Abstract
The cylindrical chaperonin GroEL and its cofactor GroES mediate ATP-dependent protein folding in Escherichia coli. Recent studies in vitro demonstrated that GroES binding to GroEL causes the displacement of unfolded polypeptide into the central volume of the GroEL cavity for folding in a sequestrated environment. Resulting native protein leaves GroEL upon GroES release, whereas incompletely folded polypeptide can be recaptured for structural rearrangement followed by another folding trial. Additionally, each cycle of GroES binding and dissociation is associated with the release of nonnative polypeptide into the bulk solution. Here we show that this loss of substrate from GroEL is prevented when the folding reaction is carried out in the presence of macromolecular crowding agents, such as Ficoll and dextran, or in a dense cytosolic solution. Thus, the release of nonnative polypeptide is not an essential feature of the productive chaperonin mechanism. Our results argue that conditions of excluded volume, thought to prevail in the bacterial cytosol, increase the capacity of the chaperonin to retain nonnative polypeptide throughout successive reaction cycles. We propose that the leakiness of the chaperonin system under physiological conditions is adjusted such that E. coli proteins are likely to complete folding without partitioning between different GroEL complexes. Polypeptides that are unable to fold on GroEL eventually will be transferred to other chaperones or the degradation machinery.
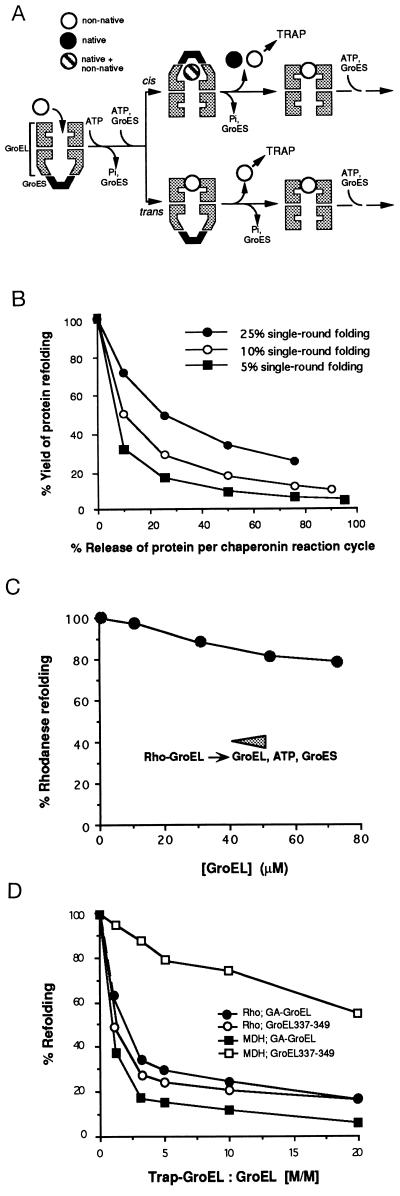
The chaperonins GroEL and GroES prevent the aggregation of newly synthesized polypeptides and promote their efficient folding in the bacterial cytosol (for review see refs. 1–5). GroEL is composed of two heptameric rings of ≈57-kDa subunits stacked back-to-back, each ring containing hydrophobic binding regions for unfolded polypeptide that face the central cavity (6). GroES is a single ring of seven ≈10-kDa subunits (7, 8). Recent studies have established the basic mechanistic principles of chaperonin action (Fig. 1A) (3, 5). Briefly, unfolded polypeptide binds to an asymmetrical GroEL–GroES complex within the cavity of the GroEL ring that is not occupied by GroES, followed by the dissociation of GroES. Rebinding of GroES to either GroEL ring together with ATP results in the encapsulation of ≈50% of bound polypeptide and the burial of the hydrophobic regions of the inner surface of GroEL. Bound polypeptide is released into an enlarged GroEL cavity (13) for folding, and the bound ATP is hydrolyzed. Within approximately 15–20 s (at 25°C), a subsequent round of ATP hydrolysis in the opposite GroEL ring triggers the dissociation of GroES from GroEL, thus opening the folding cage. Rebinding of incompletely folded polypeptide may result in the reversal of misfolded, but kinetically stable, conformations in preparation for another folding cycle. Slow-folding proteins, such as mitochondrial rhodanese, require on average up to 10 reaction cycles to complete folding in vitro (14).
Figure 1.
