Abstract
Ewing family tumors (EFTs) are prototypical primitive small round blue cell sarcomas arising in bone or extraskeletal soft tissues in children or adolescents. EFTs show fusions of EWS with a gene of the ETS family of transcription factors, either EWS-FLI1 (90 to 95%) or EWS-ERG (5 to 10%). Rare cases with fusions of EWS to other ETS family genes, such as ETV1, E1AF, and FEV, have been identified, but their clinicopathological similarity to classic EFTs remains unclear. We report four new cases of EFT-like tumors with rare EWS fusions, including two with EWS-ETV1, one with EWS-FEV, and a fourth case in which we cloned a novel EWS-SP3 fusion, the first known cancer gene fusion involving a gene of the Sp zinc finger family. Analysis of these three new cases along with data on nine previously reported cases with fusions of EWS to ETV1, E1AF, or FEV suggest a strong predilection for extraskeletal primary sites. EFT-like cases with fusions of EWS to non-ETS translocation partners are also uncommon but involve the same amino-terminal portion of EWS, which in our novel EWS-SP3 fusion is joined to the SP3 zinc-finger DNA-binding domain. As these data further support, these types of EWS fusions are associated with primitive extraskeletal small round cell sarcomas of uncertain lineage arising mainly in the pediatric population.
Classic Ewing’s sarcoma is a primitive bone sarcoma of children or adolescents composed of small round cells showing limited neural differentiation. The Ewing family of tumors (EFTs) also includes identical or very similar tumors in the soft tissues, chest wall, and visceral sites, as well as tumors previously termed peripheral neuroectodermal tumor, which show more neural features. EFTs are also defined by translocations, resulting in the fusion of the EWS gene, located at 22q12, and a gene of the ETS family of transcription factors.1,2,3 The ETS family of transcription factors is defined by the ETS domain, which interacts with DNA at sequences containing a common sequence motif.4 In 90 to 95% of cases, the gene fusion is EWS-FLI1, consisting of the N-terminal portion of EWS and the C-terminal portion of FLI1 including the ETS DNA-binding domain. An EWS-ERG fusion, where the ERG gene from 21q22 substitutes for FLI1, is found in 5 to 10% of cases.2,5 EWS is a ubiquitously expressed protein containing an RNA-binding domain in its C-terminal portion, whereas its N-terminal domain can function as a strong transactivation domain when juxtaposed to a DNA-binding domain.6 The EWS promoter is strongly and broadly activated, leading to relatively unrestricted high-level expression of the resulting fusion genes.7,8 In contrast, expression of native FLI1 is tightly regulated and lineage restricted.9 Details of the biology of native EWS and FLI1, as well as functional aspects of EWS-FLI1, are reviewed in detail elsewhere.10,11 In less than 1% of cases, variant translocations namely t(7;22)(p22;q12), t(17;22)(q12;q12), and t(2;22)(q33;q12) involving fusion of EWS gene with ETV1, E1AF (also known as ETV4), and FEV genes, respectively, have been described.12,13,14,15 ETV1, E1AF, and FEV are all three members of the ETS family of transcription factors that are highly related to FLI1 and ERG.
Although most other well-known EWS fusions have involved other transcription factor gene families and have been described in tumors that are pathologically clearly distinct from EFTs, such as clear cell sarcoma, extraskeletal myxoid chondrosarcoma, myxoid liposarcoma, and angiomatoid fibrous histiocytoma,11,16,17 there have been rare reports of unique EWS fusions in EFT-like cases that are more difficult to fit in existing categories. For instance, a single case has been reported of a small round cell tumor of the chest wall with a fusion of EWS to a zinc finger family gene, ZNF278 (also know as ZSG).18 The only other translocation fusing EWS to a zinc finger family gene is the EWS-WT1 fusion of the desmoplastic small round cell tumor (DSRCT).19,20,21 Indeed, the case bearing the EWS-ZNF278 fusion displayed some features more typical of DSRCT, such as positivity for desmin and negativity for MIC2, but lacked desmoplasia. Finally, another unique EWS fusion has recently been reported in a bone tumor, involving a POU homeodomain family gene, POU5F1.22
Studies evaluating the clinicopathological correlates of the different EWS gene fusions in EFTs have focused on cases with the common EWS-FLI1 and EWS-ERG fusion transcripts. Overall, no significant clinical or pathological differences have been found between cases with EWS-FLI1 and those with EWS-ERG.23 Among EWS-FLI1 cases, the most common type of EWS-FLI1 fusion transcript (type 1) may be associated with a somewhat more favorable prognosis compared with type 2 EWS-FLI1 and other types.24,25,26 In contrast, little or nothing has been published concerning the clinicopathological features of patients with tumors harboring rare variant EWS fusions (EWS-ETV1, EWS-E1AF, and EWS-FEV) after their initial discovery.
We report four new cases of EFT-like tumors with rare EWS fusions, including two with EWS-ETV1, one with EWS-FEV, and a fourth case in which we have cloned a novel EWS-SP3 fusion, the first known cancer gene fusion involving a gene of the Sp zinc finger family. We have also reviewed the clinical and pathological data on these cases along with other reported cases in the literature and highlight the clinicopathological features of these primitive round cell sarcomas with rare variant EWS fusions.
Materials and Methods
RNA Extraction
Total RNA was isolated from snap-frozen tissue using acid guanidium-isothiocynate followed by phenol-chloroform extraction and isopropanol precipitation. RNA was resuspended in diethylpyrocarbonate-treated water and stored at −70°C.
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
RT-PCR was performed using the Qiagen (Valencia, CA) one-step RT-PCR kit. The reaction was performed in one tube in a sequential manner, in a final volume of 50 μl. Each reaction tube contained 1 μg of total RNA diluted in 26.5 μl of RNase-free water, 10 μl of 5× buffer containing 2.5 mmol of MgCl2, 2 μl of dNTP mix (final concentration: 400 μmol/L of each dNTP), 0.5 μl of RNase inhibitor (5 U), 8 μl of primers (each primer concentration: 0.5 to 1 μmol/L), and 2 μl of one-step enzyme mix. The thermal cycler conditions were as follows: reverse transcriptase at 50°C for 30 minutes, initial PCR activation step at 95°C for 15 minutes, 35 cycles of denaturation (94°C for 30 seconds), annealing (60°C for 1 minute), and extension (72°C for 30 seconds). The final extension was performed at 72°C for 7 minutes. The primers are listed in Table 1. PCR products were electrophoresed on a 2% agarose gel containing ethidium bromide in 1× Tris acetic acid ethylenediamine tetraacetic acid (TAE) buffer.
Table 1.
Primer Sequences for EWS-ETS Fusion Transcripts
| Primers | Sequences |
|---|---|
| Forward primers | |
| EWS exon 7 (22.3) | 5′-TCCTACAGCCAAGCTCCAAGTC-3′ |
| EWS exon 7 (RACE1) | 5′-AGCAGCCTCCCACTAGTTAC-3′ |
| EWS exon 7 (22.14) | 5′-GCACCTCCATCCTACCCTCC-3′ |
| Reverse primers | |
| FLI1 exon 6 | 5′-GTTGAGGCCAGAATTCATGTTA-3′ |
| FLI1 exon 9 | 5′-ACTCCCCGTTGGTCCCTCC-3′ |
| ERG exon 9 | 5′-AAAGCTGGATCTGGCCACTG-3′ |
| ETV1 | 5′-CCGATACATTCCTGGCTCTTG-3′ |
| ETS1 (consensus for FLI1, ERG, and ETV1) | 5′-AGGAA-T/C-TGCCA-A/C-AGCTGGATCTG-3′ |
| ETS2 (consensus for ETV1 and E1AF) | 5′-CC-A/C-GTCCAGGCAAT-A/G-AAATG-3′ |
Sequencing
The RT-PCR product was purified with a Qiagen purification kit and sequenced on an Applied Biosystems (Foster City, CA) automated sequencer using the dideoxy dye terminator method.
