Abstract
We have developed a procedure for isolation of microRNA and genomic DNA in addition to total RNA from whole blood stabilized in PAXgene Blood RNA tubes. The procedure is based on automatic extraction on a BioRobot MDx and includes isolation of DNA from a fraction of the stabilized blood and recovery of small RNA species that are otherwise lost. The procedure presented here is suitable for large-scale experiments and is amenable to further automation. Procured total RNA and DNA was tested using Affymetrix Expression and single-nucleotide polymorphism GeneChips, respectively, and isolated microRNA was tested using spotted locked nucleic acid-based microarrays. We conclude that the yield and quality of total RNA, microRNA, and DNA from a single PAXgene blood RNA tube is sufficient for downstream microarray analysis.
Molecular genetic data becomes increasingly important during the process of clinical testing of new drugs. The combination of clinical and phenotypical data with expression profiles and genotyping via single-nucleotide polymorphism (SNP) analysis enables patient stratification according to, eg, treatment response or toxic effects on an individual basis. Besides parameters like efficiency and efficacy, drug metabolism has also been monitored.1,2,3 Some of the genetic data generated during drug development can often be used for the identification of biomarkers, which provide a basis for diagnostic and prognostic tests.4,5 Recently, microRNAs (miRNA) were found to be involved in disease development and progression. Their natural role in developmental processes like angiogenesis, neurogenesis, or stem cell differentiation supports their involvement in cancer progression.6,7,8,9,10 Profiling of these small regulatory RNA species may provide additional valuable information to pharmaceutical companies and diagnostic test developers. Besides testing for infectious diseases, nucleic acid-based molecular diagnostic tests for cancer or metabolic diseases have been designed.4 Regulatory agencies like the United States Food and Drug Administration encourage generation of genetic data during clinical trials to better estimate and document the effects of a drug in the human body (http://www.fda.gov/cber/gdlns/pharmdtasub.htm).
Sampling of blood is a standard procedure in most clinical trials and many diagnostic testing procedures. Pathological conditions in organs and remote tissues are often detectable in gene expression profiles from blood samples. Thus blood may serve as a surrogate tissue9 that can be collected with minimal effort and inconvenience to the patient. In contrast to classical diagnostic parameters like blood sedimentation rate or cholesterol content, which is stable in standard blood collection tubes, the nucleic acid-based information can be affected very rapidly. Postphlebotomy changes in the transcription profile caused by degradation and gene induction, leading to inconsistent results, are well documented.11,12,13,14,15,16 Consequently, there is a need for stabilization of cellular RNA species to retain the gene expression profile at the time of blood collection, and for this purpose, the PAXgene Blood RNA stabilization and isolation procedure is commonly used.17,18,19,20 This procedure is dedicated to the isolation of mRNA and large rRNA species only. Small RNAs like 5S rRNA, tRNA, snRNA, or miRNA are excluded during the isolation process, and genomic DNA (gDNA) is mechanically and enzymatically removed from the sample. To gain the complete genetic profile, additional blood tubes and isolation methods would be needed. With increased numbers of sample tubes, logistics become more complex, and the risk of failure is increased. Clinical trials work and reliable diagnosis should avoid this complication when possible.
We developed TRI-X, a new method that allows the isolation of all types of nucleic acids from one PAXgene Blood RNA tube to obtain sufficient high-quality material for the generation of microarray-based SNP, expression, and miRNA profiles. The method is suitable for clinical studies with high sample throughput because most steps can be automated on an appropriate robotic platform. The high reliability and complete documentation of all automated processes could be the starting point to develop an in vitro diagnostic compliant system.
Materials and Methods
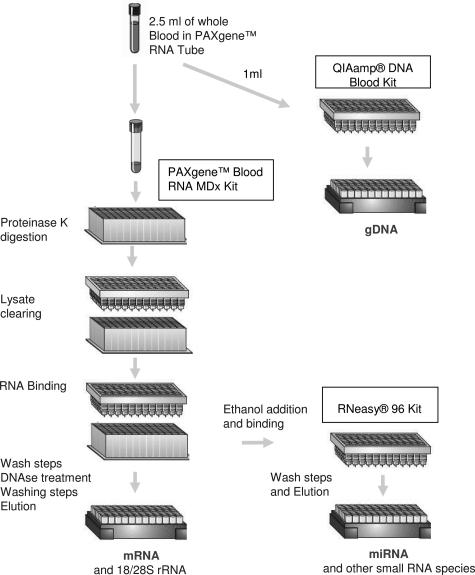
TRI-X is a combination of three isolation methods (Figure 1), starting with 2.5-ml whole blood collected in PAXgene Blood RNA tubes (PreAnalytiX, Hombrechtikon, Switzerland). Blood samples from healthy donors were collected and frozen at −80°C within 2 hours after blood withdrawal. The frozen samples were thawed for 16 hours at room temperature in batches of 48 before processing. After thawing of the tubes, 1 ml of the sample was transferred from every PAXgene Blood RNA tube into separate 2-ml tubes for gDNA isolation.