Release of native and nonnative polypeptide during cycles of chaperonin-mediated protein folding. (A) A simplified model describing the basic chaperonin mechanism. GroEL is shown as a vertical section through the cylinder outlining the three-domain structure of the subunits. In dilute solution a fraction of bound polypeptide can be readily released from the GroEL cavity in a nonnative state from both the cis topology (GroES bound to the polypeptide-containing ring of GroEL) and the trans topology (GroES and polypeptide bound to opposite GroEL rings). This polypeptide may be captured by a noncycling mutant GroEL trap-GroEL before it can rebind to GroEL to undergo another folding trial. Trapping from the cis topology recently has been demonstrated (9, 10). (B) Relationship between yields of refolding and the rate of transfer of nonnative polypeptide to trap-GroEL. Yields are modeled assuming an increasing leakiness of GroEL for nonnative polypeptide and folding efficiencies per reaction cycle of 5%, 10%, and 25%. Note how the refolding yield would increase significantly at higher folding efficiencies as they are found for most other substrate proteins of GroEL studied (see refs. 11 and 12). The folding yield per cycle for rhodanese was experimentally determined to be ≈5%. The fraction of rhodanese that is transferred from wild-type GroEL to trap-GroEL per reaction cycle was measured in single-round transfer experiments (11) to be 25% and was constant at molar ratios of trap-GroEL to GroEL from 4- to 10-fold. (C) Rhodanese refolding in concentrated chaperonin solutions. The amount of active rhodanese obtained at 0.25 μM GroEL is set to 100%. (D) Effects of trap-GroEL on GroEL/GroES-mediated protein folding. Inhibition of folding of rhodanese (•, ○) and malate dehydrogenase (MDH) (▪, □) by GA-GroEL (•, ▪) and GroEL337/349 (○, □) at the indicated molar ratios of trap-GroEL over wild-type GroEL. Enzyme activities reached in the absence of trap-GroEL are set to 100%.
While there is strong evidence for folding to occur in the chaperonin cavity (10, 11, 15–17), a fraction of bound polypeptide can be released from GroEL into solution in every reaction cycle (11, 18, 19). However, the release of nonnative polypeptide from GroEL is apparently nonproductive, and folding of this material requires rebinding to the same or another chaperonin molecule, at least in the case of rhodanese (11, 17). As shown recently, nonnative protein release can occur both when GroES binds to the GroEL ring in trans to unfolded polypeptide and upon dissociation of GroES from the cis topology in which polypeptide was enclosed within the GroEL cylinder (Fig. 1A) (9, 10). A central question in the chaperonin field is whether this release of nonnative polypeptide is an essential feature of the productive chaperonin mechanism, or whether it is a side-reaction that is favored under the experimental conditions in which the chaperonin is studied in vitro. To address this question, we used macromolecular crowding agents to mimic the excluded volume effects prevailing in the bacterial cytosol. We demonstrate that efficient chaperonin-mediated folding can occur without the premature release of nonnative polypeptide into the bulk solution.
MATERIALS AND METHODS
Chaperonin Expression and Purification.
Wild-type GroEL, the mutant GroEL337/349, and GroES were purified as described (11). GroEL internally crosslinked with glutaraldehyde (GA-GroEL) was prepared by incubation of 0.4 μM GroEL in buffer A [25 mM 3-(N-morpholino)propanesulfonic acid (Mops), pH 7.2/75 mM KCl/5 mM MgCl2/1 mM DTT] with 1.5% glutaraldehyde at 25°C for 45 min. Subsequently, glutaraldehyde action was stopped by addition of 40 mM sodium borohydride for 20 min, followed by exchange into buffer A on a NAP10 column (Pharmacia) and concentration on Centricon 100. Protein concentrations of GA-GroEL, GroEL337/349, and GroEL were determined by Bradford assay (20) and/or quantitative amino acid analysis. ATPase activity of GroEL was determined as described (14).
Protein Refolding in Concentrated Chaperonin Solutions.
Increasing concentrations of GroEL (10–73 μM) were added to 0.25 μM rhodanese-GroEL complex, formed in buffer B (see below), together with 8 mM ATP, 8 mM MgCl2, 75 μM GroES, and an ATP-regenerating system consisting of 10 mM creatine phosphate and 8.5 units/ml creatine kinase. Reactivation was terminated after 2 min at 25°C with cyclohexanediamine tetraacetate (CDTA), and the yield of native enzyme was determined enzymatically. The frequency J of encounters between a molecule of rhodanese and GroEL in solution was estimated by the equation J = 4πDGroEL(rrho + rGroEL)cGroEL, with the diffusion rate of GroEL DGroEL,20°C = 10−7 cm2·s−1, the radius of rhodanese rrho = 25 Å, the radius of GroEL rGroEL = 70 Å, and the concentration of GroEL cGroEL in units of molecules·cm−3. Assuming that only 10% of the collisions result in the formation of a rhodanese–GroEL complex (probably an underestimation), a rhodanese molecule would spend only 0.2 ms in solution before rebinding to GroEL present at 73 μM.
Refolding in the Presence of Trap-GroEL and Crowding Agents.