Rapid Amplification of cDNA Ends (RACE)
RACE was performed using the 3′ RACE system from Invitrogen (Carlsbad, CA). In brief, first-strand cDNA synthesis was initiated at the poly(A) tail of the mRNA using the adapter primer (5′-GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTT-3′). After the first-strand cDNA synthesis, the original mRNA template was destroyed with RNase H. The first-round amplification was performed using a universal amplification primer that binds the adapter sequence in the adapter primer (AUAP; 5′-GGCCACGCGTCGACTAGTAC-3′) and a forward primer that anneals to EWS exon 6 (EWS22.14; Table 1). To increase the specificity of the process, a seminested PCR was performed after the first round PCR, using the same AUAP reverse primer and a nested forward primer, EWS22.3 (Table 1), which primes from exon 7 of EWS. One μl of a 1:100 dilution of the first-round PCR product was used as the template for the second-round PCR. The products of the seminested PCR were cloned using the pGEM-T easy AT cloning system (Promega, Madison, WI), and multiple clones were sequenced as described above.
Results
We identified four new cases (Tables 2and 3, patients A to D) of variant EWS fusions. Two of these cases showed the EWS-ETV1 fusion transcript, one showed an EWS-FEV fusion, and the fourth involved a novel EWS translocation partner.
Table 2.
EFTs with Rare Variant Fusions of EWS to ETS Family Transcription Factor Genes
| Case no. (reference) | Age/sex | Presentation | Histology |
|---|---|---|---|
| EWS-ETV1 cases | |||
| No. 1 (12) | 2/F | Primary in gluteal region; lung metastases | SRCT |
| No. 2, present series, patient A | 8/M | Primary adjacent to fibula; no metastases | SRCT |
| No. 3, present series, patient B | 2/F | Primary in pelvis and thigh; lung metastases | Undifferentiated tumor with spindling (post-CT) |
| No. 4 (42,43) | 0/M | Skin and paraspinal; primary and metastatic sites unclear | Round, polygonal and spindle cells with scant cytoplasm |
| No. 5* (44) | 12/F | Primary in femur; no data on metastatic disease | NA |
| EWS-FEV cases | |||
| No. 6 (15) | 2/F | Paraspinal primary; no data on metastatic disease | SRCT |
| No. 7 (15) | 15/M | Extraosseous maxillary primary; no data on metastatic disease | SRCT |
| No. 8 (45) | 15/M | Paratesticular primary; distant metastases | Large round polygonal cells with clear cell features |
| No. 9 (46) | 24/M | Retroperitoneal primary; no metastases | SRCT with foci of pleomorphic large cells |
| No. 10, present series, patient C | 40/M | Primary in retroperitoneum and pelvis; no metastases | Poorly differentiated neoplasm |
| EWS-E1AF cases | |||
| No. 11 (13) | 0/F | Primary in cheek; no metastases | SRCT |
| No. 12 (14;47) | 10/M | Extraosseous chest wall primary; no data on metastatic disease | SRCT |
F, Female; M, male; NA, not available; SRCT, small round cell tumor; IHC, immunohistochemistry; EM, electron microscopy; CG, cytogenetics; CT, chemotherapy; RT, radiotherapy; PBSCT, peripheral blood stem cell transplant; NED, no evidence of disease; DOD, dead of disease, AWD, alive with disease.
No molecular confirmation of fusion transcript on this case.
(table continues)
Table 2.
Continued
| IHC | Cytogenetic translocation | Treatment | Outcome |
|---|---|---|---|
| + Vimentin, NSE, desmin, MSA, myoglobin | t(7;22)(p21;q12) | NA | NA |
| + Vimentin, CD99, Ki 67 >10%, S100, SMA, chromogranin, CD79A, CD34, desmin, EMA, cytokeratin | t(7;22)(p15;q11) | Resection followed by CT (see text for details) | NED at 60 months |
| + Leu-7, CD99 (focal), vimentin (weak), SMA, S100, CG, SYN, MSA, CK, EMA, NSE, GFAP, desmin, myogenin, CD34, CD31, factor VIII, CD3, CD45, CD79a | t(7;22) | Neoadjuvant CT, resection (see text for details) | DOD at 60 months |
| + S100, CD99, focal CK, CG, SYN | t(7;22)(p21;q11.2) | Surgery, CT, RT, and PBSCT | DOD at 96 months |
| NA | t(7;22)(p22;q12) | Neoadjuvant CT | DOD at 3 months |
| + CD99, desmin, NSE, LCA | t(2;21;22)(q33;q22;q12) | Resection. CT information NA | NA |
| + Vimentin, desmin, LCA, S100, NSE, CD99, chromogranin | NA | Resection. CT information NA | NA |
| + Vimentin, NSE, PGP9.5, CD99, CD57, S100, NF, CK, EMA, muscle markers | t(2;22)(q13;q22) | Excision followed by CT (vincristine, adriamycin, cisplatinum) and local RT | DOD at 28 months |
| + CD99, NSE, and weakly for vimentin. Large cells also positive for CEA, CAM 5.2, EMA, actin, desmin, CD45 | der(17)t(17;2;22;13) (q21::q11-q33::q12-q13::q14), del(22)(q12) | Partial resection. CT for 6 months | DOD at 12 months |
| + CD57, SYN, CAM5.2, CD99, CK7, CK20, EMA, PSA, PSAP, CG | NA | CT, RT (see text for details) | AWD at 42 months |
| + Vimentin, NSE, CD99, desmin, MSA, SMA, LCA, S100, NF | t(17;22)(q12;q12) | Partial resection and CT (carboplatin, etoposide, cyclophosphamide, pirarubicin, cisplatin) | DOD at 2 years |
| + CD99 | NA | Resection | NA |
Table 3.
EFT-Like Tumors with Noncanonical EWS Fusions
| Case no. (reference) | Age/sex | Presentation | Histology |
|---|---|---|---|
| No. 1 (18) | 16/M | Extraosseous chest wall primary | SRCT |
| No. 2 (22) | 39/F | Primary in pubic bone; pulmonary metastases | SRCT |
| No. 3, present series, patient D | 16/M | Disseminated disease: forehead, bones, lungs, right kidney | SRCT |
For abbreviations, see Table 2.
(table continues)
Table 3.