Figure 1.
Overview scheme for the TRI-X workflow. TRI-X is a combination of three methods, starting with 2.5-ml whole blood collected in a single PAXgene Blood RNA tube. Genomic DNA is isolated from a 1-ml aliquot with the QIAamp Blood DNA protocol. The remaining sample is extracted with the PAXgene Blood RNA MDx procedure to isolate mRNAs and large rRNA fragments. Small RNA species including miRNA are isolated from the flow through of the PAXgene Blood RNA MDx protocol binding step via a modified RNeasy procedure.
The remaining 9 ml of each sample was used for automated RNA isolation with the PAXgene Blood RNA MDx kit (PreAnalytiX) on the BioRobot MDx (Qiagen). The procedure of this RNA isolation method has been described in detail previously.12 The procedure starts with a proteinase K digest in the presence of guanidinium thiocyanate followed by a filtration step to remove cell debris and most of the gDNA. After the addition of ethanol, the cleared lysate is passed through a silica membrane to bind the large RNA species and remaining gDNA. In contrast to the original PAXgene Blood RNA MDx V1.0 protocol, we programmed a new version (PAXgene Blood RNA MDx cV17) that allows the collection of the flow through in a 96-square-well block during the RNA-binding step on the PAXgene 96 RNA plate. Immediately after the binding step, the flow through is manually removed from the robot, and the standard RNA isolation procedure is continued by the robot. This includes two washing steps, a DNase I digestion step on the membrane, three further wash steps, heat drying of the membrane, and finally elution of the RNA into a 96-well plate. The RNA samples were transferred from the elution plate into a Costar 3536 UV plate by the BioRobot MDx and absorptions at 260 and 280 nm were measured in a DTX880 plate reader (Beckman-Coulter, Fullerton, CA). In addition, the integrity of the RNA was determined with the RNA 6000 Nano LabChip on an Agilent 2100 Bioanalyzer21 (Agilent, Palo Alto, CA). The RNA was stored at −20°C until further analysis.
The small RNA fraction containing the miRNA that does not bind to the RNA-binding plate during the BioRobot MDx run was pipetted onto a new RNA-binding plate, and miRNA was procured by adjusting the binding conditions. In brief, 1.6× volumes (1.2 ml) of ethanol was added to the flow-though, mixed thoroughly, and passed through a new RNA-binding plate placed in a vacuum manifold (RNeasy 96 kit; Qiagen). The membrane was then washed according to the manufacturer’s instruction, and finally the miRNAs were eluted in 100 μl of H2O.
For gDNA isolation, the 1-ml aliquots (equivalent to 250 μl of whole blood) were processed with the QIAamp DNA Blood kit (Qiagen) according to the manufacturer’s instructions. Samples were lysed in the presence of proteinase and guanidinium hydrochloride. Ethanol was added to bind the gDNA to the QIAamp silica membrane. Nucleic acids bound to the membrane were subsequently washed in three steps and eluted from the membrane in 200 μl of H2O. The DNA concentration was determined on a NanoDrop ND-1000 UV-Vis Spectrophotometer (NanoDrop Technologies, Wilmington, DE) and stored at −20°C.
Affymetrix Genechip Human Mapping 250K SNP arrays (Affymetrix, Santa Clara, CA) were used for DNA analysis. All procedures were performed according to Affymetrix standard protocols. In brief, the DNA was vacuum dried and redissolved in 50 μl of H2O. DNA (250 ng) from 12 samples (three different samples from three donors and three control DNA samples provided with the labeling kit) was labeled and hybridized to the array. The SNP call rate and sample mismatch report was determined with GTYPE software (Affymetrix).