Unfolded bovine rhodanese (21) and porcine MDH (22) were bound to GroEL by 150-fold dilution of guanidinium chloride (GdmCl)-denatured enzyme (100 μM in 5.3 M GdmCl/1 mM DTT) either into buffer B for rhodanese (buffer A plus 10 mM DTT and 50 mM sodium thiosulfate) or into buffer A for MDH, containing in each case 0.15 μM GroEL. Formation of MDH–GroEL complexes was performed at 37°C. After reisolation of substrate–GroEL complexes by size-exclusion chromatography, the reaction mixtures were concentrated to a final GroEL concentration of 0.25 μM on Centricon 100 and divided into aliquots, which received GA-GroEL or mutant GroEL337/349 at the concentrations indicated. Refolding was initiated by adding 3 mM ATP and GroES at a 2-fold molar excess over total GroEL. After 6 min (rhodanese) or 20 min (MDH) at 25°C, 15 mM CDTA was added to stop the reaction, and enzyme activities were assayed at 25°C as described (14, 22). For refolding experiments in the presence of crowding agents, GroEL-bound rhodanese and MDH (0.25 μM final concentration) were generated in buffer B and then incubated as above, except that Ficoll 70 or dextran 70 (Sigma) were present at the concentrations (wt/vol) indicated. GroEL exhibited full ATPase activity in the presence of crowding agents and the typical inhibition of ATP hydrolytic activity by GroES. Sodium thiosulfate was omitted from the buffer solution in the reaction mixtures containing MDH. The final concentration of Ficoll 70 in the enzyme assays (8%) was without effect.
Transfer of Nonnative Rhodanese to Trap-GroEL.
A preformed complex of 3H-labeled rhodanese (21) and GroEL (0.16 μM) was incubated for 3 min in buffer B with 30% Ficoll 70 in the absence or presence of 2 mM ATP. Then, 15 mM CDTA was added and samples were diluted 5-fold with 25 mM Mops, pH 7.2/50 mM NaCl/1 mM DTT. The two forms of GroEL were then separated on a Mono Q column (Pharmacia), equilibrated in 25 mM Mops, pH 7.2/50 mM NaCl/1 mM DTT and eluted with a salt gradient from 50 to 500 mM NaCl. Fractions were analyzed by measuring A280 (GroEL) and by liquid scintillation counting ([3H]rhodanese). 3H-labeled rhodanese was functionally active and folded in a GroEL/GroES-dependent reaction.
Protein Folding in Concentrated Cytosolic Extracts.
Highly concentrated cytosol fractions from Xenopus eggs were generated as described (23). Briefly, unfertilized primed Xenopus eggs were dejellied in 2% cysteine hydrochloride (pH 7.7) and transferred into extraction buffer (50 mM sucrose/100 mM KCl/0.1 mM CaCl2/5 mM MgCl2/10 mM Hepes·KOH, pH 7.4/1 mM phenylmethanesulfonyl fluoride/10 μg/ml leupeptin/2 mM DTT). After sedimenting the eggs for 1 min at 1000 × g, surplus buffer was removed as quantitatively as possible, and the eggs were pelleted again for 15 min at 20,000 × g in a TLS55 rotor (Beckman). The concentrated lysate that was formed on top of the layer of yolk was carefully recovered and supplemented with cytochalasin B (50 μg/ml), 35 mM creatine phosphate, and 250 μg/ml creatine kinase. Yolk proteins were absent from the extract as determined by SDS/PAGE. The presence of an ATP-regenerating system was necessary to obtain full refolding efficiencies. The protein concentration of the extracts was determined by Bradford assay (Bio-Rad) with bovine IgG as standard. Concentrations of different extract batches were between 100 and 240 mg/ml, in agreement with values reported with BSA as standard (24). Refolding experiments were performed by adding concentrated preformed rhodanese-GroEL complexes (0.65 μM, final concentration) and additional reagents (10-fold molar excess of GroES and 8-fold molar excess of trap-GroEL and MgATP) to the extracts. A low background activity of rhodanese activity was subtracted. The yield of GroEL/GroES-dependent rhodanese reactivation in different batches of cytosol extracts was between 60% and 90% of that in buffer.
RESULTS AND DISCUSSION
The release of nonnative substrate polypeptide from GroEL has previously been monitored by the addition of a noncycling mutant GroEL (GroEL337/349) to the folding reaction that functions as a trap for unfolded protein (11, 18). The theoretical relationship between the transfer efficiencies for nonnative rhodanese from GroEL to trap-GroEL per reaction cycle and the final yield of folded protein in the presence of excess trap-GroEL are shown in Fig. 1B. Due to the low folding yield of only ≈5% per reaction cycle, the release of 25% of GroEL-bound rhodanese in a nonnative state per cycle (measured at up to 10-fold molar excess of trap-GroEL over GroEL) results in an 80% inhibition of refolding (see below and ref. 11). To evaluate the functional significance of nonnative protein release, we first tested whether completion of folding of nonnative intermediates in the bulk solution contributes to the overall efficiency of folding. Rhodanese refolding reactions were performed in the presence of very high concentrations of free chaperonin. Binding and rebinding of nonnative polypeptide to GroEL occur at a rate close to diffusion limited, both in the presence and in the absence of ATP (16, 25). Thus increasing the total GroEL concentration should reduce the potential of folding intermediate to reach the native state in solution. Denatured rhodanese first was bound to 0.25 μM GroEL, and then free GroEL (with GroES) was added up to a total concentration of 73 μM (Fig. 1C). This decreased the rate of rhodanese folding only slightly, perhaps due to the accumulation of higher levels of ADP that slow the GroEL ATPase. A conservative estimate suggested that under the experimental conditions nonnative rhodanese would spend less than 1 ms in solution before rebinding to GroEL at 73 μM. Similar considerations have been made for the small substrate protein barnase (16, 25). Thus the chaperonin-assisted folding of rhodanese appears to proceed exclusively in association with GroEL, consistent with the recent demonstration that rhodanese reaches the native state in the GroEL cavity with full efficiency when GroES cycling is prevented (10, 17).