Continued
| IHC | Fusion type and cytogenetic translocation | Treatment | Outcome |
|---|---|---|---|
| + SYN, desmin, CAM5.2, CD99, neurofilament | EWS-ZNF278 t(1;22)(p36.1;q12) | NA | AWD (pulmonary metastases) at 2 years |
| + vimentin, S100, NSE, focal CK, MIC2, MSA, | EWS-POU5F1 t(6;22)(p21;q12) | DOD at 6 months | |
| + BCL2, CD99 (focal), NF (focal), CK cocktail (focal), CG, SYN, S100, desmin, ALK1, EMA, CD45, CD20, CD30, CD43, CD3, CD79a, EBER | EWS-SP3 | Neoadjuvant CT, resection (kidney) irinotecan, palliative RT (see text for details) | DOD at 20 months |
Patients A, B, and C: New EWS-ETV1 and EWS-FEV Cases
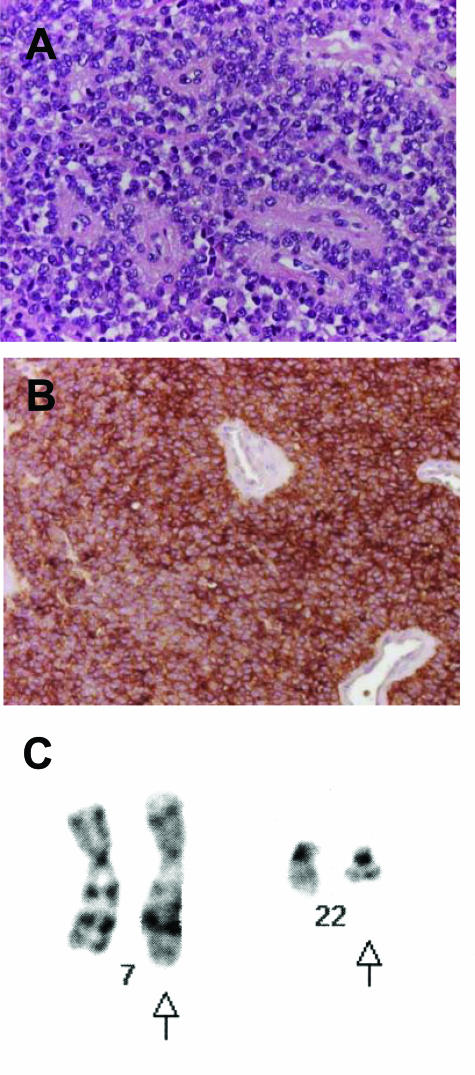
Patient A was an 8-year-old male child who had a history of trauma to his leg 1.5 years before presenting with a parosteal soft tissue tumor. The nature of the trauma was unknown, but plain films at that time failed to show any lesion. He presented with swelling in the region of trauma, and an magnetic resonance imaging showed a mass near the dorsal aspect of the left fibula. There were no other sites of disease. A resection was performed, showing a primitive tumor with small round cell morphology and diffuse positivity for CD99 by immunohistochemistry (Figure 1, A and B). The tumor karyotype was interpreted as 46,XY,t(7;22)(p15;q11) (Figure 1C). This suggested the presence of the EWS-ETV1 fusion, and this was confirmed by RT-PCR followed by direct sequencing. The fusion structure was identical to that originally reported, ie, a fusion of EWS exon 7 to the same ETV1 exon (numbered 10 or 11 based on two alternative splice forms of ETV1).12 He was treated on the CCG 7942 protocol. At 3.5 years after diagnosis, he developed pain in his right upper extremity and on plain film had a lytic lesion in the proximal humerus. On biopsy, this was found to be an osteosarcoma histologically unrelated to the previous small round cell tumor. By immunohistochemistry, the osteosarcoma showed a different staining pattern for CD99, with only scattered round and spindled cells expressing this marker. This osteosarcoma was not near the radiation field for the first tumor. He then received chemotherapy for osteosarcoma and had a limb salvage procedure with insertion of an allograft. Imaging studies of his extremities and chest do not show recurrent or metastatic disease at 5 years after initial EFT diagnosis.
Figure 1.
Patient A. A: H&E-stained section showing characteristic small round blue cell tumor morphology. B: Diffuse and strong positivity for CD99 (O13 antibody). C: Partial karyotype showing a t(7;22) translocation. Arrows indicate the derivative chromosomes.
Patient B was a 2-year-old female child who developed fever and acute right groin and right lower quadrant abdominal pain. Her mother discovered groin lumps that were mildly mobile and hard. A magnetic resonance imaging revealed a large soft tissue mass in the proximal thigh measuring 6 cm in greatest dimension and an intrapelvic mass measuring 8 cm in greatest dimension. Her initial extent of disease evaluation also showed several pulmonary metastases. A biopsy of the pelvic mass revealed a high-grade round cell sarcoma that was focally reactive for CD99 and negative for lineage markers (Table 2), and the patient was treated with five-drug chemotherapy consisting of alternating cycles of vincristine/doxorubicin/cyclophosphamide and ifosfamide/etoposide. She received 3600 cGy of radiation therapy followed by complete surgical resection of her primary tumor in the upper thigh and pelvis. The excised tumor showed spindle cell morphology (Figure 2), which was deemed to be a therapy-related change. The karyotype reportedly showed t(7;22) but further cytogenetic details were not available. RT-PCR showed the EWS-ETV1 fusion transcript. The fusion structure was identical to that originally reported,12 as described for patient A above. One year after completion of her therapy, she presented with local-regional recurrence of the tumor in her right pelvis. At that time, she underwent resection of the tumor. Several months later she began having significant progression of her pulmonary disease. Multiple types of salvage chemotherapy were given with partial responses. She died of progressive disease 5 years after diagnosis.
Figure 2.
Patient B. Undifferentiated tumor showing areas of spindling in a specimen obtained after neoadjuvant chemotherapy.
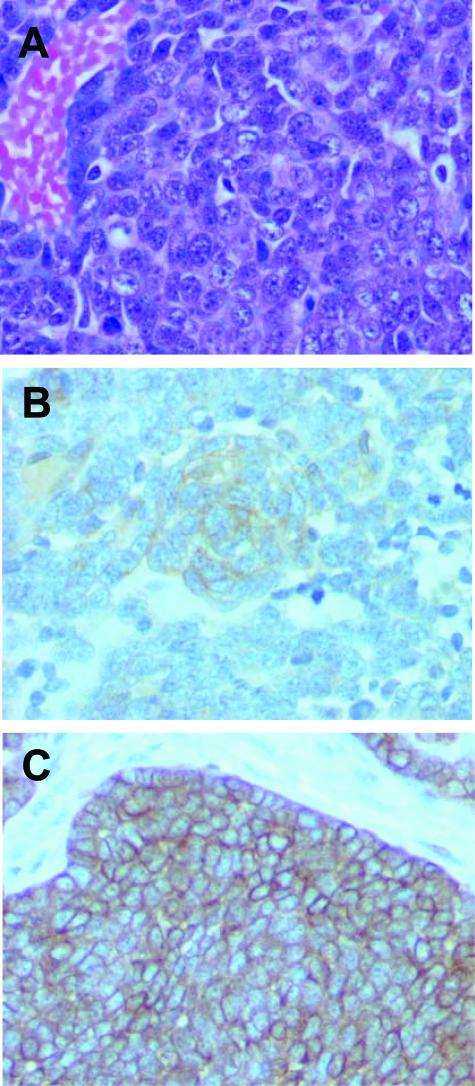
Patient C was a 40-year-old man who presented with lower abdominal pain radiating into the extremity. Imaging studies revealed a retroperitoneal and pelvic mass without skeletal involvement. A biopsy of the mass showed a poorly differentiated malignant tumor diffusely positive for CD99 and with dot-like positivity for CAM5.2 (Figure 3, A–C). The differential diagnosis was between an EFT and a high-grade neuroendocrine carcinoma. RT-PCR analysis of the fresh-frozen tumor tissue using the EWS exon 7 forward primer and the ETS1 reverse primer revealed the EWS-FEV fusion, confirming a diagnosis of an EFT. The fusion joined EWS exon 7 to FEV exon 2, as described in the original report of this fusion.15 No tissue was submitted for cytogenetics. Radiation to the primary site was given in 25 fractions to a dose of 45 Gy, along with chemotherapy (vincristine, doxorubicin, dexrazoxane, cyclophosphamide). Surgical exploration of the primary site was then performed and showed no viable tumor in extensive sampling of the residual unresectable masses. At 3.5 years after original diagnosis, he relapsed with metastatic disease in his left lung and left hilar lymph nodes and is now on second line therapy. The morphology and immunophenotype of the metastatic tumor in the lung was similar to that of the primary. In addition to strong positivity for CD99, some reactivity with AE1/AE3 antibody was also noted.