Gene expression analyses were performed using the Affymetrix HG U133 plus 2.0 Genechip (Affymetrix). RNA was concentrated by ethanol precipitation, and 1 μg was used for labeling using MessageAmp II Biotin Enhanced labeling kit (Ambion, Austin, TX). All labeling reactions were performed with or without SnX globin depletion reagent (AROS Applied Biotechnology A/S), which is a mixture of locked nucleic acid (LNA)-modified oligonucleotides that specifically hybridizes and thereby blocks the reverse transcription at the 3′-end of globin transcripts. In contrast to PNA probes as used in an Affymetrix globin depletion protocol, LNA oligonucleotides are highly soluble in water and can therefore be added to the labeling master mix. This reduces the handling processes and thereby increases the reproducibility. The scanned GeneChips were analyzed using GCOS (Affymetrix) and globally scaled to 150 U.
miRNA expression profiling was performed using LNA probes (miRCURY LNA Array Probeset V7.1; Exiqon A/S, Vedbaek, Denmark)22 spotted in duplicate on CodeLink slides (GE Healthcare, Chalfont, St. Giles, UK) using a VersArray ChipWriter Pro System (Bio-Rad Laboratories, Hercules, CA). Before spotting probes were diluted to a final concentration of 10 μmol/L using a 150 mmol/L phosphate buffer, pH 8.6, miRNA equivalent to 2 μg of total RNA isolated from the same PAXgene Blood RNA tube was used as starting material in the HY3 enzymatic labeling reaction (miRCURY LNA Array, Hy3/Hy5 labeling kit; Exiqon A/S). A pool of miRNA generated from total RNA extractions from several organs and cell lines was used as reference sample and 2 μg of reference RNA was labeled with HY5. Labeling and hybridization was performed according to the manufacturer’s instructions. TIGR Spotfinder 2.23 software (TIGR, Rockville, MD) was used after microarray scanning to generate raw-intensity data. Data were filtered to only include probes detected above background (50% background ± 2 SD). Raw expression data were Lowess-normalized using TIGR MIDAS 2.19 software, and TIGR MeV 3.1 was used for subsequent data analysis.
We used TaqMan MicroRNA assays (Applied Biosystems, Foster City, CA) for measuring the miRNA expression quantitatively. Polymerase chain reaction (PCR) assays were performed in triplicate for let-7a, miR-30b, and miR145 as described by the manufacturer using an ABI7900 PCR system (Applied Biosystems).
Results
gDNA isolation from 144 donors resulted in a yield of 1.38 ± 0.66 μg (mean ± SD) with a 260:280 nm ratio of 1.88 ± 0.38 (mean ± SD; 38 samples were excluded because they had 280 nm readings below the linear range of the spectrophotometer).
Only one sample had a yield less than 250 ng, which is the required amount for Genechip 250K SNP analysis, and only six samples gave a yield less than 500 ng (4%), which is required for 500K SNP analysis (Table 1).
Table 1.
gDNA and Total RNA Yields from TRI-X
| gDNA*
|
Total RNA†
|
|||
|---|---|---|---|---|
| Yield (ng) | N | Yield (μg) |
N
|
|
| Eluate | Conc | |||
| <250 | 1 | <1 | 0 | 6 |
| 250 to 500 | 3 | 1 to 5 | 67 | 110 |
| 500 to 1000 | 36 | >5 | 76 | 28 |
| 1000 to 1500 | 54 | |||
| 1500 to 2000 | 25 | |||
| >2000 | 25 | |||
Yield of gDNA isolated from 1-ml aliquots of blood from 144 donors.
Corresponding yield of total RNA before (Eluate) and after (Conc) ethanol concentration.
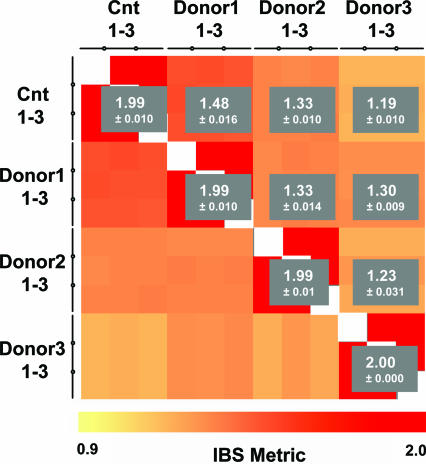
We hybridized 12 gDNA samples isolated from nine different PAXgene tubes, ie, three DNA samples from each of three donors plus three gDNA controls included in the labeling kit. The call rates achieved using donor gDNA were essentially identical to those achieved using control samples (93.2 ± 0.0090, 93.6 ± 0.0158, 94.6 ± 0.0098, and 94.4 ± 0.0158%, respectively). The gender was correctly determined in all samples, and the number of homozygotic (AA and BB) and heterozygotic (AB) calls within each group were similar (Table 2). We performed a sample mismatch analysis using the Identity By State (IBS) metric that counts the number of alleles shared by two individuals (comparison of common SNPs on two arrays). In this analysis, values of 1.95 to 2 are reported for identical gDNA, and lower values indicate increasingly unrelated donors. An IBS metric correlation heat map (Figure 2) shows that the control gDNA and the individual donor gDNA were scored as identical within the groups and different between the groups.