These observations supported the view that the release of nonnative rhodanese from GroEL represents a certain leakiness of the chaperonin system. In addition to GroEL337/349 (18), an internally glutaraldehyde-crosslinked form of trap-GroEL (GA-GroEL) (11) was employed to investigate this further. Like GroEL337/349, GA-GroEL preserved the oligomeric structure of the chaperonin and was unable to bind ATP and GroES (not shown), but exhibited a significantly higher affinity for certain polypeptide substrates (see below). Bound substrate was not released from GA-GroEL even upon prolonged incubation with ATP and GroES. When GA-GroEL was added in increasing concentrations to preformed complexes of GroEL and unfolded rhodanese or MDH, the yield of active enzyme measured upon addition of ATP and GroES decreased significantly (Fig. 1D), indicating the transfer of nonnative protein from GroEL to GA-GroEL. GroEL337/349 also inhibited rhodanese folding but had only a weak effect on the refolding of MDH (Fig. 1D), suggesting that different forms of trap-GroEL vary in their affinities for folding intermediates. Interestingly, while an ≈20% reactivation of monomeric rhodanese was observed even in the presence of a 20-fold molar excess of trap-GroEL (see above; ref. 11), high concentrations of GA-GroEL inhibited the folding/assembly of dimeric MDH almost completely. As both wild-type GroEL and the two forms of trap-GroEL interacted with denatured MDH with apparently equal affinity, it seems likely that GroEL337/349 (and wild-type GroEL), in contrast to GA-GroEL, does not recognize the prefolded subunits of MDH. GroEL is thought to release the subunits of oligomeric enzymes as monomers for spontaneous association (26). These intermediates, exposing hydrophobic surfaces, may retain a residual affinity for chaperonin. The differential behavior of GroEL337/349 and GA-GroEL suggests that some forms of trap-GroEL may recognize even late folding intermediates that may already have buried most of their hydrophobic surfaces in association with GroEL or that expose such surfaces only transiently.
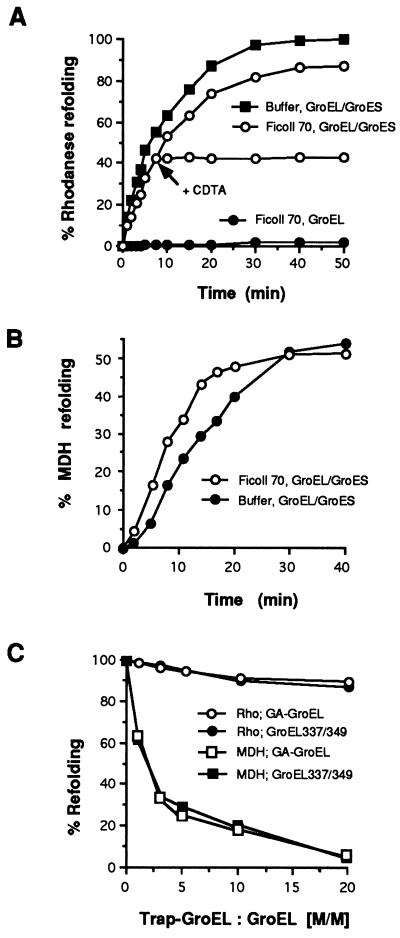
It seemed possible that under conditions of macromolecular crowding, which prevail in the bacterial cytosol, the chaperonin would function in a more tightly coupled manner. In contrast to the situation in vitro, in vivo a considerable fraction of the intracellular space is occupied by macromolecules, which reach a concentration of up to 340 g/liter in the Escherichia coli cytosol (27, 28). Excluded volume effects due to molecular crowding may significantly alter the thermodynamic equilibria and kinetics of biochemical processes (29, 30). Inert and highly soluble polymer particles, such as Ficoll or dextran, have been used to mimic the macromolecular volume occupancy of the prokaryotic cytosol and to help explain apparent discrepancies between in vivo and in vitro observations, in particular phenomena that involve macromolecular binding and assembly reactions (27, 28, 31). Indeed, we found that the release of nonnative folding intermediate from GroEL into the bulk solution was prevented by crowding agents. Chaperonin-dependent reactivation of rhodanese and MDH in a 30% solution of Ficoll 70 (average Mr 70,000) occurred with similar efficiency and kinetics as in buffer (Fig. 2 A and B). Spontaneous refolding of denatured rhodanese added directly into Ficoll solution was negligible (not shown). Refolding in Ficoll 70 required the presence of GroEL, GroES, and ATP throughout the reaction, as indicated by the immediate stop of renaturation upon addition of the Mg2+ chelator CDTA, which inhibits ATP hydrolysis and dissociates the GroEL-GroES complex (Fig. 2A) (11). However, in striking contrast to the situation in buffer, in Ficoll 70 rhodanese folding was not inhibited by increasing concentrations of either form of trap-GroEL (Fig. 2C). When, after addition of CDTA to stop refolding, the reaction mixture was diluted and wild-type GroEL was separated from excess trap-GroEL by ion-exchange chromatography, nonnative rhodanese was almost exclusively bound to wild-type GroEL (Fig. 3A), indicating that there was no increased pool of free folding intermediates in the crowded solution. It is noteworthy that the subunits of dimeric MDH were normally released from wild-type GroEL, and their assembly occurred with high efficiency in the crowded environment (Fig. 2B). Trapping of MDH was still observed in the presence of crowding agent. This was expected, because the folded subunits of MDH, unlike those of monomeric rhodanese, still will expose some hydrophobic surface until they have bound another subunit. Interestingly, not only GA-GroEL, but also GroEL337/349, effectively prevented MDH assembly under these conditions (Fig. 2C), although GroEL337/349 had only little effect in this reaction in buffer alone (Fig. 1D). We attribute this to an increased affinity of GroEL337/349 for protein substrate in the crowded solution.
