimized for use with each instrument.
In our data set, there is a great deal of variation in reported log reduction levels from laboratory to laboratory, as noted by the SD observed at all dilutions tested (Table 4). This may be a reflection of the different ways that laboratories make, calibrate, and use their standard curves and also what type of internal control is used (Tables 6and 7). Our data indicate that it is very important for each laboratory to optimize their test and ensure it achieves appropriate sensitivity and reproducibility.25 Table 3 shows that only 66% of laboratories were able to consistently detect the lowest levels of BCR-ABL1 transcripts (10−5). These data emphasize the point that ideally, all samples should be run in duplicate from the RNA extraction step because of the potential variability in results, especially at low transcript levels.19,25 However, given that this may not be practical, it may be acceptable to instead run samples in duplicate from the RT (cDNA synthesis) stage. Table 5 summarizes the results from all laboratories with regards to accuracy and sensitivity and clearly shows that although most laboratories can reproducibly detect high levels of transcript, lower levels can either be missed or inaccurately quantified. Therefore, monitoring using real-time qRT-PCR should still be undertaken in CML patients at regular intervals, even when undetectable levels of transcript are achieved.
To ensure accuracy, it is important to normalize results to an appropriate internal control. Based on the absence of pseudogenes and the level and stability of gene expression, the Europe against Cancer group showed that three genes, ABL1, B2M, and GUSB, had stable expression in different types of samples (peripheral blood, PB; bone marrow, BM; and PB stem cells, PBSCs) from patients with CML, acute myeloid leukemia, and acute lymphoblastic leukemia. However, only ABL1 gene transcript expression did not differ significantly between normal and leukemic samples at diagnosis. Europe against Cancer therefore proposed the use of the ABL1 gene as the control gene of choice for real-time qRT-PCR-based diagnosis and MRD detection in patients with CML and acute lymphoblastic leukemia.26 The primer set used to amplify the ABL1 internal control gene is located on exon a2; therefore, it also amplifies the BCR-ABL1 fusion transcript (especially when present at high levels, such as at diagnosis) and thus introduces bias for quantifying BCR-ABL1 when the proportion of BCR-ABL1-positive cells is high. This problem can be solved through the use of other internal control genes, such as BCR,10 or by changing the location of the ABL1 primers to exon 1 of the ABL1 gene. In our multilaboratory analysis, six control genes were used, including GUSB, ABL1, GAPDH, BCR, G6PD, and B2M. The laboratories (n = 4) that used BCR as their internal control seemed to achieve the most accurate and sensitive results, although because the number of laboratories is low, one cannot determine whether this is significant. The laboratories using ABL1 reported results as log reductions that were consistently lower than expected because of the undercalling of the ratio of the diagnostic sample because of overestimation of ABL1 (Table 4). When we split Table 4 into two groups (ABL1 versus all of the other internal controls) and reassessed the results, it is clear that groups using ABL1 cannot report results accurately using the log reduction method. These data have been recently verified by another group comparing the effect of different internal controls on reported real-time qRT-PCR levels in CML.27
According to the literature and published consensus guidelines, log reduction is currently the preferred method of presenting results.28 Hughes and colleagues10 proposed that a reduction in BCR-ABL1 transcript levels of at least 3 logs is used to define a major molecular response. Calculation of log reduction is based on the ratio of BCR-ABL1 transcripts/internal control transcripts in relation to the diagnostic sample ratio or to the laboratory median ratio. For a laboratory to calculate a median ratio, it should use the results from at least 30 diagnostic samples from CML patients in chronic phase.10 These samples should not include samples from patients in blast crisis or accelerated phase. A standard curve should be generated for each run unless the ΔΔCT method is being used. Because standard curves can be generated from cell lines, plasmid, cDNA, or RNA, and because the internal control used may be different, different values can