Figure 3.
Patient C. A: H&E-stained section showing small round blue cell tumor with crush artifact. B: Immunostain for CD99 showing diffuse and strong staining. C: CAM5.2 immunostain showing dot-like positivity for cytokeratin in the tumor cells.
Patient D: Identification of a Novel EWS-SP3 Fusion in an Extraskeletal SRCT
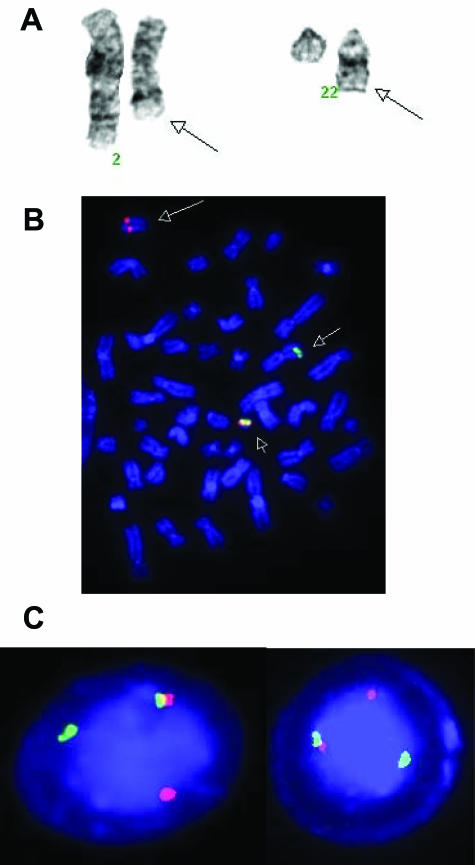
Patient D was a 16-year-old Caucasian male who presented with progressive enlargement of a right forehead mass. The initial magnetic resonance imaging and a computerized automated tomography (CT) scan revealed a heterogeneous extra-axial, extradural mass in the right frontal region measuring 5 × 3.8 × 5 cm3. In addition to the right frontotemporal mass, abnormal radiotracer accumulation was also identified in the right femoral, left clavicular, and left proximal ulnar regions by bone scan. CT scan revealed seven pulmonary nodules bilaterally in the lower lobes and a large mass in the right kidney. An excisional biopsy of the extradural mass was performed. The histopathology showed an undifferentiated small round cell tumor. Immunohistochemistry studies did not clarify the diagnosis, only showing strong positivity for BCL2 and weak and focal expression of CD99, NSE, and neurofilament (Figure 4, A–C; and see Table 3 for details). Cytogenetic analysis revealed a t(2;22)(q31;q12) (Figure 5A). Fluorescence in situ hybridization (FISH) performed with dual-color, break-apart probes flanking the EWS gene (Abbott-Vysis, Downer’s Grove, IL) showed a split in the EWS gene locus and a simple translocation of EWS gene material to 2q31 (Figure 5, B and C).
Figure 4.
Patient D. A: H&E-stained section showing small round blue cell tumor with high mitotic rate. B: Immunostain for CD99 showing focal weak staining. C: BCL2 immunostain showing strong diffuse positivity in the tumor cells.
Figure 5.
Patient D. A: Partial karyotype showing t(2;22)(q31;q12). Arrows indicate the derivative chromosomes. B: FISH using dual-color, break-apart EWS probe on a metaphase preparation. The small arrow shows the normal fusion signal (red and green signals together) on the intact chromosome 22; the large arrow indicates the der(22)t(2;22) with only red signal (EWS centromeric probe); the midsize arrow indicates the der(2)t(2;22) with the green signal (EWS telomeric probe). C: Same FISH on interphase nuclei showing one normal fusion signal and separation of the other red and green signals.
The patient was initially treated with alternating cycles of cyclophosphamide/doxorubicin/vincristine, and ifosfamide/etoposide. Because of progressive disease, he was placed on a more intensive high-dose ifosfamide/etoposide regimen with partial response. A radical nephroureterectomy was performed. He then underwent an autologous peripheral stem cells transplant as consolidation therapy. Approximately 3 months after the transplant, he relapsed with multiple bony metastases. Despite several cycles of irinotecan, the bone lesions progressed. The patient was then treated with palliative radiation therapy and palliative chemotherapy (oral etoposide). He died ∼20 months from initial diagnosis.
To address the hypothesis that a novel gene might be fused to EWS, 3′ RACE was performed using 3′ RACE kit (Invitrogen) with nested EWS forward primers (EWS22.14 for the first PCR step, EWS22.3 for the nested PCR step). Cloning and sequencing of the 3′ RACE products revealed two aberrant transcripts, both of which represented chimeric mRNAs encoding an in-frame transcript encoding a fusion of the EWS amino-terminal domain (NTD) to the zinc finger DNA-binding domain of SP3. Because the SP3 gene is located at 2q31, this was consistent with the cytogenetic and FISH data in this case. Of the two cloned chimeric EWS-SP3 transcripts, the shorter one (designated type I fusion product) represented an in-frame fusion of EWS exon 7 to nucleotide 48 of SP3 exon 6 (Figure 6A). This exon encodes the first zinc finger of SP3. The second, longer chimeric EWS-SP3 transcript (designated type II fusion product) showed an insertion of a 31-bp sequence between EWS exon 8 and nucleotide 26 of SP3 exon 6; this insertion resulted in restoration of the reading frame that would otherwise have been lost in a direct fusion between EWS exon 8 (end phase = 2) and SP3 exon 6 (start phase = 1) (Figure 6B). Sequence homology searches revealed that this additional 31-bp sequence, except the last T, is located within EWS intron 8, suggesting that it was functioning as a cryptic exon in this tumor. The extra T at the fusion point of this chimeric transcript may be attributable to error-prone nonhomologous end joining. Interestingly, the two alternative junction points within SP3 exon 6 were both at cryptic splice acceptor sites (Figure 6C). Both EWS-SP3 fusion transcripts were then confirmed by RT-PCR using EWS forward and SP3 reverse primers (Figure 7). RT-PCR assays consistently showed that the shorter fusion transcript was more prevalent. The predicted protein structure of the EWS-SP3 fusion product is schematized in Figure 8.