Figure 2.
Chaperonin-mediated protein folding in the presence of macromolecular crowding agents. (A) Chaperonin-dependent refolding of rhodanese in buffer (▪) and 30% Ficoll 70 (○, •) with GroEL and ATP in the presence (○) or absence (•) of GroES. The arrow in reaction ○ marks the addition of 15 mM CDTA after 7.5 min to stop chaperonin-mediated folding. Incubation continued for the times indicated at which enzyme activities were determined. (B) GroEL/ES-dependent refolding of MDH in buffer (•) and Ficoll 70 solution (○). (C) Refolding of rhodanese (•, ○) and MDH (▪, □) in Ficoll 70 in the presence of GA-GroEL (○, □) or GroEL337/349 (•, ▪). Preformed rhodanese-GroEL and MDH–GroEL complexes (0.25 μM) were reactivated in 30% Ficoll 70 solutions in the presence of increasing concentrations of GA-GroEL or GroEL337/349, essentially as described for A.
Figure 3.
GroES dependency of macromolecular crowding effects. (A) Absence of transfer of rhodanese from wild-type GroEL to GroEL337/349 in a 30% Ficoll 70 solution in the presence of GroES. The separation profile of the chaperonin molecules by ion-exchange chromatography and the percentage of total [3H]rhodanese associated with either wild-type or trap-GroEL is shown before and after incubation with ATP. Retention times are given in min. (B) Chaperonin-mediated rhodanese refolding in buffer and in a 30% Ficoll 70 solution in the absence or presence of a 4-fold molar excess of GA-GroEL (trap-GroEL). Rhodanese-GroEL complexes first were preincubated for 20 min either without (reactions 1 and 2) or with (reactions 3 and 4) ATP in the absence (reactions 1 and 3) or presence (reactions 2 and 4) of trap-GroEL. Folding then was initiated by adding GroES or GroES and ATP. GroEL/GroES-mediated rhodanese refolding in buffer in the absence of trap-GroEL is set to 100%.
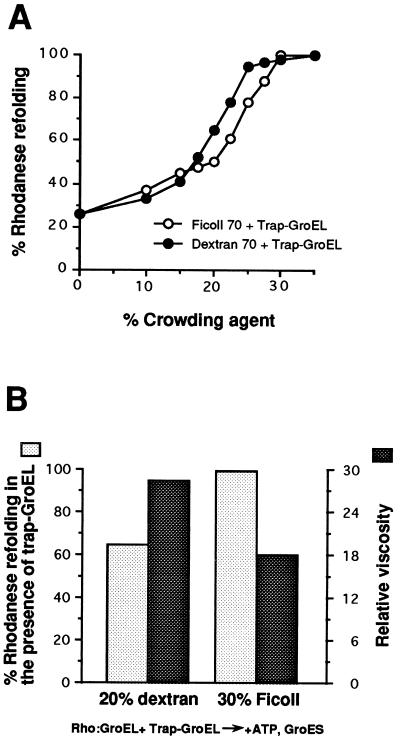
The full effect of crowding agent in preventing the release of rhodanese folding intermediate from GroEL was observed only in the presence of GroES (Fig. 3B). When GroES was initially absent, only low yields of rhodanese reactivation were observed (reaction 4). Under these conditions, nonnative rhodanese was transferred to trap-GroEL in an ATP-dependent reaction, albeit more slowly in Ficoll solution than in buffer, explaining its inability to refold upon subsequent addition of GroES. This finding confirmed that GroES is not required to promote protein release from GroEL into free solution (14) but rather to permit folding to occur in the central volume of the GroEL cavity. It also provided further evidence that the two types of trap-GroEL used were active in the presence of crowding agent. We determined that the suppression of trapping was not merely the result of the increased viscosity of the solution by analyzing the function of the chaperonin in dextran 70, a crowding agent that differs in its physicochemical properties from Ficoll 70 (32). Trapping was suppressed by both polymers in a nonlinear concentration-dependent manner (Fig. 4A), but the extent of the inhibition did not correlate with the increase in viscosity measured. For example, a 30% solution of Ficoll 70 with an 18-fold increased viscosity relative to buffer prevented rhodanese trapping completely, whereas a 20% solution of dextran 70 with 28-fold relative viscosity was only 65% efficient (Fig. 4B). Thus an increased affinity of GroEL for nonnative polypeptide due to molecular crowding most likely is responsible for the prevention of premature substrate loss from the chaperonin. In summary, crowding agents are an excellent tool to determine the functional relevance of nonnative polypeptide release from GroEL. As the kinetics and yields of GroEL-mediated folding in Ficoll are very similar to those in buffer, we conclude that nonnative release is not an integral part of the productive chaperonin cycle in folding.