Table 2.
Results from Affymetrix Human Mapping 250K SNP Genechip Analysis
| Called gender | SNP call (%) | AA call (%) | AB call (%) | BB call (%) | |
|---|---|---|---|---|---|
| Control 1 | Male | 94.48 | 38.35 | 25.43 | 36.22 |
| Control 2 | Male | 92.84 | 38.83 | 24.16 | 37.01 |
| Control 3 | Male | 95.99 | 38.22 | 25.37 | 36.41 |
| Donor 1-1 | Male | 93.54 | 38.57 | 24.52 | 36.91 |
| Donor 1-2 | Male | 93.8 | 38.77 | 24.19 | 37.04 |
| Donor 1-3 | Male | 92.13 | 38.77 | 24.69 | 36.54 |
| Donor 2-1 | Male | 92.98 | 38.61 | 25.02 | 36.37 |
| Donor 2-2 | Male | 92.45 | 39.23 | 23.08 | 37.7 |
| Donor 2-3 | Male | 95.41 | 38.12 | 25.7 | 36.18 |
| Donor 3-1 | Female | 94.15 | 37.72 | 26.16 | 36.12 |
| Donor 3-2 | Female | 93.22 | 38.38 | 25.31 | 36.31 |
| Donor 3-3 | Female | 95.17 | 37.89 | 26.27 | 35.84 |
Report data are from the GTYPE software for six SNP arrays.
AA and BB, homozygous; AB, heterozygous.
Figure 2.
Sample mismatch analysis showing IBS scores of chip to chip comparisons of 250K SNP array. Heat map showing the result of a sample mismatch analysis comparing all SNP arrays against each other. The color of each square indicates the IBS score between SNP samples ranging from 0.9 to 2. The gray boxes indicate the average IBS score and SD between each group. Cnt, control DNA.
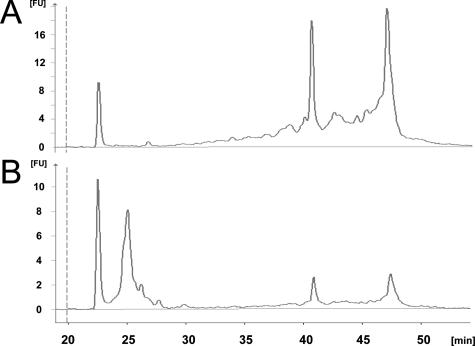
RNA isolation from 144 PAXgene Blood RNA tubes displayed a yield of 5.9 ± 3.1 μg (mean ± SD), and more than 1 μg was procured from each sample. Agilent Bioanalyzer analysis showed that RNA molecules smaller than approximately 150 nucleotides were absent from the total RNA preparations and that distinct 28S and 18S RNA were visible (Figure 3A). The RNA integrity number scores were 8.7 ± 0.47 (mean ± SD). Figure 3B shows clearly the enrichment of small RNA molecules in eluates derived from flow through of the robot binding step. For the microarray labeling reaction, the concentration of the eluted RNA was too low, and therefore an ethanol precipitation step was necessary. The recovery after ethanol precipitation was 68%, resulting in a mean yield of 4 ± 2.7 μg (mean ± SD), excluding six samples (4.2%) with yields below 1 μg (Table 1).
Figure 3.
Electropherograms from Agilent Bioanalyzer. The y axis represents fluorescence units [FU], and the x axis represents the runtime (s). A: Typical picture of a total RNA preparation with the PAXgene Blood RNA MDx protocol. The bands of the 18S and 28S rRNA fragments are clearly visible, and the RNA integrity number score is 8.7. Only very limited amounts of small RNA species are visible. B: Typical picture of a small RNA preparation isolated from the flow through from the PAXgene Blood RNA MDx binding step. Small RNAs including miRNA are clearly enriched.
The performance of the expression analysis using GeneChip U133 plus 2.0 was highly dependent on the addition of globin-blocking reagent SnX to the labeling reaction. Blocking the globin transcripts increased the number of probe sets scored as present from 7.077 (34.4%) ± 599 (2.7%) to 9.731 (43.7%) ± 347 (1.6%) and improved the glyceraldehyde-3-phosphate dehydrogenase 3′:5′ ratio from 1.87 ± 0.52 to 1.42 ± 0.31 and the total chip fluorescence signal as measured by the scaling factor from 3.7 ± 0.89 to 2.3 ± 0.34.