be produced for the transcript ratio of the same sample. If one converts these results to a log reduction, then results should be similar from laboratory to laboratory, given similar test sensitivity and specificity, regardless of the internal control used. The only exception may be with the use of ABL1 because of undercalling of the ratio at diagnosis. Another concern is whether the use of plasmid is appropriate for the generation of a standard curve because this does not take into account the analytical variability associated with RNA extraction and efficiency of cDNA synthesis.29 Thus, if the plasmid calibration standard and the cDNA template do not have equal amplification efficiencies, there is an increased potential for error.25
The use of the ΔΔCT method (rather than a standard curve) is based on the assumption that slopes are identical between BCR-ABL1 transcripts and internal control transcripts, which means that the absolute value of the difference in the slope between the target and the control gene should be less than 10%.30 With different internal controls, validation of this assumption is required. An advantage of the ΔΔCT method is that it uses only the Ct value and does not include any plasmid- or RNA-based standard curve, thus reducing the risk of contamination and batch-to-batch variation. However, recent recommendations do not support this approach for data calculation.19
More recently, it has been recommended that reporting of results should be standardized even further by using an international scale rather than log reduction.19 This international scale would call diagnostic levels as the baseline at 100% and a 3-log reduction would be at 0.1%. For all laboratories to call the same ratio 100% or 0.1%, calibration of results must be undertaken and a conversion factor derived for each laboratory. To determine the international scale conversion factor for each laboratory, laboratories would test a series of quality control samples with values established in a reference laboratory and generate a conversion factor specific to that laboratory, its testing platforms, and analytic specificities. In addition, measured BCR-ABL1 transcript levels would be calibrated using standards produced commercially in large batches to ensure their stability and reproducibility.19 Discussions regarding how to facilitate this approach are ongoing globally, in Europe, Asia, North America, and Australia.
Real-time qRT-PCR is a sensitive and accurate method that is currently used to quantitate BCR-ABL1 transcript levels. Unfortunately inter- and intralaboratory variability of quantitative analyses can occur. This can be minimized through the use of quality control (QC) samples within each run and by defining criteria for acceptance or rejection of a run.25 An international group reporting on this has suggested the use of ±2 SDs from the mean value of BCR-ABL1/control percent as a guide. A mean and range of 2 SDs is calculated by repeated analysis, and this range is used to accept or reject each real-time qRT-PCR run.31,32 Currently, our data show that there is a great deal of variability in results reported from different laboratories. It is thus important that each laboratory optimize their protocol for MRD monitoring. This will enable reliable and reproducible quantification of BCR-ABL1 transcripts regardless of specific analytic methodology. Frequent proficiency testing and the use of commercial standards (when available) will be critical in ensuring reproducible and accurate results. Furthermore, result reporting should be based on a calculation of log reduction from samples run in duplicate, using a laboratory median, until the new international reporting scale is developed. If sensitivity, specificity, and reproducibility of results are thus achieved, results should then be more comparable among different laboratories.
Acknowledgments
We thank the Association for Molecular Pathology, USA, and Novartis, Canada, for their support of these sample exchanges; and Dr. Daniel Hébert from Novartis, Canada, for the valuable role he played in the organization and facilitation of the Canadian sample exchanges.
Footnotes
Supported by the Association for Molecular Pathology and Novartis, Canada.
Standard of practice is not being defined by this article, and there may be alternatives.
In 2004 and 2005 the leadership of the Association for Molecular Pathology, Hematopathology Subdivision, consisted of Howard Ratech (Chair 2004), Suzanne Kamel-Reid (Chair-Elect 2004, Chair 2005), and Janina Longtine (Chair-Elect 2005).