Figure 4.
Comparison of the crowding effects of Ficoll 70 and dextran 70. (A) Rhodanese refolding at increasing concentrations of Ficoll 70 or dextran 70 in the presence of an 8-fold molar excess of GA-GroEL over wild-type GroEL. Rhodanese activities measured upon refolding in the presence of crowding agent and in the absence of trap-GroEL are set to 100%. The enzyme activity regained in the absence of trap-GroEL at a 30% concentration of either crowding agent was ≈80% of that measured in buffer. (B) Relationship between the viscosity of solutions containing crowding agent and their ability to suppress trapping during refolding reactions. Shown is the efficiency by which an 8-fold molar excess of trap-GroEL captures rhodanese during GroEL/GroES-mediated refolding in 30% Ficoll 70 (left) and in 20% dextran 70 (right). The relative viscosities of the crowding agents at the respective concentrations also are displayed. Viscosities relative to buffer were determined based on the Poiseuille method with an Ostwald dropping pipet at 25°C.
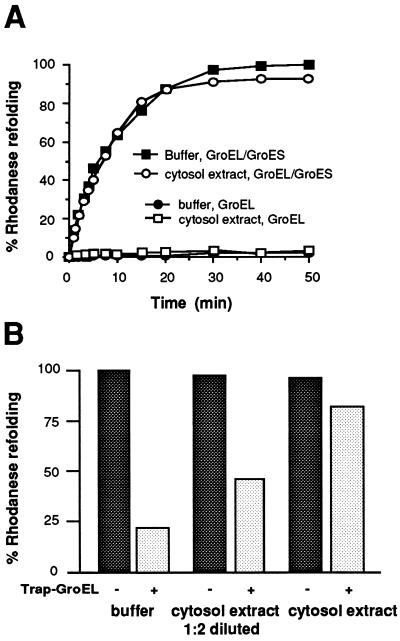
Given the known concentration dependency of molecular crowding effects (see refs. 27 and 28 and references therein), most cell lysates do not reflect the physiological conditions of excluded volume as they do not preserve the high total protein concentration of the intact cytosol (≈200–300 mg/ml in E. coli) (27, 33). Nearly undiluted extracts can, however, be prepared from unfertilized Xenopus eggs. Under optimized experimental conditions, densely packed eggs lyse during centrifugation without significant dilution. After disruption of the actin filaments these extracts are likely to have physicochemical properties similar to those of the prokaryotic cytosol. Conditions were established in which chaperonin-mediated folding of rhodanese proceeded in the egg extracts with kinetics and yields similar to those in aqueous buffer solutions or in Ficoll 70 (Fig. 5). This indicated that the concentrated extract did not contain nonnative protein that may have displaced the unfolded rhodanese from GroEL. Reactivation of GroEL-bound rhodanese was strictly dependent on GroES (Fig. 5A). When trap-GroEL was included in the extract together with a preformed rhodanese-GroEL complex, it inhibited the refolding reaction measured upon addition of GroES and ATP only marginally. As expected based on the demonstrated concentration dependency of crowding, this restriction of nonnative protein release from GroEL was observed only in undiluted extracts (≈200 mg of protein per ml) (Fig. 5B). In contrast, significant trapping was measured when the extracts were diluted 2-fold or more, similar to a recent study using extracts with a reported protein concentration of approximately 100 mg/ml (9). Both forms of trap-GroEL were functional under these conditions and did not release prebound rhodanese for folding by wild-type GroEL (not shown).
Figure 5.
Protein folding in concentrated Xenopus egg extracts. (A) Kinetics of chaperonin-mediated refolding of rhodanese in buffer and in a concentrated cytosol extract (≈200 mg of protein per ml). Experiments were performed in the presence and absence of GroES. The maximal refolding yield in buffer is set to 100%. (B) Effects of trap-GroEL on chaperonin-assisted rhodanese refolding in buffer and in cytosol extracts (original concentration of ≈200 mg/ml and 1:2 diluted). An 8-fold molar excess of trap-GroEL over wild-type GroEL was used in the experiments. Refolding reaction cycles proceeded for 15 min. The yield of rhodanese refolding in buffer in the absence of trap-GroEL is set to 100%.