Figure 6.
Patient D. A: Partial sequence of the type I EWS-SP3 fusion transcript is an in-frame junction of EWS exon 7 with SP3 exon 6 (see Results for details). The translated protein sequence is shown below, with the codon at the junction in red. B: Partial sequence of the type II EWS-SP3 transcript. The transcript has a full open reading frame and is a fusion of EWS exon 8 to SP3 exon 6 with an intervening cryptic exon. The arrowhead indicates the extra T (see Results for details). The translated protein sequence is shown below, with the codons at the junctions in red. C: Partial genomic sequence of SP3. The intron sequence is in lower case and the exon sequence is in upper case. The portion of SP3 exon 6 coding for the zinc finger is underlined. The two cryptic splice acceptor sites in SP3 exon 6 are shown in green (used in type I fusion transcript) and red (used type II fusion transcript), respectively.
Figure 7.
Patient D: RT-PCR detection of EWS-SP3 fusion. The presence of both type I and type II fusion transcripts was confirmed by RT-PCR using an EWS primer (sequence: 5′-CTGGATCCTACAGCCAAGCTCCAAGTC-3′) and an SP3 reverse primer (sequence: 5′-CAGAATGCCAACGCAGATGA-3′). Lane ptD is the RT-PCR product obtained from tumor RNA. Band A is 120 bp, and band B is 375 bp. Sequencing of A and B confirmed that A represents the type I EWS-SP3 fusion transcript, whereas B represents the type II fusion, both of which are in-frame (see Figure 6). Sequencing of band C revealed an out-of-frame EWS-SP3 fusion transcript (sequence data not shown). Lane neg is the no RNA negative control. Size marker (lane M) is φ X174/HaeIII (Invitrogen).
Figure 8.
Schematic diagrams of EWS, SP3, and the predicted EWS-SP3 protein. NTD, amino-terminal domain; AD, transactivation domain; ID, inhibitory domain; zinc finger, zinc finger DNA-binding domain.
Although we did not clone the genomic junctions of the EWS-SP3 fusion, the structure of the type II fusion product indicates that the genomic EWS breakpoint in this tumor is most likely in intron 8. Presumably, most chimeric EWS-SP3 transcripts underwent splicing that removed EWS exon 8 to produce the in-frame type I fusion product that predominated in our case. Splicing out of EWS exon 8 is also observed in EWS-FLI1 rearrangements when the genomic break is in EWS intron 8.27 As for the SP3 gene, the genomic break is most likely in intron 5, or is at least upstream of nucleotide 26 of SP3 exon 6 (based on the type I fusion product).
Discussion
Based on the patterns of associations between different translocation partners in sarcomas, the notion emerged that variant gene fusions were linked by strong similarities in the genes involved and that this in turn could be a defining feature of these specific entities. Thus, EFTs show fusions of EWS to related members of the ETS family of transcription factors (FLI1, ERG, ETV1, E1AF, and FEV). Likewise, alveolar rhabdomyosarcomas contain fusions of FKHR to members of the PAX gene family (PAX3, PAX7), clear cell sarcomas contain fusions of EWS to the CREB transcription factor family (ATF1, CREB1), in extraskeletal myxoid chondrosarcoma the TEC transcription factor is fused to a member of the TET family of RNA-binding proteins (EWS, FUS, and TAFII68), and so on.11 This has been a useful conceptual framework because tumors with related variant fusions are invariably more similar to each other than to tumors with completely different fusions. Some of the more common variant fusions have been frequent enough to allow comparisons of clinical features, including survival, within a given sarcoma entity according to fusion type. Our recent experience with three additional cases with rare variant fusions of EWS to ETS family transcription factor genes reported here prompted us to review the cumulated experience with these cases (summarized in Table 2) to begin to define their clinical and pathological phenotype.
EFTs are considered to affect primarily children and young adults, and this also holds true for EFTs with the rare variant translocations (EWS-ETV1, EWS-E1AF, EWS-FEV). Based on the data summarized in Table 2, the EWS-ETV1 and EWS-FEV fusions may be more common than the EWS-E1AF fusion. In the current study, analyzing all of the reported cases of rare variant translocations confirmed at the molecular level, the age group ranges from newborn to 40 years of age. However, 83% (10 of 12) of patients were <18 years of age at diagnosis. The two older patients had the EWS-FEV fusion. One recurrent feature of tumors with these rare variant translocations appears to be their extraskeletal location. The location in our patient A was described as adjacent to the left fibula; however, gross and microscopic examination revealed a predominantly soft tissue mass around the bone with focal periosteal involvement but no evidence of cortical bone involvement. Overall, the proportion of extraskeletal primaries is 11 of 12 cases (92%). Most cases showed a small round cell tumor on histological examination. The tumor from our patient B (EWS-ETV1 fusion transcript) showed spindle cell areas, which is unusual for EFT, but this specimen was obtained after the patient received high-dose chemotherapy. Therefore, this may reflect a therapy-related change. One case with the EWS-FEV fusion transcript (case 8 in Table 2) showed large polygonal cells with clear cell features; however, this phenotype is not uncommonly seen in EFT containing abundant glycogen. Another case of EWS-FEV fusion (case 9 in Table 2) showed a predominantly small round cell tumor with foci of large pleomorphic cells. Like most EFTs, 9 of 10 cases with immunohistochemical data for CD99/MIC2 showed positive staining, which was only focal in one case. The majority of the tumors were also positive for vimentin and variably positive for neuroendocrine markers. Other immunohistochemical data are summarized in Table 2. Clinical outcomes were quite variable, with some patients presenting with metastatic disease and progressing rapidly, whereas others have shown extended disease-free survival. Other genetic factors may account for occasional aggressive behavior. For instance, a patient whose tumor contained the EWS-FEV fusion (case 9 in Table 2) died 16 months after diagnosis, 12 months after surgery despite two courses of adjuvant chemotherapy. This case showed a p53 point mutation, which is known to have a strong negative impact on chemotherapeutic response and survival in EFT.28
Studies in NIH3T3 cells have demonstrated some functional differences between the five known EWS-ETS fusions. Introduction of EWS-FLI, EWS-ERG, or EWS-FEV into NIH3T3 leads to anchorage-independent growth, whereas EWS-ETV1 and EWS-E1AF do not.29 Intriguingly, these associations parallel the structural similarities in these ETS family members; FLI, ERG, and FEV are more closely related to each other than to ETV1 and E1AF.
We also report for the first time a sarcoma with an EWS-SP3 fusion, making this at least the 12th EWS fusion observed in this group of cancers and the first involvement of a gene encoding a SP family zinc finger protein in a cancer-associated gene fusion. Similar to all previously described fusions involving EWS, the fusion transcript encodes a predicted chimeric protein consisting of the EWS amino-terminal domain known to function as a transactivation domain in the context of EWS fusion proteins11 fused to a DNA-binding domain, in this instance the zinc finger domain from the carboxy-terminal portion of SP3 (Figure 8).