References
- Rowley JD. Letter: a new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- Shtivelman E, Lifshitz B, Gale RP, Canaani E. Fused transcript of abl and bcr genes in chronic myelogenous leukaemia. Nature. 1985;315:550–554. doi: 10.1038/315550a0. [DOI] [PubMed] [Google Scholar]
- Westbrook CA, Hooberman AL, Spino C, Dodge RK, Larson RA, Davey F, Wurster-Hill DH, Sobol RE, Schiffer C, Bloomfield CD. Clinical significance of the BCR-ABL fusion gene in adult acute lymphoblastic leukemia: a Cancer and Leukemia Group B Study (8762). Blood. 1992;80:2983–2990. [PubMed] [Google Scholar]
- Lugo TG, Pendergast AM, Muller AJ, Witte ON. Tyrosine kinase activity and transformation potency of bcr-abl oncogene products. Science. 1990;247:1079–1082. doi: 10.1126/science.2408149. [DOI] [PubMed] [Google Scholar]
- Melo JV. The molecular biology of chronic myeloid leukaemia. Leukemia. 1996;10:751–766. [PubMed] [Google Scholar]
- Schoch C, Schnittger S, Bursch S, Gerstner D, Hochhaus A, Berger U, Hehlmann R, Hiddemann W, Haferlach T. Comparison of chromosome banding analysis, interphase- and hypermetaphase-FISH, qualitative and quantitative PCR for diagnosis and for follow-up in chronic myeloid leukemia: a study on 350 cases. Leukemia. 2002;16:53–59. doi: 10.1038/sj.leu.2402329. [DOI] [PubMed] [Google Scholar]
- Mensink E, van de Locht A, Schattenberg A, Linders E, Schaap N, Geurts van Kessel A, De Witte T. Quantitation of minimal residual disease in Philadelphia chromosome positive chronic myeloid leukaemia patients using real-time quantitative RT-PCR. Br J Haematol. 1998;102:768–774. doi: 10.1046/j.1365-2141.1998.00823.x. [DOI] [PubMed] [Google Scholar]
- Merx K, Muller MC, Kreil S, Lahaye T, Paschka P, Schoch C, Weisser A, Kuhn C, Berger U, Gschaidmeier H, Hehlmann R, Hochhaus A. Early reduction of BCR-ABL mRNA transcript levels predicts cytogenetic response in chronic phase CML patients treated with imatinib after failure of interferon alpha. Leukemia. 2002;16:1579–1783. doi: 10.1038/sj.leu.2402680. [DOI] [PubMed] [Google Scholar]
- Wang L, Pearson K, Ferguson JE, Clark RE. The early molecular response to imatinib predicts cytogenetic and clinical outcome in chronic myeloid leukaemia. Br J Haematol. 2003;120:990–999. doi: 10.1046/j.1365-2141.2003.04200.x. [DOI] [PubMed] [Google Scholar]
- Hughes TP, Kaeda J, Branford S, Rudzki Z, Hochhaus A, Hensley ML, Gathmann I, Bolton AE, van Hoomissen IC, Goldman JM, Radich JP. International Randomised Study of Interferon versus STI571 (IRIS) Study Group: frequency of major molecular responses to imatinib or interferon alfa plus cytarabine in newly diagnosed chronic myeloid leukemia. N Engl J Med. 2003;349:1423–1432. doi: 10.1056/NEJMoa030513. [DOI] [PubMed] [Google Scholar]
- Branford S, Rudzki Z, Harper A, Grigg A, Taylor K, Durrant S, Arthur C, Browett P, Schwarer AP, Ma D, Seymour JF, Bradstock K, Joske D, Lynch K, Gathmann I, Hughes TP. Imatinib produces significantly superior molecular responses compared to interferon alfa plus cytarabine in patients with newly diagnosed chronic myeloid leukemia in chronic phase. Leukemia. 2003;17:2401–2409. doi: 10.1038/sj.leu.2403158. [DOI] [PubMed] [Google Scholar]