The effect of undiluted cytosol in restricting the release of nonnative rhodanese from GroEL has not been observed in experiments involving microinjection of chaperonin complexes into intact oocytes (9). Provided that mixing times in this system were sufficiently rapid (see refs. 34–36), this may be explained by the asymmetrical organization of the oocyte cytosol (37). More generally, in contrast to the bacterial cytosol (27), the eukaryotic cytosol appears to be characterized by a nonhomogeneous distribution of macromolecules between a fluid phase (equivalent to ≈110 mg of protein per ml) and a gel-like cytoskeletal network (33). Thus, extracts with a homogeneous protein distribution may more adequately reflect the macromolecular crowding effects of the bacterial cytosol.
CONCLUSIONS
The observation of nonnative protein release from GroEL initially had suggested that the chaperonin functions by ejecting unfolded polypeptide for spontaneous folding in bulk solution (18, 19). While it is now clear that proteins such as rhodanese fold in the GroES-enclosed central cavity of GroEL either to the native state or to a state that is committed to complete folding (11, 15, 17), the significance of nonnative polypeptide release from GroEL is still an issue of debate. We have shown here that folding in solution makes little, if any, contribution to the overall efficiency of rhodanese folding. We further conclude that the unproductive release of nonnative intermediate can be significantly reduced under experimental conditions of macromolecular crowding that mimic the excluded volume effects generally thought to prevail in the bacterial cytosol. We propose that GroEL has evolved to function most efficiently under conditions of crowding that appear to increase the capacity of its hydrophobic binding regions to recapture nonnative folding intermediates before they emerge into the bulk solution (4).
The inhibition of folding measured as a result of transfer of nonnative polypeptide from GroEL to trap-GroEL is most pronounced for proteins such as rhodanese that fold rather inefficiently (t1/2 ≈ 5 min, equivalent to ≈10 reaction cycles) (see Fig. 1A). Other proteins, such as ornithine transcarbamoylase, fold with very high efficiency in a single chaperonin cycle (12). We suggest that molecular crowding effects restrict the free partitioning of folding intermediates between chaperonin molecules in vivo, thus allowing substrate protein to be retained in the chaperonin cavity throughout successive chaperonin cycles, each consisting of a folding trial (upon GroES binding) followed by the structural rearrangement of kinetically trapped intermediates (upon polypeptide rebinding to GroEL when GroES dissociates) (see Fig. 1A). The extent and rate at which nonnative polypeptide is released from GroEL in vivo remain to be determined. Clearly, a certain leakiness of the system must exist to allow the partitioning of aberrant polypeptides that are unable to fold (or cannot interact productively with GroEL) to other chaperone systems or to the machinery of proteolytic degradation. Folding reactions that are as inefficient as that of rhodanese may be rare in the E. coli cytosol and may naturally be associated with a signifiant loss of protein due to degradation.
Acknowledgments
We thank Drs. Christine Schneider and Jim Denegre for providing some of the Xenopus egg extracts, and Drs. R. J. Ellis, A. Fulton, A. Minton, and S. B. Zimmerman for helpful discussion. This work was supported by the National Institutes of Health and the Howard Hughes Medical Institute.
Footnotes
Abbreviations: CDTA, cyclohexanediamine tetraacetate; MDH, malate dehydrogenase; GA-GroEL, GroEL internally crosslinked with glutaraldehyde.
References
- 1.Ellis R J, Hemmingsen S M. Trends Biochem Sci. 1989;14:339–342. doi: 10.1016/0968-0004(89)90168-0. [DOI] [PubMed] [Google Scholar]
- 2.Georgopoulos C, Welch W J. Annu Rev Cell Biol. 1993;9:601–634. doi: 10.1146/annurev.cb.09.110193.003125. [DOI] [PubMed] [Google Scholar]
- 3.Clarke A R. Curr Opin Struct Biol. 1996;6:43–50. doi: 10.1016/s0959-440x(96)80093-5. [DOI] [PubMed] [Google Scholar]
- 4.Ellis R J, Hartl F-U. FASEB J. 1996;10:20–26. doi: 10.1096/fasebj.10.1.8566542. [DOI] [PubMed] [Google Scholar]