SP3 is a member of the mammalian SP transcription factor family,30 which is ubiquitously expressed along with SP1 and is indistinguishable from SP1 in its DNA binding properties.31 Functionally, SP3 seems to be a bifunctional transcription factor, ie, it can either inhibit or activate target gene expression.32 SP family members have similar domain structures, consisting of three zinc fingers close to the C terminus and glutamine-rich activation domains in their N-terminal region.30 The last two zinc fingers are conserved classical zinc fingers, whereas the first zinc finger is less conserved. SP3 also possesses an inhibitory domain (ID) between the second glutamine-rich domain and the first zinc finger (Figure 8).30 The inhibitory domain of SP3 acts as an independent module in cis-. It can be joined to other activation domains causing them to lose their activation properties.32 The amino acid triplet KEE within its inhibitory domain has been demonstrated to be essential for repressor function of SP3.32 Splicing and alternate use of open reading frames has been postulated to affect SP3 function.33 Thus, one can speculate that a transcript that excludes the exon coding for the inhibitory domain would deprive SP3 of repressor function. In the case presented here, the chimeric protein included the EWS N-terminal domain and the SP3 zinc finger DNA-binding domain but not the inhibitory domain of SP3 (Figure 8).
It is notable that this feature of exclusion of an inhibitory domain is also observed in the two other sarcoma fusions involving EWS and a zinc finger gene, EWS-WT1 and EWS-ZNF278. WT1, the first zinc finger protein involved in a fusion with EWS,19 also acts as a transcriptional activator or repressor depending on the cellular or chromosomal context.34 WT1 protein contains a domain N-terminal to the zinc finger region, which functions as a strong transcriptional repressor.35 ZNF278 contains a POZ transcriptional repressor domain at the N terminus.18 Neither WT1’s repressor domain nor ZNF278’s POZ domain are included in their respective EWS fusion proteins. This suggests that part of the biology of these EWS-zinc finger gene fusions may involve disruption of gene expression patterns normally regulated by the alternative activator and repressor functions of these transcription factors. The clinicopathological features of these EFT-like tumors with noncanonical EWS fusions, including EWS-ZNF278, EWS-SP3, and EWS-POU5F1, are summarized in Table 3.
The existence of a growing number of rare EWS variant translocations complicates the molecular diagnosis of this group of undifferentiated small round cell sarcomas, whether by RT-PCR or FISH. To achieve a nearly definitive molecular diagnosis, the RT-PCR-based approach requires the use of multiple specific or consensus reverse primers, usually in a stepwise manner proceeding from common fusions to rare ones. As the numbers of different possible fusions rise, the RT-PCR approach is increasingly labor-intensive, making it more pressing to develop comprehensive diagnostic strategies, possibly chip-based. This may well be a continuing process because there also likely remain additional rare uncloned variant EWS fusions in these tumors, based on cytogenetic reports of cases with unusual translocations involving 22q12.36,37,38 The FISH approach based on EWS break-apart probes is comprehensive in terms of the detection of EWS rearrangements but does not identify the translocation partner and, therefore, may not provide clarification of cases with ambiguous pathological features.
Finally, to complicate further molecular diagnosis and our biological concepts of this tumor group, there are also isolated reports of EFT-like small round cell sarcomas that do not contain EWS fusions (Table 4). The recent report of a recurrent FUS-ERG gene fusion in four cases of EFT, three of which involved the chest wall,39 suggests that even a negative EWS FISH may not be sufficient to definitively exclude EFT. The existence of these FUS-ERG fusions suggests that FUS fusions with other EWS partners in EFTs may also be possible. Beyond EWS and FUS fusions, there are rare EFT-like cases with seemingly completely biologically unrelated fusions. Specifically, there are two reported cases of soft tissue sarcoma diagnosed as Ewing-like sarcoma, both with a t(4;19)(q35;q13), encoding a novel CIC-DUX4 fusion joining the CIC high-mobility group box transcription factor gene to the double-homeodomain gene DUX4.40 In addition, there has been a report of an EFT-like sarcoma harboring the BRD4-NUT fusion originally described in aggressive midline carcinomas of the chest.41 These rare non-EWS EFT-like cases are summarized in Table 4. Overall, these cases, together with the present data, highlight the growing complexity of sarcoma molecular diagnostics. Understanding the diverse or shared mechanisms underlying transformation by these distinct but often related fusions should help to clarify the biology of these aggressive cancers.
Table 4.
EFT-Like Tumors with Non-EWS Fusions
| Case no. (reference) | Age/ sex | Primary site | Histology |
|---|---|---|---|
| No. 1 (39) | 9/M | Chest wall; no data on metastatic disease | SRCT, high mitotic rate |
| No. 2 (39) | 7/F | Chest wall; no data on metastatic disease | SRCT, high mitotic rate |
| No. 3 (39) | 15/F | Chest wall; no data on metastatic disease | SRCT, high mitotic rate, pleomorphism |
| No. 4 (39) | 21/M | Femur; no data on metastatic disease | SRCT |
| No. 5 (40) | 62/F | Pelvic soft tissue mass; no metastatic disease | SRCT, glycogen granules |
| No. 6 (40) | 31/M | Shoulder soft tissue; no metastatic disease | SRCT, glycogen granules |
| No. 7 (41) | 10/M | Iliac bone; no metastatic disease | SRCT, high mitotic rate |
For abbreviations, see Table 2.
(table continues)
Table 4.
Continued
| IHC | Fusion type and cytogenetic translocation | Treatment | Outcome |
|---|---|---|---|
| + CD99, vimentin, neural markers, myogenic, lymphoid, myeloid markers | FUS-ERG t(16;21;22)(p11;q22;p11.2) | NA | DOD at 2 years |
| + CD99 (focal weak), neural, myogenic, lymphoid, myeloid markers | FUS-ERG t(16;21)(p11.2;q22.3) | NA | NA |
| + CD99, neural, myogenic, lymphoid, myeloid markers | FUS-ERG der(21)t(16;21)(p11;q22) | NA | NA |
| + CD99, other myogenic, neural, lymphoid, myeloid markers | FUS-ERG t(16;21)(p11;q22) | NA | NA |
| + CD99 (weak), vimentin | CIC-DUX4 t(4;19)(q35;q13) | No curative surgery. Palliative RT and CT. | DOD at 10 months |
| + CD99 (weak), vimentin | CIC-DUX4 t(4;19)(q35;q13) | Neoadjuvant RT and CT (adriamycin, cisplatin). Resection. | NED at 30 months |
| + CD99 (weak), CG, SYN, CD45, NSE, S100, CAM5.2, AE1/AE3, PLAP, AFP, OCT3/4, CD34 | BRD4-NUT t(15;19)(q13;p13) | No resection. RT and CT (vincristine, doxorubicin, ifosfamide, cisplatin) | NED at 13 years |
Acknowledgments
We thank Drs. N. Shukla, R. Kobos, S.W. Choi, and R.J. Mody for clinical follow-up data.
Footnotes
Supported in part by the Ewing’s Sarcoma Research Fund (M.L.).
L.W. and R.B. contributed equally to this study.