- Hughes TP, Branford S, Reynolds J, Seymour J, Taylor K, Guzzo-Pernell N, Filshie R, Arthur C, Schwarer A, Hertzberg M, Rudzki Z, Copeman M, Lynch K, Grigg A. High-dose imatinib (600m/day) with selective intensification in newly diagnosed CML patients in chronic phase: cytogenetic response rates at 12 months are superior to IRIS (abstract). Blood. 2004;104:A1001. [Google Scholar]
- Hughes TP, Branford S, Reynolds J, Koelmeyer R, Seymour J, Taylor K, Guzzo-Pernell N, Filshie R, Arthur C, Schwarer A, Hertzberg M, Copeman M, Lynch K, Grigg A. Maintenance of imatinib dose intensity in the first six months of therapy for newly diagnosed patients with CML is predictive of molecular response, independent of the ability to increase dose at a later point (abstract). Blood. 2005;106:A164. [Google Scholar]
- Hughes T, Branford S. Molecular monitoring of BCR-ABL as a guide to clinical management in chronic myeloid leukaemia. Blood Rev. 2006;20:29–41. doi: 10.1016/j.blre.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Guilhot F, O’Brien SG, Gathmann I, Kantarjian H, Gattermann N, Deininger MW, Silver RT, Goldman JM, Stone RM, Cervantes F, Hochhaus A, Powell B, Gabrilove JL, Rousselot P, Reiffers J, Cornelissen JJ, Hughes T, Agis H, Fischer T, Verhoef G, Shepherd J, Saglio G, Gratwohl A, Nielsen JL, Radich JP, Simonsson B, Taylor K, Baccarani M, So C, Letvak L, Larson RA. Five-year follow-up of patients receiving imatinib for chronic myeloid leukaemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- Mueller MC, Erben P, Saglio G, Gottardi E, Schenk T, Ernst T, Lauber S, Emig M, Hehlmann R, Hochhaus A. Harmonization of BCR-ABL mRNA quantification using an uniform control plasmid in 36 international laboratories (abstract). Blood. 2005;106:A1991. doi: 10.1038/sj.leu.2404983. [DOI] [PubMed] [Google Scholar]
- Gabert J, Beillard E, van der Velden VH, Bi W, Grimwade D, Pallisgaard N, Barbany G, Cazzaniga G, Cayuela JM, Cave H, Pane F, Aerts JL, De Micheli D, Thirion X, Pradel V, Gonzalez M, Viehmann S, Malec M, Saglio G, van Dongen JJ. Standardization and quality control studies of ‘real-time’ quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia—a Europe Against Cancer program. Leukemia. 2003;17:2318–2357. doi: 10.1038/sj.leu.2403135. [DOI] [PubMed] [Google Scholar]
- Lozzio CB, Lozzio BB. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 1975;45:321–334. [PubMed] [Google Scholar]
- Hughes T, Deininger M, Hochhaus A, Branford S, Radich JP, Kaeda J, Baccarani M, Cortes J, Cross NC, Druker BJ, Gabert J, Grimwade D, Hehlmann R, Kamel-Reid S, Lipton JH, Longtine J, Martinelli G, Saglio G, Soverini S, Stock W, Goldman JM. Monitoring CML patients responding to treatment with tyrosine kinase inhibitors—review and recommendations for ‘harmonizing’ current methodology for detecting BCR-ABL transcripts and kinase domain mutations and for expressing results. Blood. 2006;108:28–37. doi: 10.1182/blood-2006-01-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassinat B, Zassadowski F, Balitrand N, Barbey C, Rain JD, Fenaux P, Degos L, Vidaud M, Chomienne C. Quantitation of minimal residual disease in acute promyelocytic leukemia patients with t(15;17) translocation using real-time RT-PCR. Leukemia. 2000;14:324–328. doi: 10.1038/sj.leu.2401652. [DOI] [PubMed] [Google Scholar]
- Kreuzer KA, Lass U, Bohn A, Landt O, Schmidt CA. LightCycler technology for the quantitation of bcr/abl fusion transcripts. Cancer Res. 1999;59:3171–3174. [PubMed] [Google Scholar]