- 5.Hartl F-U. Nature (London) 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 6.Braig K, Otwinowski Z, Hegde R, Boisvert D C, Joachimiak A, Horwich A L, Sigler P B. Nature (London) 1994;371:578–586. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- 7.Hunt J F, Weaver A J, Landry S J, Gierasch L, Deisenhofer J. Nature (London) 1996;379:37–42. doi: 10.1038/379037a0. [DOI] [PubMed] [Google Scholar]
- 8.Mande S C, Mehra V, Bloom B, Hol W G J. Science. 1996;271:203–207. doi: 10.1126/science.271.5246.203. [DOI] [PubMed] [Google Scholar]
- 9.Burston S G, Weissman J S, Farr G W, Fenton W A, Horwich A L. Nature (London) 1996;383:96–99. doi: 10.1038/383096a0. [DOI] [PubMed] [Google Scholar]
- 10.Hayer-Hartl M K, Weber F, Hartl F-U. EMBO J. 1996;15:6111–6121. [PMC free article] [PubMed] [Google Scholar]
- 11.Mayhew M, da Silva A C R, Martin J, Erdjument-Bromage H, Tempst P, Hartl F-U. Nature (London) 1996;379:420–426. doi: 10.1038/379420a0. [DOI] [PubMed] [Google Scholar]
- 12.Weissman J S, Hohl C M, Kovalenko O, Kashi Y, Chen S, Braig K, Saibil H R, Fenton W A, Horwich A L. Cell. 1995;83:577–587. doi: 10.1016/0092-8674(95)90098-5. [DOI] [PubMed] [Google Scholar]
- 13.Chen S, Roseman A M, Hunter A S, Wood S P, Burston S G, Ranson N A, Clarke A R, Saibil H R. Nature (London) 1994;371:261–264. doi: 10.1038/371261a0. [DOI] [PubMed] [Google Scholar]
- 14.Martin J, Langer T, Boteva R, Schramel A, Horwich A L, Hartl F-U. Nature (London) 1991;352:36–42. doi: 10.1038/352036a0. [DOI] [PubMed] [Google Scholar]
- 15.Corrales F J, Fersht A R. Proc Natl Acad Sci USA. 1996;93:4509–4512. doi: 10.1073/pnas.93.9.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray T E, Fersht A R. J Mol Biol. 1993;232:1197–1207. doi: 10.1006/jmbi.1993.1471. [DOI] [PubMed] [Google Scholar]
- 17.Weissman J S, Rye H S, Fenton W A, Beechem J M, Horwich A L. Cell. 1996;84:481–490. doi: 10.1016/s0092-8674(00)81293-3. [DOI] [PubMed] [Google Scholar]
- 18.Weissman J S, Kashi Y, Fenton W A, Horwich A L. Cell. 1994;78:693–702. doi: 10.1016/0092-8674(94)90533-9. [DOI] [PubMed] [Google Scholar]
- 19.Todd M J, Viitanen P V, Lorimer G H. Science. 1994;265:659–666. doi: 10.1126/science.7913555. [DOI] [PubMed] [Google Scholar]
- 20.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 21.Hlodan R, Tempst P, Hartl F-U. Nat Struct Biol. 1995;2:587–595. doi: 10.1038/nsb0795-587. [DOI] [PubMed] [Google Scholar]
- 22.Peralta D, Hartman D J, Hoogenraad N J, Hoj P B. FEBS Lett. 1994;339:45–49. doi: 10.1016/0014-5793(94)80381-1. [DOI] [PubMed] [Google Scholar]
- 23.Matthews G, Colman A. Nucleic Acids Res. 1991;19:6405–6412. doi: 10.1093/nar/19.23.6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albritton N L, Meyer T, Stryer L. Science. 1992;258:1812–1815. doi: 10.1126/science.1465619. [DOI] [PubMed] [Google Scholar]
- 25.Corrales F J, Fersht A R. Proc Natl Acad Sci USA. 1995;92:5326–5330. doi: 10.1073/pnas.92.12.5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng X, Rosenberg L E, Kalousek F, Fenton W A. J Biol Chem. 1993;268:7489–7493. [PubMed] [Google Scholar]
- 27.Zimmerman S B, Trach S O. J Mol Biol. 1991;222:599–620. doi: 10.1016/0022-2836(91)90499-v. [DOI] [PubMed] [Google Scholar]
- 28.Zimmerman S B, Minton A P. Annu Rev Biophys Biomol Struct. 1993;22:27–65. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]
- 29.Minton A P. Mol Cell Biochem. 1983;55:119–140. doi: 10.1007/BF00673707. [DOI] [PubMed] [Google Scholar]
- 30.Minton A P. Int J Biochem. 1990;22:1063–1067. doi: 10.1016/0020-711x(90)90102-9. [DOI] [PubMed] [Google Scholar]
- 31.Lavery P E, Kowalczykowski S C. J Biol Chem. 1992;267:9307–9314. [PubMed] [Google Scholar]
- 32.Luby-Phelps K, Castele P E, Taylor D L, Lanni F. Proc Natl Acad Sci USA. 1987;84:4910–4913. doi: 10.1073/pnas.84.14.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luby-Phelps K. Curr Opin Cell Biol. 1994;6:3–9. doi: 10.1016/0955-0674(94)90109-0. [DOI] [PubMed] [Google Scholar]
- 34.Horowitz S B. J Cell Biol. 1972;54:609–625. doi: 10.1083/jcb.54.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horowitz S B, Moor L C. J Cell Biol. 1974;60:405–415. doi: 10.1083/jcb.60.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fulton A B. Cell. 1982;30:345–347. doi: 10.1016/0092-8674(82)90231-8. [DOI] [PubMed] [Google Scholar]
- 37.Klymnovsky M W, Karnovsky A. Dev Biol. 1994;165:372– 384. doi: 10.1006/dbio.1994.1260. [DOI] [PubMed] [Google Scholar]