References
- Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G, Aurias A, Thomas G. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- Zucman J, Melot T, Desmaze C, Ghysdael J, Plougastel B, Peter M, Zucker JM, Triche TJ, Sheer D, Turc-Carel C, Ambros P, Combaret V, Lenoir G, Aurias A, Thomas G, Delattre O. Combinatorial generation of variable fusion proteins in the Ewing family of tumours. EMBO J. 1993;12:4481–4487. doi: 10.1002/j.1460-2075.1993.tb06137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre O, Zucman J, Melot T, Garau XS, Zucker JM, Lenoir GM, Ambros PF, Sheer D, Turc-Carel C, Triche TJ, Aurias A, Thomas G. The Ewing family of tumors—a subgroup of small-round-cell tumors defined by specific chimeric transcripts. N Engl J Med. 1994;331:294–299. doi: 10.1056/NEJM199408043310503. [DOI] [PubMed] [Google Scholar]
- Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- Sorensen PHB, Lessnick SL, Lopez-Terrada D, Liu XF, Triche TJ, Denny CT. A second Ewing’s sarcoma translocation, t(21;22), fuses the EWS gene to another ets-family transcription factor, ERG. Nat Genet. 1994;6:146–151. doi: 10.1038/ng0294-146. [DOI] [PubMed] [Google Scholar]
- May WA, Lessnick SL, Braun BS, Klemsz M, Lewis BC, Lunsford LB, Hromas R, Denny CT. The Ewing’s sarcoma EWS/FLI-1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI-1. Mol Cell Biol. 1993;13:7393–7398. doi: 10.1128/mcb.13.12.7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman P, Panagopoulos I, Lassen C, Fioretos T, Mencinger M, Toresson H, Hoglund M, Forster A, Rabbitts TH, Ron D, Mandahl N, Mitelman F. Expression patterns of the human sarcoma-associated genes FUS and EWS and the genomic structure of FUS. Genomics. 1996;37:1–8. doi: 10.1006/geno.1996.0513. [DOI] [PubMed] [Google Scholar]
- Plougastel B, Zucman J, Peter M, Thomas G, Delattre O. Genomic structure of the EWS gene and its relationship to EWSR1, a site of tumor-associated chromosome translocation. Genomics. 1993;18:609–615. doi: 10.1016/s0888-7543(05)80363-5. [DOI] [PubMed] [Google Scholar]
- Truong AH, Ben David Y. The role of Fli-1 in normal cell function and malignant transformation. Oncogene. 2000;19:6482–6489. doi: 10.1038/sj.onc.1204042. [DOI] [PubMed] [Google Scholar]
- Ladanyi M. EWS-FLI1 and Ewing’s sarcoma: recent molecular data and new insights. Cancer Biol Ther. 2002;1:330–336. [PubMed] [Google Scholar]
- Xia SJ, Barr FG. Chromosome translocations in sarcomas and the emergence of oncogenic transcription factors. Eur J Cancer. 2005;41:2513–2527. doi: 10.1016/j.ejca.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Jeon IS, Davis JN, Braun BS, Sublett JE, Roussel MF, Denny CT, Shapiro DN. A variant Ewing’s sarcoma translocation (7;22) fuses the EWS gene to the ETS gene ETV1. Oncogene. 1995;10:1229–1234. [PubMed] [Google Scholar]
- Kaneko Y, Yoshida K, Handa M, Toyoda Y, Nishihira H, Tanaka Y, Sasaki Y, Ishida S, Higashino F, Fujinaga K. Fusion of an ETS-family gene, E1AF, to EWS by t(17;22)(q12;q12) chromosome translocation in an undifferentiated sarcoma of infancy. Genes Chromosom Cancer. 1996;15:115–121. doi: 10.1002/(SICI)1098-2264(199602)15:2<115::AID-GCC6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Urano F, Umezawa A, Hong W, Kikuchi H, Hata J. A novel chimera gene between EWS and E1A-F, encoding the adenovirus E1A enhancer-binding protein, in extraosseous Ewing’s sarcoma. Biochem Biophys Res Commun. 1996;219:608–612. doi: 10.1006/bbrc.1996.0281. [DOI] [PubMed] [Google Scholar]
- Peter M, Couturier J, Pacquement H, Michon J, Thomas G, Magdelenat H, Delattre O. A new member of the ETS family fused to EWS in Ewing tumors. Oncogene. 1997;14:1159–1164. doi: 10.1038/sj.onc.1200933. [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Nafa K, Segal NH, Dal Cin P, Ladanyi M. EWS-CREB1: A recurrent variant fusion in clear cell sarcoma. Association with gastrointestinal location and absence of melanocytic differentiation. Clin Cancer Res. 2006;12:5356–5362. doi: 10.1158/1078-0432.CCR-05-2811. [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Dal Cin P, Nafa K, Teot L, Surti U, Fletcher CDM, Ladanyi M: EWS-CREB1 is the predominant gene fusion in angiomatoid fibrous histiocytoma. Genes Chromosome Cancer, (in press) [DOI] [PubMed] [Google Scholar]
- Mastrangelo T, Modena P, Tornielli S, Bullrich F, Testi MA, Mezzelani A, Radice P, Azzarelli A, Pilotti S, Croce CM, Pierotti MA, Sozzi G. A novel zinc finger gene is fused to EWS in small round cell tumor. Oncogene. 2000;19:3799–3804. doi: 10.1038/sj.onc.1203762. [DOI] [PubMed] [Google Scholar]
- Ladanyi M, Gerald W. Fusion of the EWS and WT1 genes in the desmoplastic small round cell tumor. Cancer Res. 1994;54:2837–2840. [PubMed] [Google Scholar]
- Gerald WL, Rosai J, Ladanyi M. Characterization of the genomic breakpoint and chimeric transcripts in the EWS-WT1 gene fusion of desmoplastic small round cell tumor. Proc Natl Acad Sci USA. 1995;92:1028–1032. doi: 10.1073/pnas.92.4.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerald WL, Ladanyi M, de Alava E, Cuatrecasas M, Kushner BH, La Quaglia MP, Rosai J. Clinical, pathologic, and molecular spectrum of tumors associated with t(11;22)(p13;q12): desmoplastic small round-cell tumor and its variants. J Clin Oncol. 1998;16:3028–3036. doi: 10.1200/JCO.1998.16.9.3028. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Yamazaki Y, Ishikawa Y, Kawaguchi N, Mukai H, Nakamura T. EWSR1 is fused to POU5F1 in a bone tumor with translocation t(6;22)(p21;q12). Genes Chromosom Cancer. 2005;43:217–222. doi: 10.1002/gcc.20171. [DOI] [PubMed] [Google Scholar]
- Ginsberg JP, de Alava E, Ladanyi M, Wexler L, Kovar H, Paulussen M, Zoubek A, Dockhorn-Dworniczak BH, Juergens H, Wunder JS, Andrulis IL, Malik R, Sorensen PHB, Womer RB, Barr FG. EWS-FLI1 and EWS-ERG gene fusions are associated with similar clinical phenotypes in Ewing’s sarcoma. J Clin Oncol. 1999;17:1809–1814. doi: 10.1200/JCO.1999.17.6.1809. [DOI] [PubMed] [Google Scholar]
- Zoubek A, Dockhorn-Dworniczak B, Delattre O, Christiansen H, Niggli F, Gatterer-Menz I, Smith TL, Jurgens H, Gadner H, Kovar H. Does expression of different EWS chimeric transcripts define clinically distinct risk groups of Ewing tumor patients? J Clin Oncol. 1996;14:1245–1251. doi: 10.1200/JCO.1996.14.4.1245. [DOI] [PubMed] [Google Scholar]