Using LNA-based oligonucleotide miRNA microarrays, we identified 42 miRNAs that were detected above background in all measurements of the small RNA fraction isolated from three different PAXgene Blood RNA tubes from four donors (Table 3). The lowest signal value detected above background was 88,000 for hsa-miR-512-5p, and the highest was 1268,000 for hsa-let-30d. Three miRNA expression profiles were validated by real-time quantitative PCR on all samples. All of these miRNAs showed amplification and furthermore showed a high positive Pearson correlation to microarray data ranging between 0.74 and 0.86 (Table 3). We compared the log2 microarray data to log2 average signals.
Table 3.
miRNAs Identified Using LNA-Based Oligonucleotide Microarrays
| miRNA name | Published data* | Microarray (signal ×1000)† | qPCR‡
|
|
|---|---|---|---|---|
| Mean quantity | Correlation | |||
| hsa-let-7a
|
−
|
348 | 1329 | 0.86 |
| hsa-let-7e | − | 284 | ||
| hsa-miR-20a | * | 1034 | ||
| hsa-miR-20b | nd | 334 | ||
| hsa-miR-23a | * | 594 | ||
| hsa-miR-23b | * | 240 | ||
| hsa-miR-25 | * | 501 | ||
| hsa-miR-27a | b | 460 | ||
| hsa-miR-296 | nd | 189 | ||
| hsa-miR-29b | − | 250 | ||
| hsa-miR-29c | b | 292 | ||
| hsa-miR-30b | * | 874 | 1180 | 0.77 |
| hsa-miR-30d | * | 1268 | ||
| hsa-miR-127 | − | 237 | ||
| hsa-miR-129 | − | 281 | ||
| hsa-miR-145 | − | 295 | 1205 | 0.74 |
| hsa-miR-151 | − | 461 | ||
| hsa-miR-184 | − | 352 | ||
| hsa-miR-185 | − | 132 | ||
| hsa-miR-195 | − | 255 | ||
| hsa-miR-200b | nd | 209 | ||
| hsa-miR-202 | * | 904 | ||
| hsa-miR-214 | − | 217 | ||
| hsa-miR-223 | * | 525 | ||
| hsa-miR-320 | nd | 598 | ||
| hsa-miR-326 | nd | 140 | ||
| hsa-miR-338 | nd | 186 | ||
| hsa-miR-346 | nd | 284 | ||
| hsa-miR-370 | nd | 357 | ||
| hsa-miR-423 | nd | 285 | ||
| hsa-miR-452 | nd | 746 | ||
| hsa-miR-494 | nd | 818 | ||
| hsa-miR-498 | nd | 261 | ||
| hsa-miR-503 | nd | 177 | ||
| hsa-miR-510 | nd | 166 | ||
| hsa-miR-512-5p | nd | 88 | ||
| hsa-miR-513 | nd | 103 | ||
| hsa-miR-519d | nd | 128 | ||
| hsa-miR-519e* | nd | 146 | ||
| hsa-miR-525 | nd | 144 | ||
| hsa-miR-526c | nd | 106 | ||
| hsa-miR-527-518a-2* | nd | 653 | ||
The 42 human miRNAs detected above background in all 16 blood cell samples (four donors, four samples from each) are included.
miRNA detected in CD34+ peripheral blood stem cells and bone marrow by Georgantas et al26; asterisk, detected in both tissues; b, detected only in bone marrow; −, not detected; nd, not determined.
Mean signal intensities of the 42 miRNAs measured in single-channel microarray analysis.
Mean values of three miRNAs validated using real-time quantitative PCR (qPCR) on all 16 samples and Pearson correlation between quantitative PCR mean signal and microarray signals.
The yield of small RNA is highly influenced by even a small degree of degradation of the total RNA as the degradation products accumulate in the small RNA size range. It therefore makes little sense to use optical density measurements to determine the yield of miRNA. Because protocols for labeling miRNA from a total RNA preparation recommend using 1 to 2 μg of total RNA, we therefore used a volume of miRNA preparation equivalent to 2 μg of total RNA isolated from the same PAXgene Blood RNA tube.
Next, we evaluated the stability of RNA during long-term storage of the PAXgene tubes. A sample collection of 48 samples, of which one-half had been stored for more than 2.5 years and the other one-half for less than 5 months at −80°C, were processed using the TRI-X protocol. The yield and RNA integrity number scores of total RNA, which is the molecule that is most likely to suffer from long storage time, are summarized in Table 4. We observed no significant deterioration of total RNA during 2.5 years of storage in PAXgene tubes.
Table 4.
Effect of Long-Term Storage of PAXgene RNA Tubes on RNA Yield and Quality
| >2.5 years | <5 month | All samples | |
|---|---|---|---|
| Yield (μg) | 4.48 ± 1.42 | 4.32 ± 1.86 | 4.40 ± 1.64 |
| RNA integrity number | 8.71 ± 0.31 | 8.74 ± 0.50 | 8.73 ± 0.41 |
Twenty-four samples stored for more than 2.5 years at −80°C and 24 samples stored for less than 5 months were randomized and processed in the same batch. The table shows yield and quality of the two groups and corresponding SDs.
Discussion
A total of 144 blood samples from healthy donors were processed for isolation of RNA, miRNA, and DNA. To check the versatility of the protocols, we accessed the product for yield and quality. Yield should be sufficient for high-density microarray analysis, and quality should result in high-quality microarray data.
For the DNA microarray analysis, we chose to run one of two 250K mapping arrays from the Affymetrix 500K mapping array set. This platform requires a higher DNA quality compared with, eg, the Sentrix humanHap550 genotyping beadchip (Illumina, San Diego, CA), because this system uses restriction endonucleases in the labeling protocol. Analyses on both 250K mapping arrays require two separate labeling reactions with 250 ng of DNA for each reaction. In the present study, we achieved sufficient DNA for running both 250 SNP arrays in 96% of the 144 extracted samples and for running one of the arrays in >99% of the samples. Analysis on the Sentrix arrays requires 750 ng of input DNA, which was achieved from 125 of 144 samples. The yield of DNA can be increased by using a larger portion of the PAXgene Blood RNA tube content and still get enough RNA, because none of the samples with insufficient amounts of DNA had insufficient amounts of RNA.
Microarray expression analysis of whole-blood RNA presents a great challenge because reticulocyte transcripts may contribute up to 70% of all transcripts of which most are globin transcripts. These globin transcripts have been shown to reduce the microarray sensitivity measured as the number of probe sets present.23,24 We therefore checked the RNA quality in the microarray analysis both with and without blocking globin transcript during reverse transcription using an LNA-modified oligonucleotide mixture (SnX). The addition of SnX to the labeling reaction increased the sensitivity from 7.077 (34.4%) present calls to 9.731 (43.7%), which is highly similar to the results obtained by Affymetrix (Globin Reduction Protocol: A Method for Processing Whole Blood RNA Samples for Improved Array Results. Santa Clara, CA, Affymetrix technical note, Part no. 701497, Rev. 2) using a PNA-blocking reagent. The Affymetrix protocol, however, prescribes 3- to 10-fold higher amounts of input RNA.
In total, 42 different miRNA were detected in all samples using custom spotted miRNA microarrays. In cancer, miRNAs have been identified to have tumor suppressor and oncogene functions, and expression changes of several miRNAs may have diagnostic and prognostic significance.25 Although miRNAs have been found in all mammalian tissues examined so far, there is little published information about miRNA expression in blood.26 It is likely that miRNA expression profiles in white blood cells for blood-related diseases can be defined. However, several studies on transcriptional profiles in peripheral blood of a central nervous system disease, ie, multiple sclerosis, have identified specific expression patterns.27 Therefore, it is feasible to assume also that expression profiles of miRNAs in blood can reflect the activity in other tissue compartments and thus serve as a diagnostic and/or prognostic tool for non-blood-related diseases. Some of the 42 detected miRNA species, especially let-7a,28 mir145,29 and mir195,30 are supposed to be connected to different forms of cancer and could potentially be used as a starting point for diagnostic or prognostic assay development.
In this study, we showed that the amounts of total RNA, miRNA, and gDNA from a single PAXgene Blood RNA tube are sufficient and of an adequate quality to perform microarray experiments. As the number of genomic tests based on SNP genotyping and gene and miRNA expression grows, it would be of great importance that the material required for these tests can be isolated from a single blood sample. Furthermore, it would allow the analysis of blood samples from completed or running clinical studies where only single samples are available or for all new studies where the blood sampling is limited. The stability of RNA in PAXgene tubes stored at −80°C more than 2.5 years guarantees that the samples of longitudinal clinical studies, which often last several years, can be processed simultaneously in one or a few sets at the end of the study. Therefore, analytical “batch-effect” can also be avoided.
In some cases, donors may have low numbers of white blood cells, which is likely to result in lower yields of all three nucleic acid specimens. For diagnostic purposes, this may not be critical. Diagnostic and prognostic signatures often contain a limited number of genes, miRNAs, or SNPs, and therefore these methods often require less starting material than is used for microarrays. Moreover, labeling technologies are under constant development, and new systems that require less than 20 ng of input RNA are available, like the Ovation Whole Blood Solution kit (NuGen Technologies, Bemmel, The Netherlands).
The workflow described here for total RNA isolation is fully automated, and the concentration step before labeling is amenable for automation using MinElute 96 UF PCR Purification kit (Qiagen). We have tested this method in a pilot study and found a mean recovery of 78%, which is superior to the ethanol precipitation used in this study (data not shown). Further automation of the TRI-X procedure can be achieved by substituting the manual QiaAmp kit for gDNA isolation used in this study with the QIAamp DNA Blood MDx kit (Qiagen) designed for the BioRobot MDx. The automation of the miRNA isolation should be possible as well because similar procedures for automated total RNA isolation are already available on a comparable robot system (BioRobot Universal System; Qiagen). A completely automated isolation procedure would decrease failure rates and simplify high sample throughput logistics via sample tracking. The TRI-X procedure is readily implemented in AROS laboratories in a GLP environment that is usually required for clinical trials.
Acknowledgments
We thank Gitte Høj, Ulla Mejlvang, Lotte Gernyx, and Annette Hedegaard for excellent technical assistance.
Footnotes
Supported in part by grants from the Danish Research Council and the Nordic Centre of Excellence in Molecular Medicine (to L.D.).
TRI-X is intended for research use only. No claim or representation is intended for its use to provide information for the diagnosis, prevention or treatment of a disease.
References
- Park JS, Chang HS, Park CS, Lee JH, Lee YM, Choi JH, Park HS, Kim LH, Park BL, Choi YH, Shin HD. Association analysis of cysteinyl-leukotriene receptor 2 (CYSLTR2) polymorphisms with aspirin intolerance in asthmatics. Pharmacogenet Genomics. 2005;15:483–492. doi: 10.1097/01.fpc.0000166456.84905.a0. [DOI] [PubMed] [Google Scholar]
- Bercovich D, Friedlander Y, Korem S, Houminer A, Hoffman A, Kleinber L, Shochat C, Leitersdorf E, Meiner V. The association of common SNPs and haplotypes in the CETP and MDR1 genes with lipids response to fluvastatin in familial hypercholesterolemia. Atherosclerosis. 2006;185:97–107. doi: 10.1016/j.atherosclerosis.2005.05.025. [DOI] [PubMed] [Google Scholar]
- Puisset F, Chatelut E, Dalenc F, Busi F, Cresteil T, Aze J, Poublanc M, Hennebelle I, Lafont T, Chevreau C, Roche H. Dexamethasone as a probe for docetaxel clearance. Cancer Chemother Pharmacol. 2004;54:265–272. doi: 10.1007/s00280-004-0823-0. [DOI] [PubMed] [Google Scholar]
- Manne U, Srivastava R, Srivastava S. Recent advances in biomarkers for cancer diagnosis and treatment. Drug Discov Today. 2005;10:965–976. doi: 10.1016/S1359-6446(05)03487-2. [DOI] [PubMed] [Google Scholar]
- Gutman S, Kessler LG. The U.S. Food and Drug Administration perspective on cancer biomarker development. Nat Rev Cancer. 2006;6:565–571. doi: 10.1038/nrc1911. [DOI] [PubMed] [Google Scholar]
- Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- Mendell JT. Critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4:1179–1184. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- Tong AW. Small RNAs and non-small cell lung cancer. Curr Mol Med. 2006;6:339–349. doi: 10.2174/156652406776894554. [DOI] [PubMed] [Google Scholar]
- Slack FJ, Weidhaas JB. MicroRNAs as a potential magic bullet in cancer. Future Oncol. 2006;2:73–82. doi: 10.2217/14796694.2.1.73. [DOI] [PubMed] [Google Scholar]
- Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, Relman DA, Brown PO. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci USA. 2003;100:1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainen L, Oelmueller U, Jurgensen S, Wyrich R, Ballas C, Schram J, Herdman C, Bankaitis-Davis D, Nicholls N, Trollinger D, Tryon V. Stabilization of mRNA expression in whole blood samples. Clin Chem. 2002;48:1883–1890. [PubMed] [Google Scholar]
- Müller MC, Merx K, Weier A, Kreil S, Lahaye T, Hehlmann R, Hochhaus A. Improvement of molecular monitoring of residual disease in leukemias by bedside RNA stabilization. Leukemia. 2002;16:2395–2399. doi: 10.1038/sj.leu.2402734. [DOI] [PubMed] [Google Scholar]
- Stordeur P, Zhou L, Goldman M. Analysis of spontaneous mRNA cytokine production in peripheral blood. J Immunol Methods. 2002;261:195–713. doi: 10.1016/s0022-1759(01)00548-8. [DOI] [PubMed] [Google Scholar]
- Talwar S, Munson PJ, Barb J, Fiuza C, Cintron AP, Logun C, Tropea M, Khan S, Reda D, Shelhamer JH, Danner RL, Suffredini AF. Gene expression profiles of peripheral blood leukocytes after endotoxin challenge in humans. Physiol Genomics. 2006;25:203–215. doi: 10.1152/physiolgenomics.00192.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Robinson JF, Khan HMR, Carter DE, McKinney J, Miskie BA, Hegele RA. Optimizing RNA extraction yield from whole blood for microarray gene expression analysis. Clin Biochem. 2004;37:741–744. doi: 10.1016/j.clinbiochem.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Tan FK, Zhou X, Mayes MD, Gourh P, Guo X, Marcum C, Jin L, Arnett FC. Signatures of differentially regulated interferon gene expression and vasculotrophism in the peripheral blood cells of systemic sclerosis patients. Rheumatology. 2006;45:694–702. doi: 10.1093/rheumatology/kei244. [DOI] [PubMed] [Google Scholar]
- Vlasova TI, Stratton SL, Wells AM, Mock NI, Mock DM. Biotin deficiency reduces expression of SLC19A3, a potential biotin transporter, in leukocytes from human blood. J Nutr. 2005;135:42–47. doi: 10.1093/jn/135.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Sahni NS, Tibshirani R, Skaane P, Urdal P, Berghagen H, Jensen M, Kristiansen L, Moen C, Sharma P, Zaka A, Arnes J, Sauer T, Akslen LA, Schlichting E, Børresen-Dale AL, Lönneborg A. Early detection of breast cancer based on gene-expression patterns in peripheral blood cells. Breast Cancer Res. 2005;7:R634–R644. doi: 10.1186/bcr1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batliwalla FM, Li W, Ritchlin CT, Xiao X, Brenner M, Laragione T, Shao T, Durham R, Kemshetti S, Schwarz E, Coe R, Kern M, Baechler EC, Behrens TW, Gregersen PK, Gulko PS. Microarray analyses of peripheral blood cells identifies unique gene expression signature in psoriatic arthritis. Mol Med. 2006;11:21–29. doi: 10.2119/2006-00003.Gulko. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoldi M, Schmidt S, Benes V, Noerholm M, Kulozik AE, Hentze MW, Muckenthaler MU. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA). RNA. 2006;12:913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feezor RJ, Baker HV, Mindrinos M, Hayden D, Tannahill CL, Brownstein BH, Fay A, MacMillan S, Laramie J, Xiao W, Moldawer LL, Cobb JP, Laudanski K, Miller-Graziano CL, Maier RV, Schoenfeld D, Davis RW, Tompkins RG. Inflammation and Host Response to Injury, Large-Scale Collaborative Research Program: Whole blood and leukocyte RNA isolation for gene expression analyses. Physiol Genomics. 2004;19:247–254. doi: 10.1152/physiolgenomics.00020.2004. [DOI] [PubMed] [Google Scholar]
- Debey S, Schoenbeck U, Hellmich M, Gathof BS, Pillai R, Zander T, Schultze JL. Comparison of different isolation techniques prior gene expression profiling of blood derived cells: impact on physiological responses, on overall expression and the role of different cell types. Pharmacogenomics J. 2004;4:193–207. doi: 10.1038/sj.tpj.6500240. [DOI] [PubMed] [Google Scholar]
- Dalmay T, Edwards DR. MicroRNAs and the hallmarks of cancer. Oncogene. 2006;25:6170–6175. doi: 10.1038/sj.onc.1209911. [DOI] [PubMed] [Google Scholar]
- Georgantas RW, III, Hildreth R, Morisot S, Alder J, Li C, Heimfeld S, Calin GA, Croce CM, Civin CI. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci USA. 2007;104:2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg RLP, Kappos L. Transcriptional profiling of multiple sclerosis: towards improved diagnosis and treatment. Expert Rev Mol Diagn. 2006;6:843–855. doi: 10.1586/14737159.6.6.843. [DOI] [PubMed] [Google Scholar]
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Chang-Gong L, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard M, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA Gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinomas and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]