- Emig M, Saussele S, Wittor H, Weisser A, Reiter A, Willer A, Berger U, Hehlmann R, Cross NC, Hochhaus A. Accurate and rapid analysis of residual disease in patients with CML using specific fluorescent hybridization probes for real time quantitative RT-PCR. Leukemia. 1999;13:1825–1832. doi: 10.1038/sj.leu.2401566. [DOI] [PubMed] [Google Scholar]
- Bolufer P, Colomer D, Gomez MT, Martinez J, Gonzalez SM, Gonzalez M, Nomdedeu J, Bellosillo B, Barragan E, Lo-Coco F, Diverio D, Hermosin L, Garcia-Marco J, De Juan MD, Barros F, Romero R, Sanz MA. Quantitative assessment of PML-RARa and BCR-ABL by two real-time PCR instruments: multiinstitutional laboratory trial. Clin Chem. 2004;50:1088–1092. doi: 10.1373/clinchem.2003.028308. [DOI] [PubMed] [Google Scholar]
- Silvy M, Mancini J, Thirion X, Sigaux F, Gabert J. Evaluation of real-time quantitative PCR machines for the monitoring of fusion gene transcripts using the Europe against Cancer protocol. Leukemia. 2005;19:305–307. doi: 10.1038/sj.leu.2403590. [DOI] [PubMed] [Google Scholar]
- Branford S, Cross NCP, Hochhaus A, Radich J, Saglio G, Kaeda J, Goldman J, Hughes T. Rationale for the recommendations for harmonizing current methodology for detecting BCR-ABL transcripts in patients with chronic myeloid leukaemia. Leukemia. 2006;20:1925–1930. doi: 10.1038/sj.leu.2404388. [DOI] [PubMed] [Google Scholar]
- Beillard E, Pallisgaard N, van der Velden VH, Bi W, Dee R, van der Schoot E, Delabesse E, Macintyre E, Gottardi E, Saglio G, Watzinger F, Lion T, van Dongen JJ, Hokland P, Gabert J. Evaluation of candidate control genes for diagnosis and residual disease detection in leukemic patients using ‘real-time’ quantitative reverse-transcriptase polymerase chain reaction (RQ-PCR)—a Europe against Cancer program. Leukemia. 2003;17:2474–2486. doi: 10.1038/sj.leu.2403136. [DOI] [PubMed] [Google Scholar]
- Rulcová J, Zmekova V, Zemanova Z, Klamova H, Moravcova J. The effect of total ABL, GUS and B2M control genes on BCR-ABL monitoring by real-time RT-PCR. Leuk Res. 2007;31:483–491. doi: 10.1016/j.leukres.2006.07.021. [DOI] [PubMed] [Google Scholar]
- Laneuville P, Barnett MJ, Bélanger R, Couban S, Forrest DL, Roy DC, Lipton JH. Recommendations of the Canadian Consensus Group on the management of chronic myeloid leukaemia. Curr Oncol. 2006;13:201–221. [PMC free article] [PubMed] [Google Scholar]
- Hong KM, Nijjar H, Hawley M, Press RD. Quantitative real-time PCR with automated sample preparation for diagnosis and monitoring of cytomegalovirus infection in bone marrow transplant patients. Clin Chem. 2004;50:846–856. doi: 10.1373/clinchem.2003.026484. [DOI] [PubMed] [Google Scholar]
- Arocho A, Chen B, Ladanyi M, Pan Q. Validation of the 2−ΔΔCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn Mol Pathol. 2006;15:56–61. doi: 10.1097/00019606-200603000-00009. [DOI] [PubMed] [Google Scholar]
- Branford S, Hughes T. Diagnosis and monitoring of chronic myeloid leukaemia by qualitative and quantitative RT-PCR. Methods Mol Med. 2006;125:69–92. doi: 10.1385/1-59745-017-0:69. [DOI] [PubMed] [Google Scholar]
- Westgard JO, Barry PL, Hunt MR, Groth T. A multi-rule Shewhart chart for quality control in clinical chemistry. Clin Chem. 1981;27:493–501. [PubMed] [Google Scholar]