- de Alava E, Kawai A, Healey JH, Fligman I, Meyers P, Huvos AG, Gerald WL, Jhanwar SC, Argani P, Antonescu CR, Pardo-Mindan FJ, Ginsberg J, Womer R, Lawlor ER, Wunder J, Andrulis I, Sorensen PHB, Barr FG, Ladanyi M. EWS-FLI1 fusion transcript structure is an independent determinant of prognosis in Ewing’s sarcoma. J Clin Oncol. 1998;16:1248–1255. doi: 10.1200/JCO.1998.16.4.1248. [DOI] [PubMed] [Google Scholar]
- Aryee DN, Sommergruber W, Muehlbacher K, Dockhorn-Dworniczak B, Zoubek A, Kovar H. Variability in gene expression patterns of Ewing tumor cell lines differing in EWS-FLI1 fusion type. Lab Invest. 2000;80:1833–1844. doi: 10.1038/labinvest.3780194. [DOI] [PubMed] [Google Scholar]
- Zucman-Rossi J, Legoix P, Victor JM, Lopez B, Thomas G. Chromosome translocation based on illegitimate recombination in human tumors. Proc Natl Acad Sci USA. 1998;95:11786–11791. doi: 10.1073/pnas.95.20.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HY, Illei PB, Zhao Z, Mazumdar M, Healey JH, Huvos AG, Wexler LH, Gorlick R, Meyers PA, Ladanyi M. Ewing sarcomas with p53 mutations or p16/p14ARF homozygous deletions: a highly aggressive subset associated with poor chemoresponse. J Clin Oncol. 2005;23:548–558. doi: 10.1200/JCO.2005.02.081. [DOI] [PubMed] [Google Scholar]
- Braunreiter CL, Hancock JD, Coffin CM, Boucher KM, Lessnick SL. Expression of EWS-ETS fusions in NIH3T3 cells reveals significant differences to Ewing’s sarcoma. Cell Cycle. 2006;5:2753–2759. doi: 10.4161/cc.5.23.3505. [DOI] [PubMed] [Google Scholar]
- Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- Hagen G, Muller S, Beato M, Suske G. Cloning by recognition site screening of two novel GT box binding proteins: a family of Sp1 related genes. Nucleic Acids Res. 1992;20:5519–5525. doi: 10.1093/nar/20.21.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennig J, Beato M, Suske G. An inhibitor domain in Sp3 regulates its glutamine-rich activation domains. EMBO J. 1996;15:5659–5667. [PMC free article] [PubMed] [Google Scholar]
- Moran KM, Crusio RH, Chan CH, Grekova MC, Richert JR. Human transcription factor Sp3: genomic structure, identification of a processed pseudogene, and transcript analysis. Gene. 2004;341:235–247. doi: 10.1016/j.gene.2004.06.055. [DOI] [PubMed] [Google Scholar]
- Reddy JC, Licht JD. The WT1 Wilms’ tumor suppressor gene: how much do we really know? Biochim Biophys Acta. 1996;1287:1–28. doi: 10.1016/0304-419x(95)00014-7. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Qiu QQ, Huang J, Gurrieri M, Deuel TF. Products of alternatively spliced transcripts of the Wilms’ tumor suppressor gene, wt1, have altered DNA binding specificity and regulate transcription in different ways. Oncogene. 1995;10:415–422. [PubMed] [Google Scholar]
- Davison JM, Morgan TW, Hsi BL, Xiao S, Fletcher JA. Subtracted, unique-sequence, in situ hybridization; experimental and diagnostic applications. Am J Pathol. 1998;153:1401–1409. doi: 10.1016/S0002-9440(10)65727-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Hayes K, Van FT, Glassman A. Detection of a novel reciprocal t(16;22)(q11.2;q12) in a Ewing sarcoma. Cancer Genet Cytogenet. 2003;140:55–57. doi: 10.1016/s0165-4608(02)00637-4. [DOI] [PubMed] [Google Scholar]
- Szuhai K, Ijszenga M, Tanke HJ, Taminiau AH, de Schepper A, van Duinen SG, Rosenberg C, Hogendoorn PC. Detection and molecular cytogenetic characterization of a novel ring chromosome in a histological variant of Ewing sarcoma. Cancer Genet Cytogenet. 2007;172:12–22. doi: 10.1016/j.cancergencyto.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Shing DC, McMullan DJ, Roberts P, Smith K, Chin SF, Nicholson J, Tillman RM, Ramani P, Cullinane C, Coleman N. FUS/ERG gene fusions in Ewing’s tumors. Cancer Res. 2003;63:4568–4576. [PubMed] [Google Scholar]
- Kawamura-Saito M, Yamazaki Y, Kaneko K, Kawaguchi N, Kanda H, Mukai H, Gotoh T, Motoi T, Fukayama M, Aburatani H, Takizawa T, Nakamura T. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum Mol Genet. 2006;15:2125–2137. doi: 10.1093/hmg/ddl136. [DOI] [PubMed] [Google Scholar]
- Mertens F, Wiebe T, Adlercreutz C, Mandahl N, French CA: Successful treatment of a child with t(15;19)-positive tumor. Pediatr Blood Cancer 2006, 10.1002/pbc. 20755 [DOI] [PubMed] [Google Scholar]
- Smith LM, Adams RH, Brothman AR, Vanderhooft SL, Coffin CM. Peripheral primitive neuroectodermal tumor presenting with diffuse cutaneous involvement and 7;22 translocation. Med Pediatr Oncol. 1998;30:357–363. doi: 10.1002/(sici)1096-911x(199806)30:6<357::aid-mpo10>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Lewis TB, Coffin CM, Bernard PS. Differentiating Ewing’s sarcoma from other round blue cell tumors using a RT-PCR translocation panel on formalin-fixed paraffin-embedded tissues. Mod Pathol. 2007;20:397–404. doi: 10.1038/modpathol.3800755. [DOI] [PubMed] [Google Scholar]
- Squire J, Zielenska M, Thorner P, Tennyson S, Weitzman S, Pai KM, Yeger H, Ng YK, Weksberg R. Variant translocations of chromosome 22 in Ewing’s sarcoma. Genes Chromosom Cancer. 1993;8:190–194. doi: 10.1002/gcc.2870080309. [DOI] [PubMed] [Google Scholar]
- Llombart-Bosch A, Pellin A, Carda C, Noguera R, Navarro S, Peydro-Olaya A. Soft tissue Ewing sarcoma—peripheral primitive neuroectodermal tumor with atypical clear cell pattern shows a new type of EWS-FEV fusion transcript. Diagn Mol Pathol. 2000;9:137–144. doi: 10.1097/00019606-200009000-00003. [DOI] [PubMed] [Google Scholar]
- Navarro S, Noguera R, Pellin A, Lopez-Guerrero JA, Rosello-Sastre E, Cremades A, Llombart-Bosch A. Atypical pleomorphic extraosseous Ewing tumor/peripheral primitive neuroectodermal tumor with unusual phenotypic/genotypic profile. Diagn Mol Pathol. 2002;11:9–15. doi: 10.1097/00019606-200203000-00003. [DOI] [PubMed] [Google Scholar]
- Urano F, Umezawa A, Yabe H, Hong W, Yoshida K, Fujinaga K, Hata J. Molecular analysis of Ewing’s sarcoma: another fusion gene, EWS-E1AF, available for diagnosis. Jpn J Cancer Res. 1998;89:703–711. doi: 10.1111/j.1349-7006.1998.tb